Abstract
GS-5885 is a novel hepatitis C virus (HCV) NS5A inhibitor. In a 3-day monotherapy study in treatment-naive genotype 1a (GT1a) and GT1b HCV-infected subjects, median viral load reductions ranged from 2.3 to 3.3 log10 HCV RNA IU/ml across dosing cohorts (1, 3, 10, 30, or 90 mg once daily). Here, we report viral sequencing and phenotypic analysis of clinical isolates from this study. Detection of baseline NS5A amino acid substitutions at positions 28, 30, 31, or 93 in GT1a was associated with a reduced treatment response. In the GT1b cohort, Y93H was detected in 100% of subjects at day 4 or 14. In the Gt1a cohort, population sequencing detected NS5A resistance-associated mutations at day 4 or 14 for 3/10 subjects at the 1-mg dose and for all subjects dosed at ≥3 mg. A subset of mutants that confer a low level of reduced susceptibility to GS-5885 was not detected by population sequencing at the 30- and 90-mg doses. Subject-derived M28T, Q30R, L31M, and Y93C mutations all conferred >30-fold reductions in GS-5885 and daclatasvir susceptibilities in vitro. Site-directed NS5A mutants also showed reduced susceptibility to GS-5885. However, all NS5A mutants tested remained fully susceptible to other classes of direct-acting antivirals (DAAs), interferon alpha, and ribavirin. Importantly, the nonoverlapping resistance profile and high potency of GS-5885 support its further development with other direct-acting antivirals for the treatment of chronic HCV. (This study has been registered at ClinicalTrials.gov under registration number NCT01193478.)
INTRODUCTION
Hepatitis C virus (HCV) infection is a global health issue, with approximately 170 million people infected worldwide (1). The standard of care (SOC) has been pegylated alpha interferon and ribavirin until the more recent approval of two NS3 protease inhibitors, telaprevir and boceprevir, for use in conjunction with pegylated alpha interferon and ribavirin (1–3). The last several years have seen a great expansion of new direct-acting antivirals (DAAs) in clinical development to augment or supplant treatment with pegylated alpha interferon and ribavirin. HCV nonstructural protein 5A (NS5A) has emerged as a viable and attractive viral target for small-molecule inhibition. Although there is no known enzymatic activity for NS5A, it is essential for viral replication (4). The first NS5A replication complex inhibitor to show efficacy in the clinic was daclatasvir (BMS-790052) (5). This compound elicited rapid and profound reductions in HCV RNA and validated NS5A as a clinical target. Sequence analysis of clinical isolates following daclatasvir monotherapy identified the main resistance-associated mutations (RAMs) at NS5A amino acid positions 28, 30, 31, and 93 (6).
In addition to daclatasvir, several other NS5A replication complex inhibitors have entered the clinic, including GS-5885, PPI-461, ABT-267, and GSK2336805 (7–10). In addition, preclinical data have recently been described for several other NS5A replication complex inhibitors, including EDP-239, IDX719, MK-4882, ITMN-9959, and ACH-3102 (11–15), highlighting the increased focus on this class of HCV DAAs.
GS-5885 is a novel NS5A replication complex inhibitor with 50% effective concentrations (EC50s) of 4 and 34 pM against genotype 1b (GT1b) and GT1a replicons, respectively (7). Through in vitro resistance selection experiments, GS-5885 selected NS5A Q30E and Y93H substitutions in GT1a and Y93H in GT1b; these mutations conferred high levels of reduced susceptibility to GS-5885 (16). A multiple-ascending-dose study was conducted in which GT1a HCV chronically infected, treatment-naive subjects were treated with GS-5885 for 3 days with 1, 3, 10, 30, or 90 mg once a day. An additional cohort of GT1b HCV-infected subjects treated with 10 mg of GS-5885 once a day was also assessed. In these subjects GS-5885 treatment resulted in median maximal reductions in HCV RNA ranging from 2.3 to 3.3 log10 IU/ml. This study describes the emergence of NS5A RAMs following 3-day GS-5885 monotherapy and the impact of baseline resistance variants, detected by population or deep sequencing of NS5A, on the clinical response. Our results support the further development of GS-5885 in combination with other DAAs with distinct mechanisms of action for the treatment of GT1 chronic HCV infection.
MATERIALS AND METHODS
Compounds.
The structures of the HCV NS5A inhibitor GS-5885 (John O. Link, James G. Taylor, Lianhong Xu, Michael Mitchell, Hongyan Guo, Hongtao Liu, Darryl Kato, Thorsten Kirschberg, Jianyu Sun, Neil Squires, Jay Parrish, Terry Keller, Zheng-Yu Yang, Chris Yang, Mike Matles, Yujin Wang, Kelly Wang, Guofeng Cheng, Yang Tian, Erik Mogalian, Elsa Mondou, Melanie Cornpropst, Jason Perry, and Manoj C. Desai, submitted for publication), sofosbuvir (GS-7977) (18), GS-9451 (19), GS-9256 (20), tegobuvir (GS-9190) (21), and daclatasvir (BMS-790052) (5) have been previously described. All compounds were synthesized at Gilead Sciences, Inc.
Study design.
Samples from this study were collected from a phase 1, multicenter, randomized, double-blind, placebo-controlled, dose-escalation study that included six cohorts: five cohorts included only subjects with GT1a HCV infection, and one cohort included subjects with only GT1b HCV infection (the study was registered at ClinicalTrials.gov under registration number NCT01193478). Doses of GS-5885 in individual cohorts were as follows: 1 mg, 3 mg, 10 mg (two cohorts, GT1a and -1b), 30 mg, and 90 mg. Each cohort had 12 subjects, 10 randomly assigned to active drug and 2 assigned to placebo. Because GS-5885 has a slightly more favorable in vitro resistance profile and potency in GT1b than in GT1a, this dose escalation study has focused on GT1a with only a 10-mg dose of GS-5885 to confirm the GT1b activity. The study protocol was approved by the institutional review boards or independent ethics committees at the participating sites prior to study initiation and was performed in accordance with Good Clinical Practice guidelines outlined by the International Conference on Harmonization. A more detailed description of the clinical study was previously described (7).
