Abstract
Across all kingdoms of life, cells store energy in a specialized organelle, the lipid droplet. In general, it consists of a hydrophobic core of triglycerides and steryl esters surrounded by only one leaflet derived from the endoplasmic reticulum membrane to which a specific set of proteins is bound. We have chosen the unicellular organism Dictyostelium discoideum to establish kinetics of lipid droplet formation and degradation and to further identify the lipid constituents and proteins of lipid droplets. Here, we show that the lipid composition is similar to what is found in mammalian lipid droplets. In addition, phospholipids preferentially consist of mainly saturated fatty acids, whereas neutral lipids are enriched in unsaturated fatty acids. Among the novel protein components are LdpA, a protein specific to Dictyostelium, and Net4, which has strong homologies to mammalian DUF829/Tmem53/NET4 that was previously only known as a constituent of the mammalian nuclear envelope. The proteins analyzed so far appear to move from the endoplasmic reticulum to the lipid droplets, supporting the concept that lipid droplets are formed on this membrane.
INTRODUCTION
Fat is the ideal molecule for storing energy at low volume and weight because the triacylglycerol (TAG) molecule self-assembles due to van der Waals interactions and excludes water. Based alone on these physicochemical properties, fat would be able to form a separate organelle in the cell. However, in order to shield the hydrophobic surface from unspecific interactions, the cellular drop of fat is surrounded by one leaflet of membrane phospholipids pointing their hydrocarbon chains toward the interior and exposing their hydrophilic head groups to the aqueous cytoplasm. This surface provides the target for interactions with structural or regulatory proteins as well as metabolic enzymes.
Within the past 20 years, this simple view of the lipid droplet (LD) has been refined, and many molecular details were added, as recently reviewed (1–3). Analysis of lipid droplet structure and composition has continued to provide surprising results. Examples are the detection of proteins in the inner hydrophobic core (4, 5), the function of lipid droplets as histone storage sites in Drosophila embryogenesis (6), or the discovery that coatomer proteins (COPs), known to coat vesicles formed at membrane bilayers, mediate the translocation of enzymes to the lipid droplet, despite the fact that this organelle only bears a phospholipid monolayer (7, 8).
Proteomic analyses of lipid droplets have been conducted for various organisms such as mammals, insects, Saccharomyces cerevisiae and Yarrowia lipolytica, bacteria, microalgae, and plants (summarized in reference 9), but virtually nothing is known about lipid droplets in the otherwise well-studied model system Dictyostelium that is also evolutionarily distant from all the organisms studied until now (10). In the wild, Dictyostelium amoebae live in the forest soil, efficiently phagocytosing bacteria. After cleaving the bacterial membrane lipids, amoebae finally release complex oligosaccharides but retain the fatty acid moiety (11). Even uncommon fatty acids from the diet, such as the ones containing a cyclopropane moiety, become integrated into the predator's lipids (12).
Further work took advantage of Dictyostelium strains able to grow axenically, i.e., in a broth providing sugar, amino acids, vitamins, and trace elements but low in fatty acids. Experimental addition of polyunsaturated fatty acids to the medium impaired the subsequent progression of Dictyostelium through the developmental cycle (13), and monounsaturated fatty acids inhibited cellular growth (14). Addition of palmitic acid was tolerated best; it was incorporated into cellular lipids and even slightly enhanced cell growth (13). It is conceivable that this product is preferred because it is the common end product of endogenous de novo fatty acid synthesis in eukaryotes.
In the course of studying fatty acid activation in Dictyostelium, we discovered that the fluorescent palmitic acid analog C1-BODIPY-C12 became incorporated into small cytoplasmic dots, presumably lipid droplets (15). In the study reported here, we establish the kinetics of lipid droplet formation and degradation. This enables us to purify these organelles and to analyze their lipid structures down to the level of the fatty acid composition of the core and surrounding membrane. After proteomic analysis, we confirmed that a group of proteins moves from the endoplasmic reticulum (ER) to lipid droplets when they are formed. Among these are novel lipid droplet proteins, as well as one mammalian homologue that was previously recognized only as a constituent of the nuclear envelope.
MATERIALS AND METHODS
Internet resources for sequence analysis.
Dictyostelium DNA and protein sequences were retrieved from the fully sequenced genome (10) via http://dictybase.org (16), where they are also linked to studies of expression patterns. Transmembrane regions and domains forming coiled coils were identified at http://ch.EMBnet.org. A tool for calculating the isoelectric point of a protein according to several algorithms is found at http://isoelectric.ovh.org.
Fluorescent protein tagging.
Subsequent constructs were produced in vector 48 pDd-A15-GFP (where GFP is green fluorescent protein) without ATG (according to Gerisch et al. [17] modified by Hanakam et al. [18] to delete the start codon of the actin 15 promoter) that produced a protein using its own ATG and carrying a GFP tag on its C terminus. Alternatively, we used plasmid 68 pDNeoGFP (19), where the green fluorescent protein resides at the N terminus of the intended hybrid and the continuity of the reading frame is achieved by deleting the stop codon of the upstream open reading frame.
The Dictyostelium protein formerly called DdLSD for its homology to the Drosophila homologue is now named perilipin and abbreviated Plin according to a recent nomenclature initiative (20). The corresponding gene in Dictyostelium now bears the name plnA. For labeling the N-terminal end of perilipin with GFP, primers 159 (CGTGTCGACATGTCATCTCAAGAACAACAAAAATCAAAGC) and 160 (CGTGGATCCATCTAATTGGTTGAGTTATCATTTGAAGATGAAG) were used for PCR on the cDNA clone SLE 217 obtained from the Dictyostelium cDNA project in Japan at Tsukuba University, and the SalI/BamHI-doubly digested product was integrated into vector 68.
As a basis for further cloning steps, the coding sequence of smtA was amplified with primers 674 (CCATAGAATTCAAAATGAATACTCAACAACGTGCTATGG) and 675 (CCATAGAATTCTTAATCAGTGCTTGGTTTACGACATAATAAG) using reverse-transcribed mRNA of AX2 as the template and then ligated into vector pGem-TEasy by virtue of single A-residue overhangs to yield plasmid 845. Subsequent digestion of the PCR-engineered EcoRI sites allowed insertion of the released fragment into plasmid 68 that now expresses GFP-Smt1 (plasmid 846). The reverse construct is based on the amplification of smtA lacking its stop codon by primers 258 (CCGAATTCAAAATGAATACTCAACAACG) and 474 (CCGAATTCGATCAGTGCTTGGTTTACG) from genomic DNA and its intermediate cloning into pGEM-TEasy (plasmid 759), from where it was excised with EcoRI and transferred into vector 48 to yield 760 expressing Smt1-GFP.
The novel lipid droplet constituent encoded by ldpA was amplified with primers 302 (CGGGATCCAAAATGAATACTTCAACAACAAC) and 303 (CCGAATTCTTAATTACGTTTATTTTTTTTACC) using genomic DNA of AX2 as the template, cleaved with BamHI and EcoRI, and then ligated into vector 68 so that a GFP-Ldp hybrid protein is expressed from plasmid 581. The complementary construct 571 producing Ldp-GFP is based on vector 48 that received a PCR product from primers 304 (CCGAATTCAAAATGAATACTTCAACAACAAC) and 305 (CCGGATCCATTACGTTTATTTTTTTTACCC).
To construct a C-terminally tagged version of the Dictyostelium Net4 homologue, a gene-specific PCR was performed on total cDNA with a combination of primers 614 (GGCCGAATTCAAAATGGGTGCCCAA) and 615 (GGCCGGATCCTTTATTTTGTAATTTTTTC), purified, and cut with EcoRI and BamHI before ligation into the same sites of vector 48, resulting in plasmid 809 that serves to express Net4-GFP. A different set of primers, 618 (GGCCGTCGACATGGGTGCCCAAAAATTAC) and 619 (GGCCGAATTCTTATTTATTTTGTAAT), yielded a product suitable for insertion into plasmid 68 after digestion with SalI and EcoRI. This cloning step yielded plasmid 810 (GFP-Net4).
The above constructs were transformed into Dictyostelium discoideum AX2 vegetative cells (referred to as the wild type) by electroporation. Transformants were selected by virtue of G418 resistance, and individual clones were derived by spreading dilutions on bacterial lawns. Two or more clones originating from separate transformation events and showing the same patterns of florescence distribution were conserved. The localization of tagged proteins to the endoplasmic reticulum was confirmed by indirect immunofluorescence (21) using mouse monoclonal antibodies (MAbs) raised against the protein disulfide isomerase (PDI) (MAb 221-64-1) (22). The lipid droplet-specific dye LD540 (23) was diluted from its stock (0.5 mg/ml in ethanol) to a final concentration of 0.1 μg/ml in phosphate-buffered saline (PBS) and used to stain fixed cells for 30 min instead of using an antibody. In order to stain lipid droplets in living cells, we used the fluorescent fatty acid analogue C1-BODIPY-C12 (as described in reference 15) or replaced the growth medium by phosphate buffer containing 2 μM Nile red (from a 3 mM stock in ethanol).
In order to test the subcellular distribution of mammalian NET4, the appropriate expression plasmid encoding the GFP-tagged long splice variant (24) was transiently transfected as a complex with linear polyethyleneimine of 25 kDa (Polysciences, Warrington, PA) into COS7 or HEK293T cells growing on collagen-coated coverslips according to standard methods. Twenty-four hours after transfection the cells were challenged with bovine serum albumin (BSA)-coupled oleic acid at a concentration of 400 μM in growth medium for a further 24 h to induce lipid droplet formation. After samples were washed with PBS, lipid droplets were stained in living cells with LD540 as specified above for fixed Dictyostelium cells, washed twice with PBS, and then fixed in 3.7% formaldehyde in PBS for 20 min.
Biochemical lipid droplet analysis.
To induce the formation of lipid droplets, we add palmitic acid from a 100 mM stock dissolved at 50°C in methanol to HL5 growth medium after cooling to reach a final concentration of 200 μM. For some experiments cholesterol (soluble as a stock solution of 10 mM) was added at 100 μM.
The biochemical preparation of lipid droplets was based on the method of Fujimoto et al. (25) with the following modifications. About 5 × 108 cells from shaking culture were suspended in 1 ml of 0.25 M STKM buffer (50 mM Tris, pH 7.6, 25 mM KCl, 5 mM MgCl2, and 0.25 M sucrose), and the plasma membrane was broken by 20 passages through a cell cracker (EMBL Workshop, Heidelberg, Germany) so that the organelles remained intact. The postnuclear supernatant was adjusted to 0.8 M sucrose and loaded in the middle of a step gradient ranging from 0.1 to 1.8 M sucrose in STKM buffer and centrifuged at 180,000 × g for 2.5 h at 4°C in an SW40 rotor (Beckmann Coulter, Krefeld, Germany). Lipid droplets formed a white cushion of about 400 μl on top of the tube, which was collected by means of a microbiological inoculation loop. Seventeen further fractions of 800 μl each were taken with a pipette tip from the top to bottom of the tube.
