Abstract
SbmA is an inner membrane protein of Gram-negative bacteria that is involved in the internalization of glycopeptides and prokaryotic and eukaryotic antimicrobial peptides, as well as of peptide nucleic acid (PNA) oligomers. The SbmA homolog BacA is required for the development of Sinorhizobium meliloti bacteroids within plant cells and favors chronic infections with Brucella abortus and Mycobacterium tuberculosis in mice. Here, we investigated functional features of SbmA/BacA using the proline-rich antimicrobial peptide Bac7(1-35) as a substrate. Circular dichroism and affinity chromatography studies were used to investigate the ability of SbmA to bind the peptide, and a whole-cell transport assay with fluorescently labeled peptide allowed the determination of transport kinetic parameters with a calculated Km value of 6.95 ± 0.89 μM peptide and a Vmax of 53.91 ± 3.17 nmol/min/mg SbmA. Use of a bacterial two-hybrid system coupled to SEC-MALLS (size exclusion chromatography coupled with multiangle laser light scattering) analyses established that SbmA is a homodimer in the membrane, and treatment of the cells with arsenate or ionophores indicated that the peptide transport mediated by SbmA is driven by the electrochemical gradient. Overall, these results shed light on the SbmA-mediated internalization of peptide substrates and suggest that the transport of an unknown substrate(s) represents the function of this protein.
INTRODUCTION
SbmA is an inner membrane protein of Gram-negative bacteria found in distantly related species, such as Enterobacteriaceae and the alphaproteobacteria Sinorhizobium meliloti and Brucella abortus, and required for the establishment of symbiosis with the leguminous plant (1) or the chronic infection of the host (2, 3). It has recently been recognized as a transporter involved in the internalization of antimicrobial peptides (AMPs) of prokaryotic (4) and eukaryotic (5, 6) origin as well as of the glycopeptide antibiotic bleomycin (7–9).
SbmA is a 406-residue-long polypeptide showing seven or eight transmembrane-spanning domains (1, 10) originally identified in Escherichia coli as a consequence of the phenotype of resistance of sbmA mutants to microcin B17 (11). SbmA is involved in the uptake of the lasso peptide microcin J25 (4) and is also required for the direct uptake of the eukaryotic proline-rich antimicrobial peptides Bac7 (5) and PR-39 (12). Mutants of SbmA were recently found to be resistant to the antisense peptide phosphorodiamidate morpholino oligomers, suggesting that these molecules may be transported across the plasma membrane by SbmA (13). In addition, a role of this protein in the uptake of peptide nucleic acid (PNA) conjugates in E. coli has been proposed, indicating SbmA as the first protein identified to recognize PNA (14).
Although a physiological role of SbmA has not been found yet, BacA, the ortholog of SbmA in S. meliloti, is required for the development of S. meliloti bacteroids within plant cells (1). BacA shows 64% sequence identity to SbmA, and it is functionally interchangeable with E. coli SbmA (7). Recently, a role of BacA in the establishment of host infections has been suggested. The protein mediates protection of bacteria against host nodule-specific cysteine-rich (NCR) antimicrobial peptides, a number of host plant peptides essential to bacteroid development (15, 16). In addition, BacA is vital for the survival of B. abortus in the macrophages of the host, and it favors chronic infections in BALB/c mice (2, 3). A BacA-related protein was identified in the chronic pathogen Mycobacterium tuberculosis (3). This protein (MtBacA) was found to be important for the maintenance of chronic infection in a murine infection model (3) and, recently, to be able to partially complement the symbiotic defect of an S. meliloti BacA-deficient mutant (17). These observations suggest that the presence of SbmA/BacA may offer advantages for the survival of the bacteria in different environments.
Based on sequence analysis, SbmA has been proposed to be the transmembrane domain (TMD) of an ABC transporter (10), a widely diffused superfamily of proteins that mediate the uptake or export of a variety of substances across cell membranes (18). ABC transporters are dimers and have a common architecture consisting of a TMD and a nucleotide binding domain (NBD), which hydrolyzes ATP and drives the transport. Since the sbmA sequence does not encode an NBD (see Fig. S1 in the supplemental material), we investigated the driving force for the eukaryotic proline-rich antimicrobial peptide Bac7(1-35) uptake by SbmA. In addition, we looked at the oligomeric state of SbmA and measured its binding affinity for Bac7(1-35). The results contribute to our understanding of SbmA-mediated internalization of peptides.
MATERIALS AND METHODS
Antimicrobial peptides.
Bac7(1-35) and the dipyrromethene boron difluoride (BODIPY [BY]) fluorescently labeled derivative Bac7(1-35)-BY were prepared as previously described (19).
Bacterial strains, media, and growth conditions.
All the bacterial strains and plasmids used in this study are listed in Table 1. Bacterial cultures were grown in Luria-Bertani (LB) or Mueller-Hinton (MH) broth at 37°C under aerobic conditions with the addition, when required, of the appropriate antibiotic at the following concentrations: 100 μg ml−1 for ampicillin, 50 μg ml−1for kanamycin, and 10 μg ml−1 for tetracycline.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| BTH101 | F− cya-99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1 | Euromedex |
| BW25113 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 | Genobasea |
| JW3710 | BW25113 atpD::Kmr mutant | Genobasea |
| DH5α | supE44 ΔlacU169(ϕ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | NAc |
| JW0368-1 | F− Δ(araD-araB)567ΔlacZ4787(::rrnB-3) ΔsbmA742::kan λ− rph-1Δ(rhaD-rhaB)568 hsdR514 | Coli Genetic Stock Center collection |
| C43(DE3) | Derived from C41(DE3); C41(DE3) was derived from BL21(DE3) [E. coli F− ompT hsdSB(rB− mB−) gal dcm (DE3)] and has at least one uncharacterized mutation | 35 |
| Plasmids | ||
| pQE-9(sbmA) | Expression vector, Ampr | 5 |
| pUT18C(zip) | GCN4 leucine zipper cloned in pUT18C | Euromedex |
| pKT25(zip) | GCN4 leucine zipper cloned in pKT25 | Euromedex |
| pKNT25(sbmA) | sbmA cloned BamHI-EcoRI in pKNT25 | Provided by G. C. Walker |
| pUT18(sbmA) | sbmA cloned BamHI-EcoRI in pUT18 | Provided by G. C. Walker |
| pWaldoGFPd(sbmA) | Expression vector, Kanr, carrying sbmAGFP-His8 tag and a TEVb site as fusion | 20 |
| pWaldoGFPd(bacA) | Expression vector, Kanr, carrying bacAGFP-His8 tag and a TEV site as fusion | 20 |
Keio Collection of GenoBase (http://cgsc.biology.yale.edu/index.php).
TEV, tobacco etch virus.
NA, not available.
Cloning, expression, and purification of 6-His-tagged SbmA.
