Abstract
Corynebacterium glutamicum is equipped with abundant membrane transporters to adapt to a changing environment. Many amino acid transporters have been identified in C. glutamicum, but histidine uptake has not been investigated in detail. Here, we identified the aromatic amino acid transporter encoded by aroP as a histidine transporter in C. glutamicum by a combination of the growth and histidine uptake features. Characterization of histidine uptake showed that AroP has a moderate affinity for histidine, with a Km value of 11.40 ± 2.03 μM, and histidine uptake by AroP is competitively inhibited by the aromatic amino acids. Among the four substrates, AroP exhibits a stronger preference for tryptophan than for tyrosine, phenylalanine, and histidine. Homology structure modeling and molecular docking were performed to predict the substrate binding modes and conformational changes during substrate transport. These results suggested that tryptophan is best accommodated in the binding pocket due to shape compatibility, strong hydrophobic interactions, and the lowest binding energy, which is consistent with the observed substrate preference of AroP. Furthermore, the missense mutations of the putative substrate binding sites verified that Ser24, Ala28, and Gly29 play crucial roles in substrate binding and are highly conserved in the Gram-positive bacteria. Finally, the expression of aroP is not significantly affected by extracellular histidine or aromatic amino acids, indicating that the physiological role of AroP may be correlated with the increased fitness of C. glutamicum to assimilate extracellular amino acid for avoiding the high energy cost of amino acid biosynthesis.
INTRODUCTION
Membrane transport is vitally important to the communication of cells with the environment and allows cells to maintain internal homeostasis and adapt to environmental changes (1). Two kinds of membrane transport systems that are primary active carriers or secondary active carriers are responsible for amino acid uptake and efflux in bacteria (1). The majority of amino acid uptake systems follow a secondary transport mechanism in which transport is driven by an electrochemical ion potential across the plasma membrane (1). These transporters are integral membrane proteins with different numbers of α-helical transmembrane segments (TMs) and are classified into different families according to their sequences and structural properties, among which are the amino acid-polyamine-organocation (APC) superfamily and the ATP-binding cassette (ABC) superfamily, which represents a majority of the amino acid transporters in bacteria (2).
Histidine, as an important building block of proteins, is found especially at active sites of enzymes with crucial physiological functions in bacteria (3). De novo biosynthesis of histidine in bacteria initiates from the condensation of ATP and phosphoribosyl pyrophosphate (PRPP) and undergoes attenuation and inhibition regulation at the transcriptional and enzymatic activity levels when there is adequate histidine for growth (4–7). Histidine biosynthesis is considered to be a high-energy-cost process (8), which makes the uptake of external histidine more important for bacteria to quickly adapt to the changing environment and to assign their resources economically and rationally.
Histidine transport has been intensively studied in the Gram-negative bacteria Escherichia coli, Salmonella enterica serovar Typhimurium, and Sinorhizobium meliloti, which adopt similar ABC transporters for histidine uptake (9–11). The well-understood high-affinity ABC-type histidine transporter of E. coli and S. Typhimurium consists of four components: the histidine-binding protein HisJ, the two membrane-bound proteins HisQ and HisM, and the ATPase HisP (10). Additionally, the aromatic amino acid transporter AroP from S. Typhimurium has been found to participate in histidine transport, although it has a relatively low affinity for histidine (12). However, in Gram-positive bacteria, histidine transport has been investigated only in lactic acid bacteria, including Lactococcus lactis, Streptococcus thermophilus, and Lactobacillus hilgardii. A secondary carrier, HisP, responsible for histidine uptake, was demonstrated to be essential for L. lactis to grow on a low concentration of histidine (13). Furthermore, a histidine-histamine exchanger, HdcP, in S. thermophilus (14) and L. hilgardii (15) was found to participate in histidine decarboxylation. To date, only a few crystal structures of membrane transporters are available to provide insight into amino acid binding modes and transport mechanisms, such as the glutamate transporter (Glt) from Pyrococcus horikoshii (16), the leucine transporter (LeuT) from Aquifex aeolicus (17), the arginine-agmatine antiporter (AdiC) from E. coli (18, 19), and the proton-coupled broad-specificity amino acid transporter (ApcT) from Methanocaldococcus jannaschii (20). Due to the nonavailability of the crystal structures of histidine transporters, the molecular mechanism of histidine transport remains unclear.
Corynebacterium glutamicum, a Gram-positive soil bacterium with high G+C content, is widely used for large-scale production of amino acids and is considered to be an ideal model to study amino acid metabolism and transport (21, 22). The genome of C. glutamicum carries approximately 750 membrane proteins that could possibly function as transporters, and members of 17 transporter families are predicted to be involved in amino acid transport (1). Among them, GluABCD, LysI, BrnQ, MetQNI, MetPS, PheP, and AroP have been functionally characterized (23–28). AroP was identified as a general aromatic amino acid uptake system by gene disruption and complementation experiments (28). The kinetics of tyrosine uptake by AroP was further analyzed, and tyrosine uptake was competitively inhibited by tryptophan and phenylalanine (28). Besides AroP, another transporter (PheP) specific for phenylalanine uptake was also identified (27), confirming the presence of more than one uptake system for aromatic amino acids in C. glutamicum. As for histidine transport, an attempt to identify a histidine uptake system was mentioned only as unpublished data in a recent review (29) in which the deletion of the pheP gene rather than the aroP gene resulted in a significant reduction in the growth rate of the histidine-auxotrophic ΔhisG mutant, but no further direct evidence was provided to demonstrate the involvement of PheP in histidine transport. Thus, histidine uptake has not been investigated in detail to date, and the transport mechanism of histidine remains unknown.
In this study, we screened six putative histidine transporters and identified NCgl1062 (AroP) as a physiologically active histidine transporter in C. glutamicum. Uptake of l-[14C]histidine by C. glutamicum was performed to determine the kinetic characterization and substrate specificity of AroP. The comparison of two AroP homology models, as well as the binding modes between AroP and the four substrates, elucidated the conformational changes of AroP with and without substrate binding, as well as the molecular basis of the substrate preference of AroP. The analysis of aroP expression modes under the different conditions indicated that AroP plays an important physiological role in facilitating C. glutamicum assimilation of extracellular amino acids.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. The E. coli DH5α strain was used for gene cloning and was aerobically grown in Luria-Bertani broth at 37°C. C. glutamicum strains were aerobically grown in brain heart infusion (BHI) or in CGXII minimal medium (30) at 30°C. The histidine-auxotrophic strain RES167 ΔhisD, which was used for screening the histidine transporter, was cultivated in CGXII supplemented with 0.5 mM histidine. To evaluate the growth of C. glutamicum strains on different amino acids, 0.5 mM (each) histidine, tryptophan, tyrosine, and phenylalanine was added to CGXII as the sole nitrogen sources.
Constructions of plasmids and strains.
Total genomic DNA of C. glutamicum ATCC 13032 was extracted according to a previously described procedure (31). For gene overexpression and complementation, the shuttle vector pXMJ19 was used according to the procedure described previously (31). When needed, 1 mM isopropylthio-β-d-galactopyranoside (IPTG) was added to the culture media to induce target gene expression. The mutated aroP gene with a single codon change was generated by overlap extension PCR. The outer and inner primers used to amplify the mutants of aroP are listed in Table S1 in the supplemental material. The inner primers were designed to generate one codon change. The overlap extension PCR products were ligated to the shuttle vector pXMJ19, and the sequences of the mutated aroP were verified by DNA sequencing to confirm that only the desired mutations were generated. The pXMJ19 plasmids harboring mutated aroP were transformed to C. glutamicum by electroporation. For gene disruption, the upstream and downstream homologous fragments of the target genes were amplified with the primers listed in Table S1 in the supplemental material, ligated into the homologous-recombination integration vector pK18mobsacB, and transformed into RES167. Positive clones were selected according to the previously described procedure (32), and gene disruption was verified by PCR amplification and DNA sequencing.
