Abstract
Posttranscriptional modifications of bacterial rRNA serve a variety of purposes, from stabilizing ribosome structure to preserving its functional integrity. Here, we investigated the functional role of one rRNA modification in particular—the methylation of guanosine at position 518 (G518) of the 16S rRNA in Mycobacterium tuberculosis. Based on previously reported evidence that G518 is located 5 Å; from proline 44 of ribosomal protein S12, which interacts directly with the mRNA wobble position of the codon:anticodon helix at the A site during translation, we speculated that methylation of G518 affects protein translation. We transformed reporter constructs designed to probe the effect of functional lesions at one of the three codon positions on translational fidelity into the wild-type strain, H37Rv, and into a ΔgidB mutant, which lacks the methyltransferase (GidB) that methylates G518. We show that mistranslation occurs less in the ΔgidB mutant only in the construct bearing a lesion in the wobble position compared to H37Rv. Thus, the methylation of G518 allows mistranslation to occur at some level in order for translation to proceed smoothly and efficiently. We also explored the role of methylation at G518 in altering the susceptibility of M. tuberculosis to streptomycin (SM). Using high-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS), we confirmed that G518 is not methylated in the ΔgidB mutant. Furthermore, isothermal titration calorimetry experiments performed on 70S ribosomes purified from wild-type and ΔgidB mutant strains showed that methylation significantly enhances SM binding. These results provide a mechanistic explanation for the low-level, SM-resistant phenotype observed in M. tuberculosis strains that contain a gidB mutation.
INTRODUCTION
Posttranscriptional modification of rRNA is highly conserved and spans all kingdoms of life (1). In bacteria, ribosomal RNAs are typically posttranscriptionally modified with one of the following modifications: (i) conversion of uridine to pseudouridine; (ii) methylation of the 2′hydroxyl of the pentose moiety; and (iii) modification, typically methylation, of the base moiety at various positions (2). Such modifications expand the chemical and structural interactions and functionalities of the ribosome beyond what could be afforded by the four bases alone. Some modifications help maintain the structural integrity of the ribosome by stabilizing RNA-RNA interactions and facilitating 30S and 50S subunit assembly (1, 3–5); other modifications serve to fine-tune the functional capabilities of the ribosome (6–8). The role of modifications in maintaining the functional integrity of the ribosome is further supported when one considers that most rRNA modifications cluster in functionally important regions of the ribosome (2). Yet, despite the evidence demonstrating the importance of such modifications on a macromolecular complex that is so vital for the livelihood of the organism, the functional role of many of the known ribosomal modifications remains poorly understood (1).
In this study, we investigated the functional role of the methyl modification on the guanine base at position 518 (G518) of the 16S rRNA in Mycobacterium tuberculosis. We hypothesized that G518 methylation is indirectly involved in structurally stabilizing the 16S rRNA during protein translation via interaction with the S12 ribosomal protein, which stabilizes tRNA-mRNA interactions during aminoacyl-tRNA selection in the decoding process (9). Several lines of evidence have been reported that support this proposed role of the GidB-methylated nucleotide. First, the GidB-methylated, N7 atom of G518 is situated in close (∼5 Å) proximity to C4 of the proline residue at position 44 (P44) of the S12 ribosomal protein (Fig. 1) (9). Second, P44 interacts with the third, wobble position of the mRNA codon and helps position other 16S rRNA nucleotides into making critical contact with the first and second base pairs of the codon:anticodon helix (Fig. 1) (9, 10). And third, P44 is part of a highly conserved loop region of the S12 protein (data not shown) and a proline-to-alanine mutation is lethal in Escherichia coli, underscoring the importance of the residue (10). Using a set of misincorporation assays that monitor changes in translational fidelity at each of the three positions in a codon, we demonstrate that loss of methylation indeed results in altered translational fidelity at the wobble position.
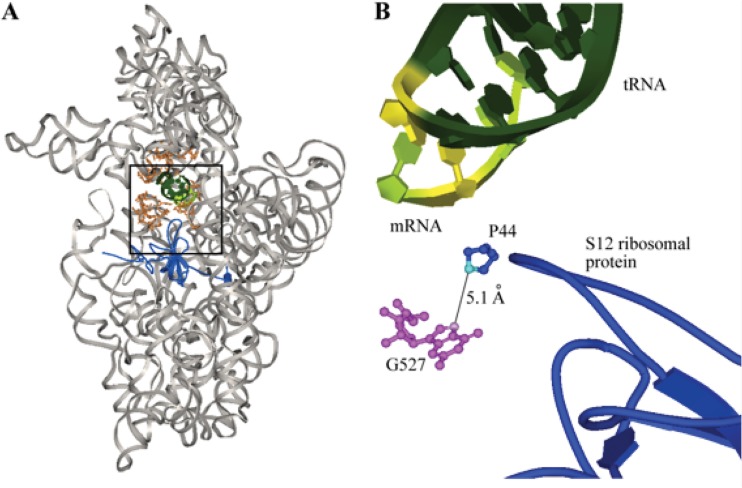
Fig 1.
(A) Crystal structure of the 30S ribosomal subunit of Thermus thermophilus complexed with mRNA and cognate tRNA in the A site (Protein Data Bank [PDB] accession no. 1IBM). 16S rRNA is shown in gray, the A site is in orange, the S12 ribosomal protein is in blue, tRNA is in dark green, and mRNA is in light green. The black box highlights the region shown in panel B. (B) The N7 atom (pink) of G527 (magenta balls and sticks), which corresponds to G518 in M. tuberculosis, is approximately 5 Å; from the C4 atom (turquoise) of proline 44 (blue balls and sticks) of the S12 ribosomal protein (blue tube). The S12 ribosomal protein interacts with the wobble position (highlighted in yellow) of the mRNA:tRNA codon:anticodon helix (dark and light green) (9).