The E. coli sbmA gene was amplified from chromosomal DNA and ligated into the expression vector pQE9 as previously described (5). The expression of the protein was induced with 1 μM isopropyl-β-d-1-thiogalactopyranoside overnight at 30°C when the bacterial suspension reached an optical density at 600 nm (OD600) of 0.7 to 0.8. The bacterial culture was harvested by centrifugation for 10 min at 2,000 × g; resuspended in ice-cold lysis buffer containing 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1% (vol/vol) Triton X-100; and left at 4°C with stirring for 10 min. The sample was then sonicated three times for 10 s each time with a Branson sonicator (Branson Sonic Power S75) at 2.5 direct current (DC) amperes. After a 20-min centrifugation at 10,000 × g, supernatants were applied to a nickel-nitrilotriacetic acid (Ni-NTA) column (Invitrogen) preequilibrated with 5 mM imidazole in 20 mM Tris-HCl, pH 8, 1 M NaCl, 5 mM β-mercaptoethanol, 1% (vol/vol) Triton X-100. The purified protein was eluted with 100 mM imidazole in the same buffer. The protein was further purified with gel filtration chromatography onto the Sephadex G-25 resin (GE Healthcare) preequilibrated with 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5% (vol/vol) Triton X-100, and eluted in the same buffer. Its concentration was determined by the bicinchoninic acid (BCA) assay kit (Pierce) using bovine serum albumin (BSA) to obtain a standard curve.
Purification of SbmA-GFP-His8 and BacA-GFP-His8.
sbmA and bacA were cloned into the pWaldoGFPd vector. Overexpression and purification were performed as previously described (20). Both proteins were purified in 0.03% (wt/vol) dodecyl-maltoside (DDM).
Affinity chromatography assay onto Bac7-functionalized resin.
The column preparation and the affinity chromatography were performed according to the manufacturer's instructions (Sulfolink resin; Pierce). The protein bound to the resin was then eluted with 12% (wt/vol) SDS, 0.4 M dithiothreitol (DTT), 30% (vol/vol) glycerol, 0.05% (wt/vol) bromophenol blue in 0.5 M Tris-HCl, pH 6.8, and by heating at 60°C for 10 min.
Anti-SbmA antibody purification.
To develop a polyclonal antibody directed against SbmA, a peptide corresponding to the sequence starting from position 269 was synthesized with an extra cysteine residue added at the C-terminal end, conjugated with the sulfhydryl-reactive keyhole limpet hemocyanin (KLH) (Pierce), and injected into the rabbit bloodstream. The polyclonal antibody was then purified from the rabbit antiserum via affinity chromatography onto a Sulfolink resin (Pierce) prepared according to the manufacturer's instructions. Briefly, the antiserum was centrifuged at 6,000 × g at 4°C, and the supernatant was filtered through a 0.2-μm filter and diluted with 1 volume of phosphate-buffered saline (PBS) (1.59 mM NaH2PO4, 8.45 mM Na2HPO4, 150 mM NaCl, pH 7.4) and incubated overnight at 4°C under constant shaking with the functionalized Sulfolink. After repeated washing steps of the resin with PBS, PBS HST (500 mM NaCl, 0.1% Triton X-100 in PBS), and PBS HS (500 mM NaCl in PBS), the antibody was eluted with 200 mM glycine-HCl, pH 2.8, and the pH of the fractions corresponding to the purified antibody was immediately adjusted to pH 7.5 with 2 M Tris-HCl, pH 8. The concentration of the purified antibody was measured by the BCA assay (Pierce), and the antibody was stored at −20°C with 30% glycerol.
Protein detection.
Upon separation by SDS-PAGE, the proteins were transferred to a nitrocellulose membrane using a semidry transfer apparatus (Bio-Rad) at 20 V for 30 min, and the membrane was stained with Ponceau Red. For the detection, the membrane was blocked overnight in 5% (wt/vol) milk powder in TBST solution (40 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.1% [vol/vol] Tween 20) at 4°C and then incubated for 90 min at room temperature with the rabbit anti-SbmA antibody at a titer of 1:2,000. The membrane was then washed and incubated for 1 h at room temperature with the goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (HRP) (GE Healthcare) at a titer of 1:2,000. The detection of HRP was performed using the ECL Plus kit (Amersham).
In vivo bacterial two-hybrid assay.
Electrocompetent E. coli BTH101 cells were cotransformed with 10 ng of plasmid pKNT25 and pUT18 carrying sbmA fused to the N terminus of the T25 subunit or to the N terminus of the T18 subunit of the adenylate cyclase, respectively. The cotransformants were plated on LB agar plates with 10 mg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 5 μM isopropyl-β-d-1-thiogalactopyranoside containing kanamycin (50 μg ml−1) and ampicillin (100 μg ml−1) and left to grow at 30°C for 48 h. Clones were considered positive when the intensity of the blue color was comparable to that of the positive control, represented by the two leucine zipper domains of the transcriptional factor GCN4 of Saccharomyces cerevisiae.
DNA sequencing and analysis.
Sequencing of the DNA inserts on both strands was performed at BMR Genomics (Padua, Italy) using the specific forward (5′-GTT-CGC-CAT-TAT-GCC-GCA-TC-3′) and reverse (5′-GGA-TGT-GCT-GCA-AGG-CGA-TT-3′) primers. Nucleotide sequence analyses were performed by using the EcoCYC database for the E. coli genome (http://www.ecocyc.org/). Database similarity searches were carried out with BLAST at the National Center for Biotechnology Information website.
β-Galactosidase activity assay.
The β-galactosidase activity was determined on the basis of the method described by Miller (21) with few modifications. Six hundred microliters (600 μl) of cells resuspended in 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.05 M β-mercaptoethanol, pH 7.0, was permeabilized for at least 20 min with 0.1% (wt/vol) SDS (24 μl) and chloroform (48 μl) and then processed as described previously (22). The reaction was conducted at 37°C, and the β-galactosidase activity was calculated in Miller units.
In vitro cross-linking assay.
Purified SbmA protein (∼6 μg) was incubated with increasing concentrations of glutaraldehyde (Sigma) (0.125, 0.25, and 0.5%, vol/vol) for 10 min at 4°C in a total reaction volume of 20 μl, then glycine was added to a final concentration of 0.125 M, and the sample was incubated for 10 min at 4°C in order to block the reaction.
Size exclusion chromatography coupled with multiangle laser light scattering (SEC-MALLS) analysis.
The oligomeric state of detergent-purified SbmA protein was assessed using a Malvern Viscotek TDAmax Tetra detection system, including static light scattering and UV and refractive index detectors, connected downstream of a Superdex-S200 10/300 gel filtration column equilibrated in 20 mM Tris, pH 8, 150 mM NaCl, and 0.03% (wt/vol) DDM. The protein concentration was adjusted to 21.5 μM. The data were analyzed using Omnisec (Malvern).
Flow cytometric analysis.
Uptake of Bac7(1-35)-BY in E. coli cells was determined by flow cytometry using a Cytomics FC500 instrument (Beckman-Coulter, Inc.) as previously described (5). Cultures of mid-log-phase bacteria were harvested, diluted to 106 CFU ml−1 in MH broth, incubated with 0.25 μM Bac7(1-35)-BY in the presence of 0.5, 1.25, and 2.5 mM 2,4-dinitrophenol (DNP) (Sigma); 0.5, 2.5, 5, and 10 mM sodium arsenate (Sigma); 5 μM nigericin (Sigma); 5 μM valinomycin (Sigma), or both at 37°C for 10 min, and analyzed immediately. All experiments were conducted in triplicate, and data were expressed as mean fluorescence intensity (MFI) + standard deviation (SD). Data analysis was performed with the FCS Express V3 software (De Novo Software, CA).
ATP determination.