For protein identification by mass spectrometry (MS), proteins were separated by polyacrylamide gels (Novex NuPAGE 4 to 12% Bis-Tris gel). Lanes were cut into 22 equally spaced pieces with an in-house made gel-cutter. The sample was digested with trypsin (sequencing grade-modified trypsin; Promega) as described previously (26), and peptides were analyzed subsequently on a hybrid triple quadrupole/linear ion-trap mass spectrometer (4000 QTRAP; Applied Biosystems/MDS Sciex) coupled to a one-dimension (1D) nano-liquid chromatography (LC) system (Eksigent). Five microliters (10% sample) was injected onto a PepMap RPC18 trap column (300-μm inside diameter [i.d.] by 5 mm; 5-μm particle size; C18 column with 100-Å pore size [Dionex]), purified, and desalted with 0.1% (vol/vol) formic acid–2% (vol/vol) CH3CN at 30 μl/min (all Biosolve). Samples were separated by gradient elution onto a PepMap C18 microcolumn (75-μm i.d. by 15 cm; 3-μm particle size; C18 column with 100-Å pore size [Dionex]) with a linear gradient of 2 to 45% (vol/vol) CH3CN–0.1% (vol/vol) formic acid at 250 nl/min. Analyst, version 1.4.1, and Bioanalyst, version 1.4.1, software programs (Applied Biosystems/MDS Sciex) were used for acquisition control. Tandem MS (MS/MS) spectra were searched against a nonredundant sequence database at www.dictybase.org (27) using MASCOT (version 2.2.05; Matrix Science). Tolerances for peptides were set to 1.5 Da and 0.5 Da for MS and MS/MS, respectively. Identified proteins were accepted with a minimum total score of 50 and at least two different peptides with a minimum peptide score of 10.
Western blotting employed the PDI antibody or antibodies recognizing GFP MAb 264-449-2 (available from Millipore), mitochondrial porin MAb 70-100-1 (28), severin MAb 42-65-11 (29), and FcsA MAb 221-457-5 (15). The work by von Löhneysen et al. (15) also describes how the mode of membrane association was determined by differential centrifugation, extraction, and subsequent Western blotting.
Lipid analysis.
To determine the TAG content of a whole-cell homogenate enzymatically, about 2.5 × 107 washed cells were resuspended in 200 μl of thin-layer chromatography (TLC) buffer, frozen in liquid nitrogen, and thawed at 37°C three times so that cells were disrupted and cellular lipids were released. A sample of 50 μl of the sample was added to 1 ml of TAG reagent (LT-SYS, Berlin, Germany) and incubated for 20 min at room temperature in a cuvette in the dark. This leads to the release of glycerol from fat, a phosphorylated intermediate, and its subsequent conversion to dihydroxyacetone phosphate and hydrogen peroxide. The latter metabolite is photometrically detected as the formation of quinoneimine, absorbing at 500 nm.
For lipid analysis by thin-layer chromatography (TLC), the classical method of Bligh and Dyer (30) was adapted as follows. About 5 × 107 washed cells were resuspended in 1 ml of TLC buffer (20 mM HEPES, 150 mM NaCl, pH 7.5), and an appropriate aliquot (according to the previously determined protein content by the bicinchoninic acid (BCA) method, per the manufacturer's instructions [Pierce]) was adjusted to 1.2 ml with TLC buffer. First, 4.5 ml of 1:2 chloroform-methanol was added and mixed for 1 min. Next, 1.5 ml of chloroform and finally 1.5 ml of double-distilled H2O (ddH2O) were added to the sample with mixing in between. Then methyl oleate (1 μg/ml) was added as a tracer to monitor possible sample loss during further preparation steps, and the mixture was centrifuged at 2,000 × g for 10 min at room temperature. The chloroform phase was collected from the bottom with a glass Pasteur pipette and transferred to a new glass tube, and the solvent was completely evaporated in a stream of nitrogen before the lipids were redissolved in 100 μl of chloroform.
Sample volumes of 20 μl were spotted with Hamilton glass syringes onto silica gel 60 plates (Merck, Darmstadt, Germany) next to a standard that contained cholesterol, cholesteryl palmitate, glyceryl trioleate, and methyl oleate (all from Sigma) at 1 μg/ml each and dried under a stream of nitrogen. Lipids were separated until the first solvent front (hexane-diethyl ether-acetic acid at 70:30:1) had reached half of the separation distance; then the plate was air dried and further developed in a second solvent system (hexane-diethyl ether at 49:1) to completion. To visualize the lipids, the plates were stained for 3 s with copper sulfate (0.3 M in 8.5% phosphoric acid) and heated to 160°C for 15 min to conduct the charring reaction.
For quantification of lipids, the fraction containing lipid droplets was extracted with 3 ml of chloroform-methanol (1:2, vol/vol) for 3 h with vigorous shaking and 4°C. After centrifugation for 10 min at 450 × g, the lower phase was stored for further processing and the upper phase was reextracted with 3 ml of chloroform as described above. Both lower phases were combined, and 2 ml of 0.45% (wt/vol) sodium chloride was added. The sample was centrifuged for 3 min at 450 × g, and then a spatula tip of sodium sulfate was added to the lower phase. The sample was centrifuged again; the upper phase was dried under streaming nitrogen and then redissolved in 0.1 ml of chloroform. After the extraction step, 1/5 of the samples were used for the TLC separation of the neutral lipids, and 2/5 were used for the separation of the phospholipids using either hexane-diethyl ether-acetic acid (80:20:1, vol/vol/vol) or chloroform-methanol-acetic acid (65:25:8, vol/vol/vol) as solvents with glass silica gel plates (silica gel 60, 20 by 20 cm; Merck, Darmstadt, Germany). Plates were sprayed with 8-anilino-1-naphthalenesulfonic acid (0.2%, wt/vol) so that lipid bands could be marked under UV light (31). Lipid spots were scraped from the TLC plate and reextracted two times with 1 ml of hexane, and defined amounts of triheptadecanoate were added for quantification. Fatty acid methyl esters were generated by transmethylation (32) and analyzed quantitatively as well as qualitatively by gas chromatography-flame ionization detection (GC-FID) (33), yielding the amount of fatty acids in the respective lipid class. To arrive at the molecular composition of lipid droplets, the amount of fatty acids was divided by 3 in the case of TAGs or by a factor of 2 for diacylglycerols (DAGs), phospholipids, and the unknown lipid (UKL), because the last is likely to contain one fatty acid linked by a nonhydrolyzable ether bond. Free sterols could not be quantified by the same method because they were lacking a fatty acid moiety. From densitometry of the TLC staining, however, it appears that that nonesterified sterols exceed the amount of DAG but are clearly below the level of free fatty acids.
RESULTS
Kinetics of lipid droplet formation and degradation.
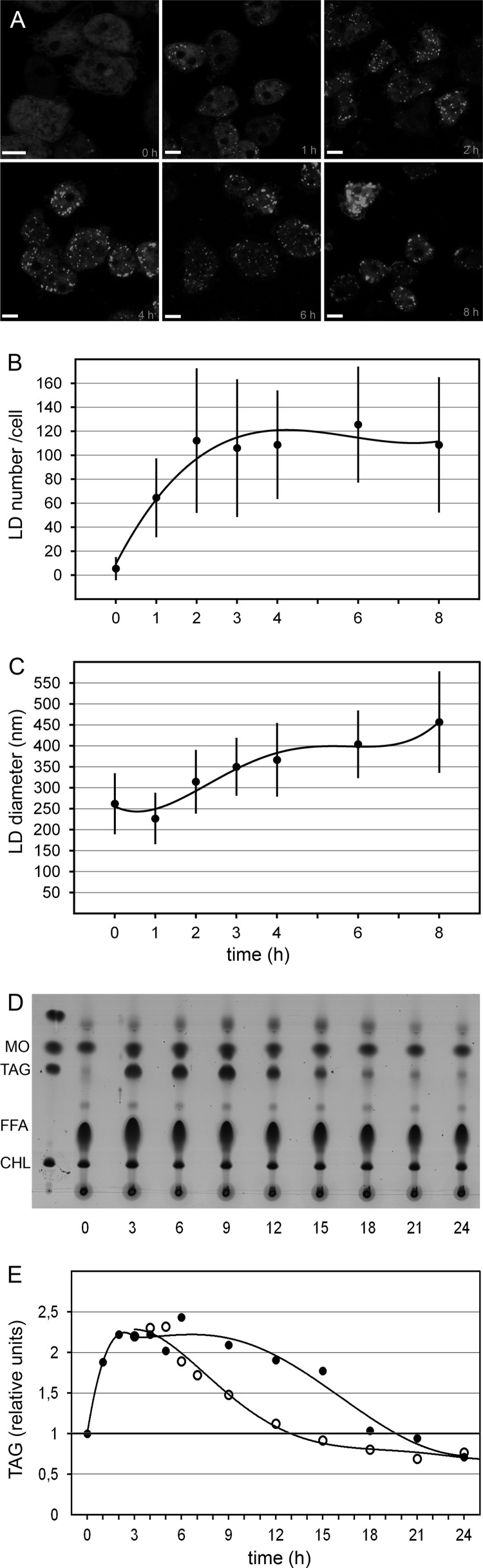
To assess the kinetics of lipid droplet (LD) formation, palmitic acid was added to a cell culture, and the well-established lipid droplet dye Nile red was used to image living cells at different times. Figure 1A shows that lipid droplet formation in Dictyostelium has some characteristics also observed in mammalian cells (34). New lipid droplets form rapidly, increasing first over 10-fold in number (Fig. 1B) and then up to 2-fold in size (Fig. 1C), with a high cell-to-cell variation at 6 or 8 h after feeding. At late time points, the lipid droplets also tend to form aggregates (Fig. 1A, 8 h).
Fig 1.
Kinetics of storage fat accumulation and utilization. (A) Wild-type cells were cultivated in the presence of palmitic acid, withdrawn at the times indicated (in hours), stained with Nile red, and photographed in a confocal microscope without prior fixation. Scale bar, 5 μm. For the experiment shown in panel B, the number of lipid droplets in one optical section was counted for at least 30 cells per time point and corrected by a factor derived from counting all lipid droplets in 20 independent stacks of sections obtained from fixed cells. (C) Over 100 lipid droplets per time point were used to determine their diameters, except at 0 h, where 30 cells were assayed. For panels B and C, the mean values are shown as closed circles connected by a fitted curve, and the bars indicate standard deviations. For the thin-layer chromatography shown in panel D, cells were cultivated in palmitic acid-containing medium, and samples were withdrawn at 3-h intervals. Lipid extracts were analyzed by TLC, where the first lane shows a standard mixture containing cholesterol (CHL), TAG, and methyl oleate (MO). The last was added to every sample to trace possible loss of material during the extraction procedure. The strong band derived from free fatty acids is labeled FFA. Panel E displays the enzymatically determined TAG values from two conditions. Wild-type cells were fed for 3 h with palmitic acid in growth medium and then washed and resuspended in normal medium (open circles) or allowed to remain in the presence of the fatty acid (filled circles). The value of 1 in the wild type is considered the background level (thick horizontal line) because of the almost undetectable level of TAG in the TLC plate (panel D) and serves as a reference for the relative units presented. The curves connect values from at least two independent experiments.