Viral sequencing.
Population sequencing of the HCV NS5A coding region was performed for all subjects at baseline (day 1 prior to dosing), on day 4, and on day 14, provided the HCV RNA level was greater than 1,000 IU/ml. All RNA isolations, amplifications by reverse transcription-PCR (RT-PCR), population sequencing, and deep sequencing were performed at Virco DBA (Virco, Belgium). Up to 1 ml of subject plasma sample was processed to isolate RNA. The full-length NS5A coding region was amplified in a nested PCR using genotype-specific primers. The pools of the PCR products were population sequenced using standard fluorescent dideoxy nucleotide sequencing methodology.
Deep sequencing was performed at baseline for the five subjects dosed at ≥3 mg of GS-5885 with less than a maximal 2.5-log10 reduction in HCV RNA IU/ml. The full-length NS5A coding region was amplified in a nested PCR using the same primers as for population sequencing. To maximize the number of input templates and to minimize variation due to PCR drift, each subject RNA sample was divided into seven aliquots, and seven parallel RT-PCRs were performed. The pool of PCR products was fragmented into smaller fragments (150 to 550 bp in length) that were pooled as equimolar concentrations and sequenced on a GS-FLX instrument according to the manufacturer's sequencing protocol (454 Life Sciences, Branford, CT).
For clonal sequencing of GT1a subjects dosed at ≥3 mg of GS-5885, day 4 amplicons from the population sequencing reactions were used as templates in amplification reaction mixtures with primers designed to amplify the first 444 bp of NS5A to ensure sufficient cloning efficiency and the maximal coverage of the diversity. The amplicons were cloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol and transformed into Escherichia coli, and individual clones were sequenced using T7 and T3 primers flanking the insertion site via a standard fluorescent dideoxy nucleotide sequencing methodology.
NS5A population sequences were aligned against respective subtype references (GT1a strain H77 [1a-H77], AF009606; GT1b strain Con1 [1b-Con1], AJ238799) to identify differences between subject and reference amino acid sequences. Analyses of the emerging amino acid changes at day 4 and day 14 compared to day 1 (baseline) were conducted and reported.
To analyze deep-sequencing data generated using the 454/Roche sequencing technology, a database-driven software package called DB454 was developed. The DB454 software was built using the Perl programming language and an Oracle 10g database and interfaced with the PyroMap sequence alignment tool (http://hivdb.stanford.edu/pages/resources.html). To eliminate the errors produced by the 454/Roche sequencing technology, algorithms were employed to exclude sequence reads with low-quality scores, homopolymer-related errors, or characteristics of primer-dimer formation. Sequence reads containing an ambiguous base call (N), any base with a quality score of <10, an average quality score of <25 across all bases, or a sequence length of <150 bp were excluded from the analysis. Sequence reads for HCV GT1a and GT1b plasmid controls were aligned to strain H77 (AF009606) and strain Con1 (AJ238799), respectively. For clinical samples, all sequence reads were aligned to the sample's baseline pretreatment sequence generated using standard population-based sequencing methods. After alignment, sequence positions with coverage of >100 reads were considered for mutation analysis. Any amino acid changes present in clinical samples in more than two reads were included in the results.
In vitro phenotypic and cross-resistance analysis.
An NS5A shuttle vector was created to phenotype clinical isolates of NS5A (see Fig. S1 in the supplemental material). To facilitate cloning of the NS5A gene from subject isolates, a silent AseI site was introduced 9 amino acids upstream from the start of the NS5A coding region. This was accomplished using site-directed mutagenesis wherein one nucleic acid base at the third position was altered but the coding amino acid was not changed. AseI was used as a 5′ cloning site for NS5A in the 1b-PI-RLuc replicon. A unique BclI site located 11 amino acids into the NS5B coding region was used as a 3′ cloning site. To generate a replication-defective parental vector, an EcoRI site was introduced 1,122 bp upstream from the 3′ end of the NS5A coding region. An endogenous EcoRI site is located in NS5A 441 bp downstream from the 5′ start of the NS5A coding region. The fragment between the two EcoRI sites was then excised, and the vector was religated to create a deletion within NS5A which rendered the replicon replication defective. Fragments containing NS5A coding amino acids at position −9 to +11 from each end of the NS5A coding regions were amplified from subject plasma samples using primers containing an AseI or BclI site. The subject isolate amplicons were digested with AseI and BclI and cloned in frame into the similarly digested NS5A shuttle vector. Successful cloning of a subject isolate restored replication competency to the NS5A shuttle vector. PCR products generated for NS5A sequencing by Janssen Diagnostics BVBA were used as templates in nested PCRs to generate gene cassettes encoding cloning sites at both ends. For HCV GT1a samples, the NS5A coding region was amplified using the forward primer 1a-5A-SV-F (5′-CAT TGG ATT AAT TCG GAG TGT ACC-3′) and the reverse primer 1a-5A-SV-R (5′-CAC GGG GTG ATC AGT GCG CCT GT-3′). For HCV GT1b samples, the forward primer 1b-5A-SV-F (5′-CCA GTG GAT TAA TGA GGA CTG CTC CA-3′) and the reverse primer 1b-5A-SV-R (5′-ATG GCG TGA TCA GGG CGC CTG TCC AT-3′) were used. Replicon RNA was transfected into Huh7-lunet cells according to previously described methods (22). Cell suspensions were seeded in 96-well plates at 10,000 cells/well and allowed to attach overnight. For EC50 determinations, compounds were serially diluted in 100% dimethyl sulfoxide (DMSO) and then added to the cells at a 1:200 dilution, achieving final concentrations of 0.5% DMSO and total volumes of 200 μl. Cells were cultured for 3 days at 37°C, after which Renilla luciferase activity was measured using a Renilla luciferase assay system (Promega, Madison, WI) with a Victor Luminometer (PerkinElmer, Waltham, MA).
NS5A resistance-associated mutants are defined as having amino acid substitutions at specific residues in the NS5A coding region that are different from those of the comparator 1a-H77 or 1b-Con1 reference sequences at specific residues. These residues are at amino acid positions 24, 30, 31, 58, 62, and 93 of the NS5A coding region. Site-directed mutants were created by gene synthesis (Genscript, Piscataway, NJ), followed by subcloning the mutant cassettes into the 1a-H77 replicon with flanking restriction sites.
