Abstract
The soil bacterium Bacillus subtilis forms biofilms on surfaces and at air-liquid interfaces. It was previously reported that these biofilms disassemble late in their life cycle and that conditioned medium from late-stage biofilms inhibits biofilm formation. Such medium contained a mixture of d-leucine, d-methionine, d-tryptophan, and d-tyrosine and was reported to inhibit biofilm formation via the incorporation of these d-amino acids into the cell wall. Here, we show that l-amino acids were able to specifically reverse the inhibitory effects of their cognate d-amino acids. We also show that d-amino acids inhibited growth and the expression of biofilm matrix genes at concentrations that inhibit biofilm formation. Finally, we report that the strain routinely used to study biofilm formation has a mutation in the gene (dtd) encoding d-tyrosyl-tRNA deacylase, an enzyme that prevents the misincorporation of d-amino acids into protein in B. subtilis. When we repaired the dtd gene, B. subtilis became resistant to the biofilm-inhibitory effects of d-amino acids without losing the ability to incorporate at least one noncanonical d-amino acid, d-tryptophan, into the peptidoglycan peptide side chain. We conclude that the susceptibility of B. subtilis to the biofilm-inhibitory effects of d-amino acids is largely, if not entirely, due to their toxic effects on protein synthesis.
INTRODUCTION
The soil bacterium Bacillus subtilis forms architecturally complex communities of cells encased in a self-produced matrix consisting of exopolysaccharide and the amyloid-like protein TasA (1, 2). These biofilms form on solid surfaces and at air-liquid interfaces (floating biofilms are known as pellicles). Whereas much is known about the mechanisms underlying biofilm formation, the mechanisms that cause cells to disperse from the biofilm and resume a planktonic existence are less well understood (3). Recently, interest has focused on the production of small molecules that inhibit biofilm formation and could therefore be involved in biofilm disassembly. Of particular interest are four d-amino acids (d-Leu, d-Met, d-Trp, and d-Tyr) that were found to be produced at late stages of biofilm development and were reported to have biofilm-inhibitory activity. Previous work from two of our laboratories suggested that the incorporation of these four d-amino acids into the peptidoglycan peptide side chain in place of d-Ala triggers the release of TasA fibers from the cell wall (4).
Here, we present evidence that indicates that the d-amino acids used (d-Leu, d-Met, d-Trp, and d-Tyr) can be growth inhibitory and that the biofilm inhibition observed when the culture medium contains these d-amino acids is largely, if not entirely, a consequence of their misincorporation into protein. This finding is in keeping with the report that d-Tyr, the most potent of the four d-amino acids, is a metabolic inhibitor of B. subtilis (5).
MATERIALS AND METHODS
Strains and growth conditions.
Bacillus subtilis NCIB3610 (hereinafter, 3610) or 168 and Escherichia coli (New England BioLabs, USA) were grown in Luria-Bertani (LB) broth (10 g tryptone per liter, 5 g yeast extract per liter, 5 g NaCl per liter) or on LB agar plates containing 1.5% Bacto agar at 37°C. When appropriate, 1 μg/ml erythromycin and 25 μg/ml lincomycin were added to liquid or solid medium. The strains used in this study are listed in Table S1 in the supplemental material.
Strain construction.
The markerless repair of dtd in B. subtilis was accomplished using the pMiniMAD protocol as described by Patrick and Kearns (6) and using primers 51 to 54, given in Table S1 in the supplemental material. Successful double-crossover events leading to the dtd+ strain were confirmed by restreaking individual clones on 1% agar plates containing LB, LB plus erythromycin and lincomycin, or LB plus 400 μM d-LMWY. Isolates which grew on LB and on LB plus d-LMWY but not on LB plus erythromycin and lincomycin were submitted for sequencing using primers 7 and 8 (see Table S1).
Luciferase reporters for epsA and tapA operon expression were constructed as described previously (7) and transferred by phage transduction into dtd+ cells (8).
Growth measurements.