We also explored the role of the G518 methyl modification in altering bacterial susceptibility to streptomycin (SM). This research was prompted by previous results from our group and others that collectively implicate the methylation status of G518 in modulating SM susceptibility. These results include crystallographic evidence demonstrating that G527 of the 16S rRNA of Thermus thermophilus, which corresponds to G518 in M. tuberculosis, interacts directly with SM (11). G527 has also been shown in Escherichia coli to be the methylation target of the 16S rRNA methyltransferase, GidB (12). Recently, we and others observed a loose but suggestive correlation between clinical isolates that contain a mutation in gidB and a low-level SM phenotype (12, 13). Upon evaluating an isogenic ΔgidB mutant constructed from the wild-type (WT) laboratory strain, H37Rv, we demonstrated the causal role of GidB in conferring low-level SM resistance in M. tuberculosis (14). In light of these findings, we hypothesized and demonstrate here that mutations in gidB cause SM resistance by disrupting the methyltransferase function of the expressed enzyme, and in turn, altering the methylation status of G518. The altered methylation status consequently disrupts binding of SM to its 16S rRNA target and ultimately gives rise to the SM resistance phenotype that has been observed in both laboratory and clinical M. tuberculosis strains (12, 13, 15).
MATERIALS AND METHODS
Mycobacterial translational fidelity assay.
A set of “gain-of-function” reporters (16; B. Javid, unpublished data) were used to sensitively measure small differences in mistranslation rates. These reporters work by expression of an enzyme—kanamycin (KM) kinase protein (Aph)—that is inactivated through mutation of a critical aspartate residue (D214) that is essential for function (17). Mistranslation of a reporter carrying a mutated copy of aph results in inadvertent reconstitution of the original (active) residue, which leads to a gain of enzymatic function that can be detected.
The aph gene was cloned into plasmid pJW3 (kind gift from Jun-Rong Wei [Harvard University, School of Public Health]). We made several constructs by site-directed mutagenesis to measure mistranslation at positions 1, 2, and 3 of the codon coding for D214 (GAT640–642). Mutation of the G640 to A (AAT, coding for Asn) would measure mistranslation at position 1 (DN reporter). Similarly, mutation of A641 to T (GTT, coding for Val) would measure mistranslation at position 2 (DV reporter). To measure wobble mistranslation, two constructs were made, T642 to A (GAA [coding for Glu], DE1 reporter) and G (GAG, DE2 reporter). An increase in mistranslation would result in active kanamycin kinase and in a measurable increase in the kanamycin MIC. However, the four reporters would be able to discriminate whether decoding errors at the ribosome occurred at position 1 (DN reporter), position 2 (DV reporter), or wobble position 3 (DE1 and DE2) of the mutated codon coding for D214.
All five reporter constructs (including the unmutated aph gene) were transformed separately into the wild-type H37Rv and ΔgidB mutant strains. Successful transformation was confirmed by direct sequencing of plasmid DNA isolated from each strain.
Cultures (5 ml) of each reporter-containing strain were grown in Middlebrook 7H9 broth (Becton, Dickinson, Franklin Lakes, NJ) supplemented with 10% albumin-dextrose-catalase (ADC), 0.05% Tween 80, 0.2% glycerol, and 25 μg/ml zeocin. Control H37Rv and ΔgidB strains containing no reporter were grown without zeocin selection. All strains were grown until the early log phase and subsequently diluted with their respective growth media to prepare a 10−3 dilution for use in the assay. Kanamycin (KM) was added to the first column of a round-bottom, 96-well plate in duplicate. A 2-fold serial dilution of the drug was created down the remaining columns, with their respective 7H9 growth media used as the diluent. Equal volumes of the cell suspensions diluted 1,000-fold were added to all drug-containing wells for a final volume in each well of 100 μl. Plates were incubated at 37°C for 14 days, after which cell growth was visually inspected.
Cells expressing mutated aph are resistant to KM only when substantial misincorporation of aspartate instead of the coded amino acid residue occurs (i.e., a gain of function due to mistranslation). The measured MIC is an indication of the relative amounts of amino acid misincorporation. Mistranslation was quantified as the KM MIC, which was defined as the lowest drug concentration that resulted in <1% growth compared to growth in no-drug control wells.
Preparation of 70S ribosomes.
The M. tuberculosis wild-type strain, H37Rv, and the mutant strain, ΔgidB, were grown in Middlebrook 7H9 broth (Becton, Dickinson, Franklin Lakes, NJ) supplemented with 10% ADC, 0.05% Tween 80, and 0.2% glycerol. Cells were harvested at the mid-log growth phase (optical density at 650 nm [OD650] of ∼1.2) and washed twice in ice-cold washing buffer (100 mM ammonium chloride, 50 mM magnesium acetate, 20 mM Tris-HCl [pH 7.5], 1.0 mM dithiothreitol [DTT], 0.5 mM EDTA). After resuspending cells in the same buffer at a 1:2 (wt/vol) ratio, 20 ml of the cell suspension was transferred to a 50-ml homogenizer chamber (Biospec Products, Bartlesville, OK) containing 15 ml of 0.1-mm-diameter zirconia/silica beads (Biospec Products, Bartlesville, OK). Cells were disrupted by three 30-s pulses in a BioSpec BeadBeater with a 5-min ice incubation after each pulse. DNase I was added to the resultant suspension to achieve a final concentration of 2 μg/ml to degrade contaminant DNA, and the reaction mixture was left to incubate at 4°C for 20 min. Cellular debris was removed by centrifuging the lysate in a Beckman Coulter Avanti J-E centrifuge fitted with the JA-12 rotor at 10,000 rpm for 8 min at 4°C followed by centrifugation at 12,000 rpm for 1 h at 4°C. The collected supernatant was then layered over a high-salt sucrose cushion (20 mM Tris-HCl [pH 7.5], 50 mM magnesium acetate, 100 mM ammonium chloride, 1 mM DTT, 0.5 mM EDTA, 1.1 M sucrose) and centrifuged at 100,000 × g in an SW Ti60 Beckman rotor for 16 h at 4°C.
70S ribosomes were further purified and fractionated using sucrose density fractionation. Samples were layered on top of a 7% to 30% linear sucrose gradient (10 mM Tris-HCl [pH 7.5], 6 mM magnesium acetate, 50 mM ammonium chloride, 1 mM DTT, 0.5 mM EDTA) and centrifuged at 85,000 × g for 4 h. After centrifugation, the gradients were fractionated and the absorbance at 260 nm was used to identify fractions containing ribosomes. Fractionated ribosomes were then pooled and recovered by centrifugation at 100,000 × g for 16 h at 4°C. Ribosomes were removed of any residual sucrose by resuspending pooled ribosomes in and dialyzing against dialysis buffer (10 mM magnesium chloride, 50 mM ammonium chloride, 5 mM β-mercaptoethanol, 10 mM HEPES-KOH, pH 7.6) in a dialysis cassette (Slide-A-Lyzer, Pierce, Rockford, IL) (10,000 molecular weight cutoff [MWCO]) for 2 days at 4°C with a change of buffer after the first day.
