Abstract
Cytoadhesion of Plasmodium falciparum parasitized red blood cells (pRBCs) has been implicated in the virulence of malaria infection. Cytoadhesive interactions are mediated by the protein family of Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1). The PfEMP1 family is under strong antibody and binding selection, resulting in extensive sequence and size variation of the extracellular domains. Here, we investigated cytoadhesion of pRBCs to CD36, a common receptor of P. falciparum field isolates, under dynamic flow conditions. Isogeneic parasites, predominantly expressing single PfEMP1 variants, were evaluated for binding to recombinant CD36 under dynamic flow conditions using microfluidic devices. We tested if PfEMP1 size (number of extracellular domains) or sequence variation affected the pRBC-CD36 interaction. Our analysis showed that clonal parasite variants varied ∼5-fold in CD36 rolling velocity despite extensive PfEMP1 sequence polymorphism. In addition, adherent pRBCs exhibited a characteristic hysteresis in rolling velocity at microvascular flow rates, which was accompanied by changes in pRBC shape and may represent important adaptations that favor stable binding.
INTRODUCTION
Cytoadhesion of Plasmodium falciparum parasitized red blood cells (pRBCs) is a major virulence determinant associated with accumulation of parasitized red blood cells in postcapillary venules of host organs, particularly the small intestines, skin, liver, lungs, and brain (1–3). Invasion by malaria parasites induces extensive morphological changes in red blood cells. Whereas healthy red blood cells have a discoid shape and possess the ability to squeeze through microvascular constrictions (1 to 2 μm in diameter) that are much smaller than the cell's diameter, pRBCs become less deformable due to parasite-induced modifications of the red blood cell surface area and volume, and reorganization of the cell membrane is critical for pRBC adhesion (4–7). The cytoadhesive phenotype enables parasites to avoid splenic clearance mechanisms and is associated with organ and system-wide disease complications (8).
The adhesion between pRBCs and vascular endothelium has many similarities to the leukocyte adhesion cascade (9) and consists of capture, rolling, and adhesion events, followed by postadhesion strengthening. CD36 is a common receptor for P. falciparum field isolates (10) and a key receptor for pRBC-endothelial binding (11–13). CD36 mediates a strong binding interaction and acts in cooperation with intercellular adhesion molecule 1 (ICAM-1) or other upstream rolling receptor interactions to firmly anchor and immobilize pRBCs to endothelial cells (14–17). The adhesion of pRBCs to CD36 on endothelial cells induces receptor clustering and dephosphorylation of an external threonine residue on CD36, which further strengthen the binding interaction and allow adherent pRBCs to withstand higher shear stress (18, 19). During adhesion, leukocytes deform from a spherical to a tear-drop shape (20). Cell deformation increases the contact patch between leukocyte and endothelial cell, increasing the number of receptor-ligand bonds and strengthening binding avidity (21). Correspondingly for P. falciparum pRBCs, variation in contact area has been predicted to affect rolling velocities in adherent parasitized cells (22, 23).
Parasite adhesion is mediated by a large and diverse family of adhesion proteins, referred to as var gene products or the P. falciparum erythrocyte membrane protein 1 (PfEMP1) family (8, 24–26). PfEMP1 members encode multiple receptor-like domains, called Duffy binding-like (DBL) and cysteine-rich interdomain region (CIDR) (26). PfEMP1 proteins are under opposing selection pressures to bind tightly to host receptors on endothelial cells and to evade host antibody responses. This has resulted in a variety of different PfEMP1 forms that differ both in sequence and size (between 2 and 9 extracellular domains) (27).
CD36 binding maps to the CIDR1 domain in the PfEMP1 head structure (28, 29) and is encoded into the majority of PfEMP1 variants (28, 30). CD36-binding CIDR1 domains are present in both small and large PfEMP1 proteins and have about 40% sequence identity (30, 31). Notably, work on the immunological synapse suggests that T cells and antigen-presenting cells align membrane surfaces with nanometer precision, and that binding partners are size optimized to maximize the alignment of specific protein domains at the adhesion synapse (32). Similarly, size differences between small and large PfEMP1 proteins could affect the alignment of CIDR1 binding domains with CD36. Consequently, differences in CD36 binding affinity may influence pRBC tropism for endothelial sites that differ in CD36 expression levels or modify the extent of endothelial or monocyte activation (33, 34). However, there has been no systematic investigation of how PfEMP1 size or CIDR sequence polymorphism affects the pRBC-CD36 binding interaction.
Standard methods to investigate P. falciparum cytoadhesion utilize static adhesion assays to characterize pRBC binding to recombinant protein, transgenic cell lines, or primary microvascular endothelial cells. Flow adhesion assays have also been applied to malaria research and have the potential for illustrating how pRBCs behave under well-controlled conditions and dynamic shear stresses that static adhesion assays tend to lack, especially during the washing step (14–16). Microfabrication and replica molding techniques can generate flow cells with dimensions that allow for approximating the physiologic flow conditions (35, 36). These microfluidic devices provide a flexible method to model pRBC binding to host receptors under the range of wall shear stresses (WSS) that occur in the microvasculature (37).
In this study, we utilized a microfluidic flow-cell assay to investigate the rolling velocities of a panel of clonally variant parasite lines to CD36 and to characterize the effect of flow rate on adherent pRBC cell shape (elongation index). This analysis indicates that clonal variants expressing different var genes exhibit a narrow range of CD36 rolling velocities and that pRBCs undergo a characteristic hysteresis in rolling velocity under microvascular flow conditions accompanied by changes in cell shape. These observations suggest the pRBC-CD36 interaction is selected for binding optimum despite extensive size and sequence polymorphisms that exist in PfEMP1 proteins.
MATERIALS AND METHODS
Malaria culture.
The isolation of clonal parasite lines used in this study was described previously (31, 38, 39). Malaria parasites were cultured under standard conditions using human type O RBCs in RPMI 1640 medium (Invitrogen) supplemented with 10% pooled A+ human serum and under a blood-gas mixture of 5% O2, 5% CO2, and 90% N2 at 37°C (40). Parasites were synchronized twice 48 h apart using 5% sorbitol in phosphate-buffered saline (PBS). Parasites were estimated to be synchronized such that invasion occurred over 6 to 10 h.
Fabrication of devices.
