Abstract
The creation of haploid gametes in yeast, termed spores, requires the de novo formation of membranes within the cytoplasm. These membranes, called prospore membranes, enclose the daughter nuclei generated by meiosis. Proper growth and closure of prospore membranes require the highly conserved Vps13 protein. Mutation of SPO71, a meiosis-specific gene first identified as defective in spore formation, was found to display defects in membrane morphogenesis very similar to those seen in vps13Δ cells. Specifically, prospore membranes are smaller than in the wild type, they fail to close, and membrane vesicles are present within the prospore membrane lumen. As in vps13Δ cells, the levels of phophatidylinositol-4-phosphate are reduced in the prospore membranes of spo71Δ cells. SPO71 is required for the translocation of Vps13 from the endosome to the prospore membrane, and ectopic expression of SPO71 in vegetative cells results in mislocalization of Vps13. Finally, the two proteins can be coprecipitated from sporulating cells. We propose that Spo71 is a sporulation-specific partner for Vps13 and that they act in concert to regulate prospore membrane morphogenesis.
INTRODUCTION
Starvation of diploid cells of the yeast Saccharomyces cerevisiae induces their differentiation into haploid spores, the equivalent of gametes in metazoans (1, 2). During meiosis, two sequential nuclear divisions produce four haploid nuclei, and each nucleus is then packaged within newly formed membranes to generate spores. As is frequently seen during differentiation in higher cells, sporulation requires rearrangement of the secretory pathway (3–5). In meiosis II, secretory vesicles are redirected from the plasma membrane to the poles of the meiotic spindle (6). At each of the four spindle pole bodies, the vesicles dock and fuse with each other to form small membrane compartments termed prospore membranes (5). As meiosis II proceeds, these membranes expand, and at the end of meiosis each prospore membrane encloses a nucleus to give rise to four immature spores. A spore wall then forms in the lumen of the prospore membrane, resulting in a mature spore (7).
Membrane closure is a type of cytokinesis, resulting in the physical separation of daughter cell and mother cell cytoplasms. Membrane closure requires removal of a protein complex that localizes to the lip of the prospore membrane termed the leading edge protein complex (LEP) (8). The LEP is composed of at least three proteins, SspI, Ady3, and Don1 (9, 10). SspI is targeted for degradation at the end of meiosis II through the action of the anaphase-promoting complex and its meiosis-specific activator, Ama1 (11). Degradation of SspI leads to release of Ady3 and Don1 from the lip of the prospore membrane and is a prerequisite for membrane closure (8, 11). An ama1 mutant, therefore, is unable to complete meiotic cytokinesis and shows persistent localization of the leading edge complex proteins at the lip of the prospore membrane.
Recently, we described a role for the VPS13 gene in promoting prospore membrane closure (12). VPS13 was originally identified by its role in vesicle traffic between the Golgi body and the vacuole (13–15). In vegetative cells, the protein is localized to the endosome, but during sporulation Vps13 relocalizes to the prospore membrane (12). vps13Δ mutants display a variety of prospore membrane defects, including small prospore membranes, the appearance of intraluminal vesicles within prospore membranes, and a failure to complete membrane closure. The small prospore membrane phenotype of the vps13Δ mutant results from reduced levels of phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] and its precursor, phosphatidylinositol-4-phosphate [PtdIns(4)P], in the prospore membrane (12). These results suggest that VPS13 is repurposed during sporulation to regulate PtdIns phosphate levels at the prospore membrane. VPS13 is highly conserved, and mutations of the human orthologs VPS13A and VPS13B give rise to the inherited disorders chorea acanthocytosis and Cohen syndrome, respectively (16, 17). Understanding the mechanism by which Vps13 influences PtdIns phosphate levels in yeast may therefore provide insight into the basis for these human diseases.
During sporulation, a highly regulated program of gene expression results in the induction of sporulation-specific proteins that can alter the function of constitutive gene products (1). For example, the sporulation-specific SNARE protein Spo20 works in concert with the constitutive SNARE molecules Sso1 and Snc1/2 to promote vesicle fusion at the prospore membrane (5, 18, 19). This work shows that Vps13 function is similarly changed during sporulation by the presence of a sporulation-specific protein encoded by SPO71. SPO71 is required for sporulation and for proper prospore membrane growth (20). Here, we report that the spo71Δ sporulation defect results from a defect in membrane closure. spo71Δ cells share several other phenotypes with vps13Δ cells as well, including small prospore membranes and intraluminal vesicles. The similarity of phenotypes in vps13Δ and spo71Δ cells suggests that they work together to promote membrane morphogenesis. We propose that meiotic induction of SPO71 triggers relocalization of Vps13 to the prospore membrane, where the complex of the two proteins functions to regulate PtdIns phosphate levels.
MATERIALS AND METHODS
Yeast strains and media.