Unless otherwise indicated, cells were grown to mid-exponential phase and diluted to a final optical density at 600 nm (OD600) of ∼0.015 into MSgg medium (8) to which amino acid or the equivalent volume of distilled water (dH2O) was added to a final volume of 20 ml. Cells were grown in shaking culture at 200 rpm at 37°C, and the OD600 was measured every hour for 7 h. Alternatively, cells grown to mid-exponential phase were diluted 1:1,000 into MSgg plus amino acids or the equivalent volume of dH2O, and 250-μl aliquots were transferred to a Costar polystyrene 96-well plate with a low-evaporation lid (Fisher Scientific, USA). The OD600 was measured every 10 min for 24 h in a BioTek Synergy 2 luminometer (BioTek, USA) with continuous slow shaking at 30°C.
Pellicle assays.
Cultures of B. subtilis 3610 or dtd+ cells were grown to early stationary phase and then diluted 1:1,000 into MSgg (9) or modified MSgg. Amino acids were stored as stock solutions of 40 mM or 60 mM in dH2O, except for d-Tyr and l-Tyr, which were stored as 10 mM stock solutions in 0.1 M HCl. Treatment solutions were diluted from stock solutions to achieve the final concentrations, and volumes were normalized using dH2O. All experiments were conducted in BD Falcon 24-well plates with low-evaporation lids (BD Biosciences, USA) in a final volume of 2 ml. Plates were incubated at 30°C or 25°C, and photographs were taken every 24 h.
Kinetic luciferase assay.
Cultures of B. subtilis 3610 or dtd+ cells were grown in LB to mid-exponential phase and diluted 1:1,000 in fresh MSgg with amino acid treatments or an equivalent volume of dH2O. Dilutions were plated in triplicate (250 μl each) in a 96-well polystyrene Costar plate (white with a clear bottom; Fisher Scientific, USA). Luciferase activity was measured on a BioTek Synergy 2 luminometer (BioTek, USA) with continuous slow shaking at 30°C. Luciferase luminescence was measured at a sensitivity setting of 200, and the culture optical density was measured at 600 nm every 10 min for 24 h. The final luciferase activity values were calculated by normalizing luciferase luminescence to culture density.
Evaluation of d-amino acid incorporation into peptidoglycan.
Analysis of B. subtilis peptidoglycan fragments by liquid chromatography-mass spectrometry (LC-MS) was performed as described by Lebar et al. (10) with the modifications detailed in the supplemental material. The extracted [M+2H]/2 ions are provided in Fig. S4 in the supplemental material.
RESULTS AND DISCUSSION
l-Amino acids but not d-alanine counteract biofilm inhibition by d-amino acids.
Kolodkin-Gal et al. (4) reported that d-Tyr at 3 μM, d-Leu at 8.5 mM, or d-Trp at 5 mM, used individually, inhibits pellicle formation by B. subtilis. These effects were prevented by the addition of d-Ala at 10 mM. Since d-Ala is present in the peptide side chain of peptidoglycan, such results suggested that biofilm inhibition by d-Leu, d-Met, d-Trp, and d-Tyr was due to their incorporation into the peptidoglycan. In our current investigation, we did not observe reversal of the biofilm-inhibitory effects of 6 μM (or 3 μM) d-Tyr by d-Ala at 10 mM (Fig. 1A; also S. A. Leiman, unpublished data). Instead, we found that l-Tyr was an effective countertreatment, even at concentrations far lower than those previously reported for d-Ala. The efficacy of the l-Tyr countertreatment was proportional to its concentration in the medium (Fig. 1A). Moreover, the effects of l-Tyr were specific. Biofilm inhibition by d-Tyr was unaffected by nonisomeric l-amino acids, such as l-Leu, l-Trp, and l-Met (Fig. 2).
Fig 1.