Uptake assays with unlabeled amino acid.
Uptake assays with unlabeled amino acids were performed according to the previously described procedure (27). C. glutamicum RES167 and its corresponding strain were grown in BHI. The cells in the exponential phase were harvested, washed twice with ammonium-free CGXII minimal medium, and resuspended in this CGXII medium supplemented with either 0.5 mM histidine for the histidine uptake assay or 0.5 mM histidine and the aromatic amino acids for substrate preference analysis, followed by incubation at 30°C. At the indicated intervals, portions of the reaction mixture were withdrawn, filtered, and analyzed by high-performance liquid chromatography (HPLC) after derivatization with 2,4-dinitrofluorobenzene. Uptake of the supplemented amino acids was indicated by concentration decreases in the culture supernatant.
Uptake assays with l-[14C]histidine.
Uptake of l-[14C]histidine was determined with a rapid filtration assay as described previously (28). C. glutamicum cells were grown to mid-exponential phase, harvested, washed with 0.1 M Tris phosphate buffer (pH 6.8), and resuspended in the buffer. The reaction mixture contained 100 μmol Tris phosphate (pH 6.8), 1 μmol MgSO4, 10 μmol glucose, and 100 μg chloramphenicol. The cell suspension was added to the reaction mixture and incubated at 30°C for 3 min to energize before the transport assay. The assay was started by the addition of l-[14C]histidine (PerkinElmer, Inc., USA) to a final concentration of 5 μM. At the indicated intervals, portions of the reaction mixture were withdrawn, vacuum filtered, and immediately washed twice with cold 0.1 M LiCl. The radioactivity retained on the filters was quantitated with a liquid scintillation counter (MicroBeta; PerkinElmer, Inc., USA). For kinetic-parameter determination, the uptake assay was performed with 0.5, 1, 1.5, 3, 6, 10, 20, and 50 μM l-[14C]histidine, and the initial linear uptake rates were used to calculate the kinetic parameters according to the Michaelis-Menten equation.
Sequence analysis and homology modeling.
Sequence comparisons and database searches were conducted with BLAST programs from the BLAST server of the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). The alignment of the secondary structure was produced with AdiC from E. coli as the template using ESPript 2.2 (http://espript.ibcp.fr/ESPript/ESPript/) with default settings. The AroP homology three-dimensional models were constructed with the automated comparative protein structure homology-modeling server SWISS-MODEL (http://swissmodel.expasy.org) (33). The crystal structures of AdiC from E. coli without and with arginine (Protein Data Bank [PDB] codes 3LRB and 3L1L) were used as templates for the two models individually (the outward-open and occluded models) (18, 19). The AroP homology model was subjected to a series of analyses to validate its consistency and reliability. Global-model quality was examined with QMEAN6, which provides a score based on six structural features using statistical potentials at the atomic level (33). The QMEAN score and Z-score of the AroP model constructed by using AdiC as the template were 0.46 and −3.669, respectively, which are higher than those of the AroP model constructed by using ApcT (PDB code 3GIA) as the template (0.32 and −7.64, respectively). DFire was used to assess the nonbonded atomic interactions in the AroP model (34). The DFire energy of the AroP model was −685.46, which is close to the −708.80 of the AdiC model, indicating that the model can represent the native conformation of AroP. The stereochemical quality of AroP was inspected with the Psi/Phi Ramachandran plot obtained from PROCHECK analysis (35). The PROCHECK results did not show significant errors in the structures based on the percentage of the residues in allowed (87.1%), additional allowed (10.3%), generously allowed (1.5%), and disallowed (1.0%) regions.
Docking analysis.
The docking analyses were performed with AutoDock tools (ADT) v1.5.4 and the AutoDock v4.2 program from the Scripps Research Institute (http://www.scripps.edu/mb/olson/doc/autodock). Aromatic amino acids and histidine were docked to the occluded model of AroP with the protein molecule considered to be a rigid body and the ligands considered to be flexible. The three-dimensional grids were created with a 60-Å grid size (x, y, and z) with a spacing of 0.375 Å. The grid maps that represent the ligand in the docking target site were calculated with AutoGrid 4.2. The search was performed with the genetic algorithm; populations of 150 individuals with a mutation rate of 0.02 were evolved for 100 generations. Automated docking was subsequently performed with AutoDock 4.2. A cluster analysis based on a root mean square (RMS) tolerance of 1.0 was subsequently performed, and the lowest energy conformation of the more populated cluster was considered to be the most trustworthy result.
RNA preparation and quantitative real-time RT-PCR.
C. glutamicum RES167 cells were grown to the exponential phase in CGXII minimal medium that was supplemented individually with histidine, tryptophan, tyrosine, or phenylalanine at final concentrations of 0.1 and 1 mM. The cells were harvested and disrupted with acid-washed glass beads (Sigma, USA). Total RNA was isolated by acid phenol-chloroform/isoamyl alcohol extraction. The reverse transcription (RT) of approximately 200 ng of RNA was performed with the specific primers listed in Table S1 in the supplemental material and the FastQuant RT Kit (Tiangen, China). Quantitative PCR was performed with Bryt Green from the GoTaq qPCR master mix (Promega, USA) and the Rotor-Gene Q Real-Time PCR Detection System (Qiagen, Germany). The C. glutamicum rpoB gene was used as the reference gene to normalize the aroP mRNA levels. Negative controls were used in each PCR run to exclude DNA and other contamination. The quantitative-PCR (qPCR) products were verified by a melting curve analysis. Data collection and analysis were facilitated by the Rotor-Gene Q Series software, version 2.0.3, according to the 2−ΔΔCT method (36).
Western blotting.
Membrane proteins were extracted with 2% Triton X-114 detergent for temperature-dependent phase separation and precipitated by ice-cold acetone (37). Samples from different culture conditions were analyzed by SDS-polyacrylamide gel electrophoresis. Specific detection of the AroP protein was performed by Western blotting. Proteins were electrotransferred to a polyvinylidene difluoride (PVDF) membrane and probed with rabbit polyclonal antibodies that were raised against a synthetic peptide analogue of the AroP protein. The blots were visualized with a peroxidase-coupled goat anti-rabbit secondary antibody and an enhanced chemiluminescence (ECL) color development reagent (GE, USA).
Phylogenetic analysis.
Sequence comparisons and protein sequence similarity searches were performed with the online BLAST programs. Pairwise and multiple protein sequence alignments were made with the CLUSTAL X program (38). The phylogenetic tree was constructed with MEGA 5.1 software according to the neighbor-joining method (39).
RESULTS
Genome data mining and screening of putative genes for histidine transport.