Yeast strains used for this study are listed in Table 1. Unless otherwise mentioned, standard yeast media and genetic techniques were used (21). JSP247 and JSP248 were segregants from a cross of AN117-4B to a strain carrying a green fluorescent protein (GFP)-tagged VPS13 allele (15). JSP290 and JSP291 were segregants from a cross of JSP247 with AN361-6A. AN361-6A is a segregant from a cross of AN117-4B and a SPO71 gene deletion strain (22). AN363 was generated by mating of AN361-6A with a second segregant from the same cross. Mating of JSP290 to JSP291 generated strain JSP294.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| AN117-4B | MATα ura3 leu2 his3ΔSK trp1::hisG arg4-NspI lys2 hoΔ::LYS2 rme1Δ::LEU2 | 23 |
| AN117-16D | MATa ura3 leu2 trp1::hisG his3ΔSK lys2 hoΔ::LYS2 | 23 |
| AN120 | MATa/MATα ura3/ura3 his3ΔSK/his3ΔSK trp1::hisG/trp1::hisG ARG4/arg4-NspI lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 leu2/leu2 | 23 |
| AN361-6A | MATa ura3 leu2 trp1::hisG his3ΔSK lys2 hoΔ::LYS2 spo71Δ::his5+ | This study |
| AN363 | MATa/MATα ura3/ura3 his3ΔSK/his3ΔSK trp1::hisG/trp1::hisG lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 leu2/leu2 ARG4/arg4-NspI spo71Δ::his5+/spo71Δ:: his5+ | This study |
| HI29 | MATa/MATα ura3/ura3 his3ΔSK/his3ΔSK trp1::hisG/trp1::hisGARG4/arg4-NspI lys2/lys hoΔ::LYS2/hoΔ::LYS2 RME1/rme1Δ::LEU2 leu2/leu2 vps13Δ::his5+/vps13Δ::his5+ | 32 |
| JSP163 | MATa/MATα ura3/ura3 TRP1/trp1 ARG4/arg4 LEU2/leu2 spo71Δ::his5+/spo71Δ::his5+ TEF2::GFP::his5+/TEF2::GFP::his5+ | This study |
| JSP247 | MATα ura3 trp1::hisG arg4-NspI lys2 hoΔ::LYS2 rme1Δ::LEU2 leu2 VPS13::GFP::his5+ | This study |
| JSP248 | MATa ura3 trp1::hisG ARG lys2 hoΔ::LYS2 leu2 VPS13::GFP::his5+ | This study |
| JSP257 | MATa/MATα ARG4/arg4-NspI LEU2/leu2 ura3/ura3 trp1/trp1VPS13::GFP::his5+/VPS13::GFP::his5+ | 12 |
| JSP290 | MATa ura3 leu2 trp1::hisG hoΔ::LYS2 arg4-NspI spo71Δ::his5+ VPS13::GFP::his5+ | This study |
| JSP291 | MATα ura3 leu2 trp1::hisG hoΔ::LYS2 spo71Δ::his5+ VPS13::GFP::his5+ | This study |
| JSP294 | MATa/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG hoΔ::LYS2/hoΔ::LYS2arg4-NspI/ATG4 spo71Δ::his5+/spo71Δ::his5+ VPS13::GFP::his5+/VPS13::GFP::his5+ | This study |
| JSP391 | MATα ura3 trp1::hisG arg4-NspI lys2 hoΔ::LYS2 rme1Δ::LEU2 leu2 his3ΔSKVPS13::3HA::his5+ | This study |
| JSP392 | MATa ura3 trp1::hisG lys2 hoΔ::LYS2 leu2 his3ΔSK VPS13::3HA::his5+ | This study |
| JSP394 | MATα ura3 trp1::hisG arg4-NspI lys2 hoΔ::LYS2 leu2 VPS13::3HA::his5+ | This study |
| JSP401 | MATa/MATα ura3/ura3 trp1::hisG/trp1::hisG hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 ARG4/arg4-NspI VPS13::3HA::his5+/VPS13::3HA::his5+ | This study |
To construct JSP163 for the fluorescence loss in photobleaching (FLIP) assay, a spo71Δ segregant was mated with a strain containing a GFP-tagged TEF2 allele (15). After tetrad dissection, segregants containing both spo71Δ::his5+ and TEF2::GFP::his5+ were mated to generate strain JSP163. This strain was transformed with pRS426-RFP-SPO2051-91 prior to the FLIP assay.
To generate JSP401, VPS13 was first tagged with 3× hemagglutinin (HA) in haploid strains AN117-4B and AN117-16D (23) by PCR-based gene tagging using primers JSO81 and JSO82, creating JSP391 and JSP392, respectively. All oligonucleotide sequences are listed in Table 2. JSP394 is a segregant from the cross of JSP391 and AN117-16D. A cross of JSP392 and JSP394 generated the diploid strain JSP401.
Table 2.
Oligonucleotides used in this study
| Name | Sequence (5′–3′) |
|---|---|
| JSO61 | CCG CTC GAG TCA CAT ATG AAA GTA TAT AC |
| JSO62 | GGG GTA CCT TTG ATC TTT AAC AGC GCA T |
| JSO64 | TGA GTA TCC TGA GAT AAA ATT TGG A |
| JSO64-r | TCC AAA TTT TAT CTC AGG ATA CTC A |
| JSO65 | CGT CAC CAA TTA AAA TAA ATC TCA G |
| JSO65-r | CTG AGA TTT ATT TTA ATT GGT GAC G |
| JSO66 | GTGTATTAACCTTCTCATCCTCATC |
| JSO66-r | GAT GAG GAT GAG AAG GTT AAT ACA C |
| JSO67 | TAG TGT ACA AAA GCG GGT ATA TAC TTT CAT ATG TGA CTC GAG CGG TCA TAG GAT AGC TTC ACA GTA CTT A |
| JSO68 | TCC CCG CGG GAA GAT GGT AGA ACG CTG TC |
| JSO69 | ATA AGA ATG CGG CCG CTT AAC TGT TCT TAA TTT TCC TTT TT |
| JSO70 | GAA AAA AGG AAA ATT AAG AAC AGT TAA GCG GCC GCA TTC TTA TAT GTT AGA GTC TTT AGC TGC TAA |
| JSO81 | ACA TCG CCA TTG CTG TTA GAG AAT ACA ATA AGT ACT GTG AAG CTA TCC TAC GGA TCC CCG GGT TAA TTA A |
| JSO82 | GAA TTA TAG CTA CAT AGT GTA CAA AAG CGG GTA TAT ACT TTC ATA TGT GAG AAT TCG AGC TCG TTT AAA C |
| JSO159 | CGG GGG ATC CAC TAG TTC TAG AGC GGC CGC CAC CGC GGT GGA GCT CCC TTT ACG GTT GGA CAG TGG |
| JSO160 | AAC TTG AAT GAA ATT ATA TTC CAA AA |
| JSO160-r | TTT TGG AAT ATA ATT TCA TTC AAG TT |
| JSO161 | CAA GCG CGC AAT TAA CCC TCA CTA AAG GGA ACA AAA GCT GGA GCT CGC TAC AGT TTC GGT CTA AGT TT |
| JSO195 | TGG CGG CCG CTC TAG AAC TAG TGG ATC CCC CGG GCT GCA GGA ATT CAT GGA TTC TAT CGT TAA TGT TGT |
| JSO196 | AGC GCA TGA ACT CCT TGA TGA CGT CCT CGG AGG AGG CCA TAA GCT TCA TAG TTT GAT TCC GTG AAT TTG |
| HT290 | AAA GAC TAA GAA GAA CTG CGT CCA CTT CAA ATT CAC GGA ATC AAA CTA TGC GGA TCC CCG GGT TAA TTA A |
| HT291 | AGT ATA CAC TAA ATT TTA TGC AAT AAT AAA AAG AAAGCA TCC CGC CAA ACG AAT TCG AGC TCG TTT AAA C |
| HT293 | GAA GAA GGT ACC AAA TGA TGA TGA AAG GAC TG |
| HT66 | GAA GAA TTC AGA TCT ATA TTA CCC TGT TAT CC |
Plasmids.