To determine the total amount of ATP in E. coli cells, the ATP Determination kit (Invitrogen) was used. Bacterial suspensions were prepared as described for the flow cytometry assays with the addition of Bac7(1-35) in place of the fluorescent Bac7(1-35)-BY. After 10 min of incubation at 37°C with the different inhibitors, 100 μl of each bacterial suspension was transferred to a fresh 2-ml tube and 900 μl of boiling buffer containing 100 mM Tris-HCl, pH 7.8, and 4 mM EDTA was added to each tube. The tubes were then incubated at 100°C for 2 min and centrifuged at 9,600 × g for 2 min. One hundred microliters of each sample was assayed in triplicate with a luminometer (Hidex Chameleon).
Time course transport assay.
E. coli cells, C43(DE3), harboring SbmA-GFP-His8, were incubated in LB at 37°C with 1 μM Bac7(1-35)-BY for the indicated time intervals. Transport was terminated after centrifugation at 20,500 × g for 1 min. Cell pellets were washed in equal volumes of PBS buffer and resuspended in 100 μl of the same buffer. Nonspecific uptake was measured under the same conditions using JW0368-1 cells that were sbmA deficient. All measurements were corrected for OD600. The fluorescence counts of internalized peptide were measured using a Spectramax spectrofluorometer at 570 nm (excitation, 544 nm). Each experiment was performed in duplicate.
Transport kinetics.
The kinetics of the Bac7(1-35)-BY transport by SbmA were measured at various concentrations of the peptide at 37°C for 60 min; the initial peptide uptake was slow and reached maximum at 60 min. The fluorescence counts of internalized peptide were measured using a Spectramax spectrofluorometer at 570 nm (excitation, 544 nm). Data were corrected for expressed SbmA-GFP-His8 by measuring the green fluorescent protein (GFP) fluorescence counts at 512 nm (excitation, 488 nm). Each experiment was performed in duplicate. The data were fitted in the Michaelis-Menten equation by nonlinear regression using GraphPad Prism.
SRCD studies.
Synchrotron radiation circular dichroism (SRCD) experiments were performed using a nitrogen-flushed Module B end-station spectrophotometer at the B23 Synchrotron Radiation CD Beamline at the Diamond Light Source, Oxfordshire, United Kingdom (23). Samples were typically prepared in 20 mM Tris-HCl, pH 7.5, and 20 mM NaCl containing 0.03% (wt/vol) DDM. Measurements were carried out at 20°C unless otherwise stated. For measurements in the far-UV range (180 to 260 nm), the concentration of SbmA and BacA employed was in the range of 10 to 25 μM. Aliquots of Bac7(1-35) stock solution in water were added to SbmA and BacA stock solutions of 10 and 24 μM, respectively, for the titrations at various molar stoichiometries. Using sample volumes of 30 μl, four scans were acquired using an integration time of 1 s, a path length of 0.02 cm, and a slit width of 0.5 mm equivalent to a 1.2-nm bandwidth. For thermal stability, both SbmA and BacA were incubated in the presence and absence of 5-fold Bac7(1-35) at 5°C and spectra were measured every 5°C over a temperature range between 5°C and 95°C with 5 min of equilibration time for each temperature. Reversibility was monitored by measuring the spectrum at 5°C after cooling from 95°C with 30 min of incubation time. For measurements in the near-UV region (250 to 340 nm), experiments were conducted in the same buffer using 100 μl of sample with concentration ranging from 10 to 25 μM at 20°C using a low-volume, small-aperture (2-mm), 1-cm-path-length cell. The spectra were reported as differential absorption ΔA, which is a differential absorption of left and right circular polarized light. The ΔA values at 223 nm were plotted against concentrations of Bac7(1-35) in the mixture. For one binding site approximation, the dissociation constant Kd was determined using the nonlinear regression analysis as reported by Siligardi et al. (24).
Statistical analysis.
The significance of differences among bacterial strains treated with increasing concentrations of sodium arsenate, DNP, nigericin, and/or valinomycin was assessed using GraphPad Prism by the Student-Newman-Keuls multiple comparison test and analysis of variance (ANOVA).
RESULTS
The SbmA-mediated transport of Bac7 is proton driven rather than ATP driven.
From the sequence analysis, sbmA does not encode an NBD (see Fig. S1 in the supplemental material). To determine the driving force of the transport, we first measured the uptake of Bac7(1-35)-BY in the presence of increasing concentrations of DNP or sodium arsenate. DNP is an ionophore which moves protons across the inner membrane and affects both the pH gradient and the membrane potential, while sodium arsenate inhibits production of ATP via substrate-level phosphorylation (25).
To correlate the uptake of Bac7(1-35)-BY with the ATP concentration of the cells, we measured the ATP level in bacterial cells treated with DNP or sodium arsenate. The uptake assay and the measurement of the ATP concentration were performed in the E. coli strain JW3710, carrying a deletion in the gene atpD encoding the β-subunit of the ATP synthase (Table 1). In this strain, ATP is produced exclusively by substrate phosphorylation, and therefore, the DNP affects only the proton gradient and not the ATP content, since the ATP synthase is impaired. By using this mutant, the effect of the DNP on the proton gradient can be evaluated separately from its indirect effect on ATP synthesis. On the other hand, the sodium arsenate should dramatically reduce the amount of ATP in the cell acting on substrate phosphorylation.
We showed that treatment with increasing concentrations of sodium arsenate did not inhibit the uptake of Bac7(1-35)-BY by the cells, despite a significant reduction in ATP cell content (Fig. 1A). In contrast, the treatment of the bacteria with increasing concentrations of DNP caused a significant decrease in the amount of internalized peptide, although the ATP content still remained high (Fig. 1B). The low reduction of the ATP content, observed after treatment of the cells with DNP, is scarcely concentration dependent and might be due to an attempt of the cell to pump out protons consuming ATP in order to counteract the DNP-mediated proton flow across the membrane. Taken together, these results highlight the predominant effect of the uncoupler on the uptake and confirm that this effect is not directly related to the ATP content.
Fig 1.
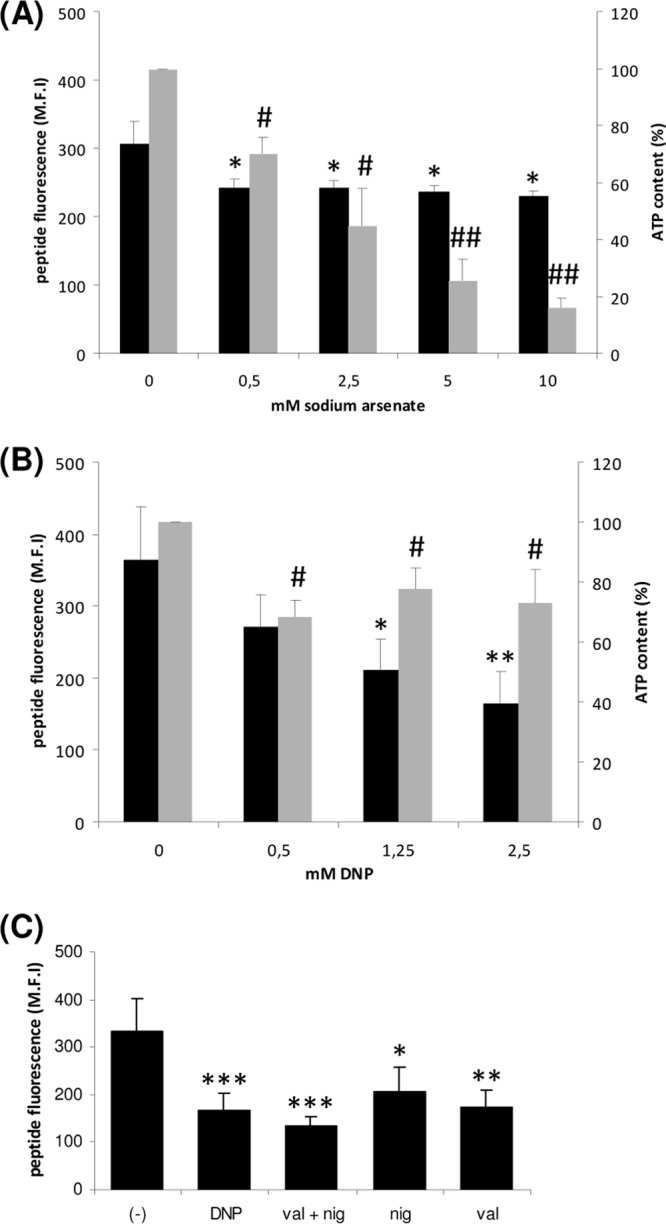
Effect of different inhibitors on the uptake of Bac7(1-35)-BY and on the ATP content of E. coli cells. Black bars represent the uptake of Bac7(1-35)-BY in E. coli JW3710 (ΔatpD) cells measured as medium fluorescence intensity (M.F.I.) in the presence of increasing concentrations of sodium arsenate (A), DNP (B), and valinomycin and/or nigericin (C). Gray bars represent the ATP content of the cells measured as luminescence produced by the firefly luciferase reaction. The level of luminescence is expressed as a percentage of luminescence after treatment with the different inhibitors with respect to the control, which represents the 100% value. *, P ≤ 0.05 versus untreated; **, P ≤ 0.01; ***, P ≤ 0.005; #, P ≤ 0.001 versus untreated; ##, P ≤ 0.0001.
