Abstract
Mannose is an important sugar in the biology of the Gram-negative bacterium Porphyromonas gingivalis. It is a major component of the oligosaccharides attached to the Arg-gingipain cysteine proteases, the repeating units of an acidic lipopolysaccharide (A-LPS), and the core regions of both types of LPS produced by the organism (O-LPS and A-LPS) and a reported extracellular polysaccharide (EPS) isolated from spent culture medium. The organism occurs at inflamed sites in periodontal tissues, where it is exposed to host glycoproteins rich in mannose, which may be substrates for the acquisition of mannose by P. gingivalis. Five potential mannosidases were identified in the P. gingivalis W83 genome that may play a role in mannose acquisition. Four mannosidases were characterized in this study: PG0032 was a β-mannosidase, whereas PG0902 and PG1712 were capable of hydrolyzing p-nitrophenyl α-d-mannopyranoside. PG1711 and PG1712 were α-1→3 and α-1→2 mannosidases, respectively. No enzyme function could be assigned to PG0973. α-1→6 mannobiose was not hydrolyzed by P. gingivalis W50. EPS present in the culture supernatant was shown to be identical to yeast mannan and a component of the medium used for culturing P. gingivalis and was resistant to hydrolysis by mannosidases. Synthesis of O-LPS and A-LPS and glycosylation of the gingipains appeared to be unaffected in all mutants. Thus, α- and β-mannosidases of P. gingivalis are not involved in the harnessing of mannan/mannose from the growth medium for these biosynthetic processes. P. gingivalis grown in chemically defined medium devoid of carbohydrate showed reduced α-mannosidase activity (25%), suggesting these enzymes are environmentally regulated.
INTRODUCTION
The Gram-negative anaerobe Porphyromonas gingivalis is an important agent in the etiology of adult periodontal disease and produces several virulence factors, which include extracellular cysteine proteases with specificities for Arg-X (Arg-gingipains [Rgps]) and Lys-X (Lys-gingipain [Kgp]) peptide bonds (1) and two lipopolysaccharides (LPSs), namely, O-LPS (2) and acidic LPS (A-LPS) (3, 4), which play important roles in the deregulation of innate and inflammatory systems in the host (1, 5).
Mannose is an important constituent of the oligosaccharide (OS) attachments to the Arg-gingipains (6), a family of five proteases derived from rgpA and rgpB. The repeating unit of A-LPS is composed of a phosphorylated branched mannan (3, 4). Monoclonal antibody (MAb) 1B5, raised against one of the isoforms of Rgp, namely, soluble monomeric RgpAcat, also cross-reacts with membrane-type Rgps (mt-Rgps) (7) and A-LPS. One of the side chains of the repeating unit of A-polysaccharide (A-PS) contains the Manα1-2Manα-1-phosphate fragment, which forms part of the epitope recognized by MAb 1B5 (3), suggesting a common biosynthetic pathway for these important macromolecules. The outer core region of O-LPS (8) occurs in two glycoforms: an “uncapped core” devoid of O-PS and a “capped core” containing the attachment site of O-PS (8). The outer region of the uncapped core is composed of a linear α-1→3-linked d-Man OS containing four or five mannopyranosyl residues (half of which are modified by phosphoethanolamine at position 6), and the “capped core” contains a three- to five-residue extension of α-1→3-linked Man residues glycosylating the outer core region at the nonreducing terminal residue (8). A mannose-containing polysaccharide has been described by Farquharson et al. (9) in the spent culture medium of P. gingivalis, which showed the presence of Man and trace amounts of Rha, Gal, Glc, and GlcNAc. The chemical shifts of the anomeric proton resonances obtained by 1H nuclear magnetic resonance (NMR) spectroscopy of this polysaccharide demonstrated that all the sugars were α-linked (9).