To characterize the lipid products that are formed upon fatty acid addition, extracts were analyzed by thin-layer chromatography (TLC) (Fig. 1D). As expected, the most prominent change is seen in the band identified as triacylglycerol (TAG) by comigration with a standard. Over the first 3 h, TAG concentration increased 23-fold (as determined by ImageJ analysis) and progressed to a plateau. This stage lasted for 6 h and was followed by a steady decline of fat levels over the next 9 h, reaching the initial value after 24 h. During this time, three cell divisions took place, explaining the gradual consumption of storage fat and its conversion into membrane lipids or metabolic energy.
As an alternative to TLC densitometry, we employed an assay developed for TAG quantification within serum samples (Fig. 1E). The TAG concentration of total cell extracts collected over 24 h paralleled the observations made by TLC (Fig. 1E, filled circles). A fatty acid washout experiment led to an immediate decline of TAG levels (Fig. 1E, open circles). One feature of this type of measurement, however, is that the values show only an apparent 2-fold increase of TAG at peak time (3 h over 0 h), which is clearly underestimated. This property is caused by the fact that the enzymatic assay indirectly determines the concentration of glycerol released from fat by lipase action. Because the endogenous content of glycerol and other possible relevant metabolites in cells from normal medium is unknown, it was arbitrarily set to 1.
One other feature we noticed during the feeding experiments is that the cells grown in medium plus fatty acid appeared smaller than those grown in axenic medium lacking fatty acids (Fig. 1A, 0-h versus 8-h panel). To measure the cell volume, we spun cells after overnight feeding (14 h) into a glass capillary made for hematocrit measurements through a funnel built from a plastic pipette tip. Indeed, the pellet volume occupied by the cells from the fatty acid-containing diet was reduced to 74% ± 3% (n = 3). Because these changes are paralleled in protein content, we corrected all quantitative data accordingly (see Materials and Methods).
Purification of lipid droplets and proteomic analysis.
About 10 years ago, Miura and coworkers had fused the sequence of the Dictyostelium homologue of perilipin (Plin) to GFP and expressed the construct in mammalian CHO cells, where it localized to lipid droplets (35). In order to obtain a marker for biochemical preparation of lipid droplets, we constructed a GFP-Plin fusion for the expression in Dictyostelium cells. This strain was fed with fatty acid, and the cells were mechanically cracked, leaving organelles intact. The preparation was loaded in the middle of a sucrose step gradient so that only fat-containing organelles were able to float to the top during ultracentrifugation. In contrast to untreated cells, the cells that were allowed to form lipid droplets had a semisolid white layer of fat on top of the gradient that was recovered with the help of a microbiological inoculation loop. Liquid fractions were taken with a pipette starting from the top, and all were separated on protein gels.
The first fraction of the fatty acid-induced cells contained protein bands that quickly decreased until fraction 5. In contrast, control cells completely lacked visible protein in the first five fractions (Fig. 2A). Indeed, Western blotting of the fractions revealed that the strong band observed at 70 kDa was GFP-Plin, which was enriched in fraction 1 (Fig. 2B), whereas it was detected only in the middle fractions if no fatty acid was added (Fig. 2C). Protein disulfide isomerase, a marker for the endoplasmic reticulum, was largely distributed over the lower half of the gradient (Fig. 2E) but gained a very small additional peak in the lipid droplet fraction (Fig. 2D). In contrast, mitochondria were most prominent in the densest fractions of the lower third of the gradient but did not reveal any association with the lipid droplet fraction, as indicated by the distribution of mitochondrial porin (Fig. 2F and G). One notable discrepancy between the fatty acid-induced samples and the untreated controls is the total amount of GFP-Plin (Fig. 2B versus C). Western blots of total cell homogenates also reveal this difference (data not shown), suggesting that, as in mammals (36, 37), perilipin is degraded if no lipid droplets are available as binding targets.
Fig 2.
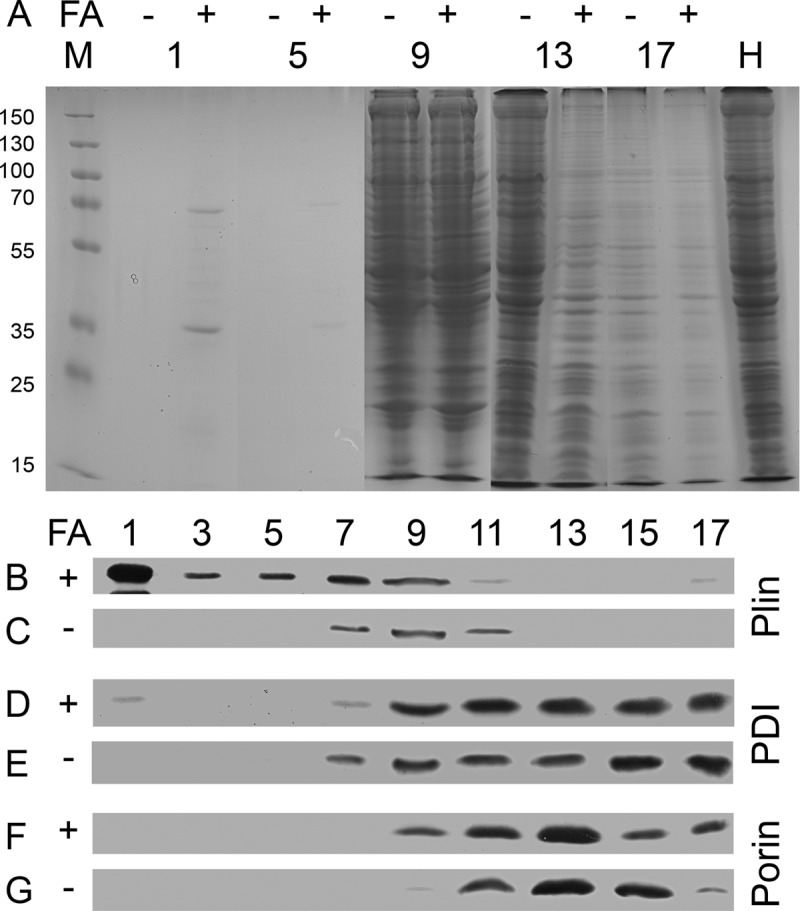
Purified lipid droplets contain a very limited set of proteins. (A) Cell homogenates from GFP-Plin-expressing untreated cells (−) or cells supplied with fatty acid (FA; +) were resolved on sucrose gradients by ultracentrifugation. Equal volumes taken from the gradient were loaded onto protein gels side by side, separated by electrophoresis, and stained by Coomassie blue. Although all 17 fractions of the gradient were analyzed on a total of three gels, only every fourth fraction (as numbered) was cut out and assembled into this panel. The assembly is flanked by a size marker (M; values in kDa) on the left and the total homogenate (H) on the right. (B to G) For Western blot analysis of the samples, every second fraction (as numbered) was taken, and GFP-perilipin (B and C), the protein disulfide isomerase (PDI) (D and E), or mitochondrial porin (F and G) was detected by the corresponding monoclonal antibody.
To achieve a broad coverage of proteins, we prepared lipid droplets under three conditions: from cells fed with fatty acid for 3 h (Table 1, first condition) and 16 h (second condition) and from cells after 5 h of fatty acid deprivation after 3 h of feeding (third condition). The protein samples from the lipid droplet preparations were subjected to a tandem mass spectrometry ion search.
Table 1.
Protein constituents of lipid droplets
| Protein group and identification no.a | MASCOT score by conditionb |
Mean MASCOT scorec | Gene name | Gene product | Presence in LDs of other cell type(s)d | ||
|---|---|---|---|---|---|---|---|
| 1st | 2nd | 3rd | |||||
| Structural LD protein | |||||||
| DDB0235170 | 930 | 968 | 2,348 | 1,416 | plnA | Perilipin | B, C, D |
| Enzymes of lipid metabolism | |||||||
| DDB0237965 | 1,608 | 480 | 1,381 | 1,157 | smtA | Putative delta-24-sterol methyltransferase Smt1 | A |
| DDB0191105 | 942 | 486 | 1,776 | 1,068 | fcsA | Long-chain fatty coenzyme A synthetase | A, B, C, D |
| DDB0304900 | 245 | 162 | 849 | 419 | Short-chain dehydrogenase/reductase family protein | ||
| DDB0185188 | 229 | 130 | 890 | 417 | comG | Putative NAD-dependent aldehyde dehydrogenase | A |
| DDB0304901 | 121 | 118 | 620 | 287 | Short-chain dehydrogenase/reductase family protein | ||
| DDB0238829 | 151 | 407 | 279 | Short-chain dehydrogenase/reductase family protein | |||
| DDB0238830 | 204 | 99 | 477 | 260 | Short-chain dehydrogenase/reductase family protein | ||
| DDB0219382 | 278 | 110 | 385 | 258 | agpC | Putative 1-acylglycerol-3-phosphate acyltransferase | A |
| DDB0233097 | 108 | 99 | 380 | 196 | dgat2 | Diacylglycerol-acyltransferase 2 | A |
| DDB0205694 | 146 | 87 | 299 | 178 | Putative acyltransferase | ||
| DDB0233059 | 50 | 36 | 240 | 109 | Putative sterol-4α-carboxylate 3-dehydrogenase | B, C, D | |
| DDB0235400 | 227 | 38 | 89 | Putative glycerol 3-phosphate acyltransferase | A, D | ||
| DDB0230057 | 59 | 94 | 51 | lip5 | Lipase family member 5 | A, B, C, D | |
| DDB0190742 | 51 | 58 | 37 | Short-chain dehydrogenase/reductase family protein | |||
| DDB0232044 | 52 | 54 | 36 | ksrA-1 | 3-Ketosphinganine reductase | A | |
| Small GTPases | |||||||
| DDB0191507 | 453 | 241 | 723 | 473 | rab7A | Rab GTPase | B, C, D |
| DDB0214821 | 393 | 243 | 732 | 456 | rab14 | Rab GTPase | D |
| DDB0191476 | 454 | 279 | 464 | 399 | rab1A | Rab GTPase | B, D |
| DDB0201663 | 213 | 215 | 436 | 288 | rasG | Ras GTPase | |
| DDB0191190 | 331 | 158 | 317 | 269 | rab11A | Rab GTPase | D |
| DDB0201639 | 164 | 90 | 393 | 216 | rab32A | Rab GTPase | |
| DDB0229398 | 178 | 149 | 308 | 212 | rab1D | Rab GTPase | |
| DDB0214820 | 173 | 146 | 288 | 203 | rab1B | Rab GTPase | D |
| DDB0214827 | 125 | 74 | 349 | 183 | rasC | Ras GTPase | |
| DDB0215409 | 50 | 44 | 394 | 163 | ranA | Ran GTPase | |
| DDB0214885 | 264 | 216 | 160 | rab8A | Rab GTPase | D | |
| DDB0185061 | 124 | 84 | 265 | 158 | rabC | Rab GTPase | |
| DDB0229409 | 123 | 87 | 222 | 144 | rab18 | Rab GTPase | C, D |
| DDB0233303 | 84 | 40 | 232 | 119 | ragC | Ras GTPase | |
| DDB0229401 | 168 | 39 | 140 | 116 | rab5A | Rab GTPase | D |
| DDB0216229 | 213 | 105 | 106 | rapA | Ras GTPase | ||
| DDB0214822 | 152 | 44 | 98 | rac1A | Rho GTPase | ||