Plasmids used in this study are listed in Table 3. To construct pRS426-VPS13, 500 bp of upstream and downstream sequence were first amplified by PCR using primer pair JSO68 and JSO69 and primer pair JSO61 and JSO62, respectively. The oligonucleotides used to amplify the upstream region (PVPS13) introduce the restriction enzyme sites SacII and NotI. After SacII and NotI double digestion, this fragment was ligated into SacII/NotI-digested pRS426 to generate pRS426-PVPS13. The downstream fragment (VPS13-Ter) was then similarly cloned into pRS426-PVPS13 using XhoI and KpnI to create pRS426-PVPS13-VPS13-Ter. Because of the large size of the VPS13 coding region (∼10 kb), this sequence was introduced into the plasmid by homologous recombination of overlapping segments in S. cerevisiae (24). Overlapping fragments of the VPS13 coding region were amplified by PCR using the primers JSO70 and JSO64-r, JSO64 and JSO65-r, JSO65 and JSO66-r, and JSO66 and JSO67. The 5′ ends of the primers JSO70 and JSO67 carry 27 and 36 nucleotides, respectively, which allow recombination with the plasmid pRS426-PVPS13-VPS13-Ter. vps13Δ mutant diploid cells (HI29) were cotransformed with the four PCR products and NotI/XhoI-linearized pRS426-PVPS13-VPS13-Ter. Transformants were screened for rescue of the vps13Δ sporulation defect, and the assembled pRS426-VPS13 was recovered from a rescued strain.
Table 3.
Plasmids used in this study
| Name | Gene(s) expressed | Source |
|---|---|---|
| pRS426-RFP-SPO2051-91 | PTEF2-RFP-SPO2051-91 | 33 |
| pRS424-DTR1-RFP | DTR1-RFP | 12 |
| pRS426-RFP-SPO2051-91-DON1-GFP | PTEF2-RFP-SPO2051-91, DON1-GFP | 12 |
| pRS426-PPRC1-GFP-PHOSH2 | GFP-PHOSH2 | 27 |
| pRS426-GFP-2 × PHPLCδ | GFP-2 × PHPLCδ | 26 |
| pRS426-VPS13 | VPS13 | This study |
| pRS424-SPO71 | SPO71 | This study |
| pRS424-SPO71-GFP | SPO71-GFP | This study |
| pRS426-PTEF2-SPO71-RFP | PTEF2-SPO71-RFP | This study |
pRS424-SPO71 was similarly constructed by homologous recombination in S. cerevisiae (24). Overlapping fragments of the SPO71 coding region including 500 bp upstream and downstream were amplified by PCR using the primers JSO159 and JSO160-r and JSO160 and JSO161. The 5′ ends of the primers JSO159 and JSO161 carry 40 nucleotides that allow recombination with the plasmid pRS424 linearized with SacI. The PCR products and linearized plasmid were cotransformed into spo71Δ mutant diploid cells (AN363), and correctly assembled plasmids were identified by complementation of the sporulation defect. pRS426-PTEF2-SPO71-RFP was constructed as pRS424-SPO71, except that pRS426-PTEF2-RFP was used as the linearized plasmid and the oligonucleotides and JSO195 and JSO196 were used in place of JSO159 and JSO161, respectively, to provide homology to the ends of the linearized plasmid. EcoRI/HindIII-digested pRS426-PTEF2-RFP and the two PCR products were introduced into spo71Δ diploid cells. pRS426-PTEF2-SPO71-RFP was recovered from a transformant whose sporulation defect was complemented. To generate pRS424-SPO71-GFP, the chromosomal copy of SPO71 was first fused to GFP by PCR-mediated transformation using HT290 and HT291 and pFA6a-yEGFP-HISMX6 as a template (10). SPO71-GFP with the SPO71 promoter was then amplified from genomic DNA of the transformant using HT293 and HT66 and cloned into pRS424.
Fluorescence microscopy.
For examination of localization of fluorescence markers to the prospore membrane, cells were replica plated from selective medium to SPO (sporulation) plates and incubated overnight at room temperature. Cells were transferred to microscopy slides, and sporulating cells at the appropriate stage were identified by the presence of prospore membranes.