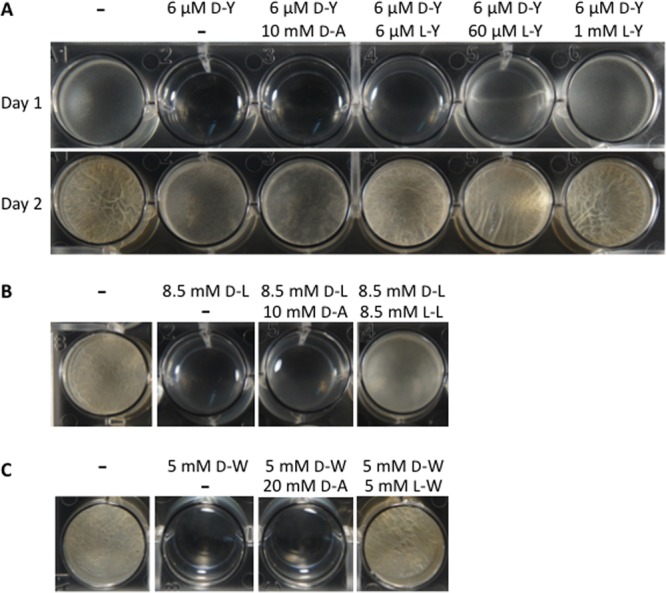
Rescue of biofilm formation by l-stereoisomers of d-amino acids. B. subtilis 3610 was treated with d-Tyr alone, d-Tyr plus d-Ala, or d-Tyr plus l-Tyr (A), with d-Leu alone, d-Leu plus d-Ala, or d-Leu plus l-Leu (B), or with d-Trp alone, d-Trp plus d-Ala, or d-Trp plus l-Trp (C). All cultures were grown in unmodified MSgg and incubated at 30°C. Photographs of d-Tyr-treated cells were taken at 24 h and at 48 h (days 1 and 2 in panel A) after treatment. Images of cells treated with d-Leu or d-Trp were taken at 48 h posttreatment.
Fig 2.
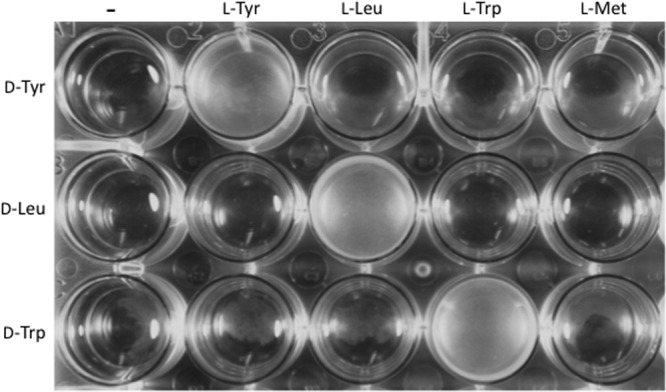
Rescue of pellicle formation by l-amino acids is specific. B. subtilis 3610 was treated with a single d-amino acid, a d-amino acid and its cognate l-amino acid, or a d-amino acid and a noncognate l-amino acid. d-Tyr was applied at 6 μM with l-Tyr, l-Leu, l-Trp, and l-Met at 600 μM each. d-Leu and all four l-amino acid countertreatments were applied at 8.5 mM. d-Trp and all four l-amino acid countertreatments were applied at 5 mM. Cultures were incubated at 30°C in unaltered MSgg and photographed at 24 h.
Similar results were obtained with other d-amino acids. We found that D-Ala did not prevent the inhibition of pellicle formation caused by 8.5 mM d-Leu or 5 mM d-Trp, whereas l-Leu and l-Trp effectively counteracted d-Leu and d-Trp, respectively (Fig. 1B and C and 2). Again, the effects of the l-amino acids were specific. Cells treated with d-Leu or d-Trp and countertreated with noncognate l-amino acids did not produce biofilms (Fig. 2).
These results indicate that competition between amino acid enantiomers influences pellicle formation. Such competition cannot occur in the peptidoglycan, as the site of noncanonical d-amino acid incorporation into the peptidoglycan peptide side chain excludes molecules with an l center (11). Rather, the specific rescue of biofilm formation by l-enantiomers suggests that d-amino acids inhibit biofilm formation by interfering with protein synthesis.