Annotation of the P. gingivalis W83 genome indicated the presence of five putative mannosidases: PG0032 was classified as a probable β-mannosidase and PG0902, PG0973, PG1711, and PG1712 as putative α-1,2-mannosidases based on homologies (10). The aim of this study was to characterize these enzymes and determine their role(s) in some or all of the biosynthetic pathways of the mannose-containing macromolecules in P. gingivalis W50.
In this study, we generated single isogenic mutants in P. gingivalis PG0032, PG0902, PG0973, PG1711, and PG1712 and assayed them against various substrates to test for α- and β-mannosidase activities. Double-isogenic mutants were made in PG1711-PG1712, and triple-isogenic mutations were made in PG0032-PG1711-PG1712, PG0902-PG1711-PG1712, and PG0973-PG1711-PG1712. The mutant strains were characterized with respect to α- and β-mannosidase activities against a variety of substrates and to the nature of their mannose-containing macromolecules.
MATERIALS AND METHODS
Materials.
DEAE-Sephacel, Sephacryl S-300HR, and PlusOne urea were purchased from GE Healthcare, Buckinghamshire, United Kingdom. A solution containing 30% acrylamide–N,N-methylenebisacrylamide (BIS) (37.5:1) and Bio-Gel P-6 was obtained from Bio-Rad Laboratories (Hercules, CA, USA). High-performance thin-layer chromatography (HPTLC) Silica Gel F254 glass plates (10 cm by 10 cm) were from Merck Chemicals Ltd., Nottingham, United Kingdom. Horseradish peroxidase-labeled mouse immunoglobulin was purchased from Dako A/S, High Wycombe, Buckinghamshire, United Kingdom. cOmplete Mini Protease inhibitor cocktail tablets and dansyl-glutamyl-glycyl-arginyl-chloromethylketone (DNS-EGR-CK) were purchased from Roche, Welwyn Garden City, Hertfordshire, United Kingdom. All other chemicals were from VWR, Lutterworth, Leicestershire, United Kingdom, or from Sigma-Aldrich Co. Ltd., Poole, Dorset, United Kingdom, and were the purest grades available. Restriction and modification enzymes were purchased from New England BioLabs, and DNA purification reagents were obtained from Qiagen.
Bacterial growth conditions.
The P. gingivalis strains used in this study (Table 1) were grown at 37°C on either blood agar plates containing 5% defibrinated horse blood or brain heart infusion (BHI) broth supplemented with hemin (5 μg ml−1) in an anaerobic atmosphere of 80% N2, 10% H2, and 10% CO2 (Don Whitley Scientific).
Table 1.
Bacterial strainsa
| Strain | Inactivated gene(s) | Relevant genotype | Source |
|---|---|---|---|
| P. gingivalis | |||
| W50 | Wild type | ATCC 53978 | |
| PG0032/2 | PG0032 | PG0032::erm | This paper |
| PG0902/5 | PG0902 | PG0902::erm | This paper |
| PG0973/2 | PG0973 | PG0973::erm | This paper |
| PG1711/3 | PG1711 | PG1711::erm | This paper |
| PG1712/2 | PG1712 | PG1712::erm | This paper |
| 1712/4 | PG1711, PG1712 (Ermr) | ΔPG1711-PG1712::erm | This paper |
| DMD | PG1711, PG1712 (Tetr) | ΔPG1711-PG1712::erm′::tetQ | This paper |
| PG0032B2 | PG0032, PG1711, PG1712 (Ermr Tetr) | ΔPG1711-PG1712::erm′::tetQ PG0032::erm | This paper |
| PG0902D1 | PG0902, PG1711, PG1712 (Ermr Tetr) | ΔPG1711-PG1712::erm′::tetQ PG0902::erm | This paper |
| PG0973E2 | PG0973, PG1711, PG1712 (Ermr Tetr) | ΔPG1711-PG1712::erm′::tetQ PG0973::erm | This paper |
| E. coli | |||
| XL-1 Blue MRF′ | Plasmid maintenance | Stratagene |
Gene notation refers to the gene ID as presented by The Institute of Genomic Research (TIGR). Ermr, macrolide-lincosamine resistance; Tetr, tetracycline resistance; nonfunctional genes are indicated with a prime.