Two fluorescence microscopes were used, a Zeiss Axioplan2 microscope (Carl Zeiss, Thornwood, NY) with a Zeiss mRM Axiocam and a Zeiss Observer.Z1 microscope with an attached Orca II ERG camera (Hamamatsu, Bridgewater, NJ). Zeiss Axiovision 4.8 software was used to acquire images. For scoring of red fluorescent protein (RFP)/GFP colocalization, only cells displaying both GFP and RFP fluorescence were included in the analysis.
Transmission electron microscopy.
Transmission electron microscopy was performed as previously described (12). Briefly, sporulating cells were fixed in glutaraldehyde and stained in potassium permanganate. Acetone was used for dehydration. The dehydrated cells were embedded in Epon. Images were collected with an AMT XR-60 camera (AMT, Danvers, MA) attached to an FEI BioTwin G2 microscope (FEI, Hillsboro, OR). To quantitate the abundance of intraluminal vesicles, every prospore membrane visible in a section was scored for the presence or absence of any intraluminal vesicles. As the presence or absence of vesicles varied between sections of the same cell, multiple sections from an individual cell were sometimes scored.
IP assays.
Sporulating cells (1 × 109 to 3 × 109) were lysed in ice-cold immunoprecipitation (IP) buffer (50 mM HEPES [pH 7.4], 150 mM KCl, 1 mM EDTA, 0.5% NP-40) containing 10 μg/ml Zymolyase, 1 mM dithiothreitol (DTT), and protease inhibitor cocktail (catalog number 04693124001, Complete Mini; Roche) using a FastPrep-24 (MP Bio) cell lysis machine. Lysates were cleared by centrifugation for 5 min at 11,000 × g, and 15 mg of cleared lysate was incubated with anti-HA (catalog number H3663; Sigma-Aldrich) at 4°C for 1 h. A 30-μl volume of Dynabeads Protein G (catalog number 100-04D; Life Technologies) was added and incubated for another hour. Protein-antibody-bead conjugates were collected by a magnet and washed twice with IP buffer containing 1 mM DTT. Proteins were eluted by the addition of 2× SDS sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 0.1 mg/ml bromophenol blue, 0.36 M β-mercaptoethanol) followed by boiling for 5 min. For precipitation of the GFP-tagged proteins, 20 mg of the lysate was incubated with anti-GFP (catalog number 632381; Clontech) and processed as for the anti-HA immunoprecipitation.
Proteins were detected by standard Western blot analysis. Since the molecular masses of Vps13-3HA and Spo71-GFP are about 365 kDa and 175 kDa, respectively, a 3 to 8% Tris-acetate gradient gel (Life Technologies) was used. For Western blot analysis, monoclonal anti-GFP or anti-HA antibodies were used at 1:1,000 dilutions.
RESULTS
SPO71 is required for membrane closure during spore formation.
We have previously described a fluorescence loss in photobleaching assay to identify mutants defective in prospore membrane closure (11). The FLIP assay differentiates whether prospore membranes are open or closed by measuring the fluorescence intensity inside the prospore membrane of a diffusible, cytoplasmic green fluorescent protein in response to repetitive photobleaching of the cytoplasm outside the prospore membrane. If the membrane is closed, then fluorescence persists within the prospore membrane but is lost in the cytoplasm of the surrounding mother cell. If closure has not occurred, GFP bleaches evenly both inside and outside the prospore membrane. Both ama1Δ and vps13Δ cells exhibit closure defects in this assay (11, 12). In addition, a significant closure defect was observed for spo71Δ cells. In postmeiotic spo71Δ cells, 72% of the prospore membranes were still open to the ascal cytoplasm, as opposed to 0% in wild-type cells at a similar stage (Table 4).
Table 4.
Comparison of prospore membrane closure in wild-type, ama1Δ, vps13Δ, and spo71Δ cells
Prospore membrane closure requires removal of the LEP from the mouth of the prospore membrane (8, 11). Don1-GFP is a convenient marker for LEP disassembly, as it is localized at the leading edge during prospore membrane growth and relocalizes throughout the spore cytoplasm coincidentally with membrane closure (8, 11). The previously identified closure ama1Δ and vps13Δ mutants differ with respect to removal of the LEP. In ama1Δ mutants, Don1-GFP persists at the leading edge in the majority of cells, whereas in vps13Δ mutants a majority of the cells display Don1-GFP redistributed to the cytoplasm (Table 4).
To determine if the closure defect of the spo71Δ mutant is upstream or downstream of LEP removal, the distribution of Don1-GFP was monitored in postmeiotic spo71Δ cells. Don1-GFP relocalizes to the spore cytoplasm in 67% of such cells, though the FLIP results suggest that the majority of prospore membranes are not yet closed (Table 4). Therefore, similar to what is seen in the vps13Δ mutant, the spo71Δ closure defect occurs after LEP removal (12).
Intraluminal vesicles accumulate in spo71Δ prospore membranes.
spo71Δ cells were examined for other vps13Δ sporulation phenotypes. In addition to the closure defects, vps13Δ mutants display a unique phenotype, the accumulation of membrane vesicles within the lumen of the prospore membrane (12). Transmission electron microscopy was performed to examine the prospore in spo71Δ cells. In wild-type cells, the luminal width between inner and outer prospore membrane is constant (Fig. 1A). In spo71Δ cells, this uniform distance is disrupted by the presence of intraluminal vesicles, just as in vps13Δ cells (Fig. 1B and C). These vesicles were visible in 37% of prospore membrane profiles in spo71Δ cells (n = 57), a percentage lower than that reported for vps13Δ cells (75% [12]) but still significant given that these vesicles were never seen in wild-type cells (n > 100). Expression of VPS13 from a high-copy-number plasmid did not suppress the sporulation defect of, or the formation of these vesicles in, the spo71Δ mutant, suggesting that VPS13 does not function downstream of SPO71 (Fig. 1D).