RESULTS
Clinical response and detection of resistance mutants.
Three-day monotherapy with GS-5885 produced rapid declines in HCV RNA levels with median maximal reductions of >3 log10 IU/ml for all dosing cohorts except those receiving 1 mg of GS-5885 (Fig. 1) (7). In the absence of a baseline RAM, emergence of a resistance mutation was detected in all subjects dosed at >1 mg of GS-5885 with successful day 4 NS5A sequence data. Individual HCV RNA levels over time are depicted in Fig. S2 in the supplemental material for each subject.
Fig 1.
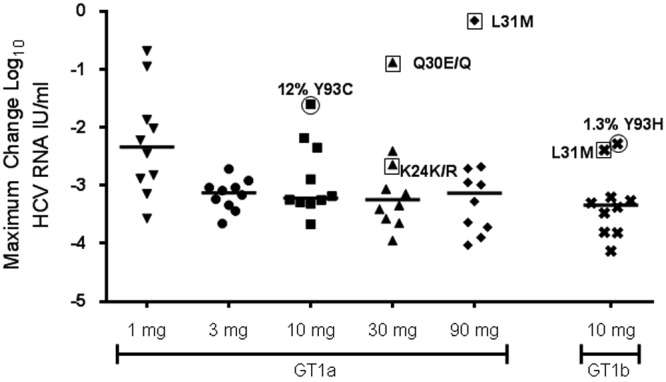
Maximal reduction in HCV RNA log10 IU/ml by cohort. The maximal change from baseline in HCV RNA is plotted for each individual subject receiving active GS-5885 in study GS-US-256-0102 (median indicated). NS5A was population sequenced for all subjects at baseline. Resistance-associated mutations detected at baseline by population sequencing are labeled and enclosed by a square. Circled subjects had resistance-associated mutations detected as minority variants by deep sequencing at baseline but not detected by population sequencing. The percentage of resistance variants present at baseline is indicated for the circled patients.
Characterization of NS5A sequence at baseline.
Viral population sequencing was performed on all baseline subject samples (day 1 prior to dosing). Subjects in the 3-, 10-, 30-, or 90-mg cohort were examined for the presence of NS5A amino acid changes previously identified as being associated with reduced susceptibility to GS-5885 (Table 1). Eight of 62 subjects randomized to receive 3, 10, 30, or 90 mg of GS-5885 or placebo had NS5A RAMs at baseline, as detected by population or deep sequencing (Table 1 and Fig. 1). The L31M variant was the most prevalent, observed in 4/8 subjects (two GT 1a and two GT 1b) with RAMs detected at baseline. Of the other four subjects with baseline RAMs, one had a Q30E/Q mixture (GT1a) by population sequencing, another had a mixture of K24K/R by population sequencing, and the other two had either 12% Y93C (GT1a) or 1.3% Y93H (GT1b) detected by deep sequencing. Among these eight subjects, six (four GT1a and two GT1b) received GS-5885, and two received placebo. All six subjects that received GS-5885 monotherapy had smaller decreases in viral load than the rest of the cohort, but two GT1b subjects with the L31M variant or 1.3% Y93H still demonstrated >2-log10 decreases in HCV RNA levels after receiving 10 mg of GS-5885 (Table 1 and Fig. 1).
Table 1.
Drug resistance mutations detected at baseline by population sequencing
| Subject | GT | Dose (mg)a | Sequencing method | Baseline RAM(s)b | Max VL reduction (log10 HCV RNA IU/ml) in:c |
|
|---|---|---|---|---|---|---|
| Subject | Cohort | |||||
| AS | 1b | 10 | Population | L31M | 2.09 | 3.31 |
| X | 1a | 30 | Population | K24K/R | 2.64 | 3.01 |
| AC | 1a | 30 | Population | Q30E/Q | 0.88 | 3.01 |
| BM | 1a | 90 | Population | L31M | 0.16 | 3.01 |
| Q | 1a | 10 | 454 | Y93C (12%) | 1.60 | 3.09 |
| AU | 1b | 10 | 454 | Y93H (1.3%) | 2.28 | 3.31 |
| AZ | 1a | PBO | Population | L31L/M | 0.25 | 0.03 |
| BH | 1b | PBO | Population | L31M | 0.15 | 0.03 |
PBO, placebo.
Resistance-associated mutations (RAMs) are defined as any substitution occurring at NS5A amino acid positions 24, 25, 28, 30, 31, 58, 62, and 93.
Max, maximum; VL, viral load.
Population sequencing of NS5A following 3 days of GS-5885 monotherapy.
Population sequencing of the NS5A coding region in GT1a-infected subjects at days 4 (approximately 24 h after the last dose of GS-5885) and 14 revealed complex mixtures of treatment-emergent NS5A RAMs (Table 2; see also Table S1 in the supplemental material). Three of 10 subjects dosed with 1 mg of GS-5885 had detectable RAMs at position 30 or 31 at day 4 or 14. One subject in the 1-mg cohort showed a loss of A25A/T from the day 4 to day 14 time point. One other subject in this cohort without a RAM detected at day 4 developed a Q30Q/R mutant mixture at day 14. No change was detected for the remaining seven subjects at day 4 or 14.
Table 2.
