Abstract
AAC(6′)-Ib-cr is a plasmid-mediated quinolone resistance mechanism described worldwide for Escherichia coli. Since it confers in vitro only a low level of resistance to ciprofloxacin, we evaluated its impact on the in vivo activity of ciprofloxacin. Isogenic strains were obtained by transferring plasmid p449, harboring aac(6′)-Ib-cr, into the quinolone-susceptible strain E. coli CFT073-RR and its D87G gyrA mutant. MICs were 0.015, 0.06, 0.25, and 0.5 μg/ml against E. coli strains CFT073-RR, CFT073-RR/p449, CFT073-RR GyrAr, and CFT073-RR GyrAr/p449, respectively. Bactericidal activity was reduced at 1× the MIC for the three resistant derivatives, while at a fixed concentration of 0.5 μg/ml, 99.9% killing was observed for all strains except E. coli CFT073-RR GyrAr/p449. In the murine model of pyelonephritis, an optimal regimen of ciprofloxacin (10 mg/kg of body weight twice a day [b.i.d.]) significantly decreased the bacterial count in the kidneys of mice infected with E. coli CFT073 (1.6 versus 4.3 log10 CFU/g of kidney compared to untreated controls; P = 0.0001), while no significant decrease was observed for E. coli CFT073-RR/p449 (2.7 versus 3.1 log10 CFU/g; P = 0.84), E. coli CFT073-RR GyrAr (4.2 versus 4.1 log10 CFU/g; P = 0.35), or E. coli CFT073-RR GyrAr/p449 (2.9 versus 3.6 log10 CFU/g; P = 0.47). While pharmacokinetic and pharmacodynamic (PK/PD) parameters accounted for ciprofloxacin failure against gyrA-containing mutants, this was not the case for the aac(6′)-Ib-cr-containing strains, suggesting an in situ hydrolysis of ciprofloxacin in the latter case.
INTRODUCTION
Fluoroquinolones are currently among the most heavily prescribed antimicrobials in the world because of their spectrum, their pharmacokinetic properties, and their generally good tolerability (1). They are particularly useful for treating urinary tract infections (UTI) due to Enterobacteriaceae (1). As a consequence of their popularity, quinolone resistance rates have increased significantly over recent years (2, 3).
Most quinolone resistance mechanisms are chromosome mediated due to mutations in the genes encoding type II topoisomerases or affecting permeability or efflux (1, 4). Plasmid-mediated quinolone resistance (PMQR) genes were described over a decade ago (5); qnr genes were first described in 1998 (6). Qnr proteins are pentapeptide repeat proteins that protect DNA gyrase and topoisomerase IV from quinolone binding. Two plasmid-mediated efflux pumps have been identified: QepA, belonging to the major facilitator superfamily (MFS) (7), and OqxAB, belonging to the resistance-nodulation-cell division superfamily (RND) (8). The last PMQR gene, aac(6′)-Ib-cr, encodes a bifunctional aminoglycoside 6′-N-acetyltransferase capable of acetylating both aminoglycosides and fluoroquinolones (5, 9). This gene is a variant of the classic aac(6′)-Ib gene, which confers resistance to certain aminoglycosides (amikacin, isepamicin, and tobramycin). The new AAC(6′)-Ib-cr enzyme includes two mutations (Trp102Arg and Asp179Tyr) that reduce aminoglycoside resistance but confer resistance to ciprofloxacin and norfloxacin. aac(6′)-Ib-cr is often found as part of a complex class 1 integron including other antibiotic resistance genes. This may explain the fact that strains of Enterobacteriaceae harboring aac(6′)-Ib-cr have been described worldwide (10). Acquisition of aac(6′)-Ib-cr genes increases ciprofloxacin MICs 2- to 4-fold; however, ciprofloxacin MICs remain below the susceptibility breakpoint according to the Clinical and Laboratory Standards Institute (1 μg/ml) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST; 0.5 μg/ml) (11, 12). The aim of the present study was to determine the in vivo impact of AAC(6′)-Ib-cr in Escherichia coli pyelonephritis on ciprofloxacin efficacy, considering that this gene confers a much lower level of resistance than qnr genes.
(Part of this work was presented at the 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, 2012.)
MATERIALS AND METHODS
Bacterial strains.
Experiments were performed with four isogenic strains: E. coli CFT073-RR, E. coli CFT073-RR/p449, E. coli CFT073-RR GyrAr, and E. coli CFT073-RR GyrAr/p449. E. coli CFT073-RR is a rifampin-resistant (Rifr) mutant of the quinolone-susceptible strain E. coli CFT073, used to set the murine model of pyelonephritis (13) and previously used in this model (14–16). E. coli CFT073-RR GyrAr is a single-step mutant harboring the gyrA D87G mutation selected as previously described (1). Plasmid p449 is a 130-kb plasmid harboring aac(6′)-Ib-cr found in the clinical strain E. coli RS449 isolated from a culture of blood from a patient with pyelonephritis. E. coli RS449 was systematically screened for PMQR genes [qnr, aac(6′)-Ib-cr, and qepA] by real-time PCR and pyrosequencing as described elsewhere (17, 18) and was shown to harbor only the aac(6′)-Ib-cr gene. Besides aac(6′)-Ib-cr, p449 carried a blaCTX-M-15 gene. Plasmid DNA was extracted using a Large-Construct Kit (Qiagen, Courtaboeuf, France) and transformed into competent E. coli DH10B cells (Invitrogen, Cergy Pontoise, France). Selection was performed on agar containing amoxicillin (100 μg/ml).