To assess which component of the proton motive force is necessary for the transport of Bac7(1-35)-BY, we evaluated its uptake in the presence of different specific ionophores (Fig. 1C). Nigericin mediates the antiport of H+ and K+ down their concentration gradients and thereby dissipates the transmembrane proton gradient (ΔpH,) leaving unaltered the transmembrane potential. In contrast, valinomycin mediates the uptake of K+ in the cell, selectively dissipating the transmembrane potential (Δψ). The treatment of the cells with nigericin or valinomycin reduced the amount of internalized peptide, but the highest reduction of internalized peptide was observed when both ionophores were present. These results indicate that both components of the electrochemical transmembrane gradient are required for transport.
Whole-cell transport assays.
The SbmA transport kinetics were measured by using E. coli whole-cell transport assays. A time course uptake assay in the presence of Bac7(1-35)-BY is shown in Fig. 2A. Cells expressing endogenous or recombinant SbmA transport larger amounts of Bac7 than do cells that are sbmA deficient, in agreement with previous studies (5). The initial peptide uptake was slow and reached maximum at 60 min. Cells expressing recombinant SbmA-GFP were used to calculate SbmA-dependent transport kinetics and displayed a Km of 6.95 ± 0.89 μM and a Vmax of 53.91 ± 3.17 nmol/min/mg SbmA (Fig. 2B).
Fig 2.
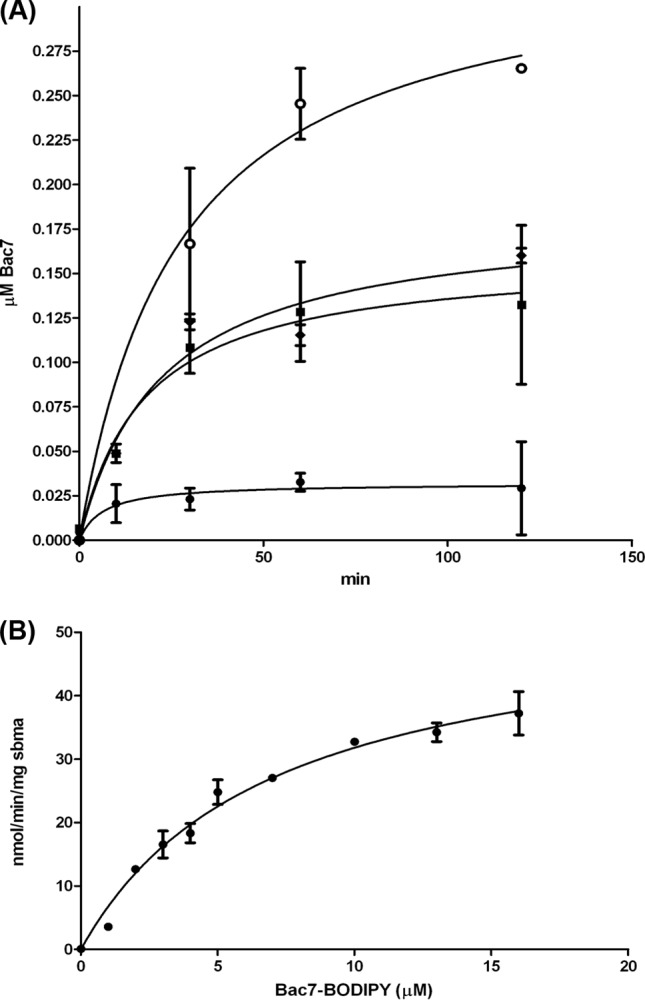
SbmA transport kinetics in whole E. coli cells. (A) Time-dependent internalization of Bac7(1-35)-BY. Cells that are sbmA deficient show a low level of nonspecific uptake (filled circles). In the presence of endogenous SbmA, the uptake increases by 6-fold (filled diamonds). Cells that are carrying the expression vector without induction show uptake levels similar to those of nontransformed cells (filled squares). Cells that are expressing recombinant SbmA-GFP-His8 show a 1.8-fold increase relative to the wild-type cells (empty circles). (B) Michaelis-Menten transport kinetics of SbmA-mediated Bac7(1-35)-BY uptake.
SbmA binds Bac7 in vitro.
A previous study has shown that SbmA is necessary for the transport of proline-rich AMPs (5). To establish whether SbmA directly binds Bac7, we analyzed the ability of the two molecules to form a complex in vitro. Purified SbmA was used in affinity chromatography on a Bac7(1-35)-functionalized resin. Two forms of recombinant SbmA were produced: one obtained after cleavage of the GFP moiety from the SbmA-GFP fusion protein and the other fused with a His tag. Both recombinant proteins were used to test the binding to Bac7 and gave similar results. Figure 3 shows that the His-tagged SbmA is retained by the Bac7(1-35) resin, even though with low efficiency.
Fig 3.
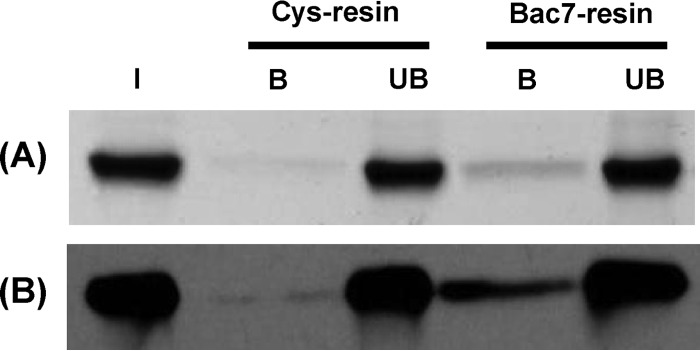
In vitro assay of the binding of SbmA to Bac7. (A) SDS-PAGE analysis of the 6-His-tagged SbmA protein bound by the Bac7(1-35)-functionalized resin. Binding of the native SbmA protein to the unfunctionalized resin (Cys-resin) has been used as a negative control. I, input SbmA; B, bound SbmA; UB, unbound SbmA. (B) Western blot analysis of the samples analyzed by SDS-PAGE using an anti-SbmA antibody.