Biofilm-inhibitory concentrations of d-amino acids inhibit growth.
If d-amino acids affect biofilm development through an indirect effect on protein synthesis, we would expect that treatment with d-amino acids would result in a growth defect. We tested this hypothesis by measuring growth rates in the absence or presence of d-Tyr, d-Leu, or d-Trp. Growth was measured in shaking cultures at 37°C in the biofilm-inducing medium MSgg. d-Tyr, d-Leu, and d-Trp markedly inhibited growth at the concentrations used to inhibit biofilm formation (6 μM, 8.5 mM, and 5 mM, respectively) (Fig. 3). d-Tyr at 3 μM caused a severe growth defect as well (S. A. Leiman, unpublished data). Moreover, growth inhibition was partially or fully reversed by the addition of an equimolar concentration of the corresponding l-enantiomer.
Fig 3.
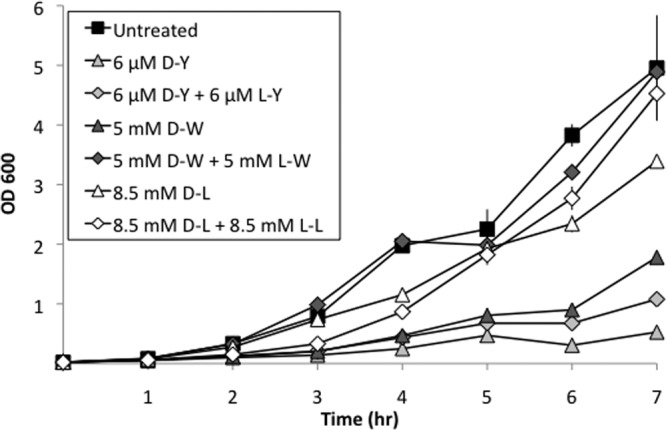
Biofilm-inhibitory concentrations of d-amino acids inhibit growth. B. subtilis 3610 was grown in shaking unmodified MSgg at 37°C. OD600 was measured every hour. Results represent the averages of duplicate experiments, and error bars show the standard deviations.
It is worth noting that at lower concentrations of d-amino acids (e.g., 700 μM d-Leu), it is possible to uncouple pellicle inhibition and growth inhibition. Under such conditions, both the corresponding l-enantiomer and, less potently, d-Ala are capable of restoring pellicle formation (H. Vlamakis and R. Kolter, unpublished results). These findings suggest that biofilm formation may be more sensitive than growth to inhibition of protein synthesis by d-amino acids.
Biofilm inhibition by a mixture of d-amino acids is reversed by l-amino acids.
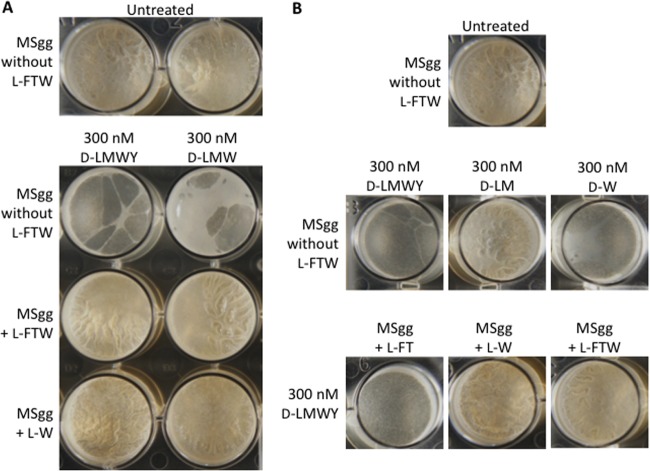
Kolodkin-Gal et al. (4) reported that a mixture of d-Leu, d-Met, d-Trp, and d-Tyr, each at 10 nM, inhibited pellicle formation in B. subtilis. We were unable to observe biofilm inhibition at this concentration, and higher concentrations sufficient to mildly inhibit pellicle formation (e.g., 500 nM) also inhibited growth (see Fig. S1 in the supplemental material). We did, however, identify conditions under which a biofilm-inhibitory concentration of d-LMWY would not inhibit growth. Biofilm inhibition occurred at a d-LMWY concentration of 300 nM each when we used a modified MSgg medium lacking the usual l-amino acid components l-phenylalanine, l-threonine, and l-tryptophan (Fig. 4A). We detected little or no growth inhibition at this concentration in shaking culture when we used a smaller-than-usual culture volume (3 ml versus 20 ml) and a larger-than-usual inoculum (OD600 0.05 versus 0.015) (see Fig. S2). Thus, there