C. glutamicum could synthesize histidine de novo from PRPP and ATP (7). Disruption of the hisD gene encoding histidinol dehydrogenase abolished the growth of RES167 in CGXII minimal medium, but growth was restored with histidine supplementation, demonstrating the existence of histidine uptake systems (see Fig. S1A in the supplemental material). Six putative histidine transporters found by BLAST searches, using the amino acid sequences of HisJQMP from E. coli, AroP from S. Typhimurium, and HisP from L. lactis as the input sequences, are summarized in Table S2 in the supplemental material. A putative ABC-type transporter encoded by the ncgl1276, ncgl1277, and ncgl1278 genes and GluABCD for glutamate (23) had moderate sequence identities to HisJQMP of E. coli (9). An unknown-substrate APC-type transporter encoded by the ncgl0453 gene, PheP for phenylalanine (27), AroP for aromatic amino acids (28), and LysI for lysine (24) also showed moderate and variable sequence identities to AroP and HisP of S. Typhimurium and L. lactis, respectively (12, 13).
The histidine-auxotrophic C. glutamicum RES167 ΔhisD was used to screen the histidine transporter, and the six candidate genes were disrupted individually in RES167 ΔhisD to investigate the effects on the growth of the mutant strains on histidine. Only the mutant with the aroP gene deleted exhibited impaired growth on histidine (see Fig. S1B in the supplemental material), indicating the relevance of the aroP gene to histidine transport. When the other five putative genes were further disrupted in RES167 ΔhisD ΔaroP, no differences in growth were observed between RES167 ΔhisD ΔaroP and its five derivative mutants (see Fig. S1C in the supplemental material). Data showing the growth properties of the mutants were listed in Table S3 in the supplemental material. These results preliminarily excluded the possibility of involvement of the other five genes in histidine transport. However, in a previous review (29), the deletion of pheP, rather than aroP, in the histidine-auxotrophic ΔhisG mutant resulted in a significant reduction in the growth rate, which is somewhat contrary to our observations. This could be attributed to the different genetic backgrounds of histidine-auxotrophic strains and the culture conditions. Considering that the growth properties of histidine-auxotrophic mutants reflect only the possible involvement of the deleted gene in l-histidine transport, direct evidence from uptake experiments with the labeled l-histidine was essential to confirm the functions of the indicated genes.
AroP is a histidine transporter.
The disruption and complementation of the aroP gene were performed in C. glutamicum RES167 to investigate the effects on growth on histidine and aromatic amino acids. Data showing the growth properties of the AroP mutants are listed in Table S3 in the supplemental material. Disruption of aroP impaired the growth of RES167 on histidine, with a maximal growth rate of 0.041 ± 0.005 h−1 and a final optical density at 600 nm (OD600) of 0.86 ± 0.04 compared to a maximal growth rate of 0.066 ± 0.004 h−1 and a final OD600 of 0.96 ± 0.01 for RES167; the genetic complementation of aroP in RES167 ΔaroP/pWYE1243 restored growth, with a maximal growth rate of 0.088 ± 0.003 h−1 and a final OD600 of 1.05 ± 0.05. The growth properties of the mutant strains on aromatic amino acids were approximately the same as those of the mutants on histidine (Fig. 1A, B, C, and D), demonstrating that aroP is involved in the uptake of histidine and aromatic amino acids. Moreover, residual growth of RES167 ΔaroP was observed, indicating that the four amino acids might be imported through additional transport systems to support the growth of RES167 ΔaroP on these amino acids. Residual uptake activities of the 14C-labeled aromatic amino acid were also found in a ΔaroP mutant in the previous study (28), suggesting the existence of additional uptake systems besides AroP. To date, only one other specific transporter for phenylalanine, PheP, has been identified and characterized (27).
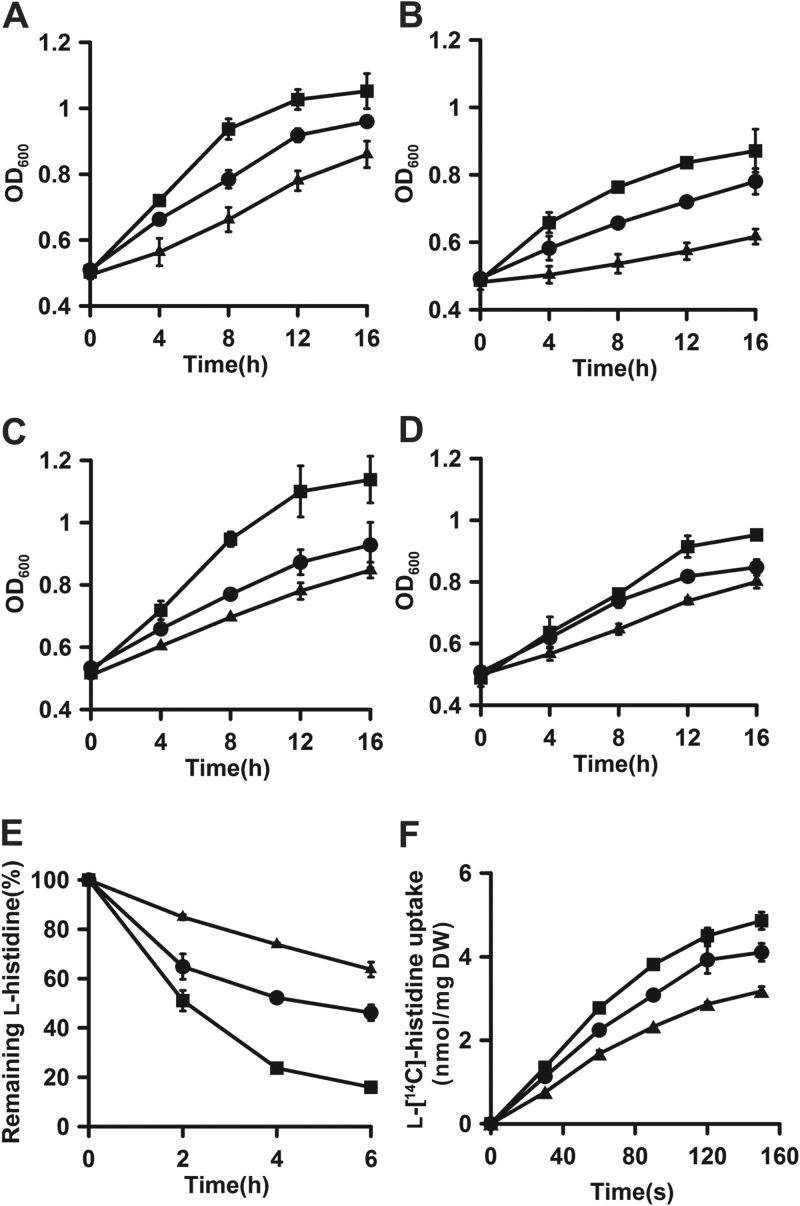
Fig 1.
Identification of AroP as a histidine transporter in C. glutamicum. (A to D) Growth of RES167 (circles), RES167 ΔaroP (triangles), and RES167 ΔaroP/pWYE1243 (squares) in CGXII minimal medium with histidine (A), tryptophan (B), tyrosine (C), and phenylalanine (D) as the sole nitrogen source. (E and F) Histidine uptake by C. glutamicum RES167 (circles), RES167ΔaroP (triangles), and RES167 ΔaroP/pWYE1243 (squares) with unlabeled l-histidine (E) and l-[14C]histidine (F). The initial concentration of the unlabeled l-histidine was 0.5 mM, which was calculated as 100%, and the unlabeled l-histidine uptake is indicated as a decrease in its concentration in the culture supernatant. The initial concentration of l-[14C]histidine was 5 μM. The error bars indicate standard deviations.