E. coli CFT073-RR GyrAr/p449 was obtained through a conjugation assay between E. coli RS449 and E. coli CFT073-RR GyrAr and selection on agar containing amoxicillin (100 μg/ml) and rifampin (250 μg/ml). E. coli CFT073-RR/p449 was obtained after a conjugation assay between E. coli DH10/p449 and E. coli CFT073-RR and selection on agar containing amoxicillin (100 μg/ml) and rifampin (250 μg/ml). The presence of aac(6′)-Ib-cr in the three CFT073 derivative strains was confirmed by PCR experiments (17).
MIC determination and time-kill experiments.
MICs were determined by the agar dilution method in accordance with EUCAST guidelines (11). Briefly, MICs of amikacin, gentamicin, tobramycin, streptomycin, nalidixic acid, ciprofloxacin, levofloxacin, moxifloxacin, norfloxacin, and ofloxacin were determined on Mueller-Hinton (MH) agar plates containing serial 2-fold-dilution of antibiotics. Plates inoculated with a Steers-type multiprong device and about 104 CFU per spot were read after incubation for 18 h at 37°C. Ciprofloxacin time-kill kinetic assays were conducted on 10 ml of MH broth at antibiotic concentrations that were 1-fold, 2-fold, 4-fold the MIC and at the fixed concentration of 0.5 μg/ml. Antimicrobial agent-free broth was evaluated in parallel as a control. Viable counts were determined by serial dilution at 0, 1, 3, 6, and 24 h of incubation and by plating 100 μl of the control test cultures or by dilution onto MH agar plates. Colony counts in log10 CFU/ml were determined after 24 h of incubation. All the in vitro experiments described above were repeated at least three times. Bactericidal effect was defined as a decrease of at least a 3-log10 reduction in CFU after 24 h of incubation compared to the initial test inoculum.
Mouse model of pyelonephritis.
As in a previous study (13), the current study used the ascending unobstructed mouse model of UTI. Animal experiments were performed in accordance with prevailing regulations regarding the care and use of laboratory animals by the European Commission. The Departmental Direction of Veterinary Services in Paris, France, approved the experimental protocol. Inocula of different strains were obtained by overnight incubation in brain heart infusion broth, washing of the cells by centrifugation at 4,000 × g for 15 min in saline, and resuspension in saline to a final inoculum of 5 × 109 CFU/ml. After general anesthesia of 8-week-old immunocompetent female CBA mice (weight, 20 to 22 g), pyelonephritis was induced by injecting 50 μl of the inoculum into the bladder through a urethral catheter.
Antimicrobial treatment.
To evaluate the effectiveness of ciprofloxacin to treat murine pyelonephritis due to AAC(6′)-Ib-cr-producing E. coli, 45 mice were inoculated with E. coli CFT073-RR, 58 mice with E. coli CFT073-RR/p449, 38 mice with E. coli CFT073-RR GyrAr, and 32 mice with E. coli CFT073-RR GyrAr/p449. For each strain, 48 h after inoculation, a start-of-treatment control group (10 to 23 mice) was sacrificed. During the following 48 h, the mice were treated or not with ciprofloxacin (2 days of treatment) and then sacrificed 18 h after the last dose of ciprofloxacin. Kidneys were aseptically sampled and homogenized in 1 ml of saline solution. One hundred microliters of this solution and its 1/10 dilution were spread onto MH agar plates and incubated for 24 h. CFU counts were enumerated and expressed as log10 CFU/g of kidney. Selection of resistant mutants after both treatments was sought by plating the latter solution onto agar containing ciprofloxacin at a concentration of 4× the MIC.
A dosing regimen for ciprofloxacin of 10 mg/kg of body weight injected subcutaneously (s.c.) twice daily was chosen from preliminary pharmacokinetic studies comparing various dosages (data not shown). This regimen provides (i) a plasma peak level in the range of that achieved in humans after an oral administration of 500 mg of ciprofloxacin (19) and (ii) peak concentration/MIC and area under the concentration-time curve from 0 to 24 h (AUC0–24)/MIC ratios against the susceptible parental strain that were above those required to achieve efficacy in humans (20).
Drug pharmacokinetics.
Single-dose pharmacokinetic studies of ciprofloxacin were performed after a single injection of 10 mg/kg s.c. in infected mice. Blood samples of 200 μl were obtained by intracardiac puncturing from three separate mice at 15, 30, 45, 60, 120, 240, 360, and 480 min after ciprofloxacin injection. Blood was centrifuged, and plasma samples were treated with a methanolic solution containing ofloxacin as an internal standard. Ciprofloxacin concentration was determined by liquid chromatography, with fluorimetric detection after deproteinization, as described previously (21). The method was linear over a concentration range from 0.1 to 40 μg/ml. Intra- and interday coefficients of variation obtained were less than 10%. The limit of quantitation was 0.05 μg/ml. The AUC0–24 was calculated using the trapezoidal rule.