Microfluidic flow cells (5 mm wide, 20 mm long, and 61 μm tall) were fabricated using standard microfabrication and replica molding techniques. Briefly, a pattern was designed in AutoCAD (AutoDesk). A transparency mask was generated by Fineline Imaging (Colorado Springs, CO). The mask was transferred to silicon wafers (Montico Silicon) using AZ 4562 thick resist. After developing the photoresist, the wafers were etched to a depth of 61 μm using a Bosche deep reactive ion etch process (Oxford Instruments ICP 380). The etch depth of 61 μm was characterized using an optical profilometer (41). The wafers were diced and mounted in aluminum molds. The microfluidic devices were generated by casting polydimethylsiloxane (Dupont Sylguard 184) replicas and cured at 140°C for 2 h. Two upstream holes and one exit hole were punched with a sharpened 16-gauge blunt syringe needle (Amazon Supply). Glass coverslips (Gold Seal 3334) and polydimethylsiloxane (PDMS) devices were then exposed to an oxygen plasma at 10 W for 40 s (Harrik plasma cleaner PDC-001) and then brought into conformal contact.
Flow experiments.
Microfluidic glass coverslips within the microfluidic devices were coated with 25 or 50 μg/ml CD36 in phosphate-buffered saline overnight at 4°C. Before flow experiments the channels were blocked with 0.5% (wt) Albumax-RPMI 1640 medium for 2 h. All experiments were performed on a Nikon TE2000 microscope (Nikon USA) with a Photometrics CoolSNAP EZ camera. All videos were recorded using a ×10 magnification lens with a camera using 2 by 2 binning for a final image of 697 by 520 pixels. Images were recorded using a 30-nm band gap filter with a peak transmission at 420 nm (Chroma) to enhance contrast between rolling pRBCs and the image background.
Microfluidic devices were assembled into an apparatus similar to one described previously, only a syringe pump (Aladdin WPI Inc.) was utilized instead of a peristaltic pump (35). Parasite cultures were loaded into the flow chamber at approximately 20% hematocrit and 6.5% ± 2.5% parasitemia, on average, as described in the supplemental material. The shear rate, τ, was calculated using the following formula, which has been widely used in different studies (14): τ = 6Qη/wh2, where Q is the volumetric flow rate, w is the chamber width, h is the chamber depth, and η is the viscosity of the suspending medium. For each flow rate, 10 movies were recorded at 10 specific positions. All of the red blood cells in the field were tracked for 5-s movies recorded at 15 frames per s.
Video processing, particle tracking, and elongation index.
All video processing was performed in MATLAB using the image-processing toolbox (Mathworks). Image-processing algorithms were developed specifically for the experiments described. Nikon nd2 video sequences were loaded using Bioformats (42). Video sequences had a threshold set manually to extract the position of cells from the background. Once cell positions in each video frame had been identified, identified cells were linked from image to image by using a bounding box. If a cell was within the bounding box in the next frame, then the two positions were identified as a cell track. Only tracks that were observed in three or more consecutive frames were retained. Identified cells were filtered according to area so that large image features, multiple cells, or background noise were excluded from the list of tracks. The pRBC elongation index was measured directly from threshold images of the tracked cells (see the supplemental material). The elongation index was calculated from the length (L) and width (W) of cells using the equation (L − W)/(L + W).
Track plotting and analysis.
Tracks identified using the image-processing software were ordered according to their rolling velocities at each flow rate. The velocities of the ordered tracks were averaged together so that roughly 50 groups of averaged cells would be plotted (see the supplemental material for details). The averaged and ordered groups were then plotted so that the cells with the lowest rolling velocity at each shear stress were assumed to be sufficiently identical to the same grouping at the next shear stress (see the supplemental material).
Quantitative PCR (qPCR).
Parasite clones had their var transcripts identified using primers and conditions described previously (31, 43). RNA was extracted using TRIzol LS (Invitrogen) from ring-stage parasites at 6 to 12 h postinvasion. The RNA was purified using RNeasy microcolumns. Prior to final elution of RNA, DNA was removed using on-column DNase I (Qiagen). cDNA was reverse transcribed from 2 μg of purified RNA starting material using Multi-Scribe reverse transcriptase (Applied Biosystems). From this mixture, an estimated 2 ng of cDNA was used per reaction against the set of known var gene primers. PCRs were performed in an ABI Prism 7500 thermocycler using optimized primer concentrations from 0.05 to 0.5 μM in 20-μl reaction volumes and using the Power-SYBR green master mix (Invitrogen). Reaction conditions were 50°C for 1 min, 95°C for 10 min, and then 40 cycles of 95°C for 15 s, 52°C for 15 s, and 60°C for 45 s. Relative transcript levels were normalized to the housekeeping gene glutaminyl-tRNA synthetase (GTS).
RESULTS
Clonal P. falciparum panel used in the binding analysis.
To investigate the role of PfEMP1 size and sequence polymorphism on the pRBC-CD36 binding interaction, a panel of isogenic clonal variants was employed to investigate CD36 binding activity (Fig. 1). Six of the clonal lines were subcloned at the same time from a clonal parasite line called A4 “long” (31), and one was independently isolated from the A4 parent clone (38). To create a more representative panel, two clonal variants (P6G2 and ItG-1E7) were included that transcribe the same 4-domain IT4var31 as the major var transcript and were independently isolated (Fig. 1; also see Table S1 in the supplemental material).
Fig 1.
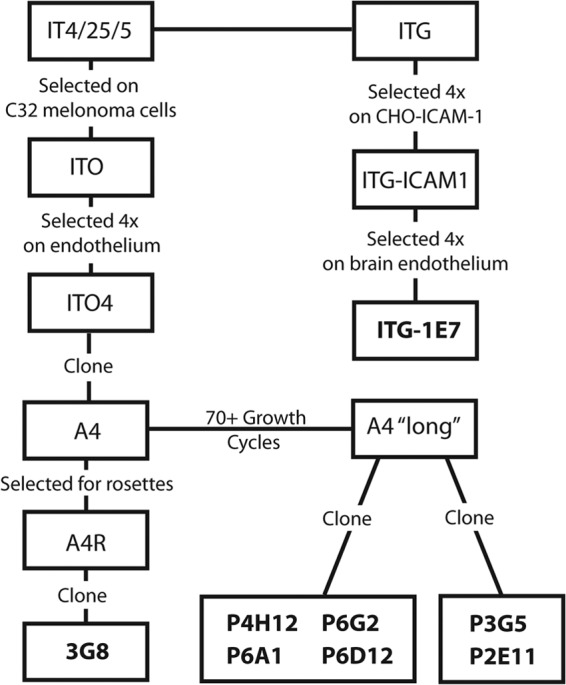
Lineage of P. falciparum clonal variants used in experiments. IT4/25/5 and ITG parasite lines are isogenically related. The eight clonal parasite variants studied were previously isolated (31, 38, 39).