Ribosomal protein extraction.
Ribosomal protein was isolated using the acid extraction method. The resuspended ribosomes were combined with a 0.1 volume of 1 M magnesium chloride and then with 2 volumes of cold glacial acetic acid and were stirred for 45 min at 4°C. The insoluble fraction containing contaminant rRNA was removed by centrifugation at 13,000 × g for 15 min at 4°C. The supernatant was then dialyzed overnight in a dialysis cassette (Slide-A-Lyzer, Pierce, Rockford, IL) (3,500 MWCO) against ammonium acetate buffer.
16S rRNA extraction.
70S ribosomes were resuspended in dissociation buffer (10 mM Tris-HCl [pH 7.5], 1 mM magnesium acetate, 100 mM ammonium chloride, 1 mM DTT, 0.5 mM EDTA) and dialyzed in the same buffer in a dialysis cassette (Slide-A-Lyzer, Pierce, Rockford, IL) (10,000 MWCO) overnight at 4°C. The retentate was layered on top of a 7% to 30% linear sucrose gradient (10 mM Tris-HCl [pH 7.5], 1 mM magnesium acetate, 100 mM ammonium chloride, 1 mM DTT, 0.5 mM EDTA) and centrifuged at 242,000 × g for 3 h in a SW Ti60 Beckman rotor at 4°C. After centrifugation, the gradients were fractionated and the absorbance at 260 nm was used to identify fractions containing the 50S and 30S subunits. Fractionated subunits were then pooled and recovered by centrifugation at 100,000 × g for 16 h at 4°C.
From the 30S ribosomal subunit, 16S rRNA was isolated using the phenol-chloroform extraction method. One A260 unit of 30S ribosomal subunit was resuspended in 300 μl of protein digestion buffer (250 mM Tris-HCl [pH 7.5], 25 mM EDTA, 0.3 M NaCl, 50 units/ml RNasin RNase inhibitor). A proteinase K solution with a final volume of 100 μl was added to the 300-μl ribosomal subunit solution at a 1:20 (WT proteinase K/WT ribosome) ratio. The resultant solution was incubated in a shaking incubator at 37°C for 45 min. One volume of 25:24:1 phenol-chloroform-isoamyl alcohol was added to the solution, vortexed for 5 min, and centrifuged at 13,000 × g for 5 min at room temperature. The top layer was removed, and the phenol-chloroform-isoamyl alcohol extraction was repeated a second time. An equal volume of cold chloroform was then added to the extracted top layer, vortexed for 5 min, and centrifuged at 13,000 × g for 5 min at 4°C. The top layer was removed, and 1 ml of cold 100% ethanol was added and mixed by inversion. After overnight incubation at −20°C, the solution was centrifuged at 13,000 × g for 30 min at 4°C. The pellet was washed twice with 70% ethanol, after which the pellet was allowed to dry for at least several hours at room temperature. The dried pellet was resuspended in 150 μl of 2.5 M ammonium acetate. Two and a half volumes of ethanol were added, and the solution was mixed by inversion. rRNA was allowed to precipitate by overnight incubation at −20°C and then recovered by centrifuging at 13,000 × g for 30 min at 4°C. After the ammonium acetate-ethanol extraction was performed a second time, 1 ml of cold 70% ethanol was added to the final rRNA pellet and the mixture was chilled at −20°C for 10 min and centrifuged at 13,000 × g for 10 min at 4°C. The supernatant was removed, and the pellet was allowed to dry overnight in vacuo.
MS of proteins.
To determine the purity of isolated ribosomes, extracted proteins from two separate biological replicates were first separated on one-dimensional (1D)-polyacrylamide gels, subjected to in-gel digestion/peptide extraction, and then analyzed by mass spectrometry (MS) using a modified version of a method described in a previous report (18). Briefly, one-dimensional liquid chromatography tandem mass spectrometry (1D LC-MS/MS) was performed using an Eksigent Nano LC 1D Proteomics high-performance LC (HPLC) system (AB Sciex, Framingham, MA) coupled online to a Thermo LTQ-XL linear ion trap mass spectrometer equipped with a nanoflow electrospray (Thermo Electron, San Jose, CA). 1D LC/MS/MS experiments were operated such that spectra were acquired for 60 min in the data-dependent mode with dynamic exclusion enabled. The top 5 peaks in the m/z 350 to 2,000 range of every MS survey scan were fragmented.
MS data files were searched against the UniProt Knowledge database (version 14.8), with the taxonomy selected for mycobacterium species using the Mascot search engine (Matrix Sciences, London, United Kingdom). Search parameters were as follows: trypsin specificity, 1 missed cleavage, carboxymethylation of cysteine as a static modification, methionine oxidation (+16 Da) as a variable modification, and +1 through +4 charge states. The precursor ion mass tolerance was ± 1.0 Da, and the fragment ion mass tolerance was ± 0.8 Da. Mascot output files were analyzed using the software MassSieve (19). MassSieve filters were adjusted to include only peptide identifications with Mascot ion scores equal to or exceeding their identity scores (corresponds to ≥95% confidence). This resulted in calculated false-positive discovery rates of 1.2% and 1.8% for the first and second biological replicates, respectively. A minimum of 2 peptide identifications were required for a protein to be considered identified.
Mass spectrometry of 16S rRNA methylation.
Identification of site-specific modification to RNA was done using RNA mass mapping and the sequencing approach using high-performance liquid chromatography–tandem mass spectrometry analysis (20). Briefly, 5 to 7 μg of purified 16S rRNA (at a 1 μg/μl concentration) was digested with RNase T1 (Roche Diagnostics; 50 U/μg of rRNA) in 10 μl of 220 mM ammonium acetate (pH 6 to 6.5). The digestion mixture was incubated at 37°C for 2 h, dried in a SpeedVac, and resuspended in 7 to 10 μl of mobile phase A (400 mM hexafluoroisopropanol [HFIP; Sigma, St. Louis, MO], 16.3 mM triethyl amine [TEA; Fisher, Fair Lawn, NJ], water [Burdick and Jackson, Bridgeport, NJ] at pH 7.0). The resuspended digestion products were subsequently analyzed by mass spectrometry.