In situ ciprofloxacin N-acetylation measurement.
To seek the potential in situ ciprofloxacin N-acetylation due to AAC(6′)-Ib-cr-producing E. coli, 13 mice were inoculated with strain E. coli CFT073-RR and 14 with E. coli CFT073-RR/p449, as described above. After 48 h of inoculation, each mouse was treated with a single injection of ciprofloxacin (10 mg/kg s.c.). Mice were sacrificed at 0, 15, and 60 min after ciprofloxacin injection. Blood and kidneys were sampled as describe above. Ciprofloxacin and N-acetyl ciprofloxacin concentrations were determined with a Waters Acquity ultraperformance liquid chromatography system (UPLC) system with a TQD tandem quadrupole mass spectrometer using electrospray ionization. The selected m/z transitions were 332.1 → 288.0, 374.2 → 242.9, and 362.2 → 318.1, with collision energies of 20, 40, and 17 eV for ciprofloxacin, N-acetyl ciprofloxacin, and ofloxacin as internal standards, respectively. Chromatography was performed on a 2.1- by 100-mm Acquity UPLC BEH C18 1.7-μm analytical column (Waters, Milford, MA). The limit of quantification was 0.01 μg/ml.
Statistical analysis.
Mean counts (log10 CFU/g) in kidneys were compared for the different groups by the Mann-Whitney U test. The proportions of sterile mice in the different groups were compared by Fisher's exact test. All statistical analyses were performed using BiostaTGV (http://marne.u707.jussieu.fr/biostatgv/). A P value of less than 0.05 was considered significant.
RESULTS
In vitro susceptibility of AAC(6′)-Ib-cr-producing strains.
MICs of selected aminoglycosides and quinolones against the E. coli isogenic strains are presented in Table 1. The resistance profiles conferred by the acquisition of p449 in E. coli CFT073-RR and E. coli CFT073-RR GyrAr were in agreement with those observed by the production of AAC(6′)-Ib-cr. Indeed, MICs of amikacin, tobramycin, ciprofloxacin, and norfloxacin observed for E. coli CFT073-RR/p449 were increased 8-fold, 64-fold, 4-fold, and 4-fold compared to those for E. coli CFT073-RR; values for E. coli CFT073-RR GyrAr/p449 we re increased 16-fold, 64-fold, 2-fold, and 4-fold compared to those for E. coli CFT073-RR GyrAr.
Table 1.
Aminoglycoside and quinolone MICs determined for the E. coli CFT073 derivative strains used in this study
| E. coli strain | MIC (μg/ml)a |
||||||
|---|---|---|---|---|---|---|---|
| AN | TM | NAL | CIP | NOR | OFX | LVX | |
| CFT073-RR | 1 | 0.25 | 2 | 0.015 | 0.06 | 0.06 | 0.03 |
| CFT073-RR/p449 | 8 | 16 | 4 | 0.06 | 0.25 | 0.06 | 0.03 |
| CFT073-RR GyrAr | 1 | 0.25 | 128 | 0.25 | 0.5 | 0.5 | 0.25 |
| CFT073-RR GyrAr/p449 | 16 | 16 | 128 | 0.5 | 2 | 0.5 | 0.125 |
MICs were determined for amikacin (AN), tobramycin (TM), nalidixic acid (NAL), ciprofloxacin (CIP), norfloxacin (NOR), ofloxacin (OFX), and levofloxacin (LVF) according to EUCAST guidelines.