Generation of P. gingivalis mutants.
Single mutants defective in PG0032, PG0902, PG0973, PG1711, and PG1712 were generated using primer pairs designed to amplify the 5′ and 3′ ends of each open reading frame (ORF) by PCR (Table 2). The strategy incorporated SstI and XbaI restriction sites at the 3′ and 5′ ends of the amplicons, respectively (Table 2). Following purification and digestion with SstI and XbaI, these amplicons were ligated to the SstI-XbaI erm cassette, retrieved from pVA2198 (11) by T4-DNA ligase. The mixture was purified and used as a template in PCR to generate an erm cassette flanked by 400 to 850 bp of the ORF in question. In all cases, this generated an amplicon with an internal deletion of the relevant gene in vitro. The products were electroporated into 6-h-grown P. gingivalis W50, and colonies were selected and screened as previously described (12). Representative isogenic mutants were further screened and were designated PG0032, PG0902, PG0973, PG1711, and PG1712.
Table 2.
Properties of oligonucleotides
| Name | Sequence (5′–3′)a | Size (bp) |
|---|---|---|
| PG0032F1 | ACACCTTCCGAACATTTCTCC | |
| PG0032R1(SstI) | atatatgagctcGTCCGTTCGGGCTGTACAG | 785 |
| PG0032F2 (XbaI) | atatattctagaGCCAAACCCTACTGCATGG | |
| PG0032R2 | GTGTCGCAGACGAATAGCC | 680 |
| PG0902F1 | TTTCTCCCCTACCCCATAGC | |
| PG0902R1 (SstI) | atatatgagctcTGCGTCGTATCGTTATTGGC | 700 |
| PG0902F2 (XbaI) | atatattctagaATCACAGAGAGTAATGCATGGC | |
| PG0902R2 | ACCAGCAATGTCCCTCGC | 609 |
| PG0973F1 | TGACGATCCGAAACTTCCTC | |
| PG0973R1 (SstI) | atatatgagctcGCCGACTCACCYGAAGTACG | 671 |
| PG0973F2 (XbaI) | atatattctagaTTATGATCCGACCAACGAGC | |
| PG0973R2 | TCGTAACGGAGGTGACCG | 434 |
| PG1711F1 | TCCTCCTTGTGCTAAGCTCTG | |
| PG1711R1 (Sst) | atatatgagctcGGCTGGTGTAGAATACCTGTCG | 839 |
| PG1711F2 (XbaI) | atatattctagaATCGCTGGGATGTACAGCAC | |
| PG1711R2 | ATAGGGATCTGCAGAAGGAGG | 631 |
| PG1712F1 | TTCTCTGCTTGTTTGTCGGC | |
| PG1712R1 (SstI) | atatatgagctcCTGCCCTTTGACTTCCGC | 814 |
| PG1712F2 (XbaI) | atatattctagaGATAGAGCTGCATTGGACACG | |
| PG1712R2 | TTACTGTGATTAGCGCGACG | 841 |
| ErmFF2 | TTCGTTTTACGGGTCAGCAC | 2,100 |
| ErmAMR2 | ACTTTGGCGTGTTTCATTGC |
Lowercase letters in boldface indicate restriction enzyme sites, and lowercase letters in lightface indicate irrelevant sequences used to facilitate digestion with restriction enzymes.