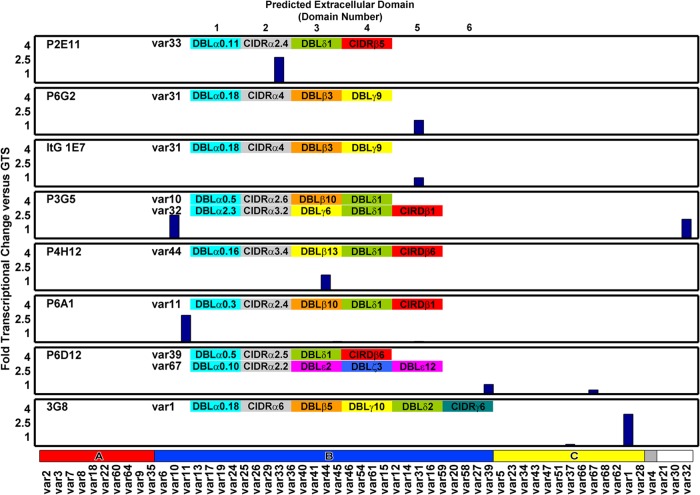
At the time of the flow assay, all of the parasite clones were confirmed by RT-qPCR to express the same major var gene transcript as that previously reported (Fig. 2) (31, 39). Six clonal lines transcribed a single primary var transcript, and two (P3G5 and P6D12) expressed a mixture of two major var transcripts. Within the panel, three parasite clones transcribed a 4-domain PfEMP1 variant, two clones transcribed a 5-domain PfEMP1 variant, one clone transcribed a 6-domain PfEMP1 variant, and two clones transcribed a mixture of a 4-domain and a 5-domain PfEMP1 variant (Fig. 2; also see Table S1 in the supplemental material).
Fig 2.
Analysis of var gene transcription in parasite lines at the time of the pRBC binding analysis. RNA was harvested from ring-stage parasites, and var gene expression profiling was performed by RT-qPCR. Most clonal variants expressed a single primary var transcript, and two of the clonal lines expressed a second var transcript. The domain structure of the dominantly expressed var gene(s) is given in each plot. Gene expression is normalized to the housekeeping control gene GTS. The var gene primer set is arranged by Ups category: UpsA (red), UpsB (blue), UpsC (yellow), UpsE (gray), and unknown (white). The predicted extracellular domain structure for each var gene transcript is shown at the top of each plot.
Rolling velocity with hysteresis.
To investigate the pRBC-CD36 binding interaction under various physiological flow conditions, trophozoite-stage clonal variants were introduced into a microfluidic device, and pRBCs were allowed to settle and adhere to CD36 under static adhesion conditions. The adherent pRBCs were then exposed to increasing wall shear stress (WSS) conditions, and rolling velocities were measured to assess the ability of CD36-adherent pRBCs to remain adherent under shear forces expected in microcirculation (Fig. 3A and C).
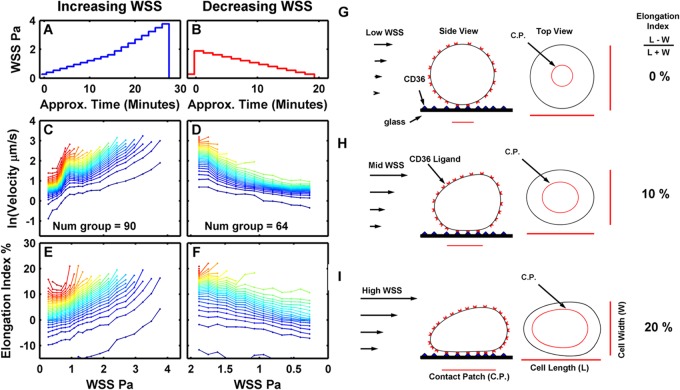
Fig 3.
Clone P6D12 binding to CD36 under increasing or decreasing shear stresses. In panels A, C, and E, adherent pRBCs were exposed to increasing shear stress, and the rolling velocity of pRBCs was tracked in 10 fields. For convenience, the total number of cells counted at each WSS time point was divided into 50 groups from lowest to highest rolling velocities. Num group is the number of pRBCs in each group, which depended on the number of pRBCs that initially adhered (e.g., a total of 4,500 pRBCs were initially adherent in panel C). Color is added to the plots to add definition between grouped pRBCs. In panels B, D, and F, the experiment was reversed. Adherent pRBCs were first exposed to a high shear stress (1.8 Pa), and then the shear stress was gradually decreased. (C) Notice a hysteresis-like effect in rolling velocities between 0.8 to 1.3 Pa observed when pRBCs were exposed to increasing shear stress. (D) This effect was largely absent from the reverse experiment with decreasing shear stress. Differences in rolling-velocity curves may be due to changes in rolling-cell footprint as cells changed shape (elongation index) under increasing (E) or decreasing (F) shear stress. A cartoon schematic illustrates how the cell contact patch (C.P.) surface area may increase with the applied low WSS (G), intermediate WSS (H), and high WSS (I). CD36 is represented as triangles on the glass surface. The CD36 ligands on the surface of the rolling cell is illustrated by the U-shaped red objects. (H and I) As the WSS increases the cell is deformed, increasing the contact area; thus, the number of bonds in contact with the CD36-glass surface increases. The larger number of bonds could alter the rolling velocity of the pRBCs.
Under increasing WSS, pRBC rolling velocity increased rapidly as the WSS increased from 0.27 to about 0.8 Pa. At approximately 0.8 Pa, the rolling velocities abruptly leveled off (and, in some cases, decreased slightly) (Fig. 3C). The rolling velocities then increased as the WSS progressed above 1.3 Pa. A similar rolling velocity-WSS pattern was observed between 0.8 and 1.3 Pa for all eight parasite clones tested and for 2 clones tested at lower CD36 concentrations of 25 μg/ml (see Fig. S1 and S2 in the supplemental material).
A potential explanation for this velocity-WSS behavior is that fluid shear forces deform and flatten the pRBCs against the glass substrate, increasing the size of the pRBC contact patch (Fig. 3G to I). Such a change in the pRBC's overall geometry would be indirectly related to the pRBC's elongation index or the relationship between the cell's length and width in the video recordings. A change in the elongation index-WSS curves (Fig. 3E) are observed at a WSS of about 0.8 Pa and above. During experiments with WSS increasing from 0.134 to 0.8 Pa (Fig. 3E), the elongation index was relatively stable and in some cases decreased slightly. From 0.8 Pa and above, the elongation index-WSS curve underwent a fairly sharp transition and then increased steadily. The change in elongation index could indicate a deformation that increased the surface area of the pRBCs' contact patch, causing a corresponding increase in the number of receptor ligand bonds at the trailing edge of the pRBC (21, 44, 45). The change in the pRBC contact patch would increase the total number of bonds contributing to pealing of the pRBC membrane away from the substrate and ultimately alter the rolling velocity of the pRBCs. Hence, the sharp transition in rolling velocity around 0.8 to 1.3 Pa may be due to a wall shear stress-induced change in the contact patch size.