Uptake assays with unlabeled l-histidine and l-[14C]histidine were performed to further verify whether AroP could function as a histidine transporter. Uptake of unlabeled l-histidine by resting cells of C. glutamicum is shown in Fig. 1E; aroP disruption resulted in an approximately 30% decrease in l-histidine uptake compared to that of RES167, and the impaired uptake activity was restored by genetic complementation of aroP in RES167 ΔaroP/pWYE1243. In the l-[14C]histidine uptake assay, the uptake rate of the l-[14C]histidine in RES167 ΔaroP was 1.67 ± 0.08 nmol/min/mg (dry weight [DW]), which decreased by approximately 25% compared to that in RES167 (2.24 ± 0.04 nmol/min/mg [DW]), and the overexpression of aroP resulted in a significant increase in the uptake activity of histidine (2.77 ± 0.20 nmol/min/mg [DW]) (Fig. 1F). Based on these results, it was concluded that AroP is a histidine transporter.
Characterization of AroP as a histidine transporter.
To characterize the kinetic properties of AroP, the uptake rates of RES167 ΔaroP/pWYE1243 at different concentrations of l-[14C]histidine were determined, and the negative-control values obtained from RES167 ΔaroP were subtracted to exclude the interference of the unknown histidine uptake systems. According to the Michaelis-Menten equation, the Km and Vmax values of AroP for histidine were 11.40 ± 2.03 μM and 3.37 ± 0.44 nmol/min/mg (DW), respectively (see Fig. S2 in the supplemental material), indicating that AroP has a much higher affinity than the low-affinity histidine transporters, such as AroP of S. Typhimurium (Km = 100 μM) (12) and TAT1 of Saccharomyces cerevisiae (Km = 300 μM) (40), and a lower affinity than the ABC-type specific histidine transporter HisJQMP of S. Typhimurium (Km = 0.08 μM) (12). Additionally, in C. glutamicum, the Km value of AroP for l-tyrosine was determined to be 3.0 ± 0.3 μM (28), which suggested a slightly higher affinity of AroP for l-tyrosine than for l-histidine. Taken together, it can be concluded that AroP is a moderate-affinity histidine transporter.
To analyze the substrate specificity of AroP, an l-[14C]histidine uptake assay was performed in RES167 ΔaroP/pWYE1243 in the presence of a 20-fold excess of unlabeled amino acids. The AroP-mediated uptake of l-[14C]histidine was not significantly affected by l-arginine, l-lysine, l-glutamate, l-valine, or l-alanine addition, whereas l-tryptophan, l-tyrosine, and l-phenylalanine resulted in 48.09%, 45.03%, and 33.04% decreases in l-histidine uptake, respectively (Fig. 2A). In combination with the results of growth assays depicted in Fig. 1, we concluded that both histidine and aromatic amino acid are transported into C. glutamicum via an aroP-encoded membrane transporter. Similarly, uptake of labeled l-tyrosine by AroP was inhibited by l-tryptophan and l-phenylalanine (28). Consequently, the functions and substrate spectrum of the AroP transporter were extended in our study on the basis of the previous study (28).
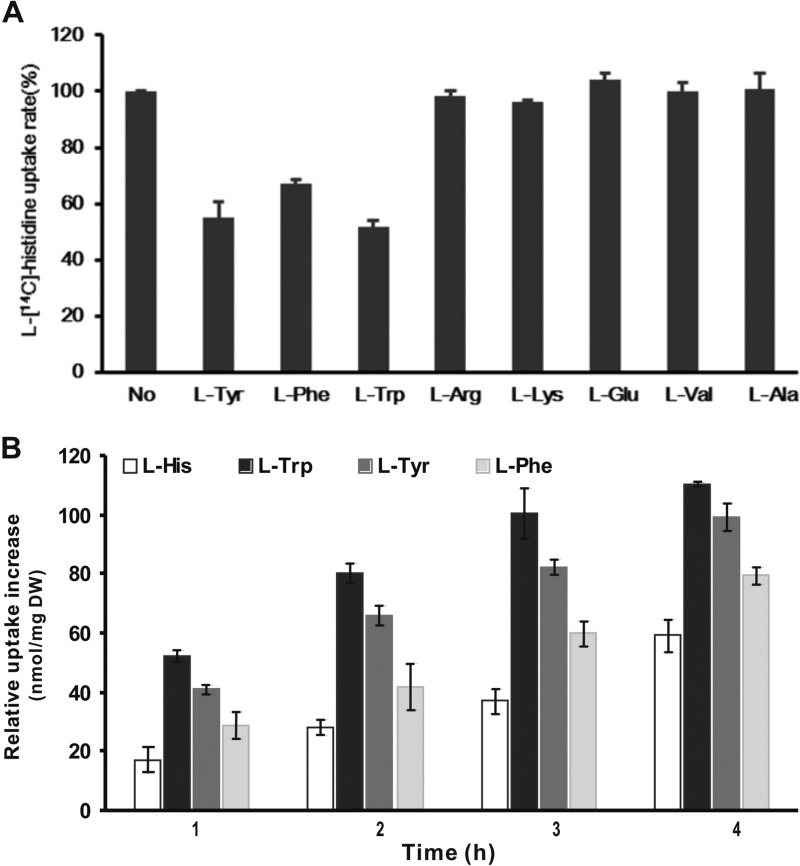
Fig 2.
Substrate specificity (A) and preferences (B) of AroP. (A) The initial concentration of l-[14C]histidine was 5 μM. The l-[14C]histidine uptake rate of C. glutamicum RES167 ΔaroP/pWYE1243 in the absence of competitors (No) was determined to be 3.20 ± 0.11 nmol/min/mg (DW) and was set as 100%. The concentration of each competitor was 100 μM. The data are shown as averages from three parallel determinations with standard deviations. (B) Uptake of l-histidine, l-tryptophan, l-tyrosine, and l-phenylalanine by AroP was analyzed with C. glutamicum RES167/pWYE1243 and RES167 in the presence of a mixture of the four amino acids. The relative uptake increase was determined as the uptake of each amino acid of C. glutamicum RES167/pWYE1243 with that of RES167 subtracted. The initial concentration of each amino acid was 0.5 mM, and the uptake of each amino acid is indicated as the decrease in the specific amino acid concentration in the culture supernatant. The data are shown as averages from three parallel determinations with standard deviations.
The broad substrate spectrum of AroP inspired us to investigate whether a substrate preference exists in transport mediated by AroP. To characterize the substrate preference, the uptake activities of AroP were analyzed in the presence of the mixture of histidine and aromatic amino acid substrates. To exclude interference from other existing aromatic amino acids and histidine uptake systems, the uptake of each amino acid by AroP was determined by the uptake of individual amino acid substrates by RES167/pWYE1243 with that of RES167 subtracted. During the first hour, the uptake rates of l-tryptophan, l-tyrosine, l-phenylalanine, and l-histidine by AroP were 0.873 ± 0.032, 0.679 ± 0.032, 0.478 ± 0.078, and 0.286 ± 0.073 nmol/min/mg (DW), respectively (Fig. 2B). The uptake trends of the four substrates at different time points were similar to those observed in the first hour. Consequently, in the simultaneous presence of the four substrates, AroP exhibits preferences in the following order: l-tryptophan > l-tyrosine > l-phenylalanine > l-histidine.
AroP structural models and the substrate binding modes.