Since PG1711 is downstream (69 bp) of and in the same transcriptional direction as PG1712, the amplicons corresponding to the 5′ end (SstI) of PG1712 and the 3′ end (XbaI) of PG1711 were ligated to the erm cassette (SstI-XbaI) in a similar manner. The representative P. gingivalis double mutant ΔPG1711-12::erm was selected for making triple mannosidase mutants. PG1711-12::erm was further manipulated to insert tetQ at the erm locus with pUCET1 (13) via electrotransformation, thereby inactivating the ermF component of erm by homologous recombination. To construct the pUCET1 integration plasmid, the erm cassette (11) from pVA2198 was initially cloned as a 2.1-kb EcoRI-HindIII fragment into the corresponding sites of pUC18 to generate pUCE. The ermF component of the erm cassette has a unique PmeI restriction site near the 3′ end of the gene (14). A 2.7-kb-HpaI-SmaI fragment of pKFT2 (15) including tetQ from pNJR12 (16) was blunted and cloned into a pUCE plasmid, described above, similarly treated and PmeI linearized. This generated pUCET1, in which the direction of tetQ is the same as the original ermF-ermAM with ermF inactivated with tetQ. Adjacent BglII-NotI sites upstream of the coding region for tetQ may be used to insert a gene expressed from its own promoter into pUCET1. Thus, the new gene tagged with tetQ and flanked by erm sequences may be used in homologous recombination to a site already possessing an erm cassette for integration of a single copy of a defined gene (13) as an insertional mutant or to replace the existing erm with tetQ. In P. gingivalis DMD, erm was replaced by a tetQ marker and selected following the usual screening procedures described above. DMD was used as the recipient of amplicons used to generate the single mutants described above in order to generate triple mutants. Triple mutants consisting of a deletion in PG1711-12 and either PG0032, PG0902, or PG0973, designated PG0032B2, PG0902D1, and PG0973E2, respectively, were generated by allelic exchange. In addition to resistance to tetracycline and/or clindamycin, purified chromosomal DNAs from these mutants were used as templates in PCRs using primer sets to the ORFs tetQ and erm and combinations of these genes to check for correct insertion.
Mannosidase assays.
α- and β-mannosidases were assayed using 4-nitrophenyl α-d-mannopyranoside or 4-nitrophenyl β-d-mannopyranoside, respectively, as the substrate. The reaction mixture (0.75 ml of 0.4 M buffer, pH 4 to 7.5, containing 4 mM Ca2+, 4 mM Co2+, or 4 mM Zn2+; 1.25 ml of 10 mM 4-nitrophenyl α [or β]-d-mannopyranoside, and 0.4 ml of distilled water) was incubated at 37°C in a water bath for 2 h. The reaction was initiated by the addition of 0.4 ml of P. gingivalis whole-cell suspension, crude cell extract, or soluble cell supernatant fraction. Aliquots (0.4 ml) were withdrawn immediately (zero time point) and at suitable time points into 0.7 ml of 0.2 M sodium carbonate solution, mixed well, and centrifuged in an Eppendorf microcentrifuge for 10 min at 4°C. The supernatant was transferred to disposable 1-ml cuvettes, and the absorbance at 450 nm was measured within 30 min. Sodium acetate buffers were used at pH 4.5, 5, 5.5, and 6.0, and Tris-HCl buffers were used at pH 6.5 and 7.0. Enzyme activity was expressed as units (nmol of 4-nitrophenol produced/h)/optical density at 600 nm (OD600) unit of cells.
α-Mannosidase assays using α-1→2-, α-1→3-, and α-1→6-linked mannobioses were performed as follows. The reaction mixtures contained 125 μg of disaccharide and 0.125 M sodium acetate buffer, pH 6.0, with 1.25 mM Ca2+ in a total volume of 30 μl. Ten microliters of reaction mixture was withdrawn and stored at 4°C, and this served as the zero time point; 10 μl of sonicated cell extracts of P. gingivalis was added to the reaction mixture and incubated at 37°C for 21 h. Aliquots of 10 μl were withdrawn and centrifuged in an Eppendorf microcentrifuge (10,000 × g) for 1 min. The supernatant was spotted onto Keiselgel F254 HPTLC glass-coated plates, and TLC was performed in a tank equilibrated for 1 h in n-butanol–ethanol–water (50:50:30, by volume). The plates were dried in air, rechromatographed in the same solvent, dried in air, sprayed with 3% sulfuric acid in methanol, air dried, and placed in an oven at 85°C to develop the spots.