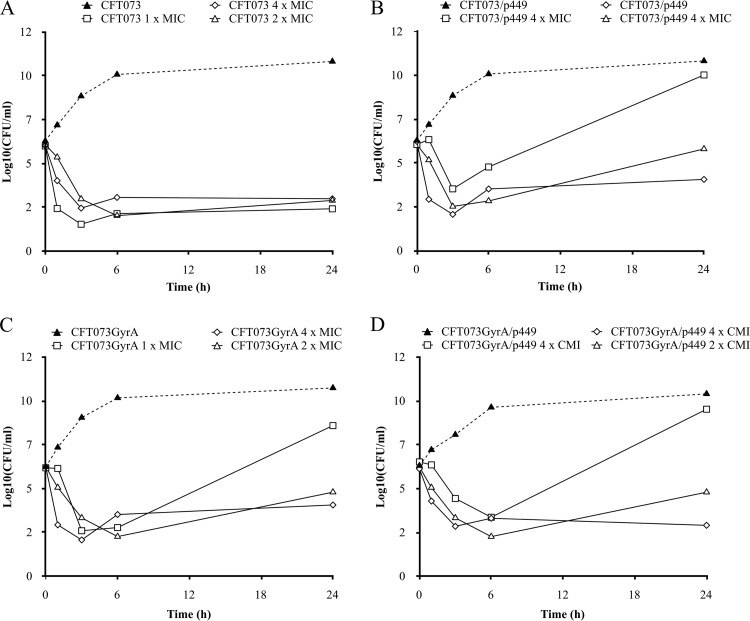
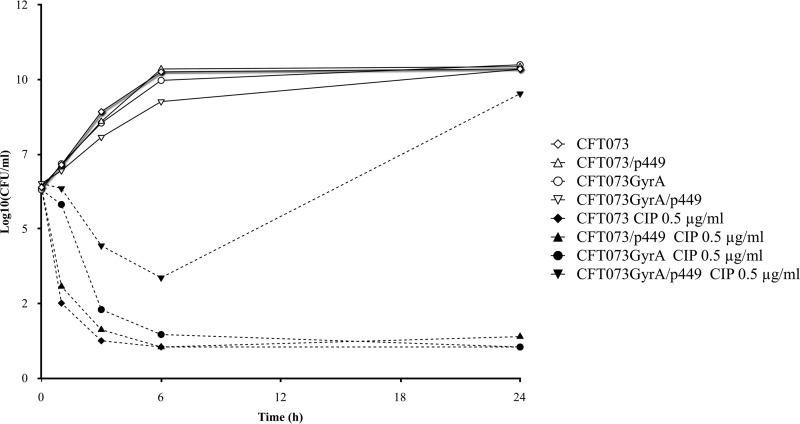
Time-kill curves of ciprofloxacin against the four isogenic strains are shown in Fig. 1 and Fig. 2 for concentrations at 1- to 4-fold the MIC and at a fixed concentration of 0.5 μg/ml, respectively. Regarding the quinolone-susceptible parental strain E. coli CFT073-RR, ciprofloxacin achieved a rapid and sustained bactericidal effect for a concentration as low as 1-fold the MIC during the 24 h of the experiment. For the three derivative strains, after a 3-log10 reduction of CFU counts at 3 h, regrowth was observed at 24 h; this regrowth was prevented only by the concentration that was 4-fold the MIC. At a ciprofloxacin concentration of 0.5 μg/ml, a rapid (3-h) and sustained (6- to 24-h) bactericidal effect was observed for all strains except E. coli CFT073-RR GyrAr/p449. For this strain, no bactericidal activity was achieved at 6 h and regrowth was observed at 24 h (P < 0.05).
Fig 1.
Viable counts of the quinolone-susceptible E. coli CFT073-RR (A) and the three isogenic derivative strains (E. coli CFT073-RR/p449 [B], E. coli CFT073-RR GyrAr [C], and E. coli CFT073-RR GyrAr/p449 [D]) in in vitro time-kill assays with ciprofloxacin concentrations at 1×, 2×, and 4× the MIC.
Fig 2.
Viable counts of the quinolone-susceptible E. coli CFT073-RR and the three isogenic derivative strains (E. coli CFT073-RR/p449, E. coli CFT073-RR GyrAr, and CFT073-RR GyrAr/p449) in in vitro time-kill assays with ciprofloxacin at the fixed concentration of 0.5 μg/ml.
In vivo ciprofloxacin efficacy in the mouse model of pyelonephritis.
With a ciprofloxacin therapeutic regimen of 10 mg/kg given subcutaneously twice a day (b.i.d.), a maximum concentration in serum (Cmax) of 1.93 ± 0.37 μg/ml (mean ± standard deviation) was obtained 15 min after the injection, and the AUC0-24 was 10.48 ± 1.96 μg · h/ml. Corresponding Cmax/MIC and AUC0-24/MIC ratios were calculated for the four isogenic strains (Table 2).
Table 2.
PK/PD parameters of the ciprofloxacin dosing regimen used in the pyelonephritis murine model
| E. coli strain | Ratio for ciprofloxacin |
|
|---|---|---|
| Cmax/MICa | AUC0–24/MICb | |
| CFT073-RR | 128.7 | 698.7 |
| CFT073-RR/p449 | 32.2 | 174.7 |
| CFT073-RR GyrAr | 7.7 | 41.9 |
| CFT073-RR GyrAr/p449 | 3.9 | 21 |
The Cmax for ciprofloxacin was 1.93 ± 0.37 μg/ml.
The AUC0–24 for ciprofloxacin was 10.48 ± 1.96 μg · ml/h.
The four strains induced pyelonephritis 48 h after inoculation in the start-of-treatment control (Table 3). Bacterial counts in kidneys were similar between the start-of-treatment groups and the end-of-treatment groups for all strains (P > 0.1). In mice infected with the quinolone-susceptible strain E. coli CFT073-RR and treated with ciprofloxacin, a significant (P < 0.05) decrease, 2.7 log10 CFU/g, of viable bacterial counts was observed in kidneys (Table 3). In contrast, no significant decrease of bacterial counts was observed after ciprofloxacin treatment of pyelonephritis due to any of the three derivatives (P > 0.1).
Table 3.