appear to be specific requirements for observing biofilm inhibition without growth inhibition. Nonetheless, under these optimized conditions, it was still possible to reverse the biofilm-inhibitory activity of 300 nM d-LMWY with 300 nM l-Trp (Fig. 4B). It is also notable that d-Trp alone at 300 nM was as effective at inhibiting biofilm formation as was the d-LMWY mixture at 300 nM, a finding that is inconsistent with previous results that suggested synergy between the four d-amino acids (4). We conclude that biofilm inhibition by nanomolar concentrations of d-amino acids happens only when the medium does not contain the corresponding l-amino acids, even under conditions that minimize growth inhibition by d-amino acids.
Fig 4.
Biofilm inhibition by low concentrations of d-amino acids requires the absence of l-amino acids and is not due to synergy. Unmodified MSgg or MSgg lacking one or more l-amino acids was inoculated with B. subtilis 3610. Cells were treated with a single d-amino acid or combinations of d-amino acids at 300 nM and were incubated at 25°C. l-Amino acids were each applied at the final concentration found in unmodified MSgg (l-Phe at 303 μM, l-Thr at 420 μM, and l-Trp at 245 μM) (A) or at 300 nM (B). The images were taken 72 h posttreatment.
Repairing dtd enhances survival and biofilm formation in the presence of d-amino acids.
We noticed that, in the strain we routinely use (NCIB3610) and in its derivative 168, the gene (dtd) for d-Tyr-tRNA deacylase is mutated, harboring a Lys codon (AAG) in place of the wild-type initiation codon (AUG). d-Tyr-tRNA deacylase was originally identified on the basis of its ability to specifically remove d-Tyr from mischarged tRNATyr but has since been shown to deacylate tRNAs mischarged with other d-amino acids (12). Thus, for simplicity, we will henceforth refer to this enzyme as d-aminoacyl-tRNA deacylase (13). If, as we hypothesize, the effects of d-amino acids are largely due to their misincorporation into protein, then repairing the mutant deacylase gene ought to confer resistance to the inhibitory effects of d-amino acids on growth and biofilm formation. To test this prediction, we repaired the dtd gene in 3610 using a protocol that left the chromosome unaltered except for the AAG-to-AUG nucleotide switch. Measurements of growth rates under standard biofilm-promoting conditions revealed that, unlike strain 3610, the dtd+ strain grew normally in the presence of d-LMWY at 3 μM each (Fig. 5). Moreover, d-LMWY at 300 μM, which was lethal for the parent strain, merely slowed the growth of the strain repaired for d-aminoacyl-tRNA deacylase activity.
Fig 5.