Fig 1.
Intraluminal vesicle accumulation within the prospore membrane. Transmission electron microscope images of prospore membranes in wild-type (AN120) cells (A), vps13Δ (HI29) cells (B), spo71Δ (AN363) cells (C), and spo71Δ cells overexpressing VPS13 (D). In all images, yellow arrows indicate prospore membranes and red arrows indicate the accumulation of vesicles within the lumen of a prospore membrane. Bars, 500 nm.
Prospore membrane PtdIns-phosphate pools are reduced in spo71Δ cells.
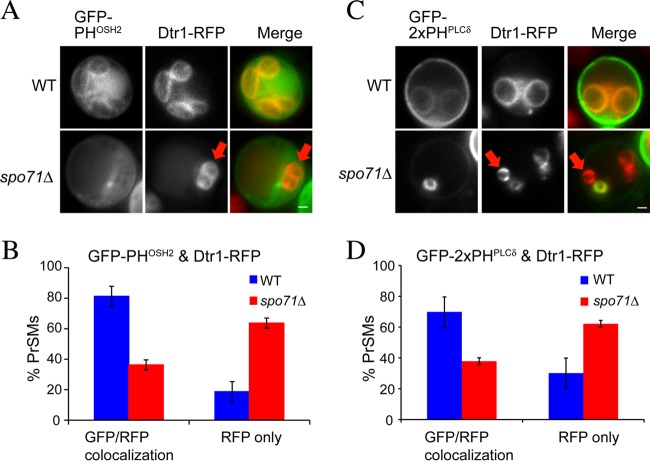
Many of the vps13Δ prospore membrane defects are caused by reduced levels of PtdIns(4)P and PtdIns(4,5)P2 within the prospore membrane (12). To investigate if these pools are reduced in spo71Δ cells, the localizations of two lipid-binding sensors—GFP-PHOSH2, a marker for PtdIns(4)P, and GFP-2xPHPLCδ, a marker for PtdIns(4,5)P2—were monitored during sporulation (25–27). In wild-type cells, both sensors colocalized with the prospore membrane marker Dtr1-RFP in >70% of the cells (Fig. 2). In spo71 cells, however, only 36% or 38% of the prospore membranes detectable by Dtr1-RFP displayed colocalization with GFP-PHOSH2 or GFP-2xPHPLCδ, respectively (Fig. 2). These results suggest that the levels of both of these lipids are reduced in the spo71Δ mutant, demonstrating another parallel between the spo71Δ and vps13Δ sporulation phenotypes.
Fig 2.
Effect of the spo71Δ mutation on prospore membrane PtdIns(4)P and PtdIns(4,5)P2 pools. (A) Sporulating wild-type (AN120) and spo71Δ (AN363) cells expressing both a PtdIns(4)P sensor (GFP-PHOSH2) and a prospore membrane marker (Dtr1-RFP). The red arrows indicate a prospore membrane without the GFP marker in the spo71Δ cell. Bar, 1 μm. (B) Quantitation of GFP-PHOSH2/Dtr1-RFP colocalization. Bars indicate standard errors of the means. More than 100 prospore membranes were scored in each of four separate experiments. (C) Sporulating wild-type (AN120) and spo71Δ (AN363) cells expressing both a PtdIns(4,5)P2 sensor (GFP-2xPHPLCδ) and a prospore membrane marker (Dtr1-RFP). The red arrows indicate a prospore membrane without the GFP marker in the spo71Δ cell. Bar, 1 μm. (D) Quantitation of GFP-PHPLCδ/Dtr1-RFP colocalization. Bars indicate standard errors of the means. More than 100 prospore membranes were scored in each of two separate experiments. The chi-square test indicates that the differences between wild-type and spo71Δ cells are significant (P < 0.001).
Spo71-GFP localizes to the prospore membrane during sporulation.
Vps13 localizes to prospore membranes during meiosis (12). If Spo71 and Vps13 work together directly, then Spo71 should also be found on these membranes. When the chromosomal copy of SPO71 was C-terminally tagged with GFP, no detectable fluorescence signal was seen in sporulating cells. Therefore, a SPO71::GFP fusion under the control of the SPO71 promoter was moved to a multicopy vector. When overexpressed, Spo71-GFP clearly colocalized with the prospore membrane marker RFP-Spo2051-91 (Fig. 3). This suggests that SPO71 may have rather direct function(s) in prospore membrane assembly.
Fig 3.
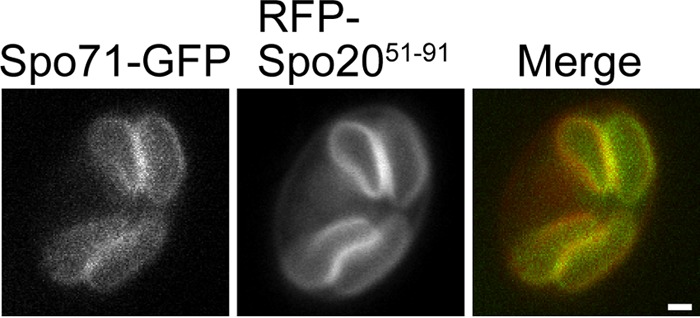
Localization of Spo71-GFP in sporulating cells. Wild-type (AN120) cells carrying pRS424-SPO71-GFP and the prospore membrane marker pRS426-RFP-Spo2051-91 were sporulated, and fluorescence was examined during prospore membrane growth. Colocalization to the prospore membrane was seen in 100% of the cells examined (n = 25). Bar, 1 μm.