A Thermo LTQ XL ion trap with a Thermo Surveyor HPLC system and a Thermo Micro AS auto sampler were used for all LC/MS analyses. Injections (5 to 10 μg) of digested 16S rRNA were made on an Xterra C18 column (Waters Milford, MA) (1.0 mm by 150 mm). A 70-min gradient elution of mobile phase A and mobile phase B (200 mM HFIP, 8.15 mM TEA, methanol [Burdick and Jackson, Bridgeport, NJ] at pH 7.0) was used at a flow rate of 40 μl/min. The gradient was as follows: 5% phase B to 20% phase B in 5 min; 20% phase B to 30% phase B in 2 min; 30% phase B to 95% phase B in 43 min; and hold at 95% phase B for 5 min followed by equilibration for another 15 min at 5% phase B.
Sheath gas, auxiliary gas, and sweep gas were set to 25, 14, and 10 arbitrary units (au), respectively. The spray voltage was 4 kV, the capillary temperature was 275°C, the capillary voltage was −23 V, and the tube lens was set to −80 V. The mass was restricted to a range of m/z 550 to 2,000 to avoid the HFIP dimer at m/z 334, with data collected in negative ion polarity. Collision-induced dissociation (CID) tandem mass spectrometry (normalized collision energy, 30 au) was used in data-dependent mode to obtain sequence information from the RNase digestion products. The data-dependent scan was recorded based on the most abundant ion, and each ion selected for CID was analyzed for up to 5 scans or 30 s before it was added to a dynamic exclusion list for 30 s. A second round of CID tandem mass spectrometery (MS/MS/MS) was performed by selecting the most abundant fragment ion from the MS/MS step and dissociating that ion using the same conditions as described above.
An in silico analysis using Mongo Oligo Mass Calculator (ver. 2.06; http://library.med.utah.edu/masspec/) was performed to identify the appropriate m/z values that would confirm methylation at G518 and to identify the number of RNase T1 digestion products that might interfere with this determination. The in silico analysis provides a complete theoretical set of the digestion products, and their corresponding m/z values, that result from RNase T1 digestion of 16S rRNA. In silico digestion was performed on the 16S rRNA sequence from locus CP003248 (GI:395136682) with and without putative methylation of G518.
ITC.
The binding interaction between streptomycin and 70S ribosomes was investigated by isothermal titration calorimetry (ITC) per a previously reported protocol (21). Briefly, 70S ribosomes purified from H37Rv and ΔgidB strains were each evaluated at a concentration of 4.5 μM in dialysis buffer (10 mM magnesium chloride, 50 mM ammonium chloride, 5 mM β-mercaptoethanol, 10 mM HEPES-KOH, pH 7.6). Streptomycin was dissolved in the respective excess dialysis buffers at 112 μM. ITC experiments were conducted at 37°C on an iTC200 isothermal titration calorimeter (GE Healthcare). Streptomycin was titrated into 4.5 μM 70S ribosomes in 3.3-μl injections in 180-s intervals. The ribosome-containing cell was stirred at 800 rpm throughout the experiment. The heats of injections were integrated using Origin 7. Data and error analyses were performed using the single-site heteroassociation model in SEDPHAT (22). Confidence interval calculations were performed as described previously (23).
RESULTS
Methyl modification on G518 affects translational fidelity via indirect interaction with the wobble position of the mRNA codon.
The functional role of the methylation of G518 in the absence of drug was evaluated in a mycobacterial translational fidelity assay that employs a gain of function to measure changes in translation error rates (see Materials and Methods). Mutations were introduced in a critical aspartate residue (D214) of the aph gene which codes for a kanamycin kinase. The mutations produce an inactive form of the kinase (17) and render the bacterium susceptible to a level of KM close to that of a bacterium containing no reporter. The mutated reporters used in this assay were designed to interrogate the effects on translation due to nucleotide mutations in the first position (DN reporter), second position (DV reporter), or wobble position (DE1 and DE2 reporters) of the mRNA codon coding for D214. All four mutated reporters and the wild-type reporter were individually transformed into H37Rv and ΔgidB strains. Translational fidelity was assessed by the level of kanamycin resistance observed in strains containing the DN, DV, DE1, or DE2 reporters. An increase in MIC between the H37Rv and ΔgidB strains can be due only to differences in protein translational fidelity, since mistranslation would result in a functional copy of the aph gene product that is expressed due to misreading of the mutated codon.
Table 1 shows the degree of mistranslation of strains H37Rv and ΔgidB, both transformed with each of the reporters, as represented by the MIC of KM. No difference between the wild-type and ΔgidB mutant strains in mistranslation was observed when the strains were transformed with the wild-type (WT), DN, or DV reporter. In both strains, however, mistranslation occurs to a greater extent in both DE reporters than in the DN and DV reporters. This observation is consistent with reports showing that translational errors due to misreading of the wobble position occur more frequently (24, 25). Moreover, of the two DE reporters, mistranslation occurred to a greater extent with the GAA version of the DE reporter than with the GAG version. This observation is also consistent with previous reports that demonstrate the codon-specific effects on misreading of the wobble position (24, 26).
Table 1.
Translational fidelity as represented by kanamycin MICa
| Reporter | MIC (μg/ml) for M. tuberculosis strain: |
|
|---|---|---|
| H37Rv | ΔgidB | |
| No pDNA | 0.98 ± 0 | 0.98 ± 0 |
| WT | >500 ± 0 | >500 ± 0 |
| DN | 0.98 ± 0 | 0.98 ± 0 |
| DV | 0.98 ± 0 | 0.98 ± 0 |
| DE1 (GAA) | 15.6 ± 0 | 3.91 ± 0 |
| DE2 (GAG) | 1.95 ± 0 | 0.98 ± 0 |
Higher MICs correspond to reduced levels of translational fidelity. Data shown are mean values and standard deviations obtained from three independent biological samples that were each tested in duplicate. pDNA, plasmid DNA.