To further investigate the hypothesis, the experiment was reversed so that adherent pRBCs were initially exposed to a high shear stress of 1.8 Pa to induce cell deformation, and then the WSS was gradually reduced to 0.134 Pa (Fig. 3B). The rolling velocities and elongation index of pRBCs in experiments with decreasing WSS (Fig. 3D and F) were compared to earlier observations for experiments under increasing WSS (Fig. 3C and E).
When pRBCs were exposed to an initial high WSS which was then gradually decreased, the rolling-velocity WSS curves were different from those of pRBCs exposed to increasing WSS. Under decreasing WSS conditions, there was less variation in rolling velocities between 0.8 and 1.3 Pa in all of the parasite clones (Fig. 3; also see Fig. S1 in the supplemental material). As with rolling velocities, the pRBC elongation index differed between the experiments with increasing and decreasing WSS. The starting elongation index was higher for pRBCs during the decreasing WSS experiments (Fig. 3E and F). With decreasing WSS, the elongation index decreased steadily and evenly without a sharp transition at 0.8 Pa. This effect was observed in all of the eight parasite clones tested (see Fig. S3). The more stable elongation index in the decreasing WSS experiments suggests that once deformed, pRBCs tend to remain deformed. This memory in deformation may account for hysteresis in rolling velocity observed between the increasing and decreasing WSS experiments.
Clonal parasite variants exhibit limited variation in rolling velocity.
To investigate the range of CD36 binding activity, the median rolling velocity of all clones was compared under decreasing WSS conditions (Fig. 4A). Multiple measurements had greater reproducibility under decreasing WSS than measurements made under increasing WSS. These decreasing WSS measurements displayed distinct differences in the median rolling velocities between the parasite clones. The parasite lines 3G8, ItG-1E7, and P3G5 consistently had the slowest rolling velocities (Fig. 4A) and also had fewer pRBCs detach under decreasing WSS (Fig. 4B). Conversely, parasite lines P6D12, P2E11, and P6A1 had the highest rolling velocities and also had a greater percentage of pRBCs detach. In general, rolling cells for all of the clonal variants stopped detaching during the time frame of the experiment at WSS below 1.3 Pa, except for the P6A1 clone, which had the highest rolling velocity and continued to detach at lower shear stresses (Fig. 4B). Overall, there was a strong positive correlation between slow median rolling velocity and the percentage of pRBCs that detached by the end of the experiment (Spearman rank correlation coefficient of 0.905, P < 0.005) (Table 1).
Fig 4.
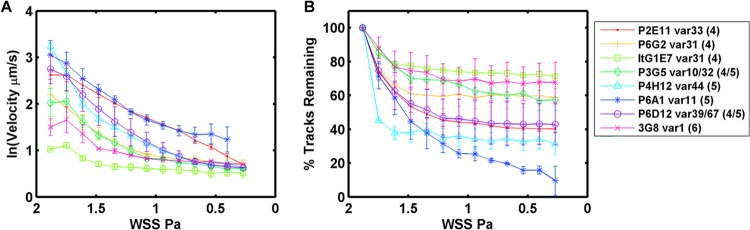
Median rolling velocity and adherence of clonal variants to CD36 under decreasing shear stress. In this experiment, adherent pRBCs were exposed to a high shear stress (1.8 Pa) and then the shear stress was gradually decreased. Error bars represent the range between two replicate experiments performed on different days. (A) The 50th percentile rolling velocity of clonal variants under decreasing shear stress. (B) The percentage of pRBCs that were tracked and remained adherent under decreasing shear stress.
Table 1.
Characteristics of clonal parasite lines binding to CD36 under flow conditions
| Parasite clone | var gene | No. of domains | % pRBCs remaining | Median rolling velocitya (5% CI) (μm s−1) | Elongation indexb (5% CI) (%) | % Remaining rankc | Rolling-velocity rankd |
|---|---|---|---|---|---|---|---|
| ItG 1E7 | var31 | 4 | 71.3 | 2.31 (2.25–2.36) | 2.0 (1.7–2.3) | 1 | 1 |
| 3G8 | var1 | 6 | 67.6 | 3.38 (3.26–3.50) | 9.3 (8.8–9.8) | 2 | 2 |
| P3G5 | var10 | 4 | 57.4 | 4.62 (4.09–5.14) | 12.9 (12.3–13.6) | 4 | 3 |
| var32 | 5 | ||||||
| P6G2 | var31 | 4 | 58.7 | 5.06 (4.90–5.23) | 9.1 (8.7–9.3) | 3 | 4 |
| P4H12 | var44 | 5 | 31.4 | 7.13 (6.24–8.02) | 9.3 (8.3–10.3) | 7 | 5 |
| P6D12 | var39 | 4 | 42.9 | 7.83 (7.59–8.06) | 11.3 (11.0–11.6) | 5 | 6 |
| var67 | 5 | ||||||
| P2E11 | var33 | 4 | 40.2 | 10.3 (10.1–10.4) | 7.6 (7.3–7.8) | 6 | 7 |
| P6A1 | var11 | 5 | 9.6 | 11.3 (11.0–11.6) | 8.9 (8.6–9.3) | 8 | 8 |
Median rolling velocity was calculated at 1.6-Pa WSS.
Elongation index was calculated at 1.6-Pa WSS.
Percent remaining rank is the percentage of adherent parasites remaining by the end of the decreasing WSS measurement. Parasites are ranked in descending order.
Rolling-velocity rank is the median rolling velocity in ascending order. The 5% confidence interval (CI) about the median is given.
To investigate if there was a relationship between rolling velocity and PfEMP1 size, the population rolling velocities of each clonal line were compared under increasing and decreasing WSS histories at a single WSS of 1.6 Pa (Fig. 5 and Table 1). The population rolling velocities were considered statistically similar if their 5% confidence intervals (CI) about the median overlapped one another. There was good agreement between the two assays, and the rank order of clonal variants did not change under increasing or decreasing WSS (Fig. 5). Overall, the median rolling velocity was similar between all parasite clones; it differed by approximately 2.5-fold among the six clonal variants that were isolated at the same time and approximately 5-fold between the more distantly related ItG-1E7 and 3G8 parasites. In addition, although the two IT4var31-expressing clones ItG-1E7 and P6G2 had statistically different rolling velocities (Fig. 5), both clonal lines were among the stronger binders and their median rolling velocities were similar (2.31 and 5.06 μm s−1) (Table 1). The number of predicted extracellular domains did not correlate with rank-order rolling velocity of the clones (Fig. 5A and B).