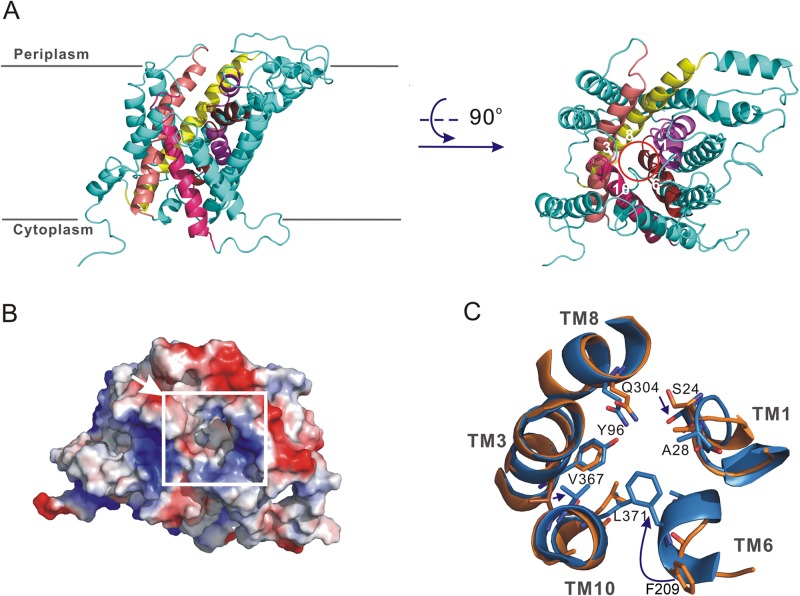
Two homology structural models of AroP with or without a binding substrate were constructed based on two of the crystal structures of E. coli AdiC (18, 19). The AroP protein contains 12 TMs and two conserved sequences, the GSG and GVESA motifs, which exist within distinct families of the APC superfamily in TM1 and TM6 (see Fig. S3 in the supplemental material). The outward-open model (without a binding substrate) of AroP shows that TM1, TM3, TM6, TM8, and TM10 are surrounded by the other 7 TMs in the outer layer to form a central cavity that opens to the periplasmic side (Fig. 3A). This cavity is potentially accessible to extracellular solutions and is ideal as a substrate binding pocket, with a volume of 297.7 Å3 (Fig. 3B) (41). A comparison of the occluded and outward-open models of AroP revealed that TM1 and TM6 moved toward TM3 and TM8 in response to substrate binding to induce the conformational changes (Fig. 3C). Along with the TM6 movement, Phe209, located at the top of the central cavity, is translocated approximately 6.2 Å in the occluded model and may function as an “extracellular gate” to block the substrate exit route to the periplasm and thus close the substrate binding pocket, as observed in the conformational transition of AdiC (19). The structural similarity of AroP to AdiC makes it likely that Tyr96 and Trp375, located at the bottom of the central cavity, could be involved in the release of substrates through opening of the intracellular gate to the cytoplasm (19).
Fig 3.
Homology structural models of AroP. (A) Two perpendicular views of the outward-open AroP from the membrane and periplasm sides. An inner layer of five TMs, marked with different colors, forms the central cavity, indicated by the red circle. (B) Electrostatic potential surface of occluded AroP. The electrostatic potential was calculated with PyMol. The central cavity is marked with a white square, and the arrow indicates what is thought to be the substrate binding site. (C) Superimposition of the central cavity of AroP with and without substrate binding. The outward-open and occluded models of AroP are shown in orange and blue, respectively. The amino acids involved in substrate binding are shown as a stick representation and colored according to the type of atom. The movement of Phe209 results in a block of the substrate binding pocket.
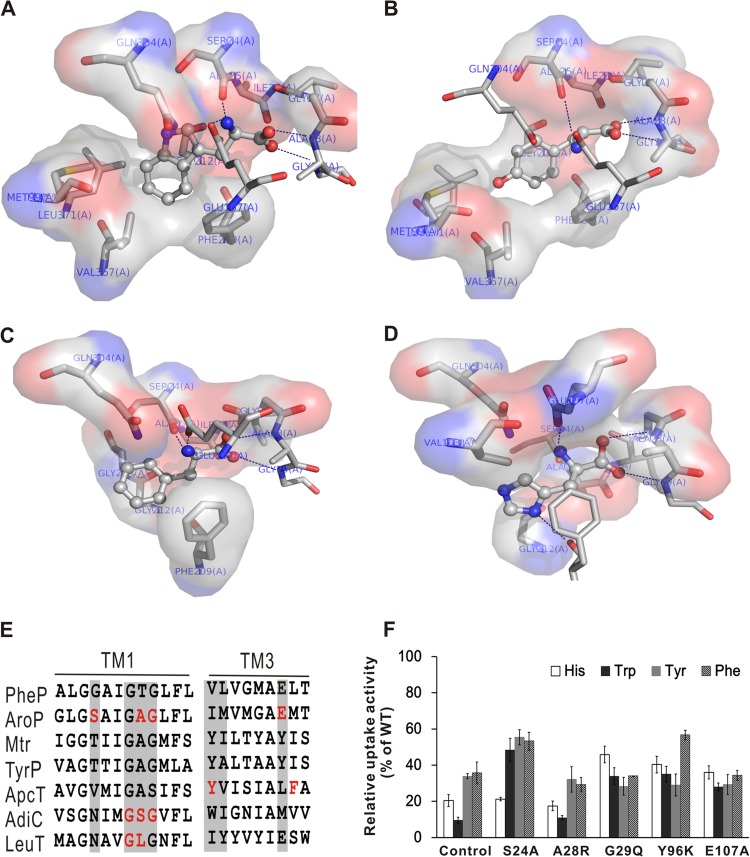
The occluded model of AroP was subjected to in silico docking calculations to investigate the binding modes of the different substrates. The best possible binding modes of the four substrates (ball and stick, colored by element) at the binding pocket of AroP are displayed in Fig. 4. The α-carboxyl group on the four amino acids accepts two hydrogen bonds from the amide nitrogens of the Ala28 and Gly29 residues located on TM1, which is in accordance with what was observed in arginine binding to AdiC (19). Sequence alignments revealed that the amino acid residues of these two sites were conserved in functional amino acid transporters (Fig. 4E), indicating that the residues are crucial for substrate binding. Furthermore, the α-amino group of the four amino acids forms two hydrogen bonds with the side chain of Ser24 in TM1 and the carboxyl group of Glu107 in TM3. The side chains of the four amino acids interact with the side chains of the hydrophobic amino acids Met99 and Val103 in TM3 and Val367 and Leu371 in TM10. Remarkably, the imidazole ring in histidine forms an additional hydrogen bond with Phe209 in TM6, with the result that the imidazole ring orientation has a relative movement toward the extracellular space. The binding free energies (ΔGb) determined by docking are −7.86 kcal/mol for tryptophan, −6.70 kcal/mol for tyrosine, −6.48 kcal/mol for phenylalanine, and −6.07 kcal/mol for histidine; thus, AroP shows higher affinity for tryptophan than for tyrosine, phenylalanine, and histidine.
Fig 4.