Frequency and spectrum of resistance-associated mutations in all subjects receiving active drug at day 4
| NS5A mutation(s) | No. of subjects with GS-5885 RAMs at day 4 by cohort and dosea |
|||||
|---|---|---|---|---|---|---|
| GT1a |
GT1b (10 mg [n = 6]) | |||||
| 1 mg (n = 10) | 3 mg (n = 10) | 10 mg (n = 8) | 30 mg (n = 8) | 90 mg (n = 9) | ||
| K24E/K | 1 (−2.6) | |||||
| A25A/T | 1 (−0.7) | |||||
| M28M/T | 1 (−3.4) | |||||
| Q30Q/R | 1 (−3.2) | 1 (−2.9) | 1 (−3.6) | 2 (−3.7, −2.7) | ||
| Q30H/Q/R | 3 (−3.1 to −3.3) | 1 (−3.2) | ||||
| E/Q30E | 1 (−0.9)c | |||||
| L31M | 2 (−1.5, −3.0)b | |||||
| L31L/M | 1 (−3.2) | 1 (−2.2) | ||||
| Y93C | 1 (−1.6) | |||||
| Y93H | 2 (−2.3, −3.2) | |||||
| Y93H/Y | 1 (−3.1) | 1 (−3.5) | ||||
| K24K/N, Q30H/Q/R | 1 (−3.0) | |||||
| K24K/R, Q30Q/R | 1 (−3.7) | |||||
| K/R24R, L31L/M | 1 (−2.6) | |||||
| M28M/T, Q30Q/R | 1 (−3.0) | |||||
| Q30Q/R, L31L/M | 1 (−3.1) | |||||
| Q30H/Q, A92A/V | 1 (−3.7) | |||||
| Q30Q/R, H58D/H | 1 (−3.6) | |||||
| Q30Q/R, Y93C/Y | 1 (−3.3) | 2 (−3.6, −3.0) | ||||
| Q30K/Q/R, Y93C/H/R/Y | 1 (−3.6) | |||||
| Q30E/G/Q/R, L31L/M | 1 (−3.3) | |||||
| L31M, Y93H | 1 (−2.4) | |||||
| L31L/M, H58H/P | 1 (−2.8) | |||||
| L31L/M, Y93C/Y | 1 (−3.3) | |||||
| L31L/M, Y93H/Y | 1 (−2.7) | |||||
| P/S58P, Y93H | 1 (−4.1) | |||||
| E/Q62E, Y93H | 1 (−3.8) | |||||
| M28M/T, Q30Q/R, L31L/M | 1 (−3.2) | |||||
| M28M/V, Q30Q/R, Y93C/Y | 1 (−2.9) | |||||
| L31M, H/P58P, D/E62D | 1 (−2.4) | |||||
| Q30Q/R, L31L/M, Y93C/Y | 1 (−3.7) | |||||
| Total (% of group) | 3 (30) | 10 (100) | 8 (100) | 8 (100) | 9 (100) | 6 (100) |
Change from baseline as determined by population sequencing. For each cohort, n is the number of subjects with available sequence data at day 4. The absence of data indicates that the mutation(s) was not detected. Except as noted, values in parentheses represent the maximum log10 change in HCV RNA.
One of these two subjects harbored L31M at baseline and on day 4.
Harbored Q30E/Q at baseline.
In contrast, emergent RAMs were detected in 100% of subjects dosed with ≥3 mg of GS-5885 with available day 4 population sequencing (Table 2). HCV variant A25T, M28T, or Q30H was not detected in any GT1a subject receiving the highest doses of GS-5885 (30 or 90 mg). The most common mutation detected at day 4 was Q30R (57.9%), followed by L31M (34.2%).
The major RAM detected at day 4 in subjects infected with GT1b HCV was Y93H. At day 4, 6/10 subjects receiving active drug were successfully sequenced, and all six of these subjects harbored the Y93H mutation. All 10 subjects infected with GT1b HCV, who received 10 mg of GS-5885, harbored Y93H following treatment (Table 2), consistent with Y93H being the primary GS-5885 resistance mutation selected in the GT1b replicon (data not shown).
Clonal sequencing of day 4 GT1a NS5A clinical isolates.
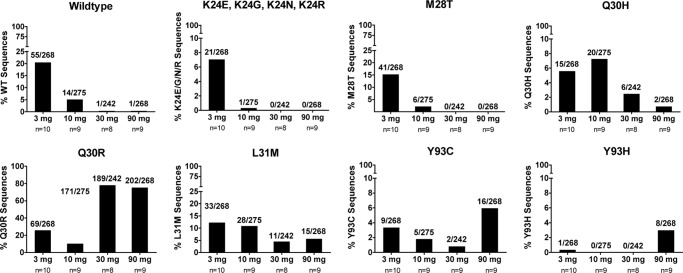
Population sequencing of NS5A following GS-5885 monotherapy often showed multiple mutations in a subject plasma sample (Table 2), yet their relative frequency with one another and their presence on the same genome of viral RNA cannot be resolved by this sequencing method. To determine the frequency of individual mutants and to assess the linkage of these mutations in a given sample, the day 4 samples with population sequence data were also analyzed by clonal sequencing for GT1a subjects dosed at ≥3 mg of GS-5885. Approximately 30 clones per sample were sequenced for every subject with available amplicons from the population-sequencing reactions. Subjects with RAMs identified by population sequencing at baseline were excluded from this analysis since the RAM frequencies following therapy were reflective of the combination of the preexisting and newly selected RAMs. Consequently, 35 subjects remained for determination of day 4 clonal mutant frequencies, for a total of 1,053 clones analyzed. The percentage of wild-type variants, defined as having no substitutions at NS5A positions 24, 25, 28, 30, 31, 58, 62, and 93 compared to the 1a-H77 NS5A sequence, declined as the dose increased. A similar trend was observed for variants having either a K24E, -G, -N, or -R substitution, as well as the M28T and L31M mutations. The frequency of Q30H mutations in clonal sequencing analysis rose slightly when doses increased from 3 to 10 mg but declined at the 30- and 90-mg doses (Fig. 2).
Fig 2.
Clonal frequency of single-substitution resistance-associated mutations in genotype 1a-infected patients at day 4 dosed at ≥3 mg of GS-5885. TOPO TA cloning and subsequent sequencing of inserts were performed from amplicons used in population-sequencing reactions. “Wildtype” indicates no mutations at NS5A positions 24, 25, 28, 30, 31, 58, 62, and 93 compared to the 1a-H77 reference strain. K24E, K24N, and K24R mutations were combined into one category. For all graphs, n = number of patients in the dosing cohort from which clonal sequences were derived.
The Q30R variant was the most frequent GT1a RAM detected by clonal sequencing. Its frequency increased over the 3- to 30-mg dosing cohorts, peaking at 89% of clones sequenced from the 30-mg cohort, and declined slightly to 75% of clones sequenced from the 90-mg cohort. In contrast, the Y93C variant was present at lower overall frequencies, ranging from 0.9% to 3.4% over the 3-, 10-, and 30-mg cohorts, and increased to 6.8% of clones in subjects dosed at 90 mg.