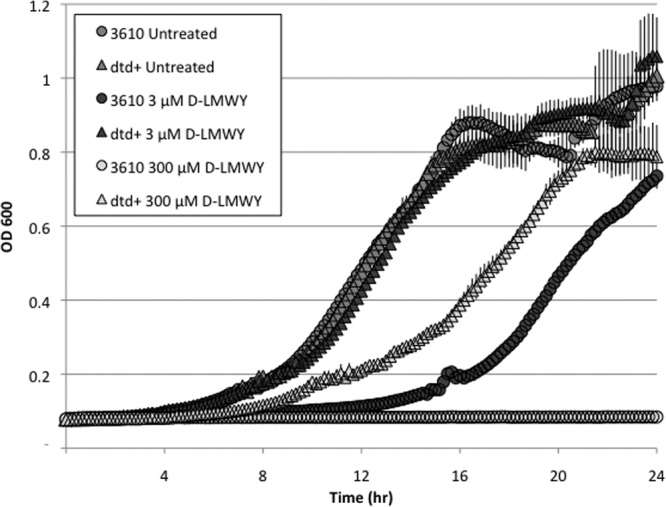
Cells repaired for the d-Tyr-tRNA deacylase gene exhibit resistance to growth inhibition by d-amino acids. The dtd mutant strain (3610) or the dtd+ repaired strain (SLH31) was grown in MSgg and either treated with an equimolar mixture of d-LMWY or left untreated. Cultures were incubated at 30°C and continuously shaken in a microplate reader in which optical density was measured at 600 nm every 10 min for 24 h. The results represent the averages of three replicates, and error bars show the standard deviations.
Importantly, whereas the parent strain produced thin biofilms in MSgg containing d-LMWY at 300 nM but lacking l-amino acids, the dtd+ strain formed robust pellicles under the same medium conditions (Fig. 6). In fact, in the absence of l-amino acids, the repaired strain resisted biofilm inhibition when treated with d-LMWY at 300 μM and was partially resistant to d-LMWY at 1 mM. These results reinforce the conclusion that the biofilm-inhibitory effects of d-amino acids in B. subtilis 3610 are an indirect consequence of their misincorporation into protein.
Fig 6.
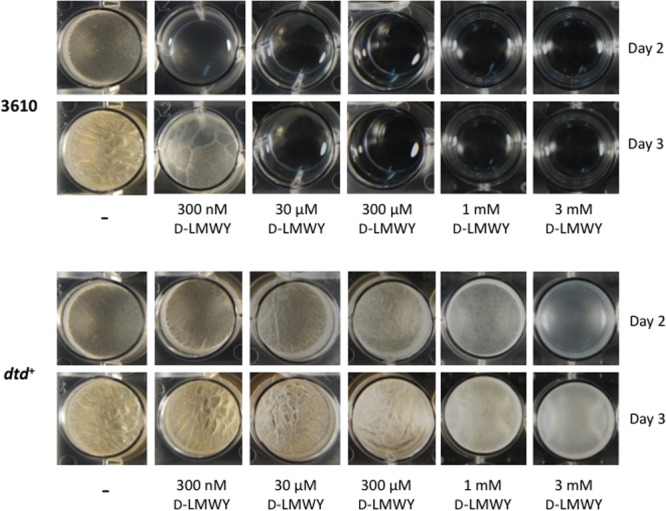
Cells repaired for the d-Tyr-tRNA deacylase gene exhibit resistance to biofilm inhibition by d-amino acids. The dtd mutant strain (3610) or the dtd+ repaired strain (SLH31) was treated with a range of concentrations of an equimolar mixture of d-LMWY. All cultures were grown in MSgg lacking l-amino acids and were incubated at 25°C. Photographs were taken after 48 and 72 h.
Matrix gene expression decreases in the presence of d-amino acids.
In light of the resistance of the dtd+ strain to d-amino acids, we revisited the effect of d-amino acids on matrix gene expression using luciferase fusions to the promoters for the epsA and tapA operons. Untreated 3610 (dtd mutant) cells, untreated dtd+ cells, and dtd+ cells treated with d-LMWY at 3 μM all expressed the luciferase fusions at comparable levels (see Fig. S3 in the supplemental material). In contrast, d-LMWY at 3 μM severely inhibited the expression of the luciferase fusions by cells of the parent strain, consistent with defects in protein synthesis. Little to no change in expression resulted from the addition of excess d-Ala; however, the expression of the luciferase fusions was partially restored when cells were treated with an equimolar mixture of d- and l-LMWY.