Studies of binding of Bac7 to SbmA and to BacA by CD spectroscopy.
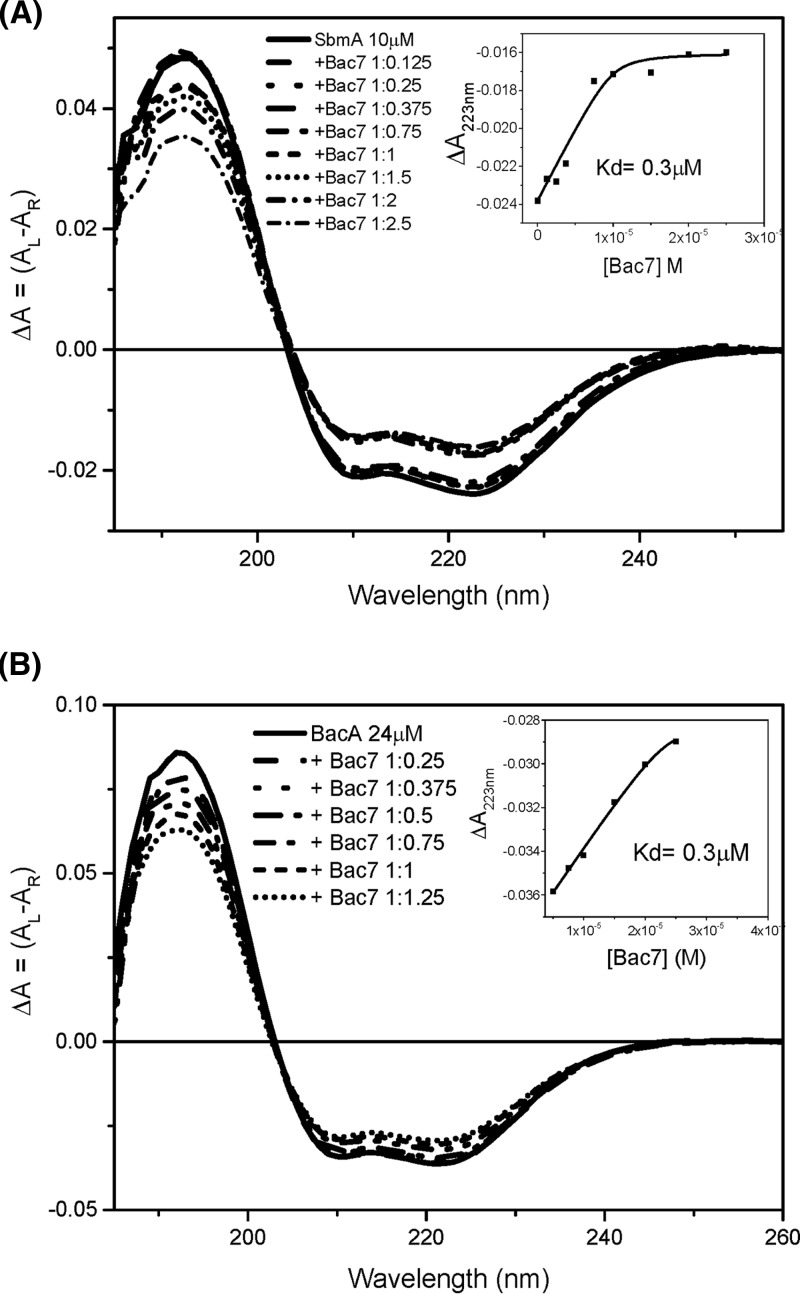
The CD spectroscopy studies provided further evidence that SbmA and BacA can interact with Bac7 in vitro. Bac7(1-35) was titrated at various molar stoichiometries to SbmA and BacA, and CD spectra in the far-UV region (180 to 260 nm) and the near-UV region (250 to 330 nm) were recorded. In the far-UV region, significant changes in the secondary structure of the mixture were observed when Bac7(1-35) was added to SbmA (Fig. 4A; see also Fig. S2A in the supplemental material) and BacA (Fig. 4B; see also Fig. S3A in the supplemental material), indicating interaction of SbmA and BacA with Bac7 accompanied by structural changes for either SbmA/BacA, Bac7, or both (Table 2). In the near-UV region, characteristic of the local tertiary structure of aromatic side chain residues, there were not significant changes, suggesting that no aromatic residues are involved in the interface between Bac7(1-35) and SbmA/BacA (see Fig. S2B and S3B). The results showed that Bac7(1-35) has a high binding affinity to both SbmA and BacA with a Kd of 0.3 μM (Fig. 4). Temperature studies were also carried out for SbmA and the mixture with Bac7 (see Fig. S2C). No significant differences between the melting temperature Tm of SbmA and that of the mixture with Bac7 (Tm = 55°C) were observed (see Fig. S2D). It is interesting that the conformational changes were not accompanied by differences in thermal stability for either SbmA or BacA (see Fig. S2D, S3C, and S3D); this could possibly be due to rearrangement of the secondary structure of the proteins, with SbmA having a loss of alpha-helix content (11%) but compensated by an increase in beta-sheet (5%) upon binding of Bac7 to SbmA, while in the case of BacA, a loss of beta-sheet (3%) is compensated by an increase in alpha-helix (5%) in the presence of Bac7, which resulted in no change of the Tm observed.
Fig 4.
Effect of Bac7 on the secondary structure of SbmA and BacA. Far-UV spectra of SbmA (A) and BacA (B) in the presence of increasing amounts of Bac7. The spectra were reported as differential absorption ΔA, which is a differential absorption of left and right circular polarized light. The insets show the ΔA values at 223 nm plotted against concentrations of Bac7(1-35) in the mixture.
Table 2.
Determination of secondary structure composition of SbmA with and without Bac7(1-35) using CONTINa
| Secondary structure | SbmA | SbmA + Bac7(1-35) | BacA | BacA+ Bac7(1-35) |
|---|---|---|---|---|
| H1, alpha-helix | 0.41 | 0.32 | 0.41 | 0.46 |
| H2, distorted alpha-helix | 0.19 | 0.17 | 0.19 | 0.19 |
| S1, β-strand | 0.04 | 0.09 | 0.06 | 0.03 |
| S2, distorted β-strand | 0.04 | 0.04 | 0.03 | 0.02 |
| T, turn | 0.12 | 0.10 | 0.12 | 0.10 |
| U, unordered | 0.21 | 0.28 | 0.20 | 0.20 |
| Spectral fit SD | 0.06 | 0.04 | 0.06 | 0.04 |
SbmA is able to form dimers.