Notably, in both DE reporters, the H37Rv strain mistranslates the GAA codon to a greater extent than the ΔgidB mutant, as indicated by the 4-fold increase in KM MIC of H37Rv containing the GAA reporter over that measured for the mutant strain. Similarly, when transformed with the GAG reporter, H37Rv experiences a 2-fold increase in KM MIC compared to the mutant strain containing the same reporter.
G518 is unmethylated in the ΔgidB strain 16S rRNA.
High-performance liquid chromatography coupled to electrospray ionization mass spectrometry was used to detect the presence of methylation on the guanine base at position 518 of the 16S rRNA. From both H37Rv and ΔgidB strains, 16S rRNA was purified and digested with RNase T1, a highly efficient endonuclease that specifically cleaves at the 3′ end of unmodified guanosine residues of single-stranded RNA. The RNA region of interest and the digestion products expected to arise after RNase T1 cleavage were determined (data not shown). In particular, the oligonucleotide digestion product from H37Rv, [516]CC[mG]CG[520] at m/z 817.6, corresponding to the doubly charged ion in negative polarity, would be detected by mass spectrometry only if G518 were modified. If the modification were absent, this specific oligonucleotide ([516]CCGCG[520]) would not be detected in the digest. RNase T1 is inhibited by the methylation at G518; thus, the absence of the methylation would result in the enzyme digesting this specific oligonucleotide into [516]CCG[518] and [519]CG[520]. By taking advantage of this specificity in RNase T1 digestion, a detected signal at m/z 817.6 would reveal the presence of methylation at G518 whereas the absence of a detected signal at this m/z would reveal the absence of methylation at this specific nucleotide residue. A complete in silico analysis of the expected RNase T1 digestion products from 16S rRNA found no other digestion products that would interfere with this analysis (see Table S1 in the supplemental material).
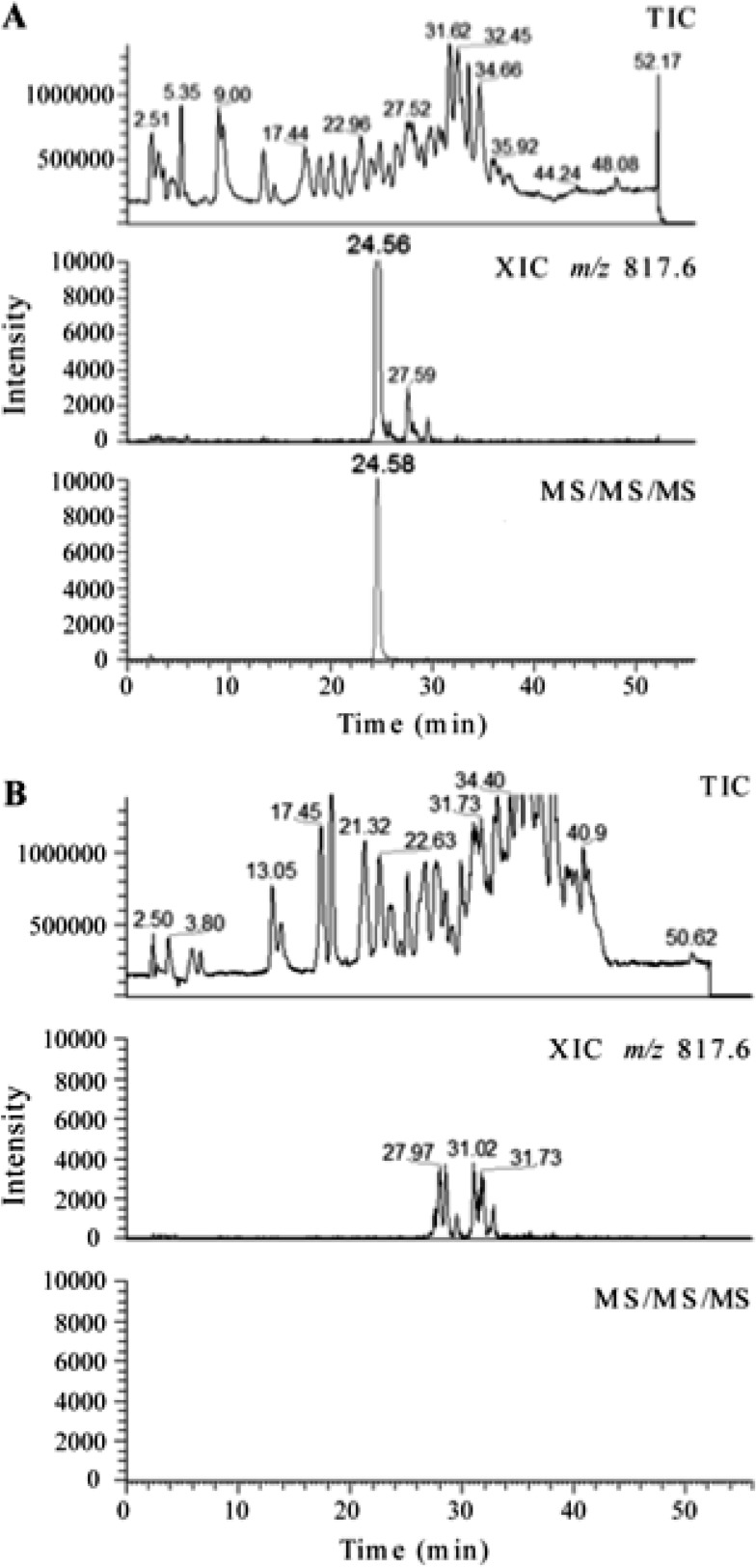
Figure 2 shows the comparative LC/MS analysis of the H37Rv wild-type and ΔgidB mutant 16S rRNA digests. Only the RNase T1-digested fragments from H37Rv exhibited a well-defined peak for m/z 817.61 in the extracted ion chromatogram (XIC) (Fig. 2A, middle chromatogram). The ΔgidB mutant sample exhibited a large number of responses in the XIC for m/z 817.6 (Fig. 2B, middle chromatogram). However, none of these responses corresponded to the correct retention time for CC[mG]CG, were all of much lower ion abundance than the response from the wild-type sample, and did not show the presence of the expected analyte ion in the mass spectrum (data not shown).