| DDB0214825 | 182 | 94 | 92 | racE | Rho GTPase | ||
| DDB0229402 | 195 | 68 | 88 | rab2B | Rab GTPase | D | |
| DDB0216191 | 143 | 78 | 74 | rab2A | Rab GTPase | C | |
| DDB0191203 | 100 | 104 | 68 | rab1C | Rab GTPase | ||
| DDB0231625 | 142 | 48 | 64 | Putative small GTPase activator | |||
| DDB0229392 | 77 | 62 | 47 | rab6 | Rab GTPase | B | |
| DDB0214824 | 76 | 61 | 46 | racB | Rho GTPase | ||
| DDB0229419 | 72 | 62 | 45 | rabF1-1 | Rab GTPase | ||
| DDB0229394 | 73 | 56 | 43 | rab5B | Rab GTPase | D | |
| DDB0201661 | 82 | 39 | 41 | rasB | Ras GTPase | ||
| DDB0191253 | 71 | 46 | 39 | rab21 | Rab GTPase | D | |
| DDB0229365 | 68 | 40 | 36 | rabQ | Rab GTPase | ||
| DDB0216195 | 57 | 47 | 35 | rasD | Ras GTPase | ||
| DDB0229413 | 40 | 65 | 35 | rabG2 | Rab GTPase | ||
| DDB0229406 | 44 | 57 | 34 | rabG1 | Rab GTPase | ||
| ER constituents | |||||||
| DDB0231364 | 143 | 85 | 444 | 224 | alg2 | Putative alpha-1,3-mannosyltransferase | A, C |
| DDB0185040 | 42 | 545 | 196 | pdi1 | Protein disulfide isomerase | A, B | |
| DDB0219977 | 54 | 298 | 118 | rplP0 | 60S ribosomal acidic ribosomal protein P0 | A | |
| DDB0231975 | 66 | 60 | 205 | 111 | ugt1 | Putative glycosyltransferase | |
| DDB0233663 | 63 | 245 | 103 | Heat shock protein 70 with ER retention signal | B, D | ||
| DDB0233863 | 77 | 160 | 79 | rtnlC | Reticulon family protein | B, C, D | |
| DDB0231366 | 53 | 158 | 71 | alg11 | Putative alpha-1,2-mannosyltransferase | A | |
| DDB0231241 | 129 | 77 | 69 | rpl4 | 60S ribosomal protein L4 | A, C | |
| DDB0231191 | 61 | 98 | 53 | rpl10 | S60 ribosomal protein L10 | A, C | |
| DDB0191150 | 101 | 48 | 50 | alg1 | Putative glycosyltransferase | B | |
| DDB0214854 | 52 | 67 | 40 | rpl19 | S60 ribosomal protein L19 | A, C | |
| Cytoskeleton components | |||||||
| DDB0191168 | 245 | 79 | 742 | 356 | hspB | Heat shock protein 70 cytoplasmic, F-actin associated | A, B, D |
| DDB0191134 | 43 | 77 | 836 | 319 | efaA11 | Elongation factor 1α, F-actin binding and bundling | C |
| DDB0185047 | 275 | 82 | 487 | 282 | hspE-1 | Heat shock protein 70, cytoplasmic, F-actin associated | A, B |
| DDB0185015 | 92 | 133 | 448 | 225 | act | Actin | B, C |
| DDB0219923 | 206 | 65 | 91 | comA | Comitin, F-actin-binding Golgi protein | ||
| DDB0305338 | 55 | 44 | 33 | Heat shock protein 70, cytoplasmic | A, B, D | ||
| Miscellaneous | |||||||
| DDB0266658 | 151 | 366 | 1,181 | 566 | aifC | Putative apoptosis inducing factor | B |
| DDB0191175 | 36 | 78 | 1,089 | 401 | cadA | Cell adhesion molecule, plasma membrane and cytosol | B |
| DDB0184006 | 124 | 68 | 726 | 306 | ldpA | Lipid droplet membrane protein | |
| DDB0238661 | 108 | 109 | 261 | 160 | netD | NET4 | |
| DDB0190526 | 96 | 43 | 171 | 104 | Putative transmembrane metallo-phosphoesterase | ||
| DDB0304659 | 59 | 67 | 42 | Contains protein phosphatase 2C-related domain | |||
| DDB0203653 | 43 | 52 | 32 | No recognizable features | |||
Proteins are grouped according to function. Rows in bold highlight proteins investigated in this work. Identification numbers are from dictyBase.
MASCOT scores of the MS/MS ion search procedure correspond to the frequency of peptide identification from three independent preparations as specified in the text.
Solely to provide a measure for sorting proteins within the groups the mean MASCOT score was determined.
To improve the chances of identifying genuine lipid droplet components, we chose only those proteins that appeared under at least two conditions with one MASCOT score being above the value of 50. In order to justify the selection, we picked the sole candidate that just fulfilled this criterion, DDB0235400, encoding a putative glycerol 3-phosphate acyltransferase. It was tagged with GFP and was verified to reside on lipid droplets (data not shown).
Our selection finally yielded 72 candidate proteins (Table 1), of which the majority were also identified in recent proteomic studies on yeast (38) and three mammalian cell types (39–41). We grouped our candidates into enzymes of lipid metabolism (15 enzymes), small GTPases (31 members), constituents of the (rough) endoplasmic reticulum (11 proteins), or cytoskeletal proteins (6 proteins). A set of seven proteins could not be classified in the above groups.
Verification of putative lipid droplet components.
To gain further support for the presence of the identified proteins on lipid droplets, we selected three candidates (shown in bold in Table 1), constructed N- and C-terminally GFP-tagged variants, and tested their lipid droplet association by microscopy.
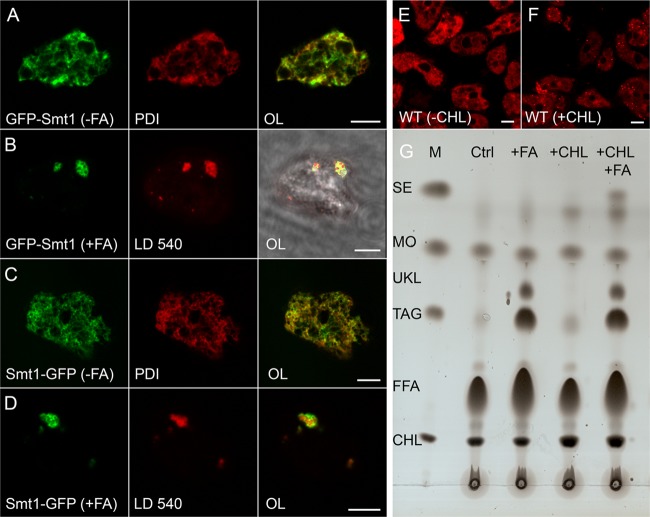
The strongest band on the protein gel (just above the 35-kDa marker in Fig. 2A) was identified as the product of the gene DDB0237965 (smtA) with homology to steryl methyltransferases (Smt) of plants and yeast. GFP-Smt1 localized at the endoplasmic reticulum in cells from axenic medium (Fig. 3A) but redistributed to lipid droplets when fatty acid was added (Fig. 3B). In an Smt1-GFP construct, where the order of protein domains was reversed, the same localization was observed (Fig. 3C and D). Since the presence of a sterol-metabolizing enzyme on lipid droplets suggested that they might contain dictyosterol, a modification of cholesterol (42) or its derivatives, we added cholesterol to the axenic culture medium, stained the cells with LD540, and indeed saw an increased number of lipid droplets (compare Fig. 3E and F). TLC analysis of these cells revealed an increase in the cholesterol band; however, only a small increase in the band of steryl esters (SEs), the form of the molecule typically stored in lipid droplets, was detectable (Fig. 3G). Because we reasoned that this might be due to limiting amounts of fatty acids, we further added palmitic acid and now observed formation of an additional band that comigrated with the marker cholesterol palmitate (Fig. 3G).
Fig 3.
Dictyostelium lipid droplets contain steryl esters. (A to D) Confocal images from fixed cells expressing steryl methyltransferase 1 (Smt1) tagged with GFP (green channel) at the N-terminal end (A and B) or at its C terminus (C and D) and incubated with (B and D) or without (A and C) fatty acid (FA). The endoplasmic reticulum was revealed by virtue of an antibody directed against PDI that appears red in panels A and C. Alternatively, lipid droplets were stained by LD540 (red in B and D). The overlaid images (OL) appear in the third column (scale bar, 5 μm), where for row B the image from transmitted light is also shown to demonstrate the outline of the otherwise barely visible cell. (E and F) Optical sections through living wild-type (WT) cells stained with LD540 (red) to reveal lipid droplets (dots in panel F) in cells fed with cholesterol (+CHL) for 3 h. In control cells (−CHL) the dye associates nonspecifically with organelle membranes such as the nuclear envelope and the closely associated Golgi apparatus (E). Scale bar, 5 μm. (G) Thin-layer chromatography of lipid samples extracted from wild-type cells grown in axenic medium without further additives (Ctrl), with 200 μM palmitic acid added (+FA), with 100 μM cholesterol (+CHL) added, or with both (+CHL +FA). Substances in the marker lane (M) are labeled as in Fig. 1D. Here, only steryl esters (SE) are relevant. An unknown lipid species (UKL) is further discussed in the text.
To obtain more quantitative information on the composition of lipid droplets, two preparations, one obtained after challenging wild-type cells with palmitic acid only and the other one isolated after feeding cells simultaneously with palmitate and cholesterol, were analyzed for their fatty acid content as well as composition (Table 2). Palmitic acid (denoted as C16:0) is readily incorporated into all lipid species. However, it is notable that the pool of free fatty acids still contains vast amounts of the major endogenous fatty acids with chain lengths of 16 or 18 carbon atoms and various degrees of unsaturation, indicating that there is no shortage in the supply of a specific acyl chain. Phospholipids building the limiting monolayer of the lipid droplet preferentially incorporate the fully saturated C18 fatty acid, whereas TAG and one unknown lipid (UKL) are rather enriched in C18:1. Lipid droplets derived from cholesterol treatment, however, show a clear increase in the amount of steryl esters with a concomitant reduction of TAG in the same order of magnitude. The added cholesterol almost completely replaces the endogenous sterol moieties in dictyosteryl esters and clionastanyl esters (Table 2, footnote c) while leaving the choice of acyl chains almost unaltered.
Table 2.