Repair of the d-aminoacyl-tRNA deacylase gene does not interfere with the incorporation of d-amino acids into peptidoglycan.
It was previously hypothesized that d-amino acids inhibit biofilms through their incorporation into peptidoglycan (4). As B. subtilis cells repaired for dtd are highly resistant to both growth inhibition and biofilm inhibition by d-amino acids, we asked whether d-amino acids were being incorporated into the peptidoglycan of dtd+ cells. Using LC-MS, we analyzed the composition of peptidoglycan from dtd+ cells grown in MSgg for 2 days in the presence of d-Trp at 50 μM or 1 mM and compared each of these samples to peptidoglycan from untreated dtd+ cells. Both untreated and treated dtd+ cultures produced robust pellicles (S. A. Leiman, unpublished data). We selected d-Trp as a representative noncanonical d-amino acid for this analysis because it gives a particularly distinctive signal.
LC-MS revealed that d-Trp was incorporated into the fifth position of the peptidoglycan peptide side chain when dtd+ cells were treated with d-Trp at 50 μM or 1 mM (Fig. 7). The elemental composition and structure of the peptidoglycan fragments containing d-Trp were confirmed by high-resolution LC-tandem mass spectrometry (MS/MS) (see Fig. S5 and S6 in the supplemental material). In contrast, we did not detect d-Trp in peptidoglycan from untreated dtd+ cells. These data indicate that repairing the dtd gene did not interfere with d-amino acid incorporation into peptidoglycan. We conclude that B. subtilis is capable of incorporating noncanonical d-amino acids, or at least d-Trp, into the peptidoglycan under conditions in which d-amino acids have little or no effect on biofilm formation or cell growth.
Fig 7.
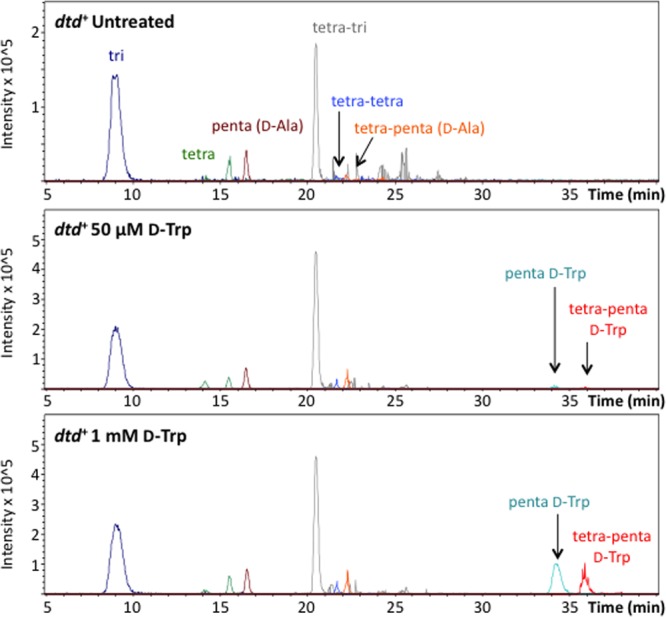
Cells repaired for the d-Tyr-tRNA deacylase gene incorporate d-tryptophan into the peptidoglycan side chain. Pellicles from dtd+ cultures left untreated or treated with d-Trp were harvested after incubation for 48 h at 25°C. Peptidoglycan from each sample was isolated, digested, and analyzed by LC-MS as depicted in Fig. S4 in the supplemental material. Peaks corresponding to peptidoglycan fragments of interest were selected by mass. Samples from all conditions were analyzed for the same eight fragment peaks.