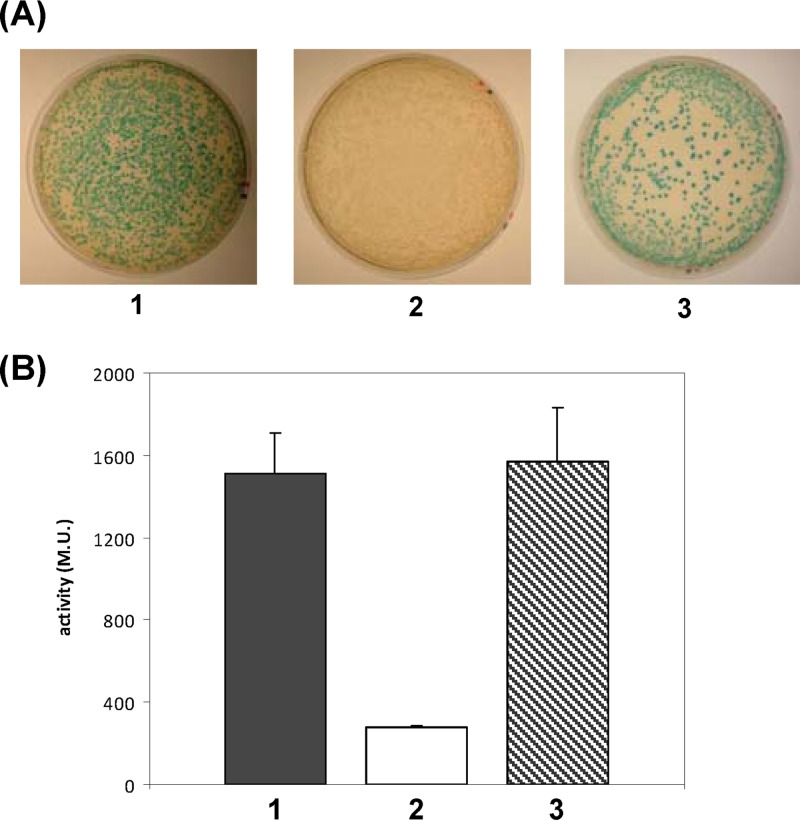
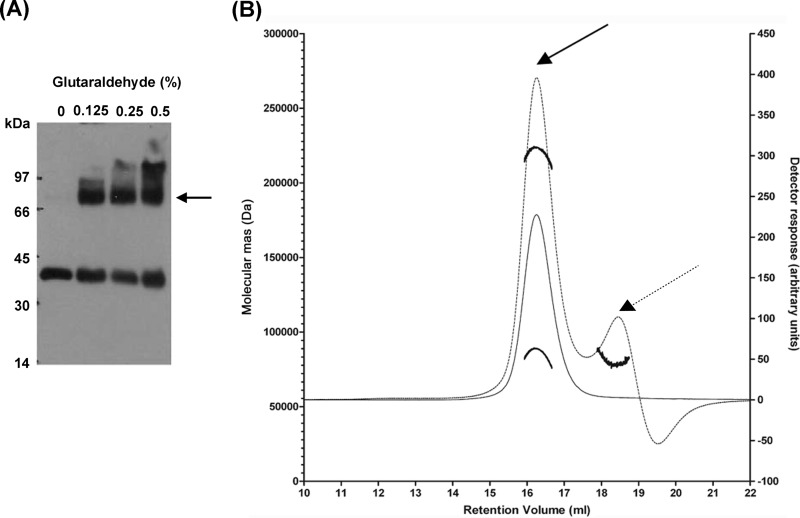
To study the oligomeric state of SbmA, we performed a bacterial two-hybrid assay expressing SbmA fused to the T18 or T25 subunit of the adenylate cyclase of Bordetella pertussis (26). Cotransformation of E. coli BTH101 with both the plasmids carrying the fusion proteins resulted in functional complementation between the two halves of the adenylate cyclase, indicating that two SbmA monomers are able to interact (Fig. 5A). We also showed that the strength of this interaction is comparable to that of the positive control (Fig. 5B). An in vitro (Fig. 6A) cross-linking assay with the purified recombinant His-tagged SbmA in the presence of increasing amounts of glutaraldehyde showed that this protein forms dimers.
Fig 5.
In vivo dimerization of SbmA. (A) Bacterial two-hybrid assay. E. coli BTH101 cells were cotransformed with pUT18C-zip and pKT25-zip vectors as a positive control (1), pKNT25-sbma and empty pUT18 (2), and pUT18-sbma and pKNT25-sbma (3). (B) β-Galactosidase assay to measure the enzymatic activity expressed as Miller units (M.U.). Bar numbers correspond to plate numbers in panel A. Results are the means of four independent experiments.
Fig 6.
In vitro dimerization of SbmA. (A) Western blot analysis of the recombinant His-tagged SbmA incubated in the presence of increasing amounts of glutaraldehyde; the dimer is indicated by the arrow. At a high percentage of cross-linking agent (0.5%, vol/vol), superaggregated forms of the protein become visible. (B) SEC-MALLS analysis of SbmA in solution. The chromatogram shows the detector readings of UV280 (solid line) and refractive index (dotted line; scale on right-hand y axis). The thick black line indicates the calculated molecular mass of the eluted protein throughout the peak (scale on left-hand y axis). Data were analyzed as in reference 27. The average molecular mass of the SbmA-DDM complex was calculated to be 223 kDa, across the refractive index detector elution peak (dotted line) (27). The average molecular mass of the SbmA assembly is 89.2 kDa. Solid and dashed arrows indicate SbmA-DDM complex and free DDM micelle, respectively.
The oligomeric state of detergent-purified SbmA was also studied using size exclusion chromatography coupled with multiangle laser light scattering (SEC-MALLS). The predicted molecular mass of SbmA from the primary sequence is 46.46 kDa. The DDM-solubilized and purified SbmA is monodisperse in solution as evaluated from the 280-nm detector. The molecular mass of the SbmA-DDM complex cannot be determined accurately from the 280-nm detector since we cannot calculate how much detergent is bound to SbmA. Therefore, the molecular mass was calculated from the refractive index detector (Fig. 6B).The elution peak was calculated to be 223 kDa, corresponding to the SbmA-DDM complex. A smaller peak at 78.5 kDa that appears only at the refractive index detector corresponds to empty detergent micelles (Fig. 6B). The average molecular mass of SbmA from the elution peak, excluding the contribution of the detergent micelle (27), was determined to be 89.2 kDa, which confirms that SbmA exists as a dimer in the DDM solution.
DISCUSSION
A number of reports have stressed the importance of the E. coli inner membrane protein SbmA in the bacterial susceptibility to some antibiotics and antibacterial peptides. However, a role for SbmA in the uptake of AMPs has been proposed only in the past 5 years (5). New insights in SbmA function came from the observation that BacA, the SbmA homolog in S. meliloti, is essential for the survival of the bacteria in the plant symbiosome and the resistance to NCR peptides released by the plant (15). In this study, we showed for the first time that (i) the energization process of the SbmA-mediated transport is based on the electrochemical transmembrane gradient rather than on ATP hydrolysis, (ii) in vitro Bac7(1-35) binds to SbmA and its homolog BacA with high affinity, (iii) Bac7(1-35) is efficiently transported across the membrane by SbmA, and (iv) SbmA is homodimeric in vitro and in vivo.
SbmA as well as BacA and BacA-related proteins has been originally proposed to be the TMD of an ABC transporter on the basis of its sequence similarity (10). However, the alignment of E. coli SbmA and S. meliloti BacA with BacA-related proteins that carry an NBD in their sequence showed low sequence identity at the TMD region (∼20%) (see Fig. S1 in the supplemental material). In addition, SbmA and its homolog BacA do not have any motif with ATPase activity in their sequence. These preliminary observations prompted us to evaluate whether ATP hydrolysis was required to translocate the proline-rich fragment Bac7(1-35) peptide across the inner membrane. A preliminary screen via a bacterial two-hybrid system to look for a possible orphan ATP binding domain and pulldown assays failed to identify any interacting NBD partners specific for SbmA (data not shown).