Fig 5.
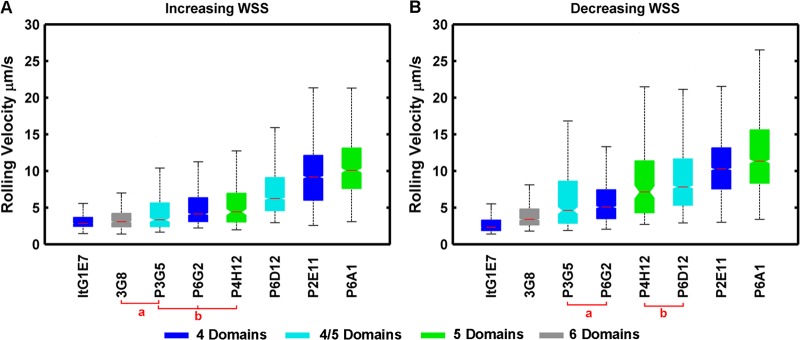
Rolling-velocity comparison and ordered rank of clonal variants at 1.6 Pa. The colors of the bars correspond to the number of extracellular domains in the predominantly expressed var product(s). Clones marked a and b are not significantly different from one another but are significantly different from other parasite clones, as indicated by overlapping confidence intervals (notches). The boxes represent the 25th and 75th percentiles, while the whiskers represent the 2.7 and 97.3 percentiles. The notches represent the 95% confidence interval about the median (horizontal line).
DISCUSSION
A common binding interaction for many PfEMP1 variants is CIDR1-CD36 (11, 12). Whereas antibody selection has led to extensive sequence diversity in CD36-binding domains (28, 30), it is not known whether CIDR1 sequence polymorphism compromises the ability of pRBCs to sequester from blood circulation or if differences in CD36 binding affinity could be selected for different microvascular niches that differ in CD36 expression levels (46, 47). Investigation of field isolates and CIDR recombinant proteins suggests there are low- and high-affinity CD36 binding variants (10, 30), but there has been limited investigation of the range of pRBC-CD36 binding affinities. A variety of factors are likely to influence pRBC adhesion and rolling velocities, including the PfEMP1 surface expression levels, the strength of the receptor-ligand interaction, the bending modulus of the pRBC membrane, and the effective hydrodynamic radius of the rolling pRBC (48–50). Recently the density of knobs on the surface has been observed to vary both as pRBCs age and between parasite isolates (51, 52). It is feasible that differences in rolling velocities are due to a combination of surface ligand density and the strength of the receptor ligand bond. Thus, rolling velocity may represent a specific phenotype of the parasites. In this study, we investigated if clonal parasite variants expressing different PfEMP1 proteins exhibit differences in CD36 rolling velocities.
Experimental strategy to study rolling velocity of different pRBC populations.
The cytoadhesion events of P. falciparum are analogous to the leukocyte cytoadhesion cascade and can be characterized into four or more distinct phases: capture, rolling, and adhesion (53), followed by postadhesion strengthening. CD36 acts in cooperation with other host receptors that tether and capture pRBCs from blood flow (14, 54). Adhesion of pRBCs to CD36 on endothelial cells has been shown to induce postadhesion signaling events that further strengthen binding by causing CD36 receptor clustering and cytoskeletal rearrangement (18, 19). Here, we used concentrations of recombinant CD36 that have previously been shown to support static adhesion of the same parasite clones (31). These concentrations of CD36 were sufficient to allow for adherent pRBCs to roll across the adsorbed protein and to measure differences in CD36-pRBC affinity. Recombinant CD36 protein was employed instead of cell platforms, because PfEMP1 proteins may bind multiple and diverse host receptors on cell surfaces, and this allowed the pRBC-CD36 interaction to be studied in isolation.
In order to minimize variables between parasite lines, we used a panel of isogenic clonal variants and compared binding over a similar age distribution between 32 and 42 h postinvasion. The 10-h window made it easier to control for the timing postinvasion in replicate experiments and to limit any subtle variations in the length of the life cycle that may exist between parasite clones. Furthermore, we developed our techniques to measure large populations of pRBCs (often greater than 1,000 individuals) from each clonal line, thereby observing the range of possible variations present even within a clonal population.
To maximize the number of rolling pRBCs observed, we utilized a method which allowed cells to first adsorb and bind to the CD36 substrate, and then we employed low flow rates to wash off unbound cells so that rolling pRBCs could be clearly observed and tracked. This approach had some advantages and disadvantages. While it is useful for observing velocities of large populations of rolling pRBCs and also the rate at which they detach from the substrate, the method described here lacks the ability to observe the rate at which pRBCs bind to the substrate surface. Measuring the pRBC adhesion rate would require continually feeding unbound pRBCs into the microfluidic flow chamber. These unbound pRBCs complicate cell tracking, because unbound cells are not easily differentiated from bound and rolling pRBCs in individual video images. Other cytoadhesion studies have utilized methods to observe both adhesion and detachment rates of cells on substrates (55–57).
Tracking an individual rolling pRBC at different WSS was not possible, because pRBCs would roll out of the field of view as the experiment progressed. Also, the number of pRBCs tracked decreased as cells detached. This complicated observing equal numbers of cells at each WSS, because at high WSS the number of cells present per field of view was significantly reduced. The solution was to assume that if the pRBC rolling velocities were ordered from least to greatest, the slowest-moving pRBCs at each WSS shared similar characteristics and could justifiably be grouped together. The grouped pRBCs at each WSS could then be related to one another according to their ordered rolling velocities (see the supplemental material). This strategy allowed generating the rolling velocity-WSS curves for large populations of pRBCs.
Determinants of variations in pRBC-CD36 cytoadhesion under flow.
Our analysis showed that although there were weaker and stronger CD36-binding parasites, clonal variants displayed a narrow 4- to 5-fold range of median rolling velocities on CD36 at 1.6-Pa WSS. This difference is relatively small, but it may still be biologically relevant for pRBC sequestration in vascular beds that differ in CD36 expression levels or other host receptors that cooperate in mediating parasite binding. In addition, there was good agreement in rolling velocities between two parasite clones (ItG-1E7 and P6G2) that expressed the same primary var transcript, even though they were independently isolated. There were no discernible trends when rolling velocity was compared to the predicted number of extracellular domains in the PfEMP1 variants. Although the strongest binding variant had 6 domains, weaker binding clones could express either 4-domain or 5-domain PfEMP1 variants. It remains a possibility that PfEMP1 size has a larger role in cell-based binding assays because endothelial cells are covered with a glycocalyx layer of proteoglycans and glycosaminoglycans (37, 58), which may shield smaller host receptors from PfEMP1 ligands. However, our findings suggest pRBCs can form stable interactions with CD36, despite extensive differences in PfEMP1 sequence. In addition, these findings suggest that CD36-binding parasite variants are under a strong binding selection to maintain a narrow CD36 rolling velocity, presumably to ensure effective pRBC sequestration and ultimately parasite transmission.