Effect of ciprofloxacin on viable organisms in kidneys of mice
| E. coli strain | Count (log10 CFU) per g (mean ± SD) of kidneys (total no. of mice) |
||
|---|---|---|---|
| Untreated mice |
Mice treated with ciprofloxacin | ||
| Start-of-treatment control | End-of-treatment control | ||
| CFT073-RR | 3.6 ± 1.7 (15) | 4.3 ± 1.7a (15) | 1.6 ± 0.3b (15) |
| CFT073-RR/p449 | 3.2 ± 1.4 (23) | 3.1 ± 1.5a (20) | 2.7 ± 1.3c (15) |
| CFT073-RR GyrAr | 4.1 ± 1.4 (13) | 4.1 ± 1.6a (13) | 4.2 ± 1.3c (12) |
| CFT073-RR GyrAr/p449 | 4.4 ± 0.9 (10) | 3.6 ± 0.9a (9) | 2.9 ± 1.4c (13) |
P > 0.1 compared with the start-of-treatment control group.
P < 0.05 compared with the end-of-treatment control group.
P > 0.1 compared with the end-of-treatment control group.
Although E. coli CFT073-RR is a pathogenic strain related to the phylogenetic group B2 (22), some of the inoculated mice presented sterile organs whether they had been treated or not (Table 3). Since we observed sterile mice in start-of-treatment groups for all the strains studied, sterile mice were included for CFU counting. The proportion of sterile mice was not different between the start-of-treatment control group and the end-of-treatment control group (P > 0.3 for each strain). For E. coli CFT073-RR, 10/15 (67%) mice were sterile in the treated group, compared to only 2/15 (13%) in the end-of-treatment control group (P < 0.05). In contrast, the proportions of sterile mice for E. coli CFT073-RR/p449, E. coli CFT073-RR GyrAr, and E. coli CFT073-RR GyrAr/p449 did not significantly differ (P > 0.1) between the end-of-treatment control group (8/20, 3/13, and 1/9) and the ciprofloxacin-treated group (6/15, 0/12, and 6/13). No resistant mutant was detected after treatment for any strain.
Ciprofloxacin N-acetylation in kidneys.
At 15 and 60 min after ciprofloxacin injection, ciprofloxacin concentrations in kidneys were higher in mice infected with E. coli CFT073-RR/p449 than in mice infected with E. coli CFT073-RR, with comparable standard deviations of concentrations at both times (0.71 versus 0.95 at 15 min and 0.26 versus 0.26 at 60 min for E. coli CFT073-RR/p449 and E. coli CFT073-RR, respectively).The N-acetyl ciprofloxacin concentration was under the limit of detection of our measuring method. Nonetheless, similar to the PK/PD data described above, the ciprofloxacin concentration in kidneys decreased between 15 and 60 min (Fig. 3A). However, this decrease was significantly more pronounced (P < 0.05) for the group of mice infected with E. coli CFT073-RR/p449 than for the group infected with E. coli CFT073-RR (Fig. 3B).
Fig 3.
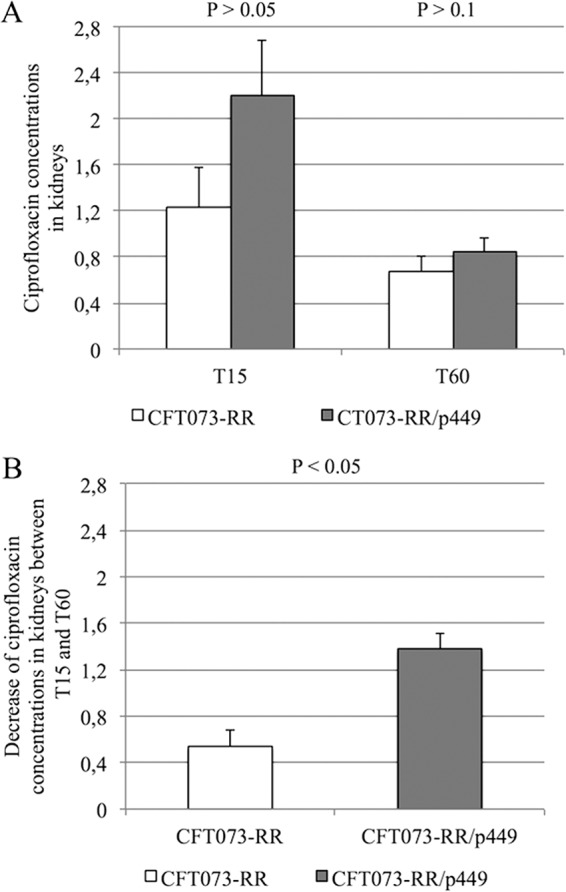
(A) Ciprofloxacin concentrations in kidneys of mice (μg/ml), infected with either E. coli CFT073-RR or E. coli CFT073-RR/p449, 15 min (T15) and 60 min (T60) and after injection of 10 mg/kg of ciprofloxacin. (B) Decrease in ciprofloxacin concentrations in kidneys of mice between 15 min and 60 min according to the study strain. The decrease in ciprofloxacin concentrations was significantly more pronounced (P < 0.05) in mice infected with E. coli CFT073-RR/p449 than in those infected with strain E. coli CFT073-RR.