Fatty acid distribution within lipid classes of isolated lipid droplets
| Condition and lipid classa | FA distributionb |
|||||||
|---|---|---|---|---|---|---|---|---|
| Total amt measured (nmol/sample) | Relative amt (%) by chain type |
Calculated amt (nmol/sample) | Mol% | |||||
| 16:0 | 16:1 | 18:0 | 18:1 | 18:2 | ||||
| +FA | ||||||||
| PL | 12.0 | 35.0 | 7.5 | 47.5 | 6.6 | 7.5 | 6.0 | 1.4 |
| DAG | 21.3 | 42.3 | 0.5 | 34.7 | 16.9 | 0.9 | 10.6 | 2.4 |
| FFA | 97.2 | 18.7 | 12.8 | 7.4 | 23.6 | 35.2 | 97.2 | 21.8 |
| TAG | 765.5 | 50.0 | 8.4 | 3.7 | 19.8 | 21.2 | 255.2 | 57.4 |
| UKL | 116.1 | 37.6 | 3.2 | 7.5 | 40.8 | 9.1 | 58.1 | 13.1 |
| SEc | 17.6 | 39.8 | 0.6 | 31.8 | 8.0 | 19.3 | 17.6 | 4.0 |
| Total | 444.7 | 100.2 | ||||||
| +FA +CHL | ||||||||
| PL | 25.5 | 34.5 | 1.2 | 56.0 | 3.1 | 4.3 | 12.8 | 3.6 |
| DAG | 20.5 | 47.8 | 2.0 | 40.5 | 8.8 | 0.5 | 10.2 | 2.9 |
| FFA | 65.1 | 27.3 | 8.8 | 16.9 | 20.6 | 26.0 | 65.1 | 18.2 |
| TAG | 516.5 | 53.4 | 6.6 | 5.0 | 18.4 | 14.1 | 172.2 | 48.2 |
| UKL | 80.4 | 44.2 | 2.5 | 14.2 | 32.7 | 6.0 | 40.2 | 11.2 |
| SEc | 57.0 | 43.0 | 4.4 | 16.3 | 8.9 | 25.3 | 57.0 | 15.9 |
| Total | 357.5 | 100.0 | ||||||
Lipid droplets were isolated under two experimental conditions, after feeding cells with palmitic acid only (+FA) or with both palmitic acid and cholesterol (+FA +CHL). The lipid classes are abbreviated as PL for phospholipids, DAG for diacylglycerol, FFA for free fatty acids, TAG for triacylglycerol, UKL for the unknown lipid, and SE for steryl esters.
Measured (total) values of fatty acids within each lipid class (nmol/sample) and relative amounts for each lipid class (%) are shown; the amounts were then calculated back according to the number of fatty acids expected in each class (nmol/sample). The relative contribution of each lipid class to the entire lipid droplet is shown as mol%.
For steryl esters, relative contributions of cholesterol, dictyosterol, clionastanol, and other sterols are as follows, in respective order: with fatty acids, 0.0, 69.3, 23.9, and 6.3%; with both fatty acids and cholesterol, 91.9, 6.0, 1.6, and 0.5%.
Next, we turned to two newly discovered proteins that do not have an obvious function in lipid metabolism. The protein encoded by the DDB0184006 gene did not bear significant homologies to any gene from other organisms. We produced N-terminal as well as C-terminal fusions of GFP to the coding region, and both hybrids changed their distribution from the ER (Fig. 4A and C) to lipid droplets upon fatty acid addition (Fig. 4B and D). Therefore, we named the protein Ldp (for lipid droplet protein). The gene is called ldpA in accordance with Dictyostelium nomenclature rules. The amino acid sequence of this protein is extremely rich in asparagine and lysine residues, resulting in an overall isoelectric point of 9.5, according to several calculation methods. The most acidic patch (pI 4.1) between residues 305 to 356 most likely participates in the formation of a coiled-coil structure (Fig. 4E, red residues). Moreover, Ldp is characterized by a high content of serine and threonine residues, opening the possibility of being phosphorylated; however, we did not detect obvious shifts in molecular masses on Western blots from samples derived from different cultivation conditions. These predominant amino acids often occur in homooligomeric repeats of up to 9 residues. Internet resources also predict the presence of three transmembrane domains (Fig. 4E, blue residues). To check the validity of this prediction, we attempted to extract Ldp-GFP with various buffers from the endoplasmic reticulum membrane and succeeded only when the detergent Triton X-100 was used (Fig. 4F). The Ldp hybrid with GFP fused to the N terminus behaved in the same way.
Fig 4.
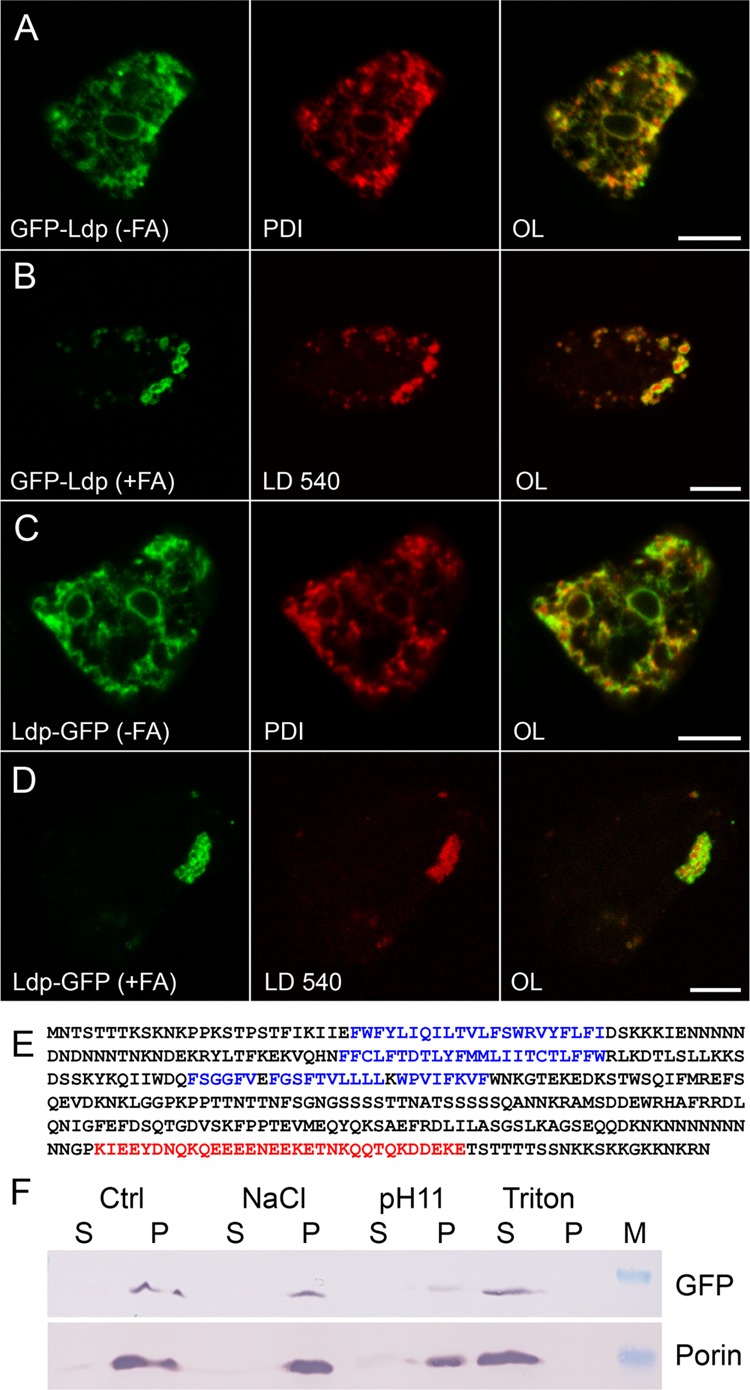
The novel protein Ldp moves from the ER to lipid droplets. (A to D) Single confocal planes through fixed cells expressing Ldp fused to GFP (green channel) at the N-terminal end (A and B) or carrying the GFP tag at the C terminus (C and D) and incubated in control medium (A and C) or in the presence of palmitic acid (B and D). The endoplasmic reticulum was revealed by immunofluorescence staining with anti-PDI (A and C), whereas lipid droplets were revealed by LD540 (B and D). The overlaid images (OL) show red and green channels. Scale bar, 5 μm. (E) Amino acid sequence of Ldp displayed in one-letter code (60 residues per line). Possible transmembrane segments are shown in blue; a region with coiled-coil character is printed in red. For other features of the protein, see the text. (F) Western blot of supernatant (S) or pellet (P) samples from separating a homogenate derived from Ldp-GFP-expressing cells incubated with homogenization buffer alone (Ctrl), 1 M NaCl, or Na2CO3 at pH 11 (pH 11) to liberate weakly or tightly associated membrane proteins, respectively. Alternatively, Triton X-100 was used to extract transmembrane proteins. The upper band is GFP-tagged LdpA detected by antibody 264−449−2; the lower band represents porin, a protein spanning the outer mitochondrial membrane.
Homologs of the third protein, encoded by the DDB0238661 gene, are found in plants, insects, and vertebrates with identities ranging between 25 and 30% only. A rather low value of conservation is also found in other Dictyostelium species such as Dictyostelium purpureum and Dictyostelium fasciculatum, which bear just 56 and 38% identical residues, respectively. The corresponding protein is best studied in mammals, where it is named DUF829 (for domains of unknown function), Tmem53 (for transmembrane protein) or, most frequently, NET4 (for nuclear envelope transmembrane protein 4). The name adopted for Dictyostelium protein is Net4, encoded by the netD locus. Indeed, this name appears suitable because both GFP fusions localize to the endoplasmic reticulum in Dictyostelium cells, with an apparent enrichment in the nuclear envelope (Fig. 5A, B, and C) as in mammals (43). When Net4-GFP-expressing cells are stimulated with fatty acids, the protein moves to lipid droplets, and the staining of endoplasmic reticulum and nuclear envelope is concomitantly reduced (Fig. 5D). The GFP-Net4 fusion, however, fails to undergo this redistribution (Fig. 5B). To test whether the mammalian NET4 protein also redistributes to lipid droplets under appropriate conditions, we transfected a plasmid expressing the human homologue tagged at its C-terminal end with GFP (24) into two mammalian cell lines and obtained a clear association with LD540-labeled structures induced by oleate feeding (Fig. 5E and F).
Fig 5.