Linkage of RAMs was also determined at day 4 by clonal sequencing. The double mutants were observed in only 13 out of 35 subjects. Among the 13 subjects with double mutants, the frequencies of double mutants were lower than those of single mutants in every subject and ranged from 3% to 13% of clones (one to four clones). The identities of these double mutations were variable, but among them, the most frequent double mutant detected was the combination of Q30R and Y93C detected at 1.8% (18/1,053).
Phenotypic analysis of clinical isolates.
To assess the impact of the identified RAMs on GS-5885 susceptibility, phenotypic analysis of clinical isolates was performed by cloning pools of full-length NS5A baseline and day 4 amplicons into an NS5A shuttle vector (see Fig. S1 in the supplemental material), transfecting into Huh7-lunet cells, and determining the GS-5885 EC50. Approximately 85% of the recombinant replicons replicated in vitro at levels sufficient for phenotypic analysis. Table 3 shows the baseline and day 4 EC50s for a group of subject isolates with RAMs identified at day 4. The day 4 resistance mutations and their frequencies detected by clonal analysis are listed to more accurately describe the composition of mutant pools being tested for each sample. Even with a variety of RAM patterns, all of the day 4 samples showed reduced susceptibility to GS-5885 compared to the pretreatment baseline sample. The fold change in day 4 GS-5885 EC50s from baseline varied from 20.4-fold to >12,500-fold (Table 3) among these subjects.
Table 3.
Phenotypic analysis of subject isolates
| GT and subject | Baseline RAM | Clonal NS5A variant(s) at day 4 (frequency)a | GS-5885 EC50 (nM)b |
Daclatasvir EC50 (nM)b |
||||
|---|---|---|---|---|---|---|---|---|
| Baseline | Day 4 | Fold change | Baseline | Day 4 | Fold change | |||
| 1a | ||||||||
| O | WT (10%), M28T (3%), Q30H (23%), Q30R (53%), L31M (3%), Y93C (7%) | 0.002 | 0.071 | 35.5 | 0.008 | >1.5 | >196 | |
| I | WT (44%), Q30R (31%), L31M (25%) | 0.005 | 0.403 | 88 | 0.007 | 2.07 | 292 | |
| BJ | Q30H (3%), Q30R (67%), Q30E (3%), H58D (3%), Y93C (7%), Y93H (7%), Q30R+Y93C (10%) | 0.017 | 4.3 | 355.6 | 0.003 | 0.8 | 267 | |
| L | WT (13%), M28T (7%), Q30H (7%), Q30R (7%), L31M (33%), Y93C (3%), Q30R+Y93C (10%) | 0.004 | >1.5 | >375 | 0.005 | >1.5 | >288 | |
| BR | Q30R (86%), L31M (4%), H58D (11%) | 0.009 | 3.5 | 389 | 0.007 | 2.1 | 300 | |
| T | WT (3%), Q30R (40%), L31M (3%), H58D (47%), Q30R+H58D (7%) | 0.006 | 0.52 | 87 | 0.003 | 0.49 | 163 | |
| BI | Q30R (87%), Q30E (3%), K24R+Q30R (10%) | 0.006 | >50 | >8,333 | 0.006 | 22.2 | 3700 | |
| BK | Q30K (7%), Q30R (66%), Y93C (14%), Y93H (7%), Q30R+Y93H (3%), Q30R+Y93C (3%) | 0.004 | >50 | >12,500 | 0.004 | >50 | >12,500 | |
| Q | 12% Y93C | M28T (3%), Q30R (26%), L31M (7%), Y93C (52%), M28V+Q30R (3%), M28T+Y93C (7%), Q30R+Y93C (3%) | 0.014 | 0.285 | 20.4 | 0.057 | 0.487 | 8.5 |
| 1b | ||||||||
| AR | Y93Hc | 0.001 | 0.994 | 994 | 0.013 | 0.12 | 9.6 | |
Clonal variants were determined by deep sequencing.
Fold change in the EC50 was calculated by dividing the day 4 EC50 by the baseline EC50.
Genotype 1b sample, day 14 population sequencing EC50.
In GT1b subjects, the Y93H mutation was detected in all subjects following 3-day 10-mg GS-5885 monotherapy. Several subject samples were unable to be analyzed by population sequencing at day 4 due to low viral loads but were successful at day 14. Phenotyping of day 4 or 14 Y93H samples from three GT1b subjects showed shifts in the EC50s of >990-fold compared to baseline or, in cases of unsuccessful baseline phenotyping, compared to wild-type 1b-Con1 EC50s (Table 3). All successfully cloned subject baseline EC50s (n = 2) were <2-fold different from the 1b-Con1 reference EC50.
Cross-resistance analysis of mutants versus other HCV inhibitors.
To determine if NS5A mutations detected at day 4 conferred reduced susceptibility to other HCV inhibitors, the EC50 was determined for baseline and day 4 subject isolates. Results are shown in Table 3 indicating that emergent RAMs detected after treatment with GS-5885 are cross-resistant to the NS5A inhibitor daclatasvir (BMS-790052). However, no cross-resistance was observed with these mutants when assayed against other classes of DAAs such as protease inhibitors (PIs), nonnucleoside inhibitors (NNIs), nucleoside inhibitors (NIs), interferon alpha, or ribavirin (data not shown).
Phenotypic analysis of subject isolates in a replicon-based assay has limitations due to the various contributions to the average EC50 value when mixtures of mutants and/or wild-type species are analyzed. To better assess the magnitude of reduced susceptibility of specific GT1a mutations, a series of NS5A site-directed mutants was created on the 1a-H77 replicon for phenotypic and cross-resistance analyses (Table 4). These NS5A mutations were identified from the population sequencing of clinical samples and were assessed for in vitro susceptibility to GS-5885, daclatasvir (Table 4), and other classes of antivirals (data not shown). With the exception of M28V, the EC50s for these mutants showed reduced susceptibilities to GS-5885 and daclatasvir. No reduced susceptibility to GS-7977 (NI), GS-9451 (PI), GS-9256 (PI), GS-910 (NNI), or ribavirin was detected with this panel of NS5A mutants (data not shown).