DISCUSSION
UTI is one of the most common bacterial infections, with a global incidence estimated at nearly 150 million cases worldwide per year (23). UTI is therefore a major public health problem given the associated morbidity and the cost of its therapeutic management (24). E. coli is the leading bacterial etiology of UTI, and reports of PMQR in E. coli are increasingly frequent (25–27). The aac(6′)-Ib-cr gene is a low-level fluoroquinolone resistance determinant. As observed with the qnr genes, strains harboring aac(6′)-Ib-cr (9) remain susceptible to fluoroquinolones according to international susceptibility breakpoint values of ciprofloxacin for Enterobacteriaceae. Most of the studies that reported a therapeutic impact of a low level of fluoroquinolone resistance in Enterobacteriaceae did not characterize the genetic determinant involved (28–31). To date, only three studies have investigated the therapeutic impact of PMQR, but only for qnr genes (14–16). Allou et al. (qnrA1 and qnrS1) and Jakobsen et al. (qnrA1, qnrB19, and qnrS1) showed that qnr genes decreased the bactericidal activity of ciprofloxacin against E. coli in a murine UTI model (14, 16). Rodríguez-Martínez et al. showed that qnrA1 decreased the efficacy of ciprofloxacin and levofloxacin in a murine model of pneumonia due to a strain of Klebsiella pneumoniae (15).
The present study investigated the bactericidal activity of ciprofloxacin against aac(6′)-Ib-cr-producing E. coli in a murine pyelonephritis model using a dose corresponding to an optimal human regimen. The main finding was that mice infected with E. coli carrying aac(6′)-Ib-cr did not show any significant reduction in bacterial counts when they were treated with ciprofloxacin, as opposed to the mice infected with the susceptible parental strain. The therapeutic impact was similar with a derivative strain harboring a single gyrA mutation with or without aac(6′)Ib-cr. Ciprofloxacin treatment significantly increased the proportion of sterile mice compared with the end-of-treatment control group only for the susceptible strain. These results were in agreement with the in vitro bactericidal findings. Indeed, at concentrations ranging from 1-fold to 4-fold the MIC, the bactericidal activity of ciprofloxacin was markedly reduced against the three resistant strains compared with the susceptible parental one. At the EUCAST clinical breakpoint concentration of 0.5 μg/ml, which is achievable in human serum during therapy with ciprofloxacin, the bactericidal activity of ciprofloxacin was achieved during the first 6 h of exposure against the derivative strains except those harboring both plasmid p449 and the gyrA D87G mutation, in comparison with the susceptible E. coli parental strain. However, the bactericidal activity of ciprofloxacin remained stable from 6 h to 24 h after exposure, except against the strain harboring both the gyrA mutation and the aac(6′)-Ib-cr gene.
The most striking result was that the therapeutic failure of ciprofloxacin in mice infected with derivative E. coli aac(6′)-cr-positive strains could not be explained by pharmacodynamic-pharmacokinetic (PK/PD) indices, in contrast to previous results reported with qnr and the gyrA mutation (14). In vivo, the parameters that best predict fluoroquinolone efficacy are the AUC0-24/MIC ratio and the Cmax/MIC ratio. Values higher than 125 and 10, respectively, have been shown to be predictive of clinical and microbiological efficacy in different foci of infection (20). In the current experiments, although the AUC0-24/MIC and Cmax/MIC ratios of ciprofloxacin were well above 125 and 10 to 12 for strain E. coli CFT073-RR/p449, we observed a failure of ciprofloxacin to decrease the bacterial counts in kidneys. Such surprising results have been previously obtained in an experimental model of rabbit endocarditis due to a K. pneumoniae strain carrying aac(6′)-Ib and treated with amikacin or isepamicin (32). This study showed that although the AAC(6′)-Ib enzyme conferred a low level of resistance to amikacin and isepamicin in vitro (MICs of amikacin and isepamicin were 4 and 0.5 μg/ml, respectively), its production compromised the two drugs' bactericidal efficacies. This observation had been explained by the possibility that aminoglycoside concentrations in cardiac vegetations were appreciably lower than their concentrations in sera. This is not the case for ciprofloxacin concentrations in the renal parenchyma, since ciprofloxacin reaches concentrations in kidneys significantly higher than in serum (33). It was therefore hypothesized that the competition between rates of drug accumulation and drug hydrolysis could explain the fact that the in vitro quinolone-susceptible but AAC(6′)-Ib-cr-producing strain led to ciprofloxacin therapeutic failure. Measurement of N-acetylation of ciprofloxacin by AAC(6′)-Ib-cr has been studied in vitro (9, 34, 35). Wachino et al. developed a disk-based method for detecting AAC(6′)-Ib-cr production in E. coli isolates based on fluoroquinolone inactivation (34). Jung et al. reported N-acetylation of norfloxacin and ciprofloxacin by an aac(6′)-Ib-cr-carrying E. coli strain from an environmental source. The authors showed, using high-performance liquid chromatography (HPLC) and mass spectrometry, that this strain transformed both ciprofloxacin and norfloxacin by N-acetylation (35). Genes present on the clinical plasmid p449, other than known resistance genes, could have affected the ability of the plasmid-containing strain to persist in kidney tissue. Nonetheless, results in the present study showing that ciprofloxacin concentration decreased more significantly (P < 0.05) when mice were infected with an AAC(6′)-Ib-cr-producing strain compared to a nonproducing strain suggest an in situ hydrolysis of ciprofloxacin that could explain why PK/PD parameters did not account for failure of ciprofloxacin against aac(6′)-Ib-cr-containing strains.