We observed that a major reduction of the uptake of Bac7(1-35) could be obtained after treatment of the bacterial cells with the uncoupler DNP, without a consistent decrease in ATP level, or with ionophores that disrupt both the chemical and electrical components of the proton motive force. These results indicate that the uptake of Bac7(1-35) requires the presence of a transmembrane electrochemical gradient rather than ATP hydrolysis to be fully accomplished.
SbmA has the notable feature of internalizing structurally unrelated bacterial peptides, including MccB17 (11), MccJ25 (4), and the nonpeptide antibiotic bleomycin (9), in addition to the eukaryotic AMP Bac7(1-35). MccJ25 is a 21-amino-acid posttranslationally modified peptide that adopts a lasso structure, whereas MccB17 and bleomycin contain thiazole heterocycles (MccB17 also contains oxazoles). Moreover, recent findings suggest the involvement of SbmA also in the internalization of antisense peptide phosphorodiamidate morpholino oligomers and PNAs (13, 14), expanding the spectrum of substrates transported by this protein. More importantly, a high level of promiscuity to export structurally unrelated peptides is a feature typical of multidrug transporters (28, 29). Interestingly, when the multidrug ABC transporter LmrA from Lactococcus lactis is deprived of its NBD, it acts as a secondary-active multidrug uptake system energized by the proton gradient (30), which suggests that the evolutionary precursor of LmrA was a secondary-active transporter that acquired an NBD to become a multidrug efflux system. Another example of switching mode of energy coupling to transport is the E. coli ArsAB transporter responsible for arsenite efflux. In vivo transport studies suggest that the ArsB protein mediates the electrochemical energy-dependent arsenite efflux in the absence of the NBD protein ArsA, while the ArsAB complex catalyzes ATP-dependent transport (31). Across the bacterial genomes, some ars operons have both arsA and arsB genes and thus encode ATP-dependent pumps, while others have only the arsB gene and encode secondary systems, thus suggesting that the acquisition of a gene for a catalytic subunit might be a recent evolutionary event. This observation suggests that SbmA may have the characteristics of an evolutionary ancestor of a multidrug ABC transporter with “import” direction of transport in the absence of an NBD. Interestingly, in E. coli there is a gene coding for a putative longer SbmA-like protein of unknown function, YddA, carrying an additional C-terminal domain with an ATPase moiety, and this protein represents a full ABC transporter. The BacA-related protein of M. tuberculosis is another example of a complete ABC transporter. The MtBacA protein is 22% identical to S. meliloti BacA and contains a fused putative ATPase domain at the C terminus of the protein. It mediates peptide uptake of the truncated bovine Bac7(1-16) in a process requiring a functional ATPase domain (17).
The possibility that SbmA may be part of a dimer or a complex was reported to explain the dominant negative character of some BacA (10) and SbmA (5) mutations. Two-hybrid screening, cross-linking assay, and size exclusion chromatography strongly indicated that SbmA forms homodimers both in vitro and in the bacterial membrane and that this is very likely the functional form of this protein. In addition, we obtained some insights on the number of transmembrane spans and of its N and C terminus localization. Given the constraints of the two-hybrid system, which can functionally reconstitute the two subunits of the adenylate cyclase only on the cytoplasmic side of the bacterial membrane, in this study we provide indirect evidence on the localization of the C terminus of SbmA inside the bacterial cell, as the two subunits fused to the C termini gave a detectable interaction. In addition, when the T25 and T18 subunits were cloned at the N terminus of SbmA, the two monomers still continued to interact, suggesting that their N terminus ends are also on the cytoplasmic side, as detailed in the accompanying article by Corbalan et al. (32). These data are in agreement with a previous study on the global topology of the E. coli inner membrane proteome (33), indicating that the C terminus of this protein is in the cytoplasm. Taken together, these data suggest that SbmA is an eight-transmembrane-span protein with both ends in the cytoplasm.
A role of SbmA in the transport of Bac7 was already proposed (5). In the present work, we calculated the Km and maximum rate of transport for the Bac7(1-35) by SbmA. Both parameters are in the same concentration range of other peptide transporters, such as the oligopeptide transport system Opp of L. lactis (34). The Opp has a transport affinity in the range of 700 μM for tetrapeptides, and the affinity increases to 8.8 μM for 11-amino-acid-long peptides. SbmA also appears to have high affinity, with a Km of 6.95 μM for the 35-amino-acid-long peptide Bac7(1-35).
The CD spectroscopy studies showed that binding of Bac7(1-35) to SbmA and also to BacA produced some significant changes in the secondary structure of one of the partners or both of them. The Bac7(1-35) did not show any structural features in the CD experiments, and the SbmA was mostly helical. The protein secondary structure estimation of SbmA showed a more pronounced unfolding with a loss of 11% of alpha-helix and a significant increase in disorder content of the protein upon binding to Bac7 (using the CONTIN algorithm) (Table 2). BacA, however, showed a mixture of a loss of 4% in β-strand and turn and a gain of 5% alpha-helix upon binding to Bac7. This contributed to the overall small change in the conformation upon binding to Bac7. In addition, the binding of SbmA and its homologous protein BacA to the antibiotic bleomycin did not show any changes in the secondary structure (data not shown). This not only supports the binding of structurally unrelated peptides to BacA and SbmA but suggests also that the Bac7 binding process involves secondary structural changes.
It is known that BacA is essential for the persistence of bacteria within plant and mammalian hosts (1), whereas the physiological function of SbmA is still unknown. Our data suggest that this protein may be involved in the uptake of certain unknown peptides, triggering a specialized function. Since SbmA is widespread in enteric bacteria, its function may be linked to a specific ecological niche represented by the gut of the host and it may contribute to the persistence of these bacteria within the intestine.
In conclusion, in this work we showed that SbmA is a secondary active transporter energized by the transmembrane electrochemical gradient and we shed light on the SbmA-mediated internalization of peptides, showing that the eukaryotic peptide Bac7(1-35) is capable of directly binding to SbmA with high affinity, causing conformational changes of the transporter. The results from this study are useful for designing novel peptides and antibiotic peptides able to penetrate efficiently into SbmA/BacA-carrying bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank Diamond Light Source for circular dichroism beam time and access. We also thank Paula Vincent for discussing the results of the bacterial two-hybrid assays and Graham C. Walker and Hajime Kobayashi for generously providing the pKNT25(sbmA) and pUT18C(sbmA) plasmids.
This study was supported by grants from the Italian Ministry for University and Research (PRIN 2008) and from the Regione Friuli Venezia Giulia grant under the LR 26/2005, art. 23, for the R3A2 network and by a Biotechnology and Biological Sciences Research Council grant (BBSRC: BB/H01778X/1) to Konstantinos Beis. Tatiana Stoilova acknowledges a fellowship from the NAM, New AntiMicrobials (PIAP-GA-2008-218191).
Footnotes
Published ahead of print 27 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00818-13.