Fig 2.
A comparative LC-MS (multiple-stage mass spectrometry) analysis of the RNase T1 digest of 16S rRNA purified from strain H37Rv (A) and strain ΔgidB (B) shown to scale. (A) The top chromatogram is the total ion chromatogram (TIC) of the RNase T1 digest of 5 μg of 16S rRNA from H37Rv. The middle chromatogram shows a well-defined peak for m/z 817.6, which corresponds to the modification of the specific oligonucleotide CC[mG]CG. The bottom chromatogram shows the peak for multiple-stage mass spectrometry of the analyte ion, where m/z 817.6 was subjected to a first stage of fragmentation (MS/MS) and the resulting most abundant fragment ion m/z 735.1 was subjected to a second stage of fragmentation (MS/MS/MS). Only an ion that generates m/z 735.1 from m/z 817.6 yields a response in this analysis. (B) The top chromatogram is the TIC of the RNase T1 digest of 7 μg of 16S rRNA from strain ΔgidB. The middle chromatogram does not show a well-defined peak for m/z 817.6. The bottom chromatogram corresponds to the multiple-stage mass spectrometry of the analyte ions as described above. The absence of any response represented in the bottom chromatogram indicates the lack of a methylguanine base loss from the oligonucleotides that yielded a response in the first stage of tandem mass spectrometry shown in the middle chromatogram.
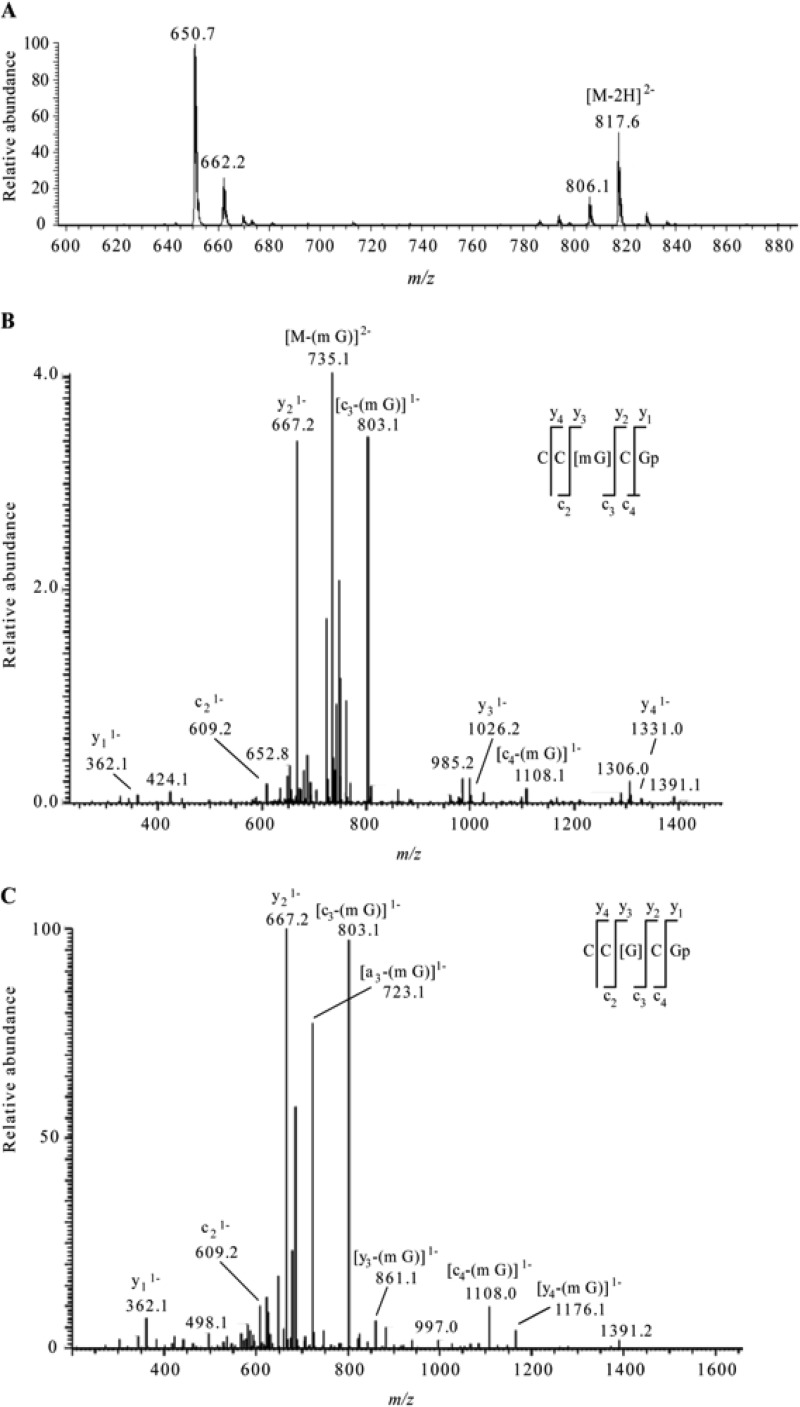
The m/z value corresponding to the peak retained at 24.56 min for the wild-type sample was confirmed to be 817.6 as shown in Fig. 3A. CID of the selected ion, m/z 817.6, yielded a fragment ion (with m/z 735.1) as the most abundant peak in the MS/MS spectrum (Fig. 3B). This observed m/z value corresponds to the loss of a methylguanine base (−164 Da) from the precursor ion. Although the oligonucleotide sequence may be reconstructed from sequence-informative fragments, their abundance was quite low in the CID spectrum due to the high level of nucleobase loss from the precursor ion. To provide additional confirmation that this m/z corresponded to the targeted oligonucleotide with methylation at G518, the most abundant fragment ion in the MS/MS analysis (m/z 735.1) was mass selected and again analyzed by CID to generate an MS/MS/MS spectrum (Fig. 3C). The sequence-informative c- and y-type ions were used to identify the sequence as CC[mG]CG, after accounting for loss of the methylated base (see above) from the sequence. These analyses confirm the presence of methylation at position 518 in the 16S rRNA of the wild-type strain, H37Rv. Multistage fragmentation was also performed on the ions observed in the XIC for m/z 817.6 for the ΔgidB mutant sample. The highly specific MS/MS/MS approach did not detect any signal, and a well-defined peak was absent at the expected retention time (Fig. 2B, bottom chromatogram).