Table 4.
GS-5885 and daclatasvir EC50s and fold shifts to NS5A resistance-associated mutations
| NS5A strain or mutant | GS-5885 |
Daclatasvir |
||
|---|---|---|---|---|
| Mean EC50 ± SD (nM)a | EC50 fold changeb | Mean EC50 ± SD (nM)a | EC50 fold changeb | |
| 1a-H77 | 0.051 ± 0.044 | 1.0 | 0.023 ± 0.017 | 1.0 |
| M28V | 0.078 ± 0.011 | 0.6 ± 0.08 | 0.02 ± 0.001 | 1.1 ± 0.13 |
| M28T | 1.801 ± 0.77 | 61 ± 8.2 | 8.85 ± 2.0 | 390 ± 125 |
| Q30H | 5.301 ± 2.55 | 183 ± 51 | 7.03 ± 0.5 | 309 ± 32.8 |
| Q30R | 12.42 ± 10.44 | 632 ± 176 | 5.73 ± 0.2 | 252 ± 15.1 |
| Q30E | 44.05 ± 14.71 | 5,458 ± 1,166 | 85.53 ± 16.8 | 3,764 ± 1,045 |
| L31M | 14.61 ± 3.97 | 554 ± 65 | 5.81 ± 1.5 | 197 ± 25 |
| H58D | 32.73 ± 8.63 | 1,127 ± 101 | 2.68 ± 0.01 | 118 ± 0.6 |
| Y93C | 48.72 ± 10.22 | 1,602 ± 372 | 12.28 ± 2.4 | 540 ± 149 |
| Y93H | 86.43 ± 21.69 | 1,677 ± 603 | 73.94 ± 28.6 | 2,082 ± 121 |
| Y93N | >500 | >14,706 | >500 | >32,000 |
| Q30R+Y93C | 56.45 ± 5.55 | 1,046 ± 391 | 229.14 ± 222.7 | 4,213 ± 646 |
The EC50 values and standard deviations (SD; n ≥ 2) for site-directed mutants in a 1a-H77 replicon were determined for GS-5885 and daclatasvir.
Fold change in EC50 is calculated relative to the baseline EC50.
DISCUSSION
These analyses were performed to identify HCV NS5A mutations potentially associated with resistance to GS-5885 from a 3-day monotherapy study and to correlate the effects of these mutations with the antiviral response to GS-5885. This monotherapy clinical trial provided a unique opportunity to examine GS-5885 resistance mutations and clinical response without the confounding effects of other antiviral agents.
In general, the majority of subjects from this study dosed at 3 mg or higher responded with >2.5-log10 reductions in their HCV RNA levels. Retrospective sequencing identified baseline RAMs by either population or deep sequencing in several subjects dosed with ≥10 mg of GS-5885. This is a potential explanation for the relatively low variability in maximal HCV RNA reduction observed in subjects in the 3-mg cohort (Fig. 1). For the other cohorts, subjects with the lower HCV RNA reductions harbored baseline RAMs, with one exception. Among the subjects with baseline RAMs, two subjects, one GT1a (subject Q) and one GT1b (subject AU), harbored RAMs detected only by deep sequencing and had maximal viral load drops of 1.9 log10 (12% Y93C variants at baseline) and 2.3 log10 (1.3% Y93H) (see Fig. S2 in the supplemental material). Our data indicate that even a minority population of resistance mutations present at 1.3% of the viral quasispecies can result in a suboptimal clinical response in a monotherapy study, provided that the resistance mutant has a high degree of reduced susceptibility.
In contrast, one subject had a 2.2-log10 HCV RNA viral load drop without a RAM detected at baseline. Drug exposure for this subject was comparable to that of others in the dosing cohort (data not shown); therefore, it is possible that this subject had baseline RAMs below 1% of the deep-sequencing cutoff but slightly higher than those of other subjects in the same cohort. Nevertheless, our data indicate that the presence of baseline RAMs correlates with a suboptimal response over a 3-day monotherapy study. Further studies are needed to determine the impact of the NS5A baseline RAMs on the treatment outcome in clinical trials investigating combination therapies of GS-5885 with other classes of DAAs. Whether there is a need for resistance testing (probably sequencing) in subjects being considered for therapy with an NS5A inhibitor would be dependent on the treatment outcome of the combination regimes in subjects with NS5A baseline RAMs.
Population sequencing of all GT1a subjects in this study at days 4 and 14 revealed that the mutation with the broadest representation across subjects and dosing cohorts is Q30R (7) (see Table S1 in the supplemental material) and was often detected with other RAMs. The resolution of population sequencing does not allow one to quantify the relative percentage of a particular mutation with respect to others in a pool of sequences, nor does it provide linkage information for mutations detected in the same sample. To address this in GT1a, clonal sequencing of the first 444 bp of NS5A at day 4 was performed and showed that the single Q30R mutation was the most frequent mutation, reaching a peak of 89.2% of clones sequenced at day 4 for the 30-mg dosing cohort. This percentage dropped to 75.4% in the 90-mg cohort and likely reflects increased drug pressure selecting for mutants with greater reduced susceptibility. This is indeed what we observed as single Y93C and Y93H mutant clones were detected at the highest frequencies in the 90-mg cohort even though these frequencies were still low (6.8% and 3.4%, respectively) compared to the frequency of the Q30R mutant.
Clonal analysis of day 4 samples revealed an inverse relationship of detection of substitutions at K24, M28T, Q30H, and L31M with increasing GS-5885 dose (Fig. 2). In contrast, the Q30R and Y93C/H clonal frequencies trended higher as the dose increased. In general, mutations with lower fold shifts in EC50s were detected more frequently at the lower doses while higher GS-5885 doses increased the frequency of higher EC50 fold shift mutations such as Y93 substitutions. Twenty-four hours after the third and final dose (day 4, 72 h) the vast majority of sequences detected by clonal analysis were single Q30R variants. Since Q30R is a common emergent mutant following treatment with GS-5885 and daclatasvir, it would be desirable to identify new NS5A inhibitors which are active against Q30R and other common RAMs.