To summarize, the principal conclusion from our work is that inhibition of biofilm formation by noncanonical d-amino acids in B. subtilis is largely, if not entirely, mediated by misincorporation into protein, presumably resulting in proteotoxicity. This conclusion rests on the following observations: (i) the biofilm-inhibitory effects of d-amino acids at concentrations previously reported and used here were not reversed by d-Ala but were reversed by their cognate l-amino acids; (ii) the concentrations of d-amino acids that inhibited biofilm formation also inhibited growth and the expression of the matrix operons epsA and tapA, although, under specific conditions, low d-amino acid concentrations did not produce a significant growth defect; and (iii) importantly, cells corrected for the mutant d-aminoacyl-tRNA deacylase gene were resistant to the biofilm-inhibitory and growth-inhibitory effects of d-amino acids while retaining the ability to incorporate d-Trp into peptidoglycan. Whether d-amino acids have any other effect on the process of biofilm formation (or disassembly) aside from interference with protein synthesis remains to be determined.
It is curious, given the toxic effects of d-amino acids, that certain strains of B. subtilis lack d-Tyr-tRNA deacylase activity. The absence of deacylase activity in the laboratory strain 168 was discovered by Calendar and Berg in 1966 (14). Interestingly, B. subtilis strain 23 is naturally resistant to d-Tyr, and from this observation, Champney and Jenson (5) inferred that strain 23 had retained d-Tyr-tRNA deacylase activity. Indeed, the presence of a wild-type dtd gene in this strain was later confirmed by DNA sequencing. d-Tyr-tRNA deacylase activity is not, however, the only basis for resistance to d-amino acids. In other work, we have found that when treated with lethal concentrations of d-LMWY, B. subtilis strain 3610 readily gives rise to resistant mutants that are nonetheless still have the mutant D-Tyr-tRNA deacylase gene (S. A. Leiman, unpublished data). The nature of these resistance mutations is the subject of ongoing research.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Foulston, H. Vlamakis, T. Norman, M. Cabeen, A. Elsholz, and N. Bradshaw for discussions. We thank G. Byrd and The Small Molecule Mass Spectrometry Facility at Harvard University for performing high-resolution LC-MS/MS.
This work was supported by NIH grants GM18568 to R.L., GM58213 and GM82137 to R.K., GM103056 to M.D.L., and GM066174 and GM076710 to D.K., as well as by NSF grant DGE-1144152 to J.M.M.
Footnotes
Published ahead of print 4 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00975-13.
REFERENCES
- 1.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59:1229–1238 [DOI] [PubMed] [Google Scholar]
- 2.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U. S. A. 107:2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. 2010. D-Amino acids trigger biofilm disassembly. Science 328:627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champney WS, Jensen RA. 1969. D-Tyrosine as a metabolic inhibitor of Bacillus subtilis. J. Bacteriol. 98:205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patrick JE, Kearns DB. 2008. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70:1166–1179 [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Cao S, Chai Y, Clardy J, Kolter R, Guo JH, Losick R. 2012. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol. Microbiol. 85:418–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55:739–749 [DOI] [PubMed] [Google Scholar]
- 9.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98:11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebar MD, Lupoli TJ, Tsukamoto H, May JM, Walker S, Kahne D. 2013. Forming cross-linked peptidoglycan from synthetic gram-negative lipid II. J. Am. Chem. Soc. 135:4632–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lupoli TJ, Tsukamoto H, Doud EH, Wang TS, Walker S, Kahne D. 2011. Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J. Am. Chem. Soc. 133:10748–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soutourina O, Soutourina J, Blanquet S, Plateau P. 2004. Formation of D-tyrosyl-tRNATyr accounts for the toxicity of D-tyrosine toward Escherichia coli. J. Biol. Chem. 279:42560–42565 [DOI] [PubMed] [Google Scholar]
- 13.Yang H, Zheng G, Peng X, Qiang B, Yuan J. 2003. D-Amino acids and D-Tyr-tRNA(Tyr) deacylase: stereospecificity of the translation machine revisited. FEBS Lett. 552:95–98 [DOI] [PubMed] [Google Scholar]
- 14.Calendar R, Berg P. 1966. Purification and physical characterization of tyrosyl ribonucleic acid synthetases from Escherichia coli and Bacillus subtilis. Biochemistry 5:1681–1690 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.