Fig 3.
(A) Electrospray mass spectrum corresponding to the peak seen in the extracted ion chromatogram for m/z 817.6 of the RNase T1 digest of 5 μg of 16S rRNA from H37Rv M. tuberculosis (Fig. 2A, middle chromatogram). The mass spectral data are consistent with the doubly charged ion that would be expected for CC[mG]CG. The other m/z values in this mass spectrum correspond to additional RNase T1 digestion products from 16S rRNA. (B) Collision-induced dissociation (CID) mass spectrum of the RNase T1 digestion product at m/z 817.6 shown in panel A. (C) CID mass spectrum of the fragment with m/z 735.1 shown in panel B. The observed sequence-informative fragments correspond to the expected fragmentation pattern of oligonucleotide CC[mG]CG, which has an mG-base loss. The absence of mG is depicted as [G] in the sequence representation. Sequence-informative fragment ions are labeled following the nomenclature of McLuckey et al. (31).
Based on the results shown in Fig. 2 and 3, we conclude that methylguanosine was present at G518 in the wild-type sample and absent in the ΔgidB mutant sample.
Streptomycin binds to strain ΔgidB ribosomes with less affinity than to strain H37Rv ribosomes.
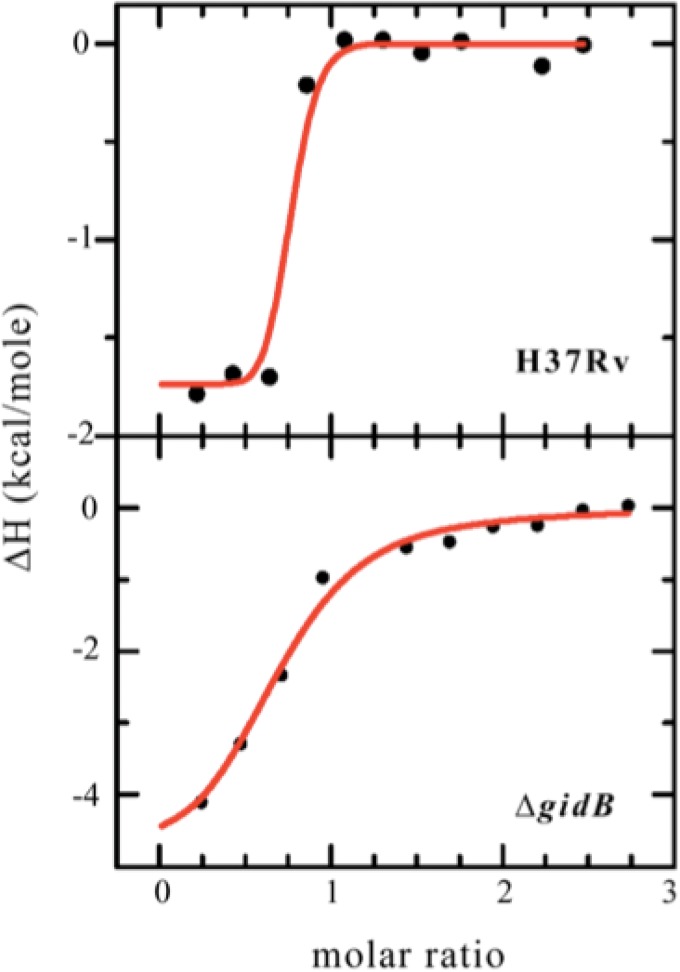
To evaluate the binding interaction between SM and the ribosome, we used isothermal titration calorimetry. The binding isotherms obtained from this analysis are shown in Fig. 4. SM binds to mutant 70S ribosomes with an affinity that is 2 orders of magnitude less than that of the binding of SM to wild-type ribosomes (strain H37Rv Kd [dissociation constant] = 0.8 nM, with a confidence interval of 4.3 nM or greater; strain ΔgidB Kd = 340 nM, with a confidence interval of 147 nM to 787 nM).
Fig 4.
Binding isotherms obtained from isothermal titration calorimetry analysis of streptomycin (SM) bound to H37Rv wild-type (top plot) or ΔgidB mutant (bottom plot) 70S ribosomes. SM binds to wild-type ribosomes with a binding affinity that is 2 orders greater than that seen with the binding of SM to mutant ribosomes. (strain H37Rv Kd = 0.8 nM, with a confidence interval of 4.3 nM or greater; strain ΔgidB Kd = 340 nM, with a confidence interval of 147 nM to 787 nM).
DISCUSSION
Posttranscriptional modifications of bacterial rRNA serve a variety of purposes, ranging from stabilizing the ribosome structure to preserving the functional integrity of the ribosome. For this report, we sought to elucidate the functional role of one rRNA modification in particular—the methyl modification of the guanine base at position 518 of the 16S rRNA in M. tuberculosis.
We provide the first biochemical evidence that G518 methylation plays a role in protein translation in normal bacterial function. Specifically, we demonstrate that the methyl modification affects translational fidelity through interaction with the mRNA wobble position, and not with the first position or second position, of the mRNA:tRNA codon:anticodon helix. The interaction is likely to be indirect and occurs via the S12 ribosomal protein since crystallographic evidence suggests that the methyl modification and the wobble position are too distant to have direct contact (11). Based on the increased mistranslation of strain H37Rv containing DE1 or DE2 reporters compared to strain ΔgidB containing the same reporters, we also show that the methyl modification actually enhances mistranslation. We speculate that this role of allowing some level of mistranslation to occur allows translation to proceed smoothly and efficiently.
We also demonstrate that, in the presence of the antibiotic SM, the methyl modification renders the bacterium vulnerable to the toxic effects of SM by enabling the drug to bind to the ribosome with high affinity and subsequently killing the bacterium. However, in the absence of the methyl modification, due to mutations in the methyltranferase (GidB) that is responsible for methylating G518, SM binds to the ribosome with less affinity, which confers low-level resistance of the bacterium to the drug.