Genetically linked double resistance mutations were detected by clonal sequencing in 13/35 subjects. This frequency of double mutants is much lower than for single mutants for every subject (Table 5). Among the linked double mutants detected at day 4, 40.9% (18/44) of them were Q30R Y93C double mutants. The observation of a much lower frequency of double mutants than single mutants is consistent with the lower probability of two nucleotide changes for double mutants than a single nucleotide change for the single mutants. It is widely accepted that the two required nucleotide changes pose a higher genetic barrier.
Table 5.
Distribution of mutations in genotype 1a patients at day 4 treated with GS-5885
| Cohort (dose) and patienta | No. of clones (%) with:b |
||
|---|---|---|---|
| Wild type | Single mutation | Double mutation | |
| 3 mg | |||
| A | 17 (53) | 15 (47) | |
| B | 3 (10) | 24 (77) | 4 (13) |
| C | 30 (100) | ||
| D | 7 (26) | 19 (70) | 1 (4) |
| E | 13 (43) | 16 (54) | 1 (3) |
| F | 5 (26) | 14 (74) | |
| G | 5 (31) | 11 (69) | |
| H | 9 (29) | 22 (71) | |
| I | 14 (44) | 18 (56) | |
| J | 4 (20) | 16 (80) | |
| 10 mg | |||
| K | 31 (100) | ||
| L | 4 (13) | 23 (77) | 3 (10) |
| M | 32 (100) | ||
| N | 3 (10) | 26 (90) | |
| O | 3 (10) | 27 (90) | |
| P | 5 (17) | 22 (73) | 3 (10) |
| R | 1 (3) | 30 (97) | |
| S | 27 (90) | 3 (10) | |
| T | 1 (3) | 27 (90) | 2 (7) |
| 30 mg | |||
| U | 29 (100) | ||
| V | 25 (89) | 3 (11) | |
| X | 32 (100) | ||
| Y | 1 (3) | 29 (97) | |
| Z | 31 (100) | ||
| AA | 32 (100) | ||
| AC | 31 (97) | 1 (3) | |
| 90 mg | |||
| BI | 31 (100) | ||
| BJ | 27 (90) | 3 (10) | |
| BK | 27 (93) | 2 (7) | |
| BL | 30 (97) | 1 (3) | |
| BN | 29 (100) | ||
| BO | 32 (100) | ||
| BP | 1 (3.5) | 26 (93) | 1 (3.5) |
| BQ | 28 (100) | ||
| BR | 28 (100) | ||
Patients with resistance-associate mutations detected at baseline by population sequencing were excluded from analysis.
No triple mutations were detected.
Phenotypic analysis of clinical isolates from this study showed reduced susceptibility to GS-5885 following 3-day monotherapy. In HCV GT1a, these samples generally consisted of mixtures of resistance mutations at multiple sites as well as some remaining wild-type variants, particularly for the subjects treated with ≤3 mg of GS-5885. In GT1b, phenotypic analysis showed reduced susceptibility of clinical isolates containing the Y93H mutation, consistent with an EC50 of 5.3 nM (1,319-fold shift in EC50 versus 1b-Con1) for a site-directed Y93H mutant (7). The magnitude of reduced susceptibility among GT1b Y93H mutants likely varies in the context of the clinical isolate sequence as well as the presence of a mixture of quasispecies in the analysis. However, the extent to which Y93H would be suppressed at 30 or 90 mg of GS-5885 is yet to be determined in GT1b HCV infections because GS-5885 was tested only at 10 mg in this subject population.
Evaluation of clinical isolates with RAMs showed cross-resistance to the NS5A replication complex inhibitor daclatasvir. These data are consistent with the resistance profile previously reported for daclatasvir (5, 23). However, no cross-resistance was observed when the same isolates were assayed against DAAs targeting the NS3 protease or NS5B polymerase. Furthermore, these isolates remained sensitive to alpha interferon and ribavirin.
Cross-resistance analysis with clinical isolates and site-directed mutant replicons showed that none of the NS5A mutants conferred resistance to sofosbuvir (formerly GS-7977), GS-9451, ribavirin, or interferon alpha and ribavirin, suggesting that GS-5885 would be a suitable candidate for combination therapy with these DAAs as well as alpha interferon and ribavirin.
Combination therapy with these other classes of HCV inhibitors would raise the resistance barrier over treating subjects with GS-5885 alone. For example, treatment with a potent NI in combination with GS-5885 could suppress the NS5A mutations driven by GS-5885 pressure. Conversely, GS-5885 would suppress the NI resistance-associated HCV variants. Clinically relevant resistance would likely be genetically linked RAMs in both NS5B and NS5A in order for the virus to overcome the drug pressure. Finally, the fitness of a dual-class mutation may be reduced, thus lowering the frequency of viral rebound during combination therapy. With high potency and the potential for combination therapy against GT1 HCV infection, GS-5885 is currently in multiple phase 2 clinical studies in combination with DAAs with distinct mechanisms of action with or without alpha interferon and ribavirin.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Gilead Sciences, Inc.
We thank the members of the clinical study team, the subjects who participated in this study, and the medicinal chemistry group for providing test compounds.
Footnotes
Published ahead of print 22 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02193-12.