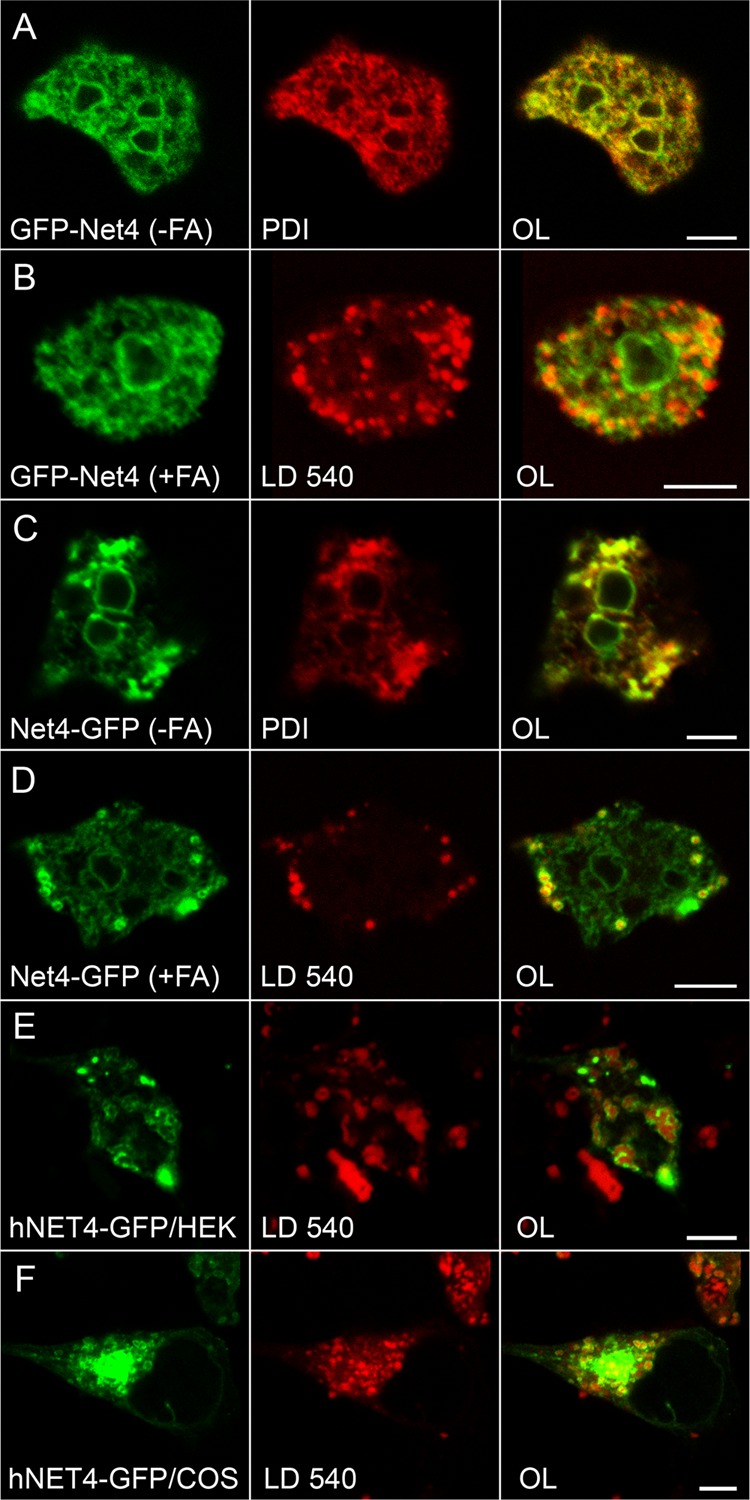
Net4 is a facultative lipid droplet protein. (A to D) Confocal images taken from fixed Dictyostelium cells expressing Net4 tagged with GFP (green channel) at its N-terminal end (A and B) or producing GFP fused to the C terminus of Net4 (C and D). The cells were incubated with (B and D) or without (A and C) fatty acid (FA), whereupon the endoplasmic reticulum was identified by virtue of an antibody directed against PDI (red in panels A and C). For panels B and D, lipid droplets were stained using LD540. Mammalian HEK293T (E) or COS7 (F) cells were transfected with a plasmid encoding the long splice variant of human NET4 fused to GFP (green) and imaged after 24 h by confocal microscopy. The formation of lipid droplets (stained with LD540; red) was stimulated with 400 μM oleic acid overnight. Cells were selected to express low levels of the hybrid protein so that the decoration of lipid droplets is visible, despite the presence of dispersed aggregates in COS7 cells or juxtanuclear accumulations in the HEK293T line. The overlaid images (OL) are shown in the third column. Scale bar, 5 μm.
DISCUSSION
Flow of fatty acids.
The natural food sources of Dictyostelium, i.e., various species of bacteria, contain fatty acids mostly in esterified form. Endosomal lipases are thought to liberate fatty acids, which can partition in the membrane vectorially. The direction of this pathway is brought about by the activity of the FcsA enzyme on the cytoplasmic face of the endosome, which adds a coenzyme A moiety to produce the activated fatty acid, which is then further funneled into metabolism (15).
In the laboratory, it is more reproducible to induce the formation of lipid droplets that store neutral fat by adding a fatty acid to the axenic medium (Fig. 1). It is interesting that added fatty acid is incorporated first into TAG and only with a delay leads to the accumulation of steryl esters (the band above methyl oleate [MO] in Fig. 1D). Depletion of the fatty acids from the medium is followed by the loss of lipid droplets and the degradation of the TAG storage molecule (Fig. 1). It is possible that the liberated fatty acids are metabolized to yield energy in mitochondria or peroxisomes, both of which contain the enzymes necessary for β oxidation (44). Peroxisomes especially are important for degrading the cyclopropane fatty acids that derive from phagocytosed bacteria (45). Alternatively, fatty acids could be incorporated into membrane lipids (46) which are required to meet the demands of the organelles that will be distributed to daughter cells during the three cell generations that occur within the 24-h cycle of lipid droplet formation and breakdown (Fig. 1D and E). Nevertheless, it is interesting that fatty acid addition and thus the presence of TAG stores do not significantly shorten the generation time of Dictyostelium amoebae (13, 14; also data not shown). Thus, the endogenous rate of de novo fatty acid synthesis appears to be fully sufficient for normal cell division. This view is further supported by two observations. First, an inhibitor of fatty acid synthase, cerulenin, completely inhibits growth of Dictyostelium cultures at a concentration of 5 μg/ml unless an exogenous fatty acid is added (data not shown). Second, amoebae growing on bacteria as a food source strongly downregulate the transcription of enzymes involved in de novo fatty acid synthesis (47). Knowledge about the path and kinetics of fatty acid flow will further support upcoming studies on the effect of therapeutically useful substances on fatty acid metabolism using Dictyostelium as a model system (48).
Composition of lipid droplets.
For experimental purposes, we have chosen to induce lipid droplet formation by the addition of palmitic acid and of cholesterol to the medium. Our quantitative analysis of lipid composition suggests that no fundamental differences exist in comparison to lipid droplets from other organisms.
By far, the major neutral lipid species in Dictyostelium lipid droplets is TAG, comprising roughly 57% of the constituent molecules. When cholesterol is given along with palmitic acid, the TAG level drops to about 48%, while steryl ester (SE) molecules increase from 4 to 16%. A similar TAG-to-SE ratio of ∼15 was seen in lipid droplets from the yeasts Yarrowia lipolytica (49) and Pichia pastoris (50) as well as in mammalian adipocytes (51). The first consequence of cholesterol addition is the appearance of a band that migrates slightly below the marker cholesteryl palmitate. Further addition of palmitate to the medium produces a second band that matches the marker perfectly (Fig. 5). Indeed, closer analysis (Table 2) reveals that 43% of this lipid is cholesteryl palmitate, apparently lacking any further modifications. Conjugated to palmitate and other acyl chains, the added cholesterol makes up 92% of the steryl esters within lipid droplets (Table 2), whereas it contributes roughly only 35% of the free sterol molecules (data not shown).
The membrane of the lipid droplet seems to be mainly composed of phospholipids, with either ethanolamine or choline as head groups in roughly equal amounts (data not shown). This composition, as well as the total amount, falls in the range of 1 to 2% as estimated for mammalian lipid droplets (52, 53) and yeast (50). The predominance of phosphatidylcholine in the limiting membrane of lipid droplets is attributed to its specific role in preventing lipid droplet coalescence within the cell (54). The amount of diacylglycerol (DAG) identified in our preparation is roughly equal to the amount of phospholipids. It is notable that the fatty acid composition of DAG more closely resembles that of phospholipids, preferentially containing stearic acid (C18:0). Thus, DAG more likely constitutes a precursor for further synthesis of membrane lipids than for TAG, which, in contrast, is enriched in unsaturated fatty acids (C18:1) in Dictyostelium as it is in yeast (38).
More frequently, biochemically prepared lipid droplet fractions from various organisms ranging from yeast and Drosophila to various mammalian cell types or organs have been analyzed by proteomic techniques. The numbers of proteins identified increase from 30 to 120 in mammals (25, 55–59) or 57 in yeast (38) to around 250 in Drosophila (60). The higher numbers do not necessarily reflect contaminations but may reveal intimate connections to specific organelles such as mitochondria (40) or point to specialized functions such as the storage of maternally provided histones in the Drosophila embryo (6).
The hallmark and most frequently used protein marker of lipid droplets is perilipin. In mammals (20) the perilipin 1 locus produces four isoforms, A to D. Moreover, four other proteins, adipose differentiation-related protein (ADRP; perilipin 2), TIP47 (perilipin 3), S3-12 (perilipin 4), and OXPAT (perilipin 5), contain the conserved PAT domain and decorate lipid droplets often at different times during their biogenesis (61) as well as serving as informative indicators for their lipid composition (62). In Drosophila, the two perilipin homologues are called LSD1 and -2 (63). Dictyostelium has a single gene (63), plnA, and Dictyostelium perilipin tagged by fluorescent proteins is a cytosolic protein until it associates with lipid droplets after induction by fatty acid feeding (Fig. 2) (35; also data not shown). Interestingly, no perilipin genes are found in Caenorhabditis and yeast (63) although both organisms produce lipid droplets for TAG storage (64, 65). In plants and microalgae, perilipin function is fulfilled by the group of oleosin and major lipid droplet proteins (MLDPs), respectively (66, 67).
Our lipid droplet preparations contain a regularly appearing set of 72 proteins (Table 1). Among the 15 lipid-metabolizing enzymes, it is interesting that overall there is a better overlap with yeast than with mammals. In yeast and Dictyostelium especially, the enzymes that add the first, second, and third fatty acid to glycerol to produce TAG are present on lipid droplets, whereas they are not consistently found in the mammalian preparations. We are also surprised by the discovery of as many as five isoforms of the short-chain dehydrogenase/reductase gene family, absent from other investigated proteomes, the role of which needs to be determined in the future.
The other large group of proteins associated with our lipid droplet preparation are small GTPases of the Rab family (Table 1). Rabs have been found in virtually all lipid droplet proteomes thus far, sometimes with as many as 25 members (40), constituting about half of the total mammalian repertoire. Although experiments with GTPγS show some specificity of association (59), only Rab18 has also been localized on lipid droplets by microscopy and seems to play a functional role there (68, 69). Some authors could not confirm the proteomically reported presence of Rabs 5, 7, and 11 in mammalian cells (69), while others succeeded at least for Rab5 and Rab11 (70). We have failed to demonstrate lipid droplet association of GFP-tagged Rab7, obtained from Rupper et al. (71) in Dictyostelium (data not shown), but this does not exclude that a subset of the identified small GTPases is involved in regulating lipid droplet metabolism directly or indirectly, as recently investigated in depth for the Drosophila system (72).
Another enzyme that was strongly enriched in our lipid droplet preparation and confirmed by GFP tagging (Fig. 3) is Smt1. This protein was also found in proteomes from phagosomes containing latex beads (73) or macropinosomes (74). We have not observed macropinosome-like distribution of the GFP-tagged Smt protein, and its presence in the phagosome preparation can possibly be explained by the flotation method used that specifically relies on the isolation of light organelles. Smt1 has strong homologies (40% identical residues) to Erg6p from yeast, which is a major constituent of yeast lipid droplets (75). It catalyzes the addition of a methyl group to the carbon atom 24 of a cholesterol-like precursor molecule to yield ergosterol, the main sterol in yeast. We assume that Smt1 catalyzes a similar reaction, but because the C-24 position of mature dictyosterol carries an ethyl rather than a methyl group (42), the second methylation could then be performed by Smt2 (DDB0307261), similar to that shown in plants (76). Interestingly, we detected Smt2 in one of our lipid droplet preparations but rejected it from the constituent list (Table 1) based on the low score.
Apart from proteins that bear homologies to lipid-metabolizing enzymes, our proteomic analysis has also turned up proteins with unclear functions. One of them, bearing no significant homologies to any other organism, is Ldp. We are confident that it is a true lipid droplet protein because fusion proteins of GFP both to the N terminus and to the C-terminal end of Ldp localize to lipid droplets (Fig. 4). Presently, we see no effect of the increased amount of Ldp on the TAG amount or lipid patterns on TLC plates (data not shown), but it will be interesting to analyze overexpressing strains or knockout mutants with techniques that provide higher-resolution analysis of lipid constituents.