Binding modes between AroP and histidine and aromatic amino acids. (A to D) Docking results for AroP protein with tryptophan (A), tyrosine (B), phenylalanine (C), and histidine (D) in the binding pocket. The substrates in the binding pockets of the occluded model of AroP are shown as ball-and-stick models. The main hydrogen bonds between the substrates and AroP are depicted as dashed blue lines. The hydrophobic interactions between the substrates and AroP are indicated by spheres. (E) Sequence alignment of the amino acids involved in substrate binding. Residues that are implicated in substrate binding are highlighted in red, and the equivalent residues in other orthologs are highlighted by gray shading. (F) Analysis of the aromatic amino acid and histidine uptake activities of the mutant AroP. Amino acid uptake activities of RES167 ΔaroP overexpressing the mutated AroP are shown as uptake activity relative to that of RES167 ΔaroP overexpressing the wild-type AroP. The uptake activity of RES167 ΔaroP was used as a control. The error bars indicate standard deviations.
To characterize the roles of the putative substrate binding sites, several missense mutations were introduced into the AroP protein, and the impacts on uptake activities were evaluated by comparing the aromatic amino acid and histidine uptake activities of the mutants with those of the wild-type AroP (Fig. 4F). All five mutants, including S24A, A28R, G29Q, Y96K, and E107A, decreased the aromatic amino acid and histidine uptake activities to different extents. The A28R mutation almost abolished the uptake activity, and G29Q reduced the activity by 60%, suggesting that the two residues play important roles in substrate binding. Mutations in the residues that interact with the α-amino group of histidine (S24A and E107A) led to a drastic decrease in the uptake activity (more than 50%), mainly due to the abrogation of the hydrogen bond interaction. The Y96K mutation, which interferes with the release of substrate, caused a significant decrease in the uptake activity. Therefore, our results verified that the central cavity was the substrate binding site and confirmed the critical roles of the specific amino acid residues in the uptake activity of AroP.
The expression of aroP is not significantly affected by extracellular amino acid substrates.
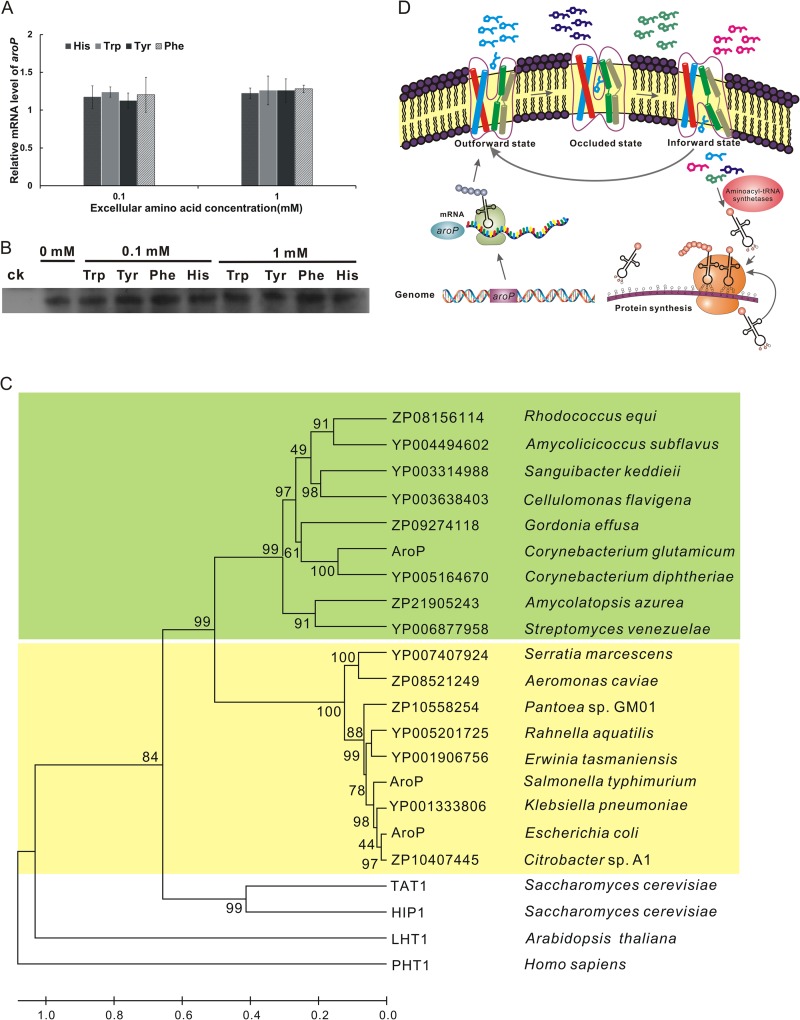
The effects of extracellular histidine and aromatic amino acids on aroP expression at the mRNA and protein levels were analyzed by quantitative RT-PCR and Western blotting, respectively. As RES167 was grown in minimal medium, the mRNA of AroP could be detected by quantitative RT-PCR. No significant differences in the quantity of AroP mRNA were observed even in the presence of 0.1 or 1 mM histidine or aromatic amino acids (Fig. 5A), suggesting that aroP transcription was not affected by increased concentrations of extracellular amino acid substrates. The same results were obtained by Western blotting, indicating that aroP expression was not affected by extracellular substrates (Fig. 5B).
Fig 5.
Working model of AroP-mediated amino acid transport. (A) Effects of histidine and aromatic amino acid addition on the transcription level of aroP. The relative fold changes of the transcriptional levels of aroP were analyzed by quantitative real-time PCR upon cultivation in CGXII minimal medium with or without histidine, tryptophan, tyrosine, or phenylalanine. The relative mRNA level represents the transcription levels of aroP in cells cultivated in the presence of 0.1 and 1 mM amino acid relative to that of cells cultivated in the absence of any of these amino acids. The data shown are mean values from three biological replicates and three technical replicates with standard deviations. (B) Effect of histidine and aromatic amino acid addition on expression of aroP analyzed by Western blotting. The RES167 ΔaroP mutant was used as a control (ck). The other lanes are samples from RES167 cultivated in minimal medium with or without histidine, tryptophan, tyrosine, or phenylalanine. (C) Phylogenetic analysis of homologous bacterial, plant, and human AroP proteins. The phylogenetic tree was generated according to the neighbor-joining method with Mega 5.1 software. The scale at the bottom indicates sequence divergence. The bootstrap confidence limits (1,000 bootstrap replicates) are indicated at the nodes. The green shading represents the AroP proteins with a moderate affinity for histidine from Gram-positive bacteria, and the yellow shading indicates the AroP proteins with a low affinity for histidine from Gram-negative bacteria. Histidine transporters from A. thaliana and Homo sapiens were used as the outgroup. (D) Model of AroP-mediated amino acid transport. AroP, constitutively expressed and located on the membrane, implements the transport function through a reciprocal transition among three conformations. In the absence of substrates, AroP is outward-open, with the binding pocket accessible to the extracellular solution. Upon substrate binding, AroP undergoes transition to the occluded conformation and then an inward-open conformation accompanied by release of substrate into the cytoplasm to participate in metabolic processes.