REFERENCES
- 1. Shepard CW, Finelli L, Alter MJ. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558–567 [DOI] [PubMed] [Google Scholar]
- 2. Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R, George J, Rizzetto M, Shouval D, Sola R, Terg RA, Yoshida EM, Adda N, Bengtsson L, Sankoh AJ, Kieffer TL, George S, Kauffman RS, Zeuzem S. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 364:2405–2416 [DOI] [PubMed] [Google Scholar]
- 3. Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, DiNubile MJ, Sniukiene V, Brass CA, Albrecht JK, Bronowicki JP. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun JH, O'Boyle DR, II, Lemm JA, Wang C, Knipe JO, Chien C, Colonno RJ, Grasela DM, Meanwell NA, Hamann LG. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fridell RA, Wang C, Sun JH, O'Boyle DR, II, Nower P, Valera L, Qiu D, Roberts S, Huang X, Kienzle B, Bifano M, Nettles RE, Gao M. 2011. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54:1924–1935 [DOI] [PubMed] [Google Scholar]
- 7. Lawitz EJ, Gruener D, Hill JM, Marbury T, Moorehead L, Mathias A, Cheng G, Link JO, Wong KA, Mo H, McHutchison JG, Brainard DM. 2012. A phase 1, randomized, placebo-controlled, three-day, dose-ranging study of GS-5885, an NS5A inhibitor, in patients with genotype 1 hepatitis C. J. Hepatol. 57:24–31 [DOI] [PubMed] [Google Scholar]
- 8. Huang Q, Huang N, Lau M, Bencsik M, Huq A, Peng E, Agarwal K, Lalezari J, Vig P, Brown N, Colonno R. 2011. Resistance monitoring of HCV patients treated for three days with the NS5A inhibitor PPI-461 reveals rapid emergence of resistant HCV variants. Hepatology 54(Suppl 1):536A [Google Scholar]
- 9. Spreen W, Wilfret DA, Bechtel J, Adkinson KK, Lou Y, Jones L, Willsie SK, Glass SJ, Roberts CD. 2011. GSK2336805 HCV NS5A inhibitor demonstrates potent antiviral activity in chronic genotype 1 infection; results from a first time in human (FTIH) single and repeat dose study. Hepatology 54(Suppl 1):400–401A [Google Scholar]
- 10. Dumas EO, Lawal A, Menon RM, Podsadecki T, Awni W, Dutta S, Williams L. 2011. Pharmacokinetics, safety and tolerability of the HCV NS5A inhibitor ABT-267 following single and multiple doses in healthy adult volunteers. J. Hepatol. 54(Suppl 1):S475–S476 [Google Scholar]
- 11. Jiang LJ, Liu S, Phan T, Owens C, Brasher B, Polemeropoulos A, Luo X, Hoang K, Wang M, Peng X, Cao H, Qiu Y, Or YS. 2011. A highly potent HCV NS5A inhibitor EDP-239 with favorable preclinical pharmacokinetics. J. Hepatol. 54(Suppl 1):S479 [Google Scholar]
- 12. Dousson CB, Chapron C, Standring D, Bilello JP, McCarville J, La Colla M, Seifer M, Parsy C, Dukhan D, Pierra C, Surleraux P. 2011. Idenix NS5A HCV replication inhibitors with low picomolar, pan-genotypic in vitro antiviral activity. J. Hepatol. 54(Suppl 1):S326–S327 [Google Scholar]
- 13. Ludmerer S, Fandozzi CM, Huang Q, Kargman S, Liu R, Meinke P, Olsen DB, Coburn C. 2011. Discovery of MK-4882, a novel inhibitor of HCV NS5A with an attractive pre-clinical profile. Hepatology 54(Suppl 1):531A–532A [Google Scholar]
- 14. Nicholas JB, Buckman BO, Serebryany V, Schaefer CJ, Kossen K, Ruhrmund D, Hooi L, Aleskovski N, Arfsten A, Lim SR, Pan L, Huang L, Rajagopalan R, Misialek S, Seiwert S. 2011. Characterization of novel, highly potent NS5A inhibitors with QD dosing potential and robust activity in an HCV chimeric animal model. J. Hepatol. 54(Suppl 1):S485 [Google Scholar]
- 15. Yang G, Zhao Y, Patel D, Fabrycki JL, Marlor C, Rivera J, Stauber K, Wiles J, Gadhachanda V, Hashimoto A, Chen D, Wang Q, Pais G, Wang X, Deshpande A, Huang M. 2011. Novel hepatitis C virus NS5A inhibitors with improved potency against genotype-1a replicons and replicons carrying mutations associated with viral resistance to 1st generation NS5A inhibitors. Hepatology 54(Suppl 1):537A [Google Scholar]
- 16. Cheng G, Peng B, Corsa A, Yu M, Nash M, Lee YJ, Xu Y, Kirschberg T, Tian Y, Taylor J, Link J, Delaney W. 2012. Antiviral activity and resistance profile of the novel HCV NS5A inhibitor GS-5885. J. Hepatol. 56(Suppl 2):S464 [Google Scholar]
- 17.Reference deleted.
- 18. Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang P, Zhang HR, Bansal S, Espiritu C, Keilman M, Lam AM, Steuer HM, Niu C, Otto MJ, Furman PA. 2010. Discovery of a beta-d-2′-deoxy-2′-alpha-fluoro-2′-beta-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J. Med. Chem. 53:7202–7218 [DOI] [PubMed] [Google Scholar]
- 19. Sheng XC, Appleby T, Butler T, Cai R, Chen X, Cho A, Clarke MO, Cottell J, Delaney WE, IV, Doerffler E, Link J, Ji M, Pakdaman R, Pyun HJ, Wu Q, Xu J, Kim CU. 2012. Discovery of GS-9451: an acid inhibitor of the hepatitis C virus NS3/4A protease. Bioorg. Med. Chem. Lett. 22:2629–2634 [DOI] [PubMed] [Google Scholar]
- 20. Sheng XC, Casarez A, Cai R, Clarke MO, Chen X, Cho A, Delaney WE, IV, Doerffler E, Ji M, Mertzman M, Pakdaman R, Pyun HJ, Rowe T, Wu Q, Xu J, Kim CU. 2012. Discovery of GS-9256: a novel phosphinic acid derived inhibitor of the hepatitis C virus NS3/4A protease with potent clinical activity. Bioorg. Med. Chem. Lett. 22:1394–1396 [DOI] [PubMed] [Google Scholar]
- 21. Shih IH, Vliegen I, Peng B, Yang H, Hebner C, Paeshuyse J, Purstinger G, Fenaux M, Tian Y, Mabery E, Qi X, Bahador G, Paulson M, Lehman LS, Bondy S, Tse W, Reiser H, Lee WA, Schmitz U, Neyts J, Zhong W. 2011. Mechanistic characterization of GS-9190 (Tegobuvir), a novel nonnucleoside inhibitor of hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 55:4196–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 23. Fridell RA, Qiu D, Wang C, Valera L, Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 54:3641–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.