The other protein, Net4, localizes to the endoplasmic reticulum in the absence of added fatty acids and shows a distinct enrichment at the nuclear envelope compared to other ER markers (Fig. 5). This distribution is similar to the mammalian NET4 protein, which is known to preferentially reside in the outer nuclear membrane (43). The function ascribed to mammalian NET4 so far is based on small interfering RNA (siRNA) studies, which indicate that loss of NET4 slows down the cell cycle, even leading to premature senescence, depending on the cell type studied (24). Because Dictyostelium Net4 is found on lipid droplets when the medium is supplemented with fatty acid (Fig. 5D), we also tested the localization for the human NET4 protein and, indeed, found this property conserved from amoebae to humans (Fig. 5E and F).
Dual localization of lipid droplet proteins.
Looking at internet resources for the expression of the genes we have confirmed above as lipid droplet components of Dictyostelium, we find that all of them are expressed in vegetatively growing cells, i.e., in the absence of fatty acid addition. This was further supported by our reverse transcription-PCR (RT-PCR) experiments (data not shown). Because there are almost no detectable lipid droplets under these conditions, it was possible that the proteins localized elsewhere within the cell.
Indeed, Smt1, Ldp, and Net4 are all found in the endoplasmic reticulum in the absence of fatty acids, i.e., when lipid droplets are absent (Fig. 3, 4, and 5). Quite a number of ER-resident proteins relocalize to lipid droplets upon their formation. Examples from mammalian cells are UBXD8, AAM-B (77), DGAT2 (34), caveolin, ALDI (78), and ACSL3 (79). A previously mentioned example from yeast is Erg6p (75). Conversely, in a yeast strain unable to form lipid droplets, all typical lipid droplet-resident proteins localize to the ER (80). The large number of common proteins shared by these organelles is not surprising because it is widely accepted that lipid droplets are derived from the ER (81) although the precise mechanism of their formation is still under debate.
The dual localization of proteins also raises a topological problem because the ER membrane is a typical biological phospholipid bilayer, whereas the triglyceride core of the lipid droplet is surrounded by a monolayer only. Thus, the mode of protein binding is theoretically restricted to lipid anchors, amphipathic helices, or hairpin structures, whereas proteins with transmembrane stretches followed by hydrophilic tails cannot be accommodated (1) unless one assumes that excess membrane could form local wrinkles of bilayer, as proposed earlier (82).
Topological studies were recently started for some lipid-synthesizing enzymes (79), and the mode of membrane insertion was also investigated for caveolin (83). Preliminary biochemical experiments suggest that LpdA and Net4 behave like transmembrane proteins in the ER (Fig. 4F and data not shown). Given the observation that both GFP fusions of LdpA show the same localization behaviors, future experiments could address the question of whether the ends of this protein face the cytoplasm or the ER lumen and compare these topological results with data obtained from the Ldp protein residing on lipid droplets.
ACKNOWLEDGMENTS
We thank Carmen Demme for production of monoclonal antibodies from hybridoma cell lines. We are grateful to Petra Fey (Northwestern University) for suggestions on the gene and protein names and for conducting the annotation at dictybase.org. Christoph Thiele (Bonn, Germany) generously provided the lipid droplet-specific probe LD540, and Eric Schirmer (Edinburgh, United Kingdom) made the mammalian NET4 plasmids available. The perilipin cDNA clone was received from Hideko Urushihara (Tsukuba, Japan).
This work was supported by the European Union FP7 Health Programme (241481 Affinomics to F.W.H.).
Footnotes
Published ahead of print 13 September 2013
REFERENCES
- 1. Thiele C, Spandl J. 2008. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20:378–385 [DOI] [PubMed] [Google Scholar]
- 2. Brasaemle DL, Wolins NE. 2012. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J. Biol. Chem. 287:2273–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walther TC, Farese RV., Jr 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81:687–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robenek H, Buers I, Hofnagel O, Robenek MJ, Troyer D, Severs NJ. 2009. Compartmentalization of proteins in lipid droplet biogenesis. Biochim. Biophys. Acta 1791:408–418 [DOI] [PubMed] [Google Scholar]
- 5. Robenek H, Buers I, Robenek MJ, Hofnagel O, Ruebel A, Troyer D, Severs NJ. 2011. Topography of lipid droplet-associated proteins: insights from freeze-fracture replica immunogold labeling. J. Lipids 2011:409371. 10.1155/2011/409371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cermelli S, Guo Y, Gross SP, Welte MA. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16:1783–1795 [DOI] [PubMed] [Google Scholar]
- 7. Beller M, Sztalryd C, Southall N, Bell M, Jäckle H, Auld DS, Oliver B. 2008. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6:e292. 10.1371/journal.pbio.0060292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS. 2009. Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 122:1834–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, Ma Y, Liu P. 2012. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J. Lipid Res. 53:1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eichinger L, Pachebat JA, Glöckner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, Tunggal B, Kummerfeld S, Madera M, Konfortov BA, Rivero F, Bankier AT, Lehmann R, Hamlin N, Davies R, Gaudet P, Fey P, Pilcher K, Chen G, Saunders D, Sodergren E, Davis P, Kerhornou A, Nie X, Hall N, Anjard C, Hemphill L, Bason N, Farbrother P, Desany B, Just E, Morio T, Rost R, Churcher C, Cooper J, Haydock S, van Driessche N, Cronin A, Goodhead I, Muzny D, Mourier T, Pain A, Lu M, Harper D, Lindsay R, Hauser H, James K, Quiles M, Madan Babu M, Saito T, Buchrieser C, Wardroper A, Felder M, Thangavelu M, Johnson D, Knights A, Loulseged H, Mungall K, Oliver K, Price C, Quail MA, Urushihara H, Hernandez J, Rabbinowitsch E, Steffen D, Sanders M, Ma J, Kohara Y, Sharp S, Simmonds M, Spiegler S, Tivey A, Sugano S, White B, Walker D, Woodward J, Winckler T, Tanaka Y, Shaulsky G, Schleicher M, Weinstock G, Rosenthal A, Cox EC, Chisholm RL, Gibbs R, Loomis WF, Platzer M, Kay RR, Williams J, Dear PH, Noegel AA, Barrell B, Kuspa A. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malchow D, Lüderitz O, Westphal O, Gerisch G, Riedel V. 1967. Polysaccharides of vegetative and aggregationally competent amoebae of the strain Dictyostelium discoideum. 1. In vivo degradation of bacterial lipopolysaccharides. Eur. J. Biochem. 2:469–479 (In German.) [DOI] [PubMed] [Google Scholar]
- 12. Long BH, Coe EL. 1977. Fatty acid compositions of lipid fractions from vegetative cells and mature sorocarps of the cellular slime mold Dictyostelium discoideum. Lipids 12:414–417 [DOI] [PubMed] [Google Scholar]
- 13. Weeks G. 1976. The manipulation of the fatty acid composition of Dictyostelium discoideum and its effect on cell differentiation. Biochim. Biophys. Acta 450:21–32 [DOI] [PubMed] [Google Scholar]
- 14. Rai A, Nothe H, Tzvetkov N, Korenbaum E, Manstein DJ. 2011. Dictyostelium dynamin B modulates cytoskeletal structures and membranous organelles. Cell. Mol. Life Sci. 68:2751–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. von Löhneysen K, Pawolleck N, Rühling H, Maniak M. 2003. A Dictyostelium long chain fatty acyl coenzyme A synthetase mediates fatty acid retrieval from endosomes. Eur. J. Cell Biol. 82:505–514 [DOI] [PubMed] [Google Scholar]
- 16. Gaudet P, Fey P, Basu S, Bushmanova YA, Dodson R, Sheppard KA, Just EM, Kibbe WA, Chisholm RL. 2011. dictyBase update 2011: web 2.0 functionality and the initial steps towards a genome portal for the Amoebozoa. Nucleic Acids Res. 39:D620–D624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerisch G, Albrecht R, Heizer C, Hodgkinson S, Maniak M. 1995. Chemoattractant-controlled accumulation of coronin at the leading edge of Dictyostelium cells monitored using a green fluorescent protein-coronin fusion protein. Curr. Biol. 5:1280–1285 [DOI] [PubMed] [Google Scholar]
- 18. Hanakam F, Albrecht R, Eckerskorn C, Matzner M, Gerisch G. 1996. Myristoylated and non-myristoylated forms of the pH sensor protein hisactophilin II: intracellular shuttling to plasma membrane and nucleus monitored in real time by a fusion with green fluorescent protein. EMBO J. 15:2935–2943 [PMC free article] [PubMed] [Google Scholar]
- 19. Jenne N, Rauchenberger R, Hacker U, Kast T, Maniak M. 1998. Targeted gene disruption reveals a role for vacuolin B in the late endocytic pathway and exocytosis. J. Cell Sci. 111:61–70 [DOI] [PubMed] [Google Scholar]
- 20. Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. 2010. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 51:468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Humbel BM, Biegelmann E. 1992. A preparation protocol for postembedding electron microscopy of Dictyostelium discoideum cells with monoclonal antibodies. Scanning Microsc. 6:817–825 [Google Scholar]
- 22. Monnat J, Hacker U, Geissler H, Rauchenberger R, Neuhaus E, Maniak M, Soldati T. 1997. Dictyostelium discoideum protein disulfide isomerase, an endoplasmic reticulum resident enzyme lacking a KDEL-type retrieval signal. FEBS Lett. 418:357–362 [DOI] [PubMed] [Google Scholar]
- 23. Spandl J, White DJ, Peychl J, Thiele C. 2009. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic 10:1579–1584 [DOI] [PubMed] [Google Scholar]
- 24. Korfali N, Srsen V, Waterfall M, Batrakou DG, Pekovic V, Hutchison CJ, Schirmer EC. 2011. A flow cytometry-based screen of nuclear envelope transmembrane proteins identifies NET4/Tmem53 as involved in stress-dependent cell cycle withdrawal. PLoS One 6:e18762. 10.1371/journal.pone.0018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. 2004. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta 1644:47–59 [DOI] [PubMed] [Google Scholar]
- 26. Bertinetti D, Schweinsberg S, Hanke SE, Schwede F, Bertinetti O, Drewianka S, Genieser HG, Herberg FW. 2009. Chemical tools selectively target components of the PKA system. BMC Chem. Biol. 9:3. 10.1186/1472-6769-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fey P, Gaudet P, Curk T, Zupan B, Just EM, Basu S, Merchant SN, Bushmanova YA, Shaulsky G, Kibbe WA, Chisholm RL. 2009. dictyBase—a Dictyostelium bioinformatics resource update. Nucleic Acids Res. 37:D515–D519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Troll H, Malchow D, Müller-Taubenberger A, Humbel B, Lottspeich F, Ecke M, Gerisch G, Schmid A, Benz R. 1992. Purification, functional characterization and cDNA sequencing of mitochondrial porin from Dictyostelium discoideum. J. Biol. Chem. 267:21072–21079 [PubMed] [Google Scholar]
- 29. Schleicher M, Gerisch G, Isenberg G. 1984. New actin-binding proteins from Dictyostelium discoideum. EMBO J. 3:2095–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 31. Heftmann E. 2004. Chromatography: fundamentals and applications of chromatography and related differential migration methods. Elsevier, San Diego, CA [Google Scholar]