The fitness costs associated with the acquisition of the various mutations that confer SM resistance are complex and not well understood. Interestingly, in this case, deletion of gidB, which induces low-level SM resistance, was found to induce a higher level of translational fidelity. Estimates of the intrinsic error frequency of amino acid misincorporation range from 10−5 to 10−3, depending on how the measurements were performed and details of the experimental methods used for the analysis (26–28). Under some conditions, mistranslation can be beneficial for the cell, for example, under growth conditions where amino acids are in limited supply, by allowing the cell to continue to produce proteins with a more limited repertoire of precursors (29). The ribosome has evolved in the face of pressure to maximize speed while achieving reasonable accuracy (30). Our experiments suggest the possibility that the organism normally methylates G518 at a lower level of translational accuracy at the wobble position, possibly to improve translational speed, although we have not yet formally tested that. The long-term selective pressure leading to the fixation of methylation appears to be relatively modest, and further experimentation is required to establish the fitness costs to M. tuberculosis for acquisition of low-level SM resistance.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this work was provided by the Intramural Research Program of the NIH, by NIAID, and by NIH grant GM58843 to P.A.L.
Footnotes
Published ahead of print 7 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00905-13.
REFERENCES
- 1. Grosjean H. 2005. Modification and editing of RNA: historical overview and important facts to remember, 1st ed. Springer-Verlag, New York, NY [Google Scholar]
- 2. Decatur WA, Fournier MJ. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27:344–351 [DOI] [PubMed] [Google Scholar]
- 3. Connolly K, Rife JP, Culver G. 2008. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 70:1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu Z, O'Farrell HC, Rife JP, Culver GM. 2008. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat. Struct. Mol. Biol. 15:534–536 [DOI] [PubMed] [Google Scholar]
- 5. Green R, Noller HF. 1996. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA 2:1011–1021 [PMC free article] [PubMed] [Google Scholar]
- 6. Green R, Noller HF. 1999. Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA. Biochemistry 38:1772–1779 [DOI] [PubMed] [Google Scholar]
- 7. Kimura S, Suzuki T. 2010. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 38:1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krzyzosiak W, Denman R, Nurse K, Hellmann W, Boublik M, Gehrke CW, Agris PF, Ofengand J. 1987. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome. Biochemistry 26:2353–2364 [DOI] [PubMed] [Google Scholar]
- 9. Ogle JM, Brodersen DE, Clemons WM, Tarry MJ, Carter AP, Ramakrishnan V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897–902 [DOI] [PubMed] [Google Scholar]
- 10. Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R. 2007. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J. Mol. Biol. 374:1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348 [DOI] [PubMed] [Google Scholar]
- 12. Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, Suzuki Y, Ochi K. 2007. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 63:1096–1106 [DOI] [PubMed] [Google Scholar]
- 13. Via LE, Cho SN, Hwang S, Bang H, Park SK, Kang HS, Jeon D, Min SY, Oh T, Kim Y, Kim YM, Rajan V, Wong SY, Shamputa IC, Carroll M, Goldfeder L, Lee SA, Holland SM, Eum S, Lee H, Barry CE. 2010. Polymorphisms associated with resistance and cross-resistance to aminoglycosides and capreomycin in Mycobacterium tuberculosis isolates from South Korean patients with drug-resistant tuberculosis. J. Clin. Microbiol. 48:402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wong SY, Lee JS, Kwak HK, Via LE, Boshoff HIM, Barry CE., III 2011. Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55:2515–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spies FS, da Silva PEA, Ribeiro MO, Rossetti ML, Zaha A. 2008. Identification of mutations related to streptomycin resistance in clinical isolates of Mycobacterium tuberculosis and possible involvement of efflux mechanism. Antimicrob. Agents Chemother. 52:2947–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruan B, Palioura S, Sabina J, Marvin-Guy L, Kochhar S, Larossa RA, Soll D. 2008. Quality control despite mistranslation caused by an ambiguous genetic code. Proc. Natl. Acad. Sci. U. S. A. 105:16502–16507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boehr DD, Thompson PR, Wright GD. 2001. Molecular mechanism of aminoglycoside antibiotic kinase APH(3′)-IIIa: roles of conserved active site residues. J. Biol. Chem. 276:23929–23936 [DOI] [PubMed] [Google Scholar]
- 18. Strader MB, Costantino N, Elkins CA, Chen CY, Patel I, Makusky AJ, Choy JS, Court DL, Markey SP, Kowalak JA. 2011. A proteomic and transcriptomic approach reveals new insight into beta-methylthiolation of Escherichia coli ribosomal protein S12. Mol. Cell. Proteomics 10:M110.005199. 10.1074/mcp.M110.005199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Slotta DJ, McFarland MA, Markey SP. 2010. MassSieve: panning MS/MS peptide data for proteins. Proteomics 10:3035–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mandal D, Kohrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Soll D, RajBhandary UL. 2010. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc. Natl. Acad. Sci. U. S. A. 107:2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gregory ST, Carr JF, Dahlberg AE. 2009. A signal relay between ribosomal protein S12 and elongation factor EF-Tu during decoding of mRNA. RNA 15:208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Houtman JC, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE, Schuck P. 2007. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 16:30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson ML. 1992. Why, when, and how biochemists should use least squares. Anal. Biochem. 206:215–225 [DOI] [PubMed] [Google Scholar]
- 24. Dix DB, Thompson RC. 1989. Codon choice and gene expression: synonymous codons differ in translational accuracy. Proc. Natl. Acad. Sci. U. S. A. 86:6888–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parker J, Johnston TC, Borgia PT, Holtz G, Remaut E, Fiers W. 1983. Codon usage and mistranslation. In vivo basal level misreading of the MS2 coat protein message. J. Biol. Chem. 258:10007–10012 [PubMed] [Google Scholar]
- 26. Kramer EB, Farabaugh PJ. 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13:87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Drummond DA, Wilke CO. 2009. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 10:715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parker J. 1989. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 53:273–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reynolds NM, Lazazzera BA, Ibba M. 2010. Cellular mechanisms that control mistranslation. Nat. Rev. Microbiol. 8:849–856 [DOI] [PubMed] [Google Scholar]
- 30. Wohlgemuth I, Pohl C, Mittelstaet J, Konevega AL, Rodnina MV. 2011. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:2979–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McLuckey SA, Van Berkel GJ, Glish GL. 1992. Tandem mass-spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 3:60–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.