DISCUSSION
Amino acids can be directly transported into cells through specific or general uptake systems. General amino acid uptake systems often have broad substrate specificities and can transport groups of amino acids with similar structures or properties, such as the aromatic amino acid transporter (AroP) of E. coli (42), the branched-chain amino acid and methionine transporter (BrnFE) of C. glutamicum (43), the acidic amino acid transporter (BztABCD) of Rhodobacter capsulatus (44), and the nearly universal protein-amino acid transporter (GAP1) of S. cerevisiae (45) and AapJQMP of Rhizobium leguminosarum (46). In this study, we identified the aromatic amino acid transporter AroP as a physiologically active histidine transporter in C. glutamicum. As a general transporter, AroP showed preferences among the four substrates. Similarly, substrate preferences of transport systems involved in osmotic regulation for proline, betaine, and ectoine have been observed in C. glutamicum (47). The substrate binding modes of AroP determined by docking analysis provide new insights into the substrate preferences. Optimal substrate binding and the formation of the occluded state require a complementary shape, and this interaction is best satisfied by tryptophan. With regard to the capacity of the binding pocket, tryptophan (228.2 Å3) could better adapt to the space than tyrosine (196.8 Å3), phenylalanine (191.9 Å3), or histidine (158.3 Å3) (41). The indole ring of tryptophan is better accommodated in the binding pocket than the aromatic rings of phenylalanine or tyrosine and can engage in strong hydrophobic interactions with the adjacent nonpolar residues. In the case of histidine binding, the electrostatic repulsive forces due to the positive charge of the imidazole ring interfered with the hydrophobic interaction, leading to decreased affinity (see Fig. S4 in the supplemental material). Moreover, the occluded state of the tryptophan-AroP complex is more stable than those of the other complexes due to the low binding energy of tryptophan.
Bacteria are often equipped with at least two uptake systems for substances with crucial physiological roles; these systems can possess different affinities, capacities, specificities, and regulation mechanisms to satisfy different physiological needs in the changing environment (48). As for aromatic amino acids, more than one uptake system was demonstrated to exist in C. glutamicum (28). In addition to AroP, a second phenylalanine transporter, PheP, was recently identified, and the residual phenylalanine uptake activity of the ΔaroP ΔpheP mutant indicated the existence of a third phenylalanine transporter (27). Similarly, the residual histidine uptake activity of the ΔaroP mutant was observed in our study, suggesting the presence of at least one unknown transporter involved in histidine uptake. Studies on histidine transporters in E. coli showed that different histidine uptake systems usually employ distinct regulation mechanisms. The specific histidine transporter HisJQMP is not only repressed by the ArgR protein with arginine and histidine as cofactors at the transcriptional level (9), but also responds to nitrogen limitation induction (49). However, transcription of the general transporter AroP is repressed by the TyrR protein and the cofactors tryptophan, tyrosine, and phenylalanine (50). In this study, AroP expression was not affected by extracellular histidine or aromatic amino acids, in contrast to that observed in E. coli. For some amino acids, such as histidine and tryptophan, which are biosynthesized at high energy cost, uptake of these amino acids from the environment is more economical for bacteria than de novo biosynthesis (13). Therefore, it suggested that AroP, which follows an ion symport mechanism and functions without ATP, is a frequently used transporter to facilitate C. glutamicum assimilation of external histidine and aromatic amino acids to avoid the high energy cost of the biosynthesis of these amino acids. Furthermore, the additional histidine transporter might be subjected to a different regulatory mechanism and compensate for the function of AroP to some extent.
The phylogenetic-tree analysis showed that the AroP-homologous proteins in bacteria could be classified into two groups (Fig. 5C). One group consists of AroP proteins with low affinity for histidine from Gram-negative bacteria, such as AroP of S. Typhimurium. The AroP with a moderate affinity for histidine is the first identified histidine transporter in C. glutamicum and belongs to the other group of AroP-homologous proteins from Gram-positive bacteria. Five amino acid residues predicted to be involved in substrate binding and transport of AroP were validated by the missense mutation in our study. Among them, the critical residues Ser24 and Ala28, responsible for binding of the α-amino group and the α-carboxyl group of amino acid substrates, respectively, are conserved in AroP homologues in Gram-positive bacteria and Arabidopsis thaliana, whereas Gly29 is highly conserved in AroP homologues in bacteria, S. cerevisiae, and A. thaliana due to the strong interaction with the α-carboxyl group of the amino acid substrate. Consequently, our structural models and the interaction mechanism of AroP and its substrate could be a representative of and reference for the function and molecular mechanism of the AroP homologues.
In conclusion, a working model of AroP-mediated amino acid transport is presented in Fig. 5D. In this model, the aroP gene is always transcribed and expressed as a transmembrane protein with transporter function in the presence or absence of extracellular histidine or aromatic amino acids. In the absence of substrate binding, AroP exhibits an outward-facing conformation with a binding pocket that is accessible to the cell wall. As the substrate binds to AroP, the movement of TM1 and TM6 results in the formation of the occluded conformation. It could be deduced that the subsequent conformational changes permit AroP to form an inward-open conformation. Consequently, the bound histidine or aromatic amino acid is released into the cytoplasm for direct involvement in protein synthesis or other metabolic processes. Our findings provide new insights into the molecular basis of the amino acid transport process and contribute to a deeper understanding of the physiological roles of general amino acid transporters in bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Shuwen Liu and Qian Liu for critical reading of the manuscript.
This work was supported by grants from the Ministry of Science and Technology of China (2008BAI63B01, 2008ZX09401-05, and 2010ZX09401-403) and the Beijing Natural Science Foundation (5112023).
Footnotes
Published ahead of print 20 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00971-13.
REFERENCES
- 1.Marin K, Krämer R. 2007. Amino acid transport systems in biotechnologically relevant bacteria, p 290–325 In Wendisch V. (ed), Microbiology monographs, vol 5 Springer-Verlag GmbH, Berlin, Germany [Google Scholar]
- 2.Saier MH. 2000. Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology 146:1775–1795 [DOI] [PubMed] [Google Scholar]
- 3.Waldron KJ, Robinson NJ. 2009. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7:25–35 [DOI] [PubMed] [Google Scholar]
- 4.Kasai T. 1974. Regulation of the expression of the histidine operon in Salmonella typhimurium. Nature 249:523–527 [DOI] [PubMed] [Google Scholar]
- 5.Artz SW, Broach JR. 1975. Histidine regulation in Salmonella typhimurium: an activator attenuator model of gene regulation. Proc. Natl. Acad. Sci. U. S. A. 72:3453–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung S, Chun JY, Yim SH, Cheon CI, Song E, Lee SS, Lee MS. 2009. Organization and analysis of the histidine biosynthetic genes from Corynebacterium glutamicum. Genes Genom. 31:315–323 [Google Scholar]
- 7.Zhang Y, Shang XL, Deng AH, Chai X, Lai SJ, Zhang GQ, Wen TY. 2012. Genetic and biochemical characterization of Corynebacterium glutamicum ATP phosphoribosyltransferase and its three mutants resistant to feedback inhibition by histidine. Biochimie 94:829–838 [DOI] [PubMed] [Google Scholar]
- 8.Brenner M, Ames BN. 1971. The histidine operon and its regulation, p 349–387 In Greenberg DM. (ed), Metabolic pathways. Academic Press, New York, NY [Google Scholar]