- 32. op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, Nater M, Apel K. 2003. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15:2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aggarwal S, Yurlova L, Snaidero N, Reetz C, Frey S, Zimmermann J, Pahler G, Janshoff A, Friedrichs J, Muller DJ, Goebel C, Simons M. 2011. A size barrier limits protein diffusion at the cell surface to generate lipid-rich myelin-membrane sheets. Dev. Cell 21:445–456 [DOI] [PubMed] [Google Scholar]
- 34. Kuerschner L, Moessinger C, Thiele C. 2008. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic 9:338–352 [DOI] [PubMed] [Google Scholar]
- 35. Miura S, Gan JW, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. 2002. Functional conservation for lipid storage droplet association among perilipin-, ADRP-, and TIP 47-related proteins in mammals, Drosophila, and Dictyostelium. J. Biol. Chem. 277:32253–32257 [DOI] [PubMed] [Google Scholar]
- 36. Xu G, Sztalryd C, Lu X, Tansey JT, Gan J, Dorward H, Kimmel AR, Londos C. 2005. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J. Biol. Chem. 280:42841–42847 [DOI] [PubMed] [Google Scholar]
- 37. Xu G, Sztalryd C, Londos C. 2006. Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim. Biophys. Acta 1761:83–90 [DOI] [PubMed] [Google Scholar]
- 38. Grillitsch K, Connerth M, Kofeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G. 2011. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim. Biophys. Acta 1811:1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bouchoux J, Beilstein F, Pauquai T, Guerrera IC, Chateau D, Ly N, Alqub M, Klein C, Chambaz J, Rousset M, Lacorte JM, Morel E, Demignot S. 2011. The proteome of cytosolic lipid droplets isolated from differentiated Caco-2/TC7 enterocytes reveals cell-specific characteristics. Biol. Cell 103:499–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang H, Wang Y, Li J, Yu J, Pu J, Li L, Zhang S, Peng G, Yang F, Liu P. 2011. Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein A-I. J. Proteome Res. 10:4757–4768 [DOI] [PubMed] [Google Scholar]
- 41. Larsson S, Resjo S, Gomez MF, James P, Holm C. 2012. Characterization of the lipid droplet proteome of a clonal insulin-producing β-cell line (INS-1 832/13). J. Proteome Res. 11:1264–1273 [DOI] [PubMed] [Google Scholar]
- 42. Nes WD, Norton RA, Crumley FG, Madigan SJ, Katz ER. 1990. Sterol phylogenesis and algal evolution. Proc. Natl. Acad. Sci. U. S. A. 87:7565–7569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Malik P, Korfali N, Srsen V, Lazou V, Batrakou DG, Zuleger N, Kavanagh DM, Wilkie GS, Goldberg MW, Schirmer EC. 2010. Cell-specific and lamin-dependent targeting of novel transmembrane proteins in the nuclear envelope. Cell. Mol. Life Sci. 67:1353–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paschke P, Pawolleck N, Haenel F, Otto H, Rühling H, Maniak M. 2012. The isoform B of the Dictyostelium long-chain fatty-acyl-coenzyme A synthetase is initially inserted into the ER and subsequently provides peroxisomes with an activity important for efficient phagocytosis. Eur. J. Cell Biol. 91:717–727 [DOI] [PubMed] [Google Scholar]
- 45. Matsuoka S, Saito T, Kuwayama H, Morita N, Ochiai H, Maeda M. 2003. MFE1, a member of the peroxisomal hydroxyacyl coenzyme A dehydrogenase family, affects fatty acid metabolism necessary for morphogenesis in Dictyostelium spp. Eukaryot. Cell 2:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Long BH, Coe EL. 1974. Changes in neutral lipid constituents during differentiation of the cellular slime mold, Dictyostelium discoideum. J. Biol. Chem. 249:521–529 [PubMed] [Google Scholar]
- 47. Sillo A, Bloomfield G, Balest A, Balbo A, Pergolizzi B, Peracino B, Skelton J, Ivens A, Bozzaro S. 2008. Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium. BMC Genomics 9:291. 10.1186/1471-2164-9-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Elphick LM, Pawolleck N, Guschina IA, Chaieb L, Eikel D, Nau H, Harwood JL, Plant NJ, Williams RS. 2012. Conserved valproic acid-induced lipid droplet formation in Dictyostelium and human hepatocytes identifies structurally active compounds. Dis. Model Mech. 5:231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Athenstaedt K, Jolivet P, Boulard C, Zivy M, Negroni L, Nicaud JM, Chardot T. 2006. Lipid particle composition of the yeast Yarrowia lipolytica depends on the carbon source. Proteomics 6:1450–1459 [DOI] [PubMed] [Google Scholar]
- 50. Ivashov VA, Grillitsch K, Koefeler H, Leitner E, Baeumlisberger D, Karas M, Daum G. 2013. Lipidome and proteome of lipid droplets from the methylotrophic yeast Pichia pastoris. Biochim. Biophys. Acta 1831:282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Suzuki M, Shinohara Y, Ohsaki Y, Fujimoto T. 2011. Lipid droplets: size matters. J. Electron Microsc. 60(Suppl. 1):S101–S116 [DOI] [PubMed] [Google Scholar]
- 52. Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. 2007. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 48:837–847 [DOI] [PubMed] [Google Scholar]
- 53. Blouin CM, Le Lay S, Eberl A, Koefeler HC, Guerrera IC, Klein C, Le Liepvre X, Lasnier F, Bourron O, Gautier JF, Ferre P, Hajduch E, Dugail I. 2010. Lipid droplet analysis in caveolin-deficient adipocytes: alterations in surface phospholipid composition and maturation defects. J. Lipid Res. 51:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, Farese RV, Jr, Walther TC. 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14:504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. 2004. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279:3787–3792 [DOI] [PubMed] [Google Scholar]
- 56. Brasaemle DL, Dolios G, Shapiro L, Wang R. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 279:46835–46842 [DOI] [PubMed] [Google Scholar]
- 57. Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R. 2004. Association of stomatin with lipid bodies. J. Biol. Chem. 279:23699–23709 [DOI] [PubMed] [Google Scholar]
- 58. Turro S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernandez MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A. 2006. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic 7:1254–1269 [DOI] [PubMed] [Google Scholar]
- 59. Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6:3256–3265 [DOI] [PubMed] [Google Scholar]
- 60. Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jäckle Kühnlein HRP. 2006. Characterization of the Drosophila lipid droplet subproteome. Mol. Cell. Proteomics 5:1082–1094 [DOI] [PubMed] [Google Scholar]
- 61. Wolins NE, Brasaemle DL, Bickel PE. 2006. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 580:5484–5491 [DOI] [PubMed] [Google Scholar]
- 62. Hsieh K, Lee YK, Londos C, Raaka BM, Dalen KT, Kimmel AR. 2012. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 125:4067–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lu X, Gruia-Gray J, Copeland NG, Gilbert DJ, Jenkins NA, Londos C, Kimmel AR. 2001. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm. Genome 12:741–749 [DOI] [PubMed] [Google Scholar]
- 64. Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421:268–272 [DOI] [PubMed] [Google Scholar]
- 65. Czabany T, Athenstaedt K, Daum G. 2007. Synthesis, storage and degradation of neutral lipids in yeast. Biochim. Biophys. Acta 1771:299–309 [DOI] [PubMed] [Google Scholar]
- 66. Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. 2006. The accumulation of oleosins determines the size of seed oil bodies in Arabidopsis. Plant Cell 18:1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Moellering ER, Benning C. 2010. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9:97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. 2005. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118:2601–2611 [DOI] [PubMed] [Google Scholar]
- 69. Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. 2005. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280:42325–42335 [DOI] [PubMed] [Google Scholar]
- 70. Liu P, Bartz R, Zehmer JK, Ying YS, Zhu M, Serrero G, Anderson RG. 2007. Rab-regulated interaction of early endosomes with lipid droplets. Biochim. Biophys. Acta 1773:784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rupper A, Grove B, Cardelli J. 2001. Rab7 regulates phagosome maturation in Dictyostelium. J. Cell Sci. 114:2449–2460 [DOI] [PubMed] [Google Scholar]
- 72. Wang C, Liu Z, Huang X. 2012. Rab32 is important for autophagy and lipid storage in Drosophila. PLoS One 7:e32086. 10.1371/journal.pone.0032086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gotthardt D, Blancheteau V, Bosserhoff A, Ruppert T, Delorenzi M, Soldati T. 2006. Proteomics fingerprinting of phagosome maturation and evidence for the role of a Gα during uptake. Mol. Cell. Proteomics 5:2228–2243 [DOI] [PubMed] [Google Scholar]
- 74. Journet A, Klein G, Brugiere S, Vandenbrouck Y, Chapel A, Kieffer S, Bruley C, Masselon C, Aubry L. 2012. Investigating the macropinocytic proteome of Dictyostelium amoebae by high-resolution mass spectrometry. Proteomics 12:241–245 [DOI] [PubMed] [Google Scholar]
- 75. Natter K, Leitner P, Faschinger A, Wolinski H, McCraith S, Fields S, Kohlwein SD. 2005. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol. Cell. Proteomics 4:662–672 [DOI] [PubMed] [Google Scholar]
- 76. Bouvier-Nave P, Husselstein T, Benveniste P. 1998. Two families of sterol methyltransferases are involved in the first and the second methylation steps of plant sterol biosynthesis. Eur. J. Biochem. 256:88–96 [DOI] [PubMed] [Google Scholar]
- 77. Zehmer JK, Bartz R, Bisel B, Liu P, Seemann J, Anderson RG. 2009. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J. Cell Sci. 122:3694–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ingelmo-Torres M, Gonzalez-Moreno E, Kassan A, Hanzal-Bayer M, Tebar F, Herms A, Grewal T, Hancock JF, Enrich C, Bosch M, Gross SP, Parton RG, Pol A. 2009. Hydrophobic and basic domains target proteins to lipid droplets. Traffic 10:1785–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Poppelreuther M, Rudolph B, Du C, Grossmann R, Becker M, Thiele C, Ehehalt R, Füllekrug J. 2012. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J. Lipid Res. 53:888–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sorger D, Athenstaedt K, Hrastnik C, Daum G. 2004. A yeast strain lacking lipid particles bears a defect in ergosterol formation. J. Biol. Chem. 279:31190–31196 [DOI] [PubMed] [Google Scholar]
- 81. Walther TC, Farese RV., Jr 2009. The life of lipid droplets. Biochim. Biophys. Acta 1791:459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ohsaki Y, Cheng J, Suzuki M, Shinohara Y, Fujita A, Fujimoto T. 2009. Biogenesis of cytoplasmic lipid droplets: from the lipid ester globule in the membrane to the visible structure. Biochim. Biophys. Acta 1791:399–407 [DOI] [PubMed] [Google Scholar]
- 83. Aoki S, Thomas A, Decaffmeyer M, Brasseur R, Epand RM. 2010. The role of proline in the membrane re-entrant helix of caveolin-1. J. Biol. Chem. 285:33371–33380 [DOI] [PMC free article] [PubMed] [Google Scholar]