SPO71 regulates Vps13 localization to the prospore membrane during spore formation.
To determine the genetic requirements for prospore membrane localization of Vps13 and Spo71, the localization of each protein was examined in the absence of the other gene. The prospore membrane localization of Spo71-GFP was unchanged in the absence of VPS13 (Fig. 4A). In contrast, Vps13-GFP failed to localize to prospore membranes in spo71Δ cells (Fig. 4B); only about 4% of prospore membranes contained Vps13-GFP (n > 150 in each of three independent experiments). Reintroduction of SPO71 on a plasmid restored prospore membrane localization of Vps13-GFP, confirming that SPO71 is required for proper localization of Vps13 during sporulation (85%, n > 230 in each of three independent experiments) (Fig. 4B).
Fig 4.
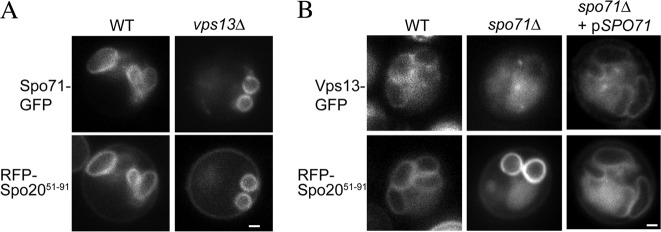
Vps13-GFP localization during sporulation in wild-type and spo71Δ cells. (A) Wild-type (AN120) and vps13Δ (HI29) cells carrying pRS424-SPO71-GFP and pRS426-RFP-Spo2051-91 were sporulated, and fluorescence was examined during prospore membrane growth. Colocalization to the prospore membrane was seen in 99% of the prospore membranes examined (n > 150). Bar, 1 μm. (B) VPS13-GFP SPO71 (JSP257) and VPS13-GFP spo71Δ (JSP294) cells carrying the prospore membrane marker RFP-Spo2051-91 were sporulated, and cells containing prospore membranes (as identified by the RFP-Spo2051-91 marker) were examined for colocalization of Vps13-GFP. Bar, 1 μm.
Ectopically expressed SPO71 alters Vps13-GFP localization in mitotic cells.
In mitotic cells, where Spo71 is not present, Vps13 is found on endosomes (15). If SPO71 is a regulator of Vps13 localization during sporulation, expression of SPO71 in mitotic cells might alter Vps13 localization. To test this hypothesis, SPO71::RFP was ectopically expressed under the control of the constitutive TEF2 promoter in mitotic cells that also expressed VPS13-GFP. The SPO71::RFP fusion is functional as shown by its ability to rescue the sporulation defect of a spo71Δ mutant (J.-S. Park, unpublished observation). In the absence of SPO71::RFP expression, Vps13-GFP foci are observed in the cytoplasm, likely at endosomes as previously reported (Fig. 5). Expression of SPO71::RFP caused a redistribution of the Vps13-GFP signal. Rather than cytoplasmic foci, GFP fluorescence was diffuse in the cytosol, with some concentration at the plasma membrane and bud neck (Fig. 5). Vps13-GFP signal at the plasma membrane was not seen in wild-type cells (Fig. 5).
Fig 5.
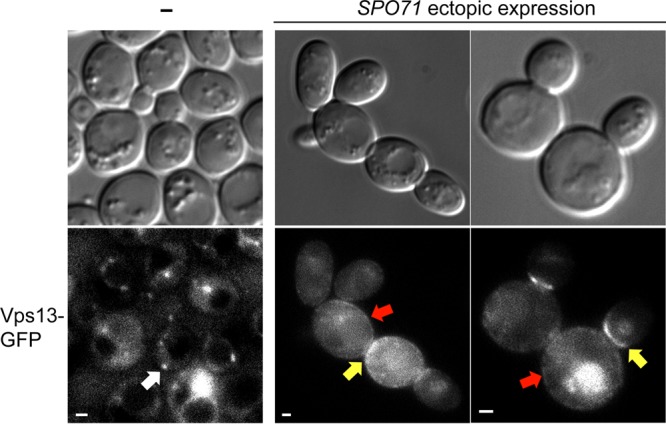
Effect of ectopically expressed SPO71 on Vps13-GFP localization in mitotic cells. VPS13-GFP (JSP257) cells with (right panels) or without (left panel) pRS426-PTEF2-SPO71-RFP were examined in mid-log-phase growth. The white arrow indicates Vps13-GFP localization to cytoplasmic foci, likely to be endosomes. Red and yellow arrows indicate Vps13-GFP localization to plasma membranes and bud necks, respectively. Bars, 1 μm. Plasma membrane or bud neck localization of Vps13-GFP was seen in 97% of the SPO71-overexpressing cells in two independent experiments, and at least 120 cells were examined in each.
Ectopically expressed SPO71 has been reported to localize to the plasma membrane in mitotic cells (20). This suggests that induction of Spo71-RFP might lead to colocalization with Vps13 at the plasma membrane and bud neck. However, we were unable to test this prediction, as the RFP fluorescence from our Spo71-RFP was seen uniformly throughout the cell, suggesting that the RFP moiety may be cleaved from Spo71 in vivo. Nonetheless, these results demonstrate that expression of SPO71::RFP is sufficient to alter Vps13 localization.
Spo71 physically interacts with Vps13 during sporulation.