Of interest, all of the clonal variants exhibited a hysteresis in rolling velocity that was dependent on the WSS history. Specifically, when starting from a low WSS, the pRBC rolling velocity increased with increasing shear force and then leveled off or, in some cases, was slightly reduced before increasing again. The velocity-WSS pattern was not observed when pRBCs were exposed to an initially high WSS that was then gradually decreased. Hence, a WSS-dependent rolling velocity hysteresis was observed.
These features in the WSS-rolling velocity curves appeared at around 0.8 to 1.3 Pa and correspond to an upper-limit WSS value of 1.6 Pa, which correspond to fluid shear rates of 1 to 3 Pa (10 to 30 dynes per cm2) observed in postcapillary venules (37). We do note that the concentration of CD36 utilized in this study was arbitrarily chosen; therefore, the relationship may be coincidental.
Possible origins of rolling velocity hysteresis.
In principle, the features in rolling velocity observed around 0.8 to 1.3 Pa could be created by a catch-like bond strengthening as the force applied to the bonds increased. However, the same velocity-shear stress pattern was not observed when adherent pRBCs were first exposed to a high WSS which was then gradually decreased (Fig. 4A and B). These WSS-velocity patterns were superficially identical for all parasite clones observed. Atomic force microscopy measurements of CD36 and pRBCs describe a slip-type bond interaction (55, 59). Consequently, the change in pRBC-CD36 rolling velocity from 0.8 to 1.3 Pa is unlikely to be due to catch-bond interaction.
The more likely explanation of the differing velocity profiles under increasing versus decreasing WSS is due to the size of the cell contact patch (21, 44). Cell flattening has been shown for leukocyte binding (20) and is predicted to affect rolling velocities in adherent parasitized cells (22, 23, 55), but rolling-velocity measurements with the WSS resolution depicted here have not been previously shown. When pRBCs first bind to CD36 within the microfluidic device the contact patch is likely to be small, but when the WSS increases the cell is pressed against the glass coverslip by the fluid shear forces. These forces cause the cell to deform and flatten against the coverslip, altering the elongation index of the cell and increasing the size of the cell contact patch. This increases the number of bond-ligand interactions between the cell and CD36 substrate and ultimately reduces the rolling velocity of the cell.
The present results agree well with complementary work from another study. Recent measurements of the pRBC contact patch by total internal reflectance microscopy show that the pRBC contact patch size increases from 0.1 to 0.3 Pa but then remains relatively constant at WSS above 0.5 Pa (55). In that study (55), pRBC rolling velocities were not measured, but taken together, the different approaches are in good agreement that the sudden increase and then leveling out of rolling velocity under increasing WSS may be due to an increase in size of the contact patch under microvascular shear stress, followed by the contact patch size remaining relatively constant and stabilizing the pRBC rolling velocity. During the decreasing WSS experiments, the cell is initially deformed by the high shear stress, so we suspect that the cell contact patch is initially larger and changes more gradually across the measurement. Hence, we observed hysteresis, since the same WSS-velocity profiles are not observed under increasing and decreasing WSS experiments.
The dependence of the pRBC rolling velocity on the cell contact patch illustrates how cell geometry and mechanics relate to gross cell behavior. Examining the mechanics of rolling cells reveals that for rolling to occur, the pRBC host membrane must be bent as it is peeled away from a substrate (48). Micropipette and optical trap studies have shown that the pRBC membrane's elastic modulus has increased over normal RBCs (60, 61). Perhaps the changes in membrane stiffness help stabilize the rolling velocities of pRBCs by increasing the energy needed to peel the cell membrane away from the substrate. If true, then the development of a stiffer membrane and stabilizing of rolling velocity under microvascular shear stresses represent adaptations that increase the tendency of pRBCs to slow on endothelial cells, followed by postadhesive CD36 signaling events that lead to stable binding (18, 19).
In summary, we have developed a new method for observing the rolling behavior of pRBCs under various wall shear stress and applied these tools to understand whether PfEMP1 sequence variation alters adherent pRBC-CD36 rolling characteristics under different sheer stress levels. Our analysis suggests that CD36-binding clonal variants exhibit a narrow rolling velocity under physiological flow conditions despite extensive differences in PfEMP1 size and sequence, and that shear stresses encountered in microvessels cause a hysteresis effect in rolling velocities. This hysteresis was accompanied by changes in pRBC shape and may represent adaptations that favor stable pRBC binding.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the U.S. NIH/NIAID International Center of Excellence for Malaria Research for South Asia under NIH grants U19AI089688 (P.K.R.) and R01 AI047953 (J.D.S.).
Footnotes
Published ahead of print 6 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00148-13.