In conclusion, low-level fluoroquinolone resistance conferred by aac(6′)-Ib-cr seems to be associated with reduced bactericidal activity of ciprofloxacin in vivo and to lead to ciprofloxacin therapeutic failure in pyelonephritis. In situ N-acetylation of ciprofloxacin may explain the reduction in bactericidal activity. aac(6′)-Ib-cr is the most prevalent PMQR gene detected in clinical isolates, especially extended-spectrum β-lactamase (ESBL)-producing strains (36). Regarding in vivo data on qnr (14–16) and considering the present data on aac(6′)-Ib-cr, we believe that care should be taken in clinical practice when PMQR is detected. However, since the level of resistance conferred is low, it is often difficult to detect the gene's presence based on the observed phenotype on a routine basis in the laboratory. Molecular detection of aac(6′)-Ib-cr among clinical bacterial isolates would be of clinical interest.
ACKNOWLEDGMENTS
We are indebted to Janick Madoux and Sara Dion for their excellent technical assistance. We have no conflict of interest to declare. We thank Tim Greacen for his help with the English language.
This work was supported by annual grants from the University of Reims Champagne-Ardenne and from University Paris-Diderot.
Footnotes
Published ahead of print 9 September 2013
REFERENCES
- 1. Hooper D. 2000. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31(Suppl 2):S24–S28. 10.1086/314056 [DOI] [PubMed] [Google Scholar]
- 2. Goossens H. 2009. Antibiotic consumption and link to resistance. Clin. Microbiol. Infect. 15(Suppl 3):12–15 [DOI] [PubMed] [Google Scholar]
- 3. Garau J, Xercavins M, Rodríguez-Carballeira M, Gómez-Vera JR, Coll I, Vidal D, Llovet T, Ruíz-Bremón A. 1999. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob. Agents Chemother. 43:2736–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drlica K, Malik M, Kerns RJ, Zhao X. 2008. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52:385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martínez-Martínez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799 [DOI] [PubMed] [Google Scholar]
- 7. Périchon B, Courvalin P, Galimand M. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 51:2464–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sørensen AH, Hansen LH, Johannesen E, Sørensen SJ. 2003. Conjugative plasmid conferring resistance to olaquindox. Antimicrob. Agents Chemother. 47:798–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83–88 [DOI] [PubMed] [Google Scholar]
- 10. Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y, Lü D, Huang L, Zhang Y, Liu J, Wang M. 2009. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob. Agents Chemother. 53:519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Dieases (ESCMID) 2000. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Clin. Microbiol. Infect. 6:509–515 [DOI] [PubMed] [Google Scholar]
- 12. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement M100-S21. CLSI, Wayne, PA [Google Scholar]
- 13. Labat F, Pradillon O, Garry L, Peuchmaur M, Fantin B, Denamur E. 2005. Mutator phenotype confers advantage in Escherichia coli chronic urinary tract infection pathogenesis. FEMS Immunol. Med. Microbiol. 44:317–321 [DOI] [PubMed] [Google Scholar]
- 14. Allou N, Cambau E, Massias L, Chau F, Fantin B. 2009. Impact of low-level resistance to fluoroquinolones due to qnrA1 and qnrS1 genes or a gyrA mutation on ciprofloxacin bactericidal activity in a murine model of Escherichia coli urinary tract infection. Antimicrob. Agents Chemother. 53:4292–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodríguez-Martínez JM, Pichardo C, García I, Pachón-Ibañez ME, Docobo-Pérez F, Pascual A, Pachón J, Martínez-Martínez L. 2008. Activity of ciprofloxacin and levofloxacin in experimental pneumonia caused by Klebsiella pneumoniae deficient in porins, expressing active efflux and producing QnrA1. Clin. Microbiol. Infect. 14:691–697 [DOI] [PubMed] [Google Scholar]
- 16. Jakobsen L, Cattoir V, Jensen KS, Hammerum AM, Nordmann P, Frimodt-Moller N. 2012. Impact of low-level fluoroquinolone resistance genes qnrA1, qnrB19 and qnrS1 on ciprofloxacin treatment of isogenic Escherichia coli strains in a murine urinary tract infection model. J. Antimicrob. Chemother. 67:2438–2444 [DOI] [PubMed] [Google Scholar]
- 17. Guillard T, Duval V, Moret H, Brasme L, Vernet-Garnier V, de Champs C. 2010. Rapid detection of aac(6′)-Ib-cr quinolone resistance gene by pyrosequencing. J. Clin. Microbiol. 48:286–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guillard T, Moret H, Brasme L, Carlier A, Vernet-Garnier V, Cambau E, de Champs C. 2011. Rapid detection of qnr and qepA plasmid-mediated quinolone resistance genes using real-time PCR. Diagn. Microbiol. Infect. Dis. 70:253–259 [DOI] [PubMed] [Google Scholar]