To test for a physical interaction between Vps13 and Spo71, coimmunoprecipitation experiments were performed. Extracts made from sporulating cells expressing SPO71::GFP, VPS13::3xHA, or both fusions were used for immunoprecipitations with both anti-HA and anti-GFP antibodies. A Western blot of the anti-HA immunoprecipitates with anti-GFP antibodies revealed that Spo71-GFP precipitated only when coexpressed with Vps13-3xHA (Fig. 6). Reciprocally, pulldown of Spo71-GFP was able to specifically coprecipitate Vps13-3xHA. Thus, consistent with the similar phenotypes of the mutants, Spo71 forms a sporulation-specific complex with Vps13. The anti-HA pulldown reveals extensive degradation of Vps13-3xHA in the extracts. In contrast, only the full-length Vps13-3xHA protein is seen in the anti-GFP precipitates. As the HA epitopes are fused to the C-terminal end of Vps13, this observation suggests that the N terminus of Vps13 is essential for interaction with Spo71.
Fig 6.
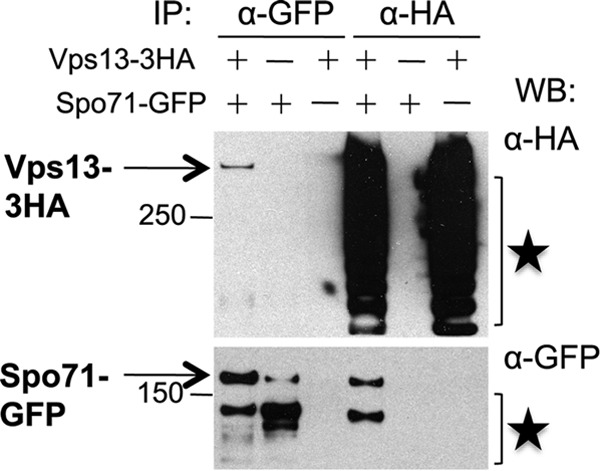
Spo71 interacts with Vps13 during sporulation. VPS13 (AN120) and VPS13-3HA (JSP401) cells carrying pRS424-SPO71-GFP or an empty vector were sporulated and then lysed, and immunoprecipitations were performed using anti-HA or anti-GFP antibodies. Precipitates were then probed by Western blotting using anti-HA and anti-GFP antibodies. Stars indicate Vps13-HA and Spo71-GFP degradation products. Molecular size markers are given in kilodaltons.
DISCUSSION
The data presented provide a model for how constitutive secretory pathway functions can be repurposed during differentiation. The Spo71 and Vps13 proteins form a sporulation-specific complex that regulates PtdIns phosphate levels in the prospore membrane. While VPS13 is constitutively expressed, SPO71 is sporulation specific in its expression. Our data suggest that induction of SPO71 when cells progress out of meiotic prophase leads to Spo71 protein binding to Vps13. In turn, interaction with Spo71 leads to release of Vps13 from the endosome, and both proteins then localize to the prospore membrane and influence PtdIns phosphate levels.
The spo71Δ prospore membrane phenotypes described are similar to those of vps13Δ cells; however, in all cases—e.g., membrane size, the percentage of membranes that fail to close, the fraction of membranes displaying intraluminal vesicles—the phenotype of the spo71Δ mutant is quantitatively less severe than that of the vps13Δ mutant. This suggests the possibility that the critical role of Spo71 is simply to recruit VPS13 to the prospore membrane, where Vps13 would be responsible for stimulating PtdIns phosphate levels. If so, overexpression of VPS13 might be expected to rescue some of the spo71 phenotypes. As overexpression of VPS13 does not bypass spo71, we prefer a model in which the two proteins act in concert to regulate PtdIns phosphates.
An important issue that remains to be resolved is whether Vps13 performs the same molecular function in two different locations, the endosome or the prospore membrane, at different stages of the life cycle, or if the function of Vps13 at the prospore membrane is different from its vegetative function in endosomal-Golgi body retrograde transport. While the answer to this question will require identifying the precise molecular function of Vps13 in both processes, the fact that SPO71 is required at the prospore membrane but not at the endosome may indicate that Vps13 has two independent functions.
Another critical question is how the Vps13/Spo71 complex affects PtdIns phosphate levels. As the levels of these lipids are based, ultimately, on the rate of their synthesis and degradation, Vps13/Spo71 likely acts either by inhibition of PtdIns-4-phosphate phosphatases or by activation of PtdIns-4 kinases. For the kinases, the likely targets would be either Pik1, responsible for the Golgi body pool of PtdIns(4)P in vegetative cells, or Stt4, responsible for the plasma membrane pool (28, 29). A previous study has implicated PIK1 in contributing to PtdIns phosphates in the prospore membrane (30). However, these effects could be indirect, as Pik1-generated PtdIns(4)P in the Golgi body could be delivered to the prospore membrane via vesicular transport. For example, initial levels of PtdIns(4)P might be from a Pik1-generated Golgi body pool delivered by vesicular transport, while maintenance of PtdIns(4)P levels might require Stt4 activity and the latter activity could be influenced by Vps13/Spo71. Consistent with this possibility, Stt4 localizes to the prospore membrane (J.-S. Park and H. Tachikawa, unpublished observations). To date, however, we have been unable to observe a physical interaction of Vps13 with either Pik1 or Stt4; however, it is possible that any effect on enzymatic activity of the kinases may not be mediated through direct binding.
VPS13 is highly conserved in eukaryotic organisms, including four paralogs in human cells (31). Mutations in two of these paralogs result in different genetic syndromes (16, 17). It is of interest, therefore, whether the Spo71/Vps13 complex or its function is conserved in higher organisms. While SPO71 is not as highly conserved as VPS13—putative SPO71 orthologs can be found in many fungi, but are absent from metazoans—the protein contains domains present in higher cells. The Spo71 protein contains 2 pleckstrin homology (PH) domains and a third conserved domain, which sequence alignments suggest may be a third, degenerate PH domain (20, 27; A. M. Neiman, unpublished observation). While no direct orthologs of SPO71 are present in metazoans, there are many PH domain-containing proteins. It is possible that one or more of these mammalian PH domain proteins interact with a Vps13 ortholog similarly to Spo71.