REFERENCES
- 1. Seydel KB, Milner DA, Jr, Kamiza SB, Molyneux ME, Taylor TE. 2006. The distribution and intensity of parasite sequestration in comatose Malawian children. J. Infect. Dis. 194:208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. MacPherson GG, Warrell MJ, White NJ, Looareesuwan S, Warrell DA. 1985. Human cerebral malaria. A quantitative ultrastructural analysis of parasitized erythrocyte sequestration. Am. J. Pathol. 119:385–401 [PMC free article] [PubMed] [Google Scholar]
- 3. Pongponratn E, Riganti M, Punpoowong B, Aikawa M. 1991. Microvascular sequestration of parasitized erythrocytes in human falciparum malaria: a pathological study. Am. J. Trop. Med. Hyg. 44:168–175 [DOI] [PubMed] [Google Scholar]
- 4. Maier AG, Cooke BM, Cowman AF, Tilley L. 2009. Malaria parasite proteins that remodel the host erythrocyte. Nat. Rev. Microbiol. 7:341–354 [DOI] [PubMed] [Google Scholar]
- 5. An X, Mohandas N. 2010. Red cell membrane and malaria. Transfus. Clin. Biol. 17:197–199 [DOI] [PubMed] [Google Scholar]
- 6. Herricks T, Seydel KB, Molyneux M, Taylor T, Rathod PK. 2012. Estimating physical splenic filtration of Plasmodium falciparum-infected red blood cells in malaria patients. Cell. Microbiol. 14:1880–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Herricks T, Antia M, Rathod PK. 2009. Deformability limits of Plasmodium falciparum-infected red blood cells. Cell. Microbiol. 11:1340–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miller LH, Baruch DI, Marsh K, Doumbo OK. 2002. The pathogenic basis of malaria. Nature 415:673–679 [DOI] [PubMed] [Google Scholar]
- 9. Ley K, Laudanna C, Cybulsky MI, Nourshargh S. 2007. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7:678–689 [DOI] [PubMed] [Google Scholar]
- 10. Newbold C, Warn P, Black G, Berendt A, Craig A, Snow B, Msobo M, Peshu N, Marsh K. 1997. Receptor-specific adhesion and clinical disease in Plasmodium falciparum. Am. J. Trop. Med. Hyg. 57:389–398 [DOI] [PubMed] [Google Scholar]
- 11. Ockenhouse CF, Tandon NN, Magowan C, Jamieson GA, Chulay JD. 1989. Identification of a platelet membrane glycoprotein as a falciparum malaria sequestration receptor. Science 243:1469–1471 [DOI] [PubMed] [Google Scholar]
- 12. Barnwell JW, Asch AS, Nachman RL, Yamaya M, Aikawa M, Ingravallo P. 1989. A human 88-kD membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum-infected erythrocytes. J. Clin. Investig. 84:765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oquendo P, Hundt E, Lawler J, Seed B. 1989. CD36 directly mediates cytoadherence of Plasmodium falciparum parasitized erythrocytes. Cell 58:95–101 [DOI] [PubMed] [Google Scholar]
- 14. Cooke BM, Berendt AR, Craig AG, MacGregor J, Newbold CI, Nash GB. 1994. Rolling and stationary cytoadhesion of red blood cells parasitized by Plasmodium falciparum: separate roles for ICAM-1, CD36 and thrombospondin. Br. J. Haematol. 87:162–170 [DOI] [PubMed] [Google Scholar]
- 15. Gray C, McCormick C, Turner G, Craig A. 2003. ICAM-1 can play a major role in mediating P. falciparum adhesion to endothelium under flow. Mol. Biochem. Parasitol. 128:187–193 [DOI] [PubMed] [Google Scholar]
- 16. Yipp BG, Anand S, Schollaardt T, Patel KD, Looareesuwan S, Ho M. 2000. Synergism of multiple adhesion molecules in mediating cytoadherence of Plasmodium falciparum-infected erythrocytes to microvascular endothelial cells under flow. Blood 96:2292–2298 [PubMed] [Google Scholar]
- 17. Ho M, Hickey MJ, Murray AG, Andonegui G, Kubes P. 2000. Visualization of Plasmodium falciparum-endothelium interactions in human microvasculature: mimicry of leukocyte recruitment. J. Exp. Med. 192:1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davis SP, Amrein M, Gillrie MR, Lee K, Muruve DA, Ho M. 2012. Plasmodium falciparum-induced CD36 clustering rapidly strengthens cytoadherence via p130CAS-mediated actin cytoskeletal rearrangement. FASEB J. 26:1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ho M, Hoang HL, Lee KM, Liu N, MacRae T, Montes L, Flatt CL, Yipp BG, Berger BJ, Looareesuwan S, Robbins SM. 2005. Ectophosphorylation of CD36 regulates cytoadherence of Plasmodium falciparum to microvascular endothelium under flow conditions. Infect. Immun. 73:8179–8187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dong C, Lei XX. 2000. Biomechanics of cell rolling: shear flow, cell-surface adhesion, and cell deformability. J. Biomech. 33:35–43 [DOI] [PubMed] [Google Scholar]
- 21. Pappu V, Doddi SK, Bagchi P. 2008. A computational study of leukocyte adhesion and its effect on flow pattern in microvessels. J. Theor. Biol. 254:483–498 [DOI] [PubMed] [Google Scholar]
- 22. Fedosov DA, Caswell B, Karniadakis GE. 2011. Wall shear stress-based model for adhesive dynamics of red blood cells in malaria. Biophys. J. 100:2084–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fedosov DA, Caswell B, Suresh S, Karniadakis GE. 2011. Quantifying the biophysical characteristics of Plasmodium-falciparum-parasitized red blood cells in microcirculation. Proc. Natl. Acad. Sci. U. S. A. 108:35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. 1995. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82:77–87 [DOI] [PubMed] [Google Scholar]
- 25. Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82:101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89–100 [DOI] [PubMed] [Google Scholar]
- 27. Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T. 2010. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes–divide and conquer. PLoS Comput. Biol. 6:e1000933. 10.1371/journal.pcbi.1000933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baruch DI, Ma XC, Singh HB, Bi X, Pasloske BL, Howard RJ. 1997. Identification of a region of PfEMP1 that mediates adherence of Plasmodium falciparum infected erythrocytes to CD36: conserved function with variant sequence. Blood 90:3766–3775 [PubMed] [Google Scholar]
- 29. Smith JD, Kyes S, Craig AG, Fagan T, Hudson-Taylor D, Miller LH, Baruch DI, Newbold CI. 1998. Analysis of adhesive domains from the A4VAR Plasmodium falciparum erythrocyte membrane protein-1 identifies a CD36 binding domain. Mol. Biochem. Parasitol. 97:133–148 [DOI] [PubMed] [Google Scholar]
- 30. Robinson BA, Welch TL, Smith JD. 2003. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Mol. Microbiol. 47:1265–1278 [DOI] [PubMed] [Google Scholar]
- 31. Janes JH, Wang CP, Levin-Edens E, Vigan-Womas I, Guillotte M, Melcher M, Mercereau-Puijalon O, Smith JD. 2011. Investigating the host binding signature on the Plasmodium falciparum PfEMP1 protein family. PLoS Pathog. 7:e1002032. 10.1371/journal.ppat.1002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dustin ML, Colman DR. 2002. Neural and immunological synaptic relations. Science 298:785–789 [DOI] [PubMed] [Google Scholar]