- 19. Fantin B, Duval X, Massias L, Alavoine L, Chau F, Retout S, Andremont A, Mentré F. 2009. Ciprofloxacin dosage and emergence of resistance in human commensal bacteria. J. Infect. Dis. 200:390–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schentag JJ. 2000. Clinical pharmacology of the fluoroquinolones: studies in human dynamic/kinetic models. Clin. Infect. Dis. 31(Suppl 2):S40–S44. 10.1086/314059 [DOI] [PubMed] [Google Scholar]
- 21. Rubinstein E, St Julien L, Ramon J, Dautrey S, Farinotti R, Huneau JF, Carbon C. 1994. The intestinal elimination of ciprofloxacin in the rat. J. Infect. Dis. 169:218–221 [DOI] [PubMed] [Google Scholar]
- 22. Johnson JR, Clermont O, Menard M, Kuskowski MA, Picard B, Denamur E. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141–1150 [DOI] [PubMed] [Google Scholar]
- 23. Stamm WE, Norrby SR. 2001. Urinary tract infections: disease panorama and challenges. J. Infect. Dis. 183(Suppl 1):S1–S4. 10.1086/318850 [DOI] [PubMed] [Google Scholar]
- 24. Ronald AR, Nicolle LE, Stamm E, Krieger J, Warren J, Schaeffer A, Naber KG, Hooton TM, Johnson J, Chambers S, Andriole V. 2001. Urinary tract infection in adults: research priorities and strategies. Int. J. Antimicrob. Agents 17:343–348 [DOI] [PubMed] [Google Scholar]
- 25. Yang H, Chen H, Yang Q, Chen M, Wang H. 2008. High prevalence of plasmid-mediated quinolone resistance genes qnr and aac(6′)-Ib-cr in clinical isolates of Enterobacteriaceae from nine teaching hospitals in China. Antimicrob. Agents Chemother. 52:4268–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones GL, Warren RE, Skidmore SJ, Davies VA, Gibreel T, Upton M. 2008. Prevalence and distribution of plasmid-mediated quinolone resistance genes in clinical isolates of Escherichia coli lacking extended-spectrum beta-lactamases. J. Antimicrob. Chemother. 62:1245–1251 [DOI] [PubMed] [Google Scholar]
- 27. Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Endimiani A, Luzzaro F, Perilli M, Lombardi G, Colì A, Tamborini A, Amicosante G, Toniolo A. 2004. Bacteremia due to Klebsiella pneumoniae isolates producing the TEM-52 extended-spectrum beta-lactamase: treatment outcome of patients receiving imipenem or ciprofloxacin. Clin. Infect. Dis. 38:243–251 [DOI] [PubMed] [Google Scholar]
- 29. Fuursted K, Schumacher H. 2002. Significance of low-level resistance to ciprofloxacin in Klebsiella pneumoniae and the effect of increased dosage of ciprofloxacin in vivo using the rat granuloma pouch model. J. Antimicrob. Chemother. 50:421–424 [DOI] [PubMed] [Google Scholar]
- 30. McCarron B, Love WC. 1997. Acalculous nontyphoidal salmonellal cholecystitis requiring surgical intervention despite ciprofloxacin therapy: report of three cases. Clin. Infect. Dis. 24:707–709 [DOI] [PubMed] [Google Scholar]
- 31. Vasallo FJ, Martín-Rabadán P, Alcalá L, García-Lechuz JM, Rodríguez-Créixems M, Bouza E. 1998. Failure of ciprofloxacin therapy for invasive nontyphoidal salmonellosis. Clin. Infect. Dis. 26:535–536 [DOI] [PubMed] [Google Scholar]
- 32. Caulin E, Coutrot A, Carbon C, Collatz E. 1996. Resistance to amikacin and isepamicin in rabbits with experimental endocarditis of an aac(6′)-Ib-bearing strain of Klebsiella pneumoniae susceptible in vitro. Antimicrob. Agents Chemother. 40:2848–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bergan T. 1990. Extravascular penetration of ciprofloxacin: a review. Diagn. Microbiol. Infect. Dis. 13:103–114 [DOI] [PubMed] [Google Scholar]
- 34. Wachino JI, Yamane K, Arakawa Y. 2011. Practical disk-based method for detection of Escherichia coli clinical isolates producing the fluoroquinolone-modifying enzyme AAC(6′)-Ib-cr. J. Clin. Microbiol. 49:2378–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jung CM, Heinze TM, Strakosha R, Elkins CA, Sutherland JB. 2009. Acetylation of fluoroquinolone antimicrobial agents by an Escherichia coli strain isolated from a municipal wastewater treatment plant. J. Appl. Microbiol. 106:564–571 [DOI] [PubMed] [Google Scholar]
- 36. Karah N, Poirel L, Bengtsson S, Sundqvist M, Kahlmeter G, Nordmann P, Sundsfjord A, Samuelsen Ø, Norwegian Study Group on PMQR 2010. Plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in Escherichia coli and Klebsiella spp. from Norway and Sweden. Diagn. Microbiol. Infect. Dis. 66:425–431 [DOI] [PubMed] [Google Scholar]