ACKNOWLEDGMENTS
We thank Nancy Hollingsworth and members of the Neiman lab for helpful discussions and comments on the manuscript. We are grateful to Susan Van Horn in the Stony Brook Center for Microscopy for assistance with electron microscopy.
This work was supported by the National Institutes of Health grant RO1 GM072540 to A.M.N.
Footnotes
Published ahead of print 13 September 2013
REFERENCES
- 1. Neiman AM. 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189:737–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neiman AM. 2005. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:565–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Byers B. 1981. Cytology of the yeast life cycle, p 59–96 In Srathern JN, Jones EW, Broach JR. (ed), The molecular biology of the yeast Saccharomyces. 1. Life cycle and inheritance. Cold Spring Harbor Monograph Archive, vol. 11 Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 4. Moens PB. 1971. Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. Can. J. Microbiol. 17:507–510 [DOI] [PubMed] [Google Scholar]
- 5. Neiman AM. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakanishi H, Morishita M, Schwartz CL, Coluccio A, Engebrecht JA, Neiman AM. 2006. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 119:1406–1415 [DOI] [PubMed] [Google Scholar]
- 7. Lynn RR, Magee PT. 1970. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J. Cell Biol. 44:688–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maier P, Rathfelder N, Finkbeiner MG, Taxis C, Mazza M, Le Panse S, Haguenauer-Tsapis R, Knop M. 2007. Cytokinesis in yeast meiosis depends on the regulated removal of Ssp1p from the prospore membrane. EMBO J. 26:1843–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moreno-Borchart AC, Strasser K, Finkbeiner MG, Shevchenko A, Shevchenko A, Knop M. 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20:6946–6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nickas ME, Neiman AM. 2002. Ady3p links spindle pole body function to spore wall synthesis in Saccharomyces cerevisiae. Genetics 160:1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diamond AE, Park JS, Inoue I, Tachikawa H, Neiman AM. 2009. The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol. Biol. Cell 20:134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park JS, Neiman AM. 2012. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 125:3004–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bankaitis VA, Johnson LM, Emr SD. 1986. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. U. S. A. 83:9075–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brickner JH, Fuller RS. 1997. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139:23–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691 [DOI] [PubMed] [Google Scholar]
- 16. Kolehmainen J, Black G, Saarinen A, Chandler K, Clayton-Smith J, Träskelin AL, Perveen R, Kivitie-Kallio S, Norio R, Warburg M. 2003. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 72:1359–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rampoldi L, Dobson-Stone C, Rubio JP, Danek A, Chalmers RM, Wood NW, Verellen C, Ferrer X, Malandrini A, Fabrizi GM. 2001. A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet. 28:119–120 [DOI] [PubMed] [Google Scholar]
- 18. Jäntti J, Aalto MK, Öyen M, Sundqvist L, Keränen S, Ronne H. 2002. Characterization of temperature-sensitive mutations in the yeast syntaxin 1 homologues Sso1p and Sso2p, and evidence of a distinct function for Sso1p in sporulation. J. Cell Sci. 115:409–420 [DOI] [PubMed] [Google Scholar]
- 19. Yang H-J, Nakanishi H, Liu S, McNew JA, Neiman AM. 2008. Binding interactions control SNARE specificity in vivo. J. Cell Biol. 183:1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parodi EM, Baker CS, Tetzlaff C, Villahermosa S, Huang LS. 2012. SPO71 mediates prospore membrane size and maturation in Saccharomyces cerevisiae. Eukaryot. Cell 11:1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rose MD, Fink GR. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22. Rabitsch KP, Tóth A, Gálová· M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M. 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11:1001–1009 [DOI] [PubMed] [Google Scholar]
- 23. Neiman AM, Katz L, Brennwald PJ. 2000. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 155:1643–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma H, Kunes S, Schatz PJ, Botstein D. 1987. Plasmid construction by homologous recombination in yeast. Gene 58:201–216 [DOI] [PubMed] [Google Scholar]
- 25. Roy A, Levine TP. 2004. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 279:44683–44689 [DOI] [PubMed] [Google Scholar]
- 26. Stefan CJ, Audhya A, Emr SD. 2002. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4, 5)-bisphosphate. Mol. Biol. Cell 13:542–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. 2004. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell 13:677–688 [DOI] [PubMed] [Google Scholar]
- 28. Audhya A, Emr SD. 2002. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2:593–605 [DOI] [PubMed] [Google Scholar]
- 29. Walch-Solimena C, Novick P. 1999. The yeast phosphatidylinositol-4-OH kinase Pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1:523–525 [DOI] [PubMed] [Google Scholar]
- 30. Rudge SA, Sciorra VA, Iwamoto M, Zhou C, Strahl T, Morris AJ, Thorner J, Engebrecht JA. 2004. Roles of phosphoinositides and of Spo14p (phospholipase D)-generated phosphatidic acid during yeast sporulation. Mol. Biol. Cell 15:207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Velayos-Baeza A, Vettori A, Copley RR, Dobson-Stone C, Monaco A. 2004. Analysis of the human VPS13 gene family. Genomics 84:536–549 [DOI] [PubMed] [Google Scholar]
- 32. Nakanishi H, Suda Y, Neiman AM. 2007. Erv14 family cargo receptors are necessary for ER exit during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 120:908–916 [DOI] [PubMed] [Google Scholar]
- 33. Nakanishi H, De Los Santos P, Neiman AM. 2004. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 15:1802–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]