- 33. Chakravorty SJ, Carret C, Nash GB, Ivens A, Szestak T, Craig AG. 2007. Altered phenotype and gene transcription in endothelial cells, induced by Plasmodium falciparum-infected red blood cells: pathogenic or protective? Int. J. Parasitol. 37:975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chakravorty SJ, Hughes KR, Craig AG. 2008. Host response to cytoadherence in Plasmodium falciparum. Biochem. Soc. Trans. 36:221–228 [DOI] [PubMed] [Google Scholar]
- 35. Herricks T, Seydel KB, Turner G, Molyneux M, Heyderman R, Taylor T, Rathod PK. 2011. A microfluidic system to study cytoadhesion of Plasmodium falciparum infected erythrocytes to primary brain microvascularendothelial cells. Lab. Chip 11:2994–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. D'Amico Oblak T, Root P, Spence DM. 2006. Fluorescence monitoring of ATP-stimulated, endothelium-derived nitric oxide production in channels of a poly(dimethylsiloxane)-based microfluidic device. Anal. Chem. 78:3193–3197 [DOI] [PubMed] [Google Scholar]
- 37. Lipowsky HH. 2005. Microvascular rheology and hemodynamics. Microcirculation 12:5–15 [DOI] [PubMed] [Google Scholar]
- 38. Horrocks P, Pinches R, Christodoulou Z, Kyes SA, Newbold CI. 2004. Variable var transition rates underlie antigenic variation in malaria. Proc. Natl. Acad. Sci. U. S. A. 101:11129–11134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Avril M, Tripathi AK, Brazier AJ, Andisi C, Janes JH, Soma VL, Sullivan DJ, Jr, Bull Stins PCMF, Smith JD. 2012. A restricted subset of var genes mediates adherence of Plasmodium falciparum-infected erythrocytes to brain endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 109:E1782–E1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675 [DOI] [PubMed] [Google Scholar]
- 41. Yen GS, Fujimoto BS, Schneider T, Huynh DT, Jeffries GD, Chiu DT. 2011. A rapid and economical method for profiling feature heights during microfabrication. Lab. Chip 11:974–977 [DOI] [PubMed] [Google Scholar]
- 42. Linkert M, Rueden CT, Allan C, Burel JM, Moore W, Patterson A, Loranger B, Moore J, Neves C, Macdonald D, Tarkowska A, Sticco C, Hill E, Rossner M, Eliceiri KW, Swedlow JR. 2010. Metadata matters: access to image data in the real world. J. Cell Biol. 189:777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Viebig NK, Levin E, Dechavanne S, Rogerson SJ, Gysin J, Smith JD, Scherf A, Gamain B. 2007. Disruption of var2csa gene impairs placental malaria associated adhesion phenotype. PLoS One 2:e910. 10.1371/journal.pone.0000910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pappu V, Bagchi P. 2008. 3D computational modeling and simulation of leukocyte rolling adhesion and deformation. Comput. Biol. Med. 38:738–753 [DOI] [PubMed] [Google Scholar]
- 45. Efremov A, Cao J. 2011. Bistability of cell adhesion in shear flow. Biophys. J. 101:1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eppihimer MJ, Wolitzky B, Anderson DC, Labow MA, Granger DN. 1996. Heterogeneity of expression of E- and P-selectins in vivo. Circ. Res. 79:560–569 [DOI] [PubMed] [Google Scholar]
- 47. Turner GD, Morrison H, Jones M, Davis TM, Looareesuwan S, Buley ID, Gatter KC, Newbold CI, Pukritayakamee S, Nagachinta B, White NJ, Berendt AR. 1994. An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am. J. Pathol. 145:1057–1069 [PMC free article] [PubMed] [Google Scholar]
- 48. Hodges SR, Jensen OE. 2002. Spreading and peeling dynamics in a model of cell adhesion. J. Fluid Mech. 460:381–409 [Google Scholar]
- 49. Ortiz V, Nielsen SO, Discher DE, Klein ML, Lipowsky R, Shillcock J. 2005. Dissipative particle dynamics simulations of polymersomes. J. Phys. Chem. B 109:17708–17714 [DOI] [PubMed] [Google Scholar]
- 50. Reboux S, Richardson G, Jensen OE. 2008. Bond tilting and sliding friction in a model of cell adhesion. Proc. R. Soc. A Math. Phys. Eng. Sci. 464:447–467 [Google Scholar]
- 51. Quadt KA, Barfod L, Andersen D, Bruun J, Gyan B, Hassenkam T, Ofori MF, Hviid L. 2012. The density of knobs on Plasmodium falciparum-infected erythrocytes depends on developmental age and varies among isolates. PLoS One 7:e45658. 10.1371/journal.pone.0045658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nagao E, Kaneko O, Dvorak JA. 2000. Plasmodium falciparum-infected erythrocytes: qualitative and quantitative analyses of parasite-induced knobs by atomic force microscopy. J. Struct. Biol. 130:34–44 [DOI] [PubMed] [Google Scholar]
- 53. Ho M, White NJ. 1999. Molecular mechanisms of cytoadherence in malaria. Am. J. Physiol. 276:C1231–C1242 [DOI] [PubMed] [Google Scholar]
- 54. Udomsangpetch R, Reinhardt PH, Schollaardt T, Elliott JF, Kubes P, Ho M. 1997. Promiscuity of clinical Plasmodium falciparum isolates for multiple adhesion molecules under flow conditions. J. Immunol. 158:4358–4364 [PubMed] [Google Scholar]
- 55. Xu X, Efremov AK, Li A, Lai L, Dao M, Lim CT, Cao J. 2013. Probing the cytoadherence of malaria infected red blood cells under flow. PLoS One 8:e64763. 10.1371/journal.pone.0064763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thomas WE, Nilsson LM, Forero M, Sokurenko EV, Vogel V. 2004. Shear-dependent ‘stick-and-roll' adhesion of type 1 fimbriated Escherichia coli. Mol. Microbiol. 53:1545–1557 [DOI] [PubMed] [Google Scholar]
- 57. Hammer DA, Brunk DK. 1999. Measuring receptor-mediated cell adhesion under flow: cell-free systems. Methods Mol. Med. 18:543–552 [DOI] [PubMed] [Google Scholar]
- 58. Vink H, Duling BR. 1996. Identification of distinct luminal domains for macromolecules, erythrocytes, and leukocytes within mammalian capillaries. Circ. Res. 79:581–589 [DOI] [PubMed] [Google Scholar]
- 59. Li A, Lim TS, Shi H, Yin J, Tan SJ, Li Z, Low BC, Tan KS, Lim CT. 2011. Molecular mechanistic insights into the endothelial receptor mediated cytoadherence of Plasmodium falciparum-infected erythrocytes. PLoS One 6:e16929. 10.1371/journal.pone.0016929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Glenister FK, Coppel RL, Cowman AF, Mohandas N, Cooke BM. 2002. Contribution of parasite proteins to altered mechanical properties of malaria-infected red blood cells. Blood 99:1060–1063 [DOI] [PubMed] [Google Scholar]
- 61. Mills JP, Diez-Silva M, Quinn DJ, Dao M, Lang MJ, Tan KS, Lim CT, Milon G, David PH, Mercereau-Puijalon O, Bonnefoy S, Suresh S. 2007. Effect of plasmodial RESA protein on deformability of human red blood cells harboring Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 104:9213–9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.