REFERENCES
- 1.Glazebrook J, Ichige A, Walker GC. 1993. A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 7:1485–1497 [DOI] [PubMed] [Google Scholar]
- 2.LeVier K, Phillips RW, Grippe VK, Roop RM, II, Walker GC. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287:2492–2493 [DOI] [PubMed] [Google Scholar]
- 3.Domenech P, Kobayashi H, LeVier K, Walker GC, Barry CE., III 2009. BacA, an ABC transporter involved in maintenance of chronic murine infections with Mycobacterium tuberculosis. J. Bacteriol. 191:477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salomon RA, Farias RN. 1995. The peptide antibiotic microcin 25 is imported through the TonB pathway and the SbmA protein. J. Bacteriol. 177:3323–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattiuzzo M, Bandiera A, Gennaro R, Benincasa M, Pacor S, Antcheva N, Scocchi M. 2007. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 66:151–163 [DOI] [PubMed] [Google Scholar]
- 6.Marlow VL, Haag AF, Kobayashi H, Fletcher V, Scocchi M, Walker GC, Ferguson GP. 2009. Essential role for the BacA protein in the uptake of a truncated eukaryotic peptide in Sinorhizobium meliloti. J. Bacteriol. 191:1519–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichige A, Walker GC. 1997. Genetic analysis of the Rhizobium meliloti bacA gene: functional interchangeability with the Escherichia coli sbmA gene and phenotypes of mutants. J. Bacteriol. 179:209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wehmeier S, Arnold MF, Marlow VL, Aouida M, Myka KK, Fletcher V, Benincasa M, Scocchi M, Ramotar D, Ferguson GP. 2010. Internalization of a thiazole-modified peptide in Sinorhizobium meliloti occurs by BacA-dependent and -independent mechanisms. Microbiology 156:2702–2713 [DOI] [PubMed] [Google Scholar]
- 9.Yorgey P, Lee J, Kordel J, Vivas E, Warner P, Jebaratnam D, Kolter R. 1994. Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc. Natl. Acad. Sci. U. S. A. 91:4519–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeVier K, Walker GC. 2001. Genetic analysis of the Sinorhizobium meliloti BacA protein: differential effects of mutations on phenotypes. J. Bacteriol. 183:6444–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavina M, Pugsley AP, Moreno F. 1986. Identification, mapping, cloning and characterization of a gene (sbmA) required for microcin B17 action on Escherichia coli K12. J. Gen. Microbiol. 132:1685–1693 [DOI] [PubMed] [Google Scholar]
- 12.Pranting M, Negrea A, Rhen M, Andersson DI. 2008. Mechanism and fitness costs of PR-39 resistance in Salmonella enterica serovar Typhimurium LT2. Antimicrob. Agents Chemother. 52:2734–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puckett SE, Reese KA, Mitev GM, Mullen V, Johnson RC, Pomraning KR, Mellbye BL, Tilley LD, Iversen PL, Freitag M, Geller BL. 2012. Bacterial resistance to antisense peptide phosphorodiamidate morpholino oligomers. Antimicrob. Agents Chemother. 56:6147–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosal A, Vitali A, Stach JE, Nielsen PE. 2013. Role of SbmA in the uptake of peptide nucleic acid (PNA)-peptide conjugates in E. coli. ACS Chem. Biol. 8:360–367 [DOI] [PubMed] [Google Scholar]
- 15.Haag AF, Baloban M, Sani M, Kerscher B, Pierre O, Farkas A, Longhi R, Boncompagni E, Herouart D, Dall'angelo S, Kondorosi E, Zanda M, Mergaert P, Ferguson GP. 2011. Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol. 9:e1001169. 10.1371/journal.pbio.1001169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z, Farkas A, Mikulass K, Nagy A, Tiricz H, Satiat-Jeunemaitre B, Alunni B, Bourge M, Kucho K, Abe M, Kereszt A, Maroti G, Uchiumi T, Kondorosi E, Mergaert P. 2010. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327:1122–1126 [DOI] [PubMed] [Google Scholar]
- 17.Arnold MF, Haag AF, Capewell S, Boshoff HI, James EK, McDonald R, Mair I, Mitchell AM, Kerscher B, Mitchell TJ, Mergaert P, Barry CE, III, Scocchi M, Zanda M, Campopiano DJ, Ferguson GP. 2013. Partial complementation of Sinorhizobium meliloti bacA mutant phenotypes by the Mycobacterium tuberculosis BacA protein. J. Bacteriol. 195:389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidson AL, Chen J. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241–268 [DOI] [PubMed] [Google Scholar]
- 19.Scocchi M, Mattiuzzo M, Benincasa M, Antcheva N, Tossi A, Gennaro R. 2008. Investigating the mode of action of proline-rich antimicrobial peptides using a genetic approach: a tool to identify new bacterial targets amenable to the design of novel antibiotics. Methods Mol. Biol. 494:161–176 [DOI] [PubMed] [Google Scholar]
- 20.Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. 2006. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 3:303–313 [DOI] [PubMed] [Google Scholar]
- 21.Miller JH. (ed). 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22.Griffith KL, Wolf RE., Jr 2002. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem. Biophys. Res. Commun. 290:397–402 [DOI] [PubMed] [Google Scholar]
- 23.Javorfi T, Hussain R, Myatt D, Siligardi G. 2010. Measuring circular dichroism in a capillary cell using the b23 synchrotron radiation CD beamline at diamond light source. Chirality 22(Suppl 1):E149–E153 [DOI] [PubMed] [Google Scholar]
- 24.Siligardi G, Panaretou B, Meyer P, Singh S, Woolfson DN, Piper PW, Pearl LH, Prodromou C. 2002. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J. Biol. Chem. 277:20151–20159 [DOI] [PubMed] [Google Scholar]
- 25.Cascales E, Gavioli M, Sturgis JN, Lloubes R. 2000. Proton motive force drives the interaction of the inner membrane TolA and outer membrane pal proteins in Escherichia coli. Mol. Microbiol. 38:904–915 [DOI] [PubMed] [Google Scholar]
- 26.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slotboom DJ, Duurkens RH, Olieman K, Erkens GB. 2008. Static light scattering to characterize membrane proteins in detergent solution. Methods 46:73–82 [DOI] [PubMed] [Google Scholar]
- 28.Putman M, van Veen HW, Konings WN. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulsen IT, Brown MH, Skurray RA. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venter H, Shilling RA, Velamakanni S, Balakrishnan L, Van Veen HW. 2003. An ABC transporter with a secondary-active multidrug translocator domain. Nature 426:866–870 [DOI] [PubMed] [Google Scholar]
- 31.Kuroda M, Dey S, Sanders OI, Rosen BP. 1997. Alternate energy coupling of ArsB, the membrane subunit of the Ars anion-translocating ATPase. J. Biol. Chem. 272:326–331 [DOI] [PubMed] [Google Scholar]
- 32.Corbalan N, Runti G, Adler C, Covaceuszach S, Ford RC, Lamba D, Beis K, Scocchi M, Vincent PA. 2013. Functional and structural study of the dimeric inner membrane protein SbmA. J. Bacteriol. 195:5352–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daley DO, Rapp M, Granseth E, Melen K, Drew D, von Heijne G. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321–1323 [DOI] [PubMed] [Google Scholar]
- 34.Detmers FJ, Kunji ER, Lanfermeijer FC, Poolman B, Konings WN. 1998. Kinetics and specificity of peptide uptake by the oligopeptide transport system of Lactococcus lactis. Biochemistry 37:16671–16679 [DOI] [PubMed] [Google Scholar]
- 35.Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289–298 [DOI] [PubMed] [Google Scholar]
- 36.Provencher SW, Glockner J. 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20:33–37 [DOI] [PubMed] [Google Scholar]
- 37.Sreerama N, Woody RW. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287:252–260 [DOI] [PubMed] [Google Scholar]
- 38.van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC. 1990. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal. Biochem. 191:110–118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.