- 9.Caldara M, Le Minh PN, Bostoen S, Massant J, Charlier D. 2007. ArgR-dependent repression of arginine and histidine transport genes in Escherichia coli K-12. J. Mol. Biol. 373:251–267 [DOI] [PubMed] [Google Scholar]
- 10.Ames GF-L, Nikaido K. 1978. Identification of a membrane protein as a histidine transport component in Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 75:5447–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boncompagni E, Dupont L, Mignot T, Østeräs M, Lambert A, Poggi MC, Le Rudulier D. 2000. Characterization of a Sinorhizobium meliloti ATP-binding cassette histidine transporter also involved in betaine and proline uptake. J. Bacteriol. 182:3717–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ames GF-L, Roth JR. 1968. Histidine and aromatic permeases of Salmonella typhimurium. J. Bacteriol. 96:1742–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trip H, Mulder NL, Lolkema JS. 2013. Cloning, expression, and functional characterization of secondary amino acid transporters of Lactococcus lactis. J. Bacteriol. 195:340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trip H, Mulder NL, Rattray FP, Lolkema JS. 2011. HdcB, a novel enzyme catalysing maturation of pyruvoyl-dependent histidine decarboxylase. Mol. Microbiol. 79:861–871 [DOI] [PubMed] [Google Scholar]
- 15.Lucas PM, Wolken WAM, Claisse O, Lolkema JS, Lonvaud-Funel A. 2005. Histamine-producing pathway encoded on an unstable plasmid in Lactobacillus hilgardii 0006. Appl. Environ. Microbiol. 71:1417–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yernool D, Boudker O, Jin Y, Gouaux E. 2004. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 431:811–818 [DOI] [PubMed] [Google Scholar]
- 17.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. 2005. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437:215–223 [DOI] [PubMed] [Google Scholar]
- 18.Gao X, Lu F, Zhou L, Dang S, Sun L, Li X, Wang J, Shi Y. 2009. Structure and mechanism of an amino acid antiporter. Science 324:1565–1568 [DOI] [PubMed] [Google Scholar]
- 19.Gao X, Zhou L, Jiao X, Lu F, Yan C, Zeng X, Wang J, Shi Y. 2010. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 463:828–832 [DOI] [PubMed] [Google Scholar]
- 20.Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. 2009. Structure and mechanism of a Na+-independent amino acid transporter. Science 325:1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leuchtenberger W, Huthmacher K, Drauz K. 2005. Biotechnological production of amino acids and derivatives: current status and prospects. Appl. Microbiol. Biotechnol. 69:1–8 [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa S, Suda M, Uematsu K, Natsuma Y, Hiraga K, Jojima T, Inui M, Yukawa H. 2013. Engineering of Corynebacterium glutamicum for high-yield l-valine production under oxygen deprivation conditions. Appl. Environ. Microbiol. 79:1250–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krämer R, Lambert C, Hoischen C, Ebbighausen H. 1990. Uptake of glutamate in Corynebacterium glutamicum. 1 Kinetic properties and regulation by internal pH and potassium. Eur. J. Biochem. 194:929–935 [DOI] [PubMed] [Google Scholar]
- 24.Seep-Feldhaus AH, Kalinowski J, Pühler A. 1991. Molecular analysis of the Corynebacterium glutamicum lysI gene involved in lysine uptake. Mol. Microbiol. 5:2995–3005 [DOI] [PubMed] [Google Scholar]
- 25.Tauch A, Hermann T, Burkovski A, Krämer R, Pühler A, Kalinowski J. 1998. Isoleucine uptake in Corynebacterium glutamicum ATCC 13032 is directed by the brnQ gene product. Arch. Microbiol. 169:303–312 [DOI] [PubMed] [Google Scholar]
- 26.Trötschel C, Follmann Nettekoven M, Mohrbach JA, Forrest T, Burkovski LR, Marin A, Krm̈er R. 2008. Methionine uptake in Corynebacterium glutamicum by MetQNI and by MetPS, a novel methionine and alanine importer of the NSS neurotransmitter transporter family. Biochemistry 47:12698–12709 [DOI] [PubMed] [Google Scholar]
- 27.Zhao Z, Ding JY, Li T, Zhou NY, Liu SJ. 2011. The ncgl1108 (PhePCg) gene encodes a new l-Phe transporter in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 90:2005–2013 [DOI] [PubMed] [Google Scholar]
- 28.Wehrmann A, Morakkabati S, Krämer R, Sahm H, Eggeling L. 1995. Functional analysis of sequences adjacent to dapE of Corynebacterium glutamicum reveals the presence of aroP, which encodes the aromatic amino acid transporter. J. Bacteriol. 177:5991–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulis-Horn RK, Persicke M, Kalinowski J. 2013. Histidine biosynthesis, its regulation and biotechnological application in Corynebacterium glutamicum. Microb. Biotechnol. 10.1111/1751-7915.12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keilhauer C, Eggeling L, Sahm H. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Shang XL, Lai SJ, Zhang GQ, Liang Y, Wen TY. 2012. Development and application of an arabinose-inducible expression system by facilitating inducer uptake in Corynebacterium glutamicum. Appl. Environ. Microbiol. 78:5831–5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schäfer A, Tauch A, Jäger Kalinowski WJ, Thierbach G, Pühler A. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 33.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 34.Benkert P, Biasini M, Schwede T. 2011. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 36.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 37.Zhang LJ, Xie JY, Wang X, Liu XH, Tang XK, Cao R, Hu WJ, Nie S, Fan CM, Liang SP. 2005. Proteomic analysis of mouse liver plasma membrane: use of differential extraction to enrich hydrophobic membrane proteins. Proteomics 5:4510–4524 [DOI] [PubMed] [Google Scholar]
- 38.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403–405 [DOI] [PubMed] [Google Scholar]
- 39.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bajmoczi M, Sneve M, Eide DJ, Drewes LR. 1998. TAT1 encodes a low-affinity histidine transporter in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 243:205–209 [DOI] [PubMed] [Google Scholar]
- 41.Tsai JTR, Chothia C, Gerstein M. 1999. The packing density in proteins: standard radii and volumes. J. Mol. Biol. 290:253–266 [DOI] [PubMed] [Google Scholar]
- 42.Chye ML, Guest JR, Pittard J. 1986. Cloning of the aroP gene and identification of its product in Escherichia coli K-12. J. Bacteriol. 167:749–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennerknecht N, Sahm H, Yen M-R, Pátek M, Saier J, Milton H, Eggeling L. 2002. Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J. Bacteriol. 184:3947–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng S, Haselkorn R. 1996. A glutamate/glutamine/aspartate/asparagine transport operon in Rhodobacter capsulatus. Mol. Microbiol. 20:1001–1011 [DOI] [PubMed] [Google Scholar]
- 45.Jauniaux JC, Grenson M. 1990. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190:39–44 [DOI] [PubMed] [Google Scholar]
- 46.Hosie AHF, Allaway D, Galloway CS, Dunsby HA, Poole PS. 2002. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J. Bacteriol. 184:4071–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peter H, Weil B, Burkovski A, Krämer R, Morbach S. 1998. Corynebacterium glutamicum is equipped with four secondary carriers for compatible solutes: Identification, sequencing, and characterization of the proline/ectoine uptake system, ProP, and the ectoine/proline/glycine betaine carrier, EctP. J. Bacteriol. 180:6005–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frossard SM, Khan AA, Warrick EC, Gately JM, Hanson AD, Oldham ML, Sanders DA, Csonka LN. 2012. Identification of a third osmoprotectant transport system, the OsmU system, in Salmonella enterica. J. Bacteriol. 194:3861–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitz G, Durre P, Mullenbach G, Ames GF-L. 1987. Nitrogen regulation of transport operons: analysis of promoters argTr and dhuA. Mol. Genet. Genomics 209:403–407 [DOI] [PubMed] [Google Scholar]
- 50.Wang PX, Yang J, Ishihama A, Pittard AJ. 1998. Demonstration that the TyrR protein and RNA polymerase complex formed at the divergent P3 promoter inhibits binding of RNA polymerase to the major promoter, P1, of the aroP gene of Escherichia coli. J. Bacteriol. 180:5466–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.