Arg-gingipain and Lys-gingipain enzyme assays.
Arg-X and Lys-X protease activities were measured at 30°C with Nα-benzoyl dl-arginine-4-nitroanilide (dl-BRpNA) (500 μM) or N-α-acetyl-l-lysine-4-nitroanilide (AcLyspNA) (250 μM) as the substrate in spectrophotometric assays, as previously described (17). The activities of Arg-X and Lys-X proteases in culture supernatants or whole cultures are expressed as the change in A405 units per minute at 30°C. The cultures were adjusted to the same OD600 prior to the assays by dilution with BHI broth.
Purification of PS: EPS from P. gingivalis W50 grown in BHI broth.
To 1.7 liters of culture supernatant from 24-h cultures of P. gingivalis W50 grown in BHI broth supplemented with hemin (5 mg/liter), 8.5 g of NaCl, followed by 3.4 liters of absolute ethanol, was added, mixed, and left at −20°C for 16 h to effect the precipitation of PS. The suspension was centrifuged at 10,000 × g (Sorvall RC5C; SLA3000) for 45 min at 4°C. The pellet was dissolved in water, dialyzed exhaustively against distilled water for 2 to 3 days at 4°C, and freeze-dried (765 mg). The residue was treated with RNase A, DNase I, and proteinase as described previously (6), dialyzed exhaustively against distilled water at 4°C, and freeze-dried (450 mg).
The freeze-dried residue was dissolved in 75 ml of 50 mM Tris-HCl, pH 6.5, and applied to a column (2.6 cm by 11 cm) of DEAE-Sephacel previously equilibrated in buffer. The column effluent was monitored using a refractive index (RI) detector (Knauer Wellchrom K-2400; Wissenschaftliche Geratebau Dr. Ing. Herbert Knauer GmbH, Berlin, Germany), and fractions (5 ml) were collected. The column was washed with buffer until there was no change in the RI value. Fractions containing neutral EPS were combined, dialyzed exhaustively against distilled water at 4°C, and freeze-dried (125 mg).
Further purification of EPS was achieved by gel filtration chromatography on a Sephacryl S-300HR column (1.6 cm by 65 cm) in 50 mM ammonium bicarbonate. The yield of EPS was 46 mg (27 mg/liter); it was used in structural analysis and is referred to here as EPS (W50).
PS from the BHI growth medium was purified exactly as described for EPS (W50) from spent culture medium and is referred to here as PS (BHI).
NMR spectroscopy.
One-dimensional (1D) and 2D NMR spectra were recorded on a Bruker Dax-500 spectrometer for solutions in 99.99% D2O at 40°C using acetone as the internal standard (δH 2.225; δC 31.45). The standard XWINNMR software versions 2.5 and 3.1 were used for carrying out 2D NMR pulse programs as follows: 2D total correlation spectroscopy (TOCSY) (homonuclear Hartmann Hahn [HOHAHA]) (18) with presaturation during relaxation delay and MLEV-17 sequence for mixing; nuclear Overhauser effect spectroscopy (NOESY) (19) using time-proportional phase incrementation (TPPI) with presaturation and during relaxation delay and mixing time, 1H (13C) heteronuclear multiple-quantum coherence (HMQC) (20) using TPPI, presaturation during relaxation delay, and globally optimized alternating-phase rectangular pulse (GARP) decoupling during acquisition; 1H (13C) heteronuclear single-quantum coherence (HSQC)-TOCSY (21); and 1H (13C) HMQC-NOESY with WALTZ 16 decoupling during acquisition. For NOESY and TOCSY experiments, mixing delays of 0.12 s and 0.075 s, respectively, were used.
Composition and methylation analysis.
The composition of EPS (W50) was determined by methanolysis according to the method of Altman et al. (22), as described previously (3). Methyl glycosides were converted to O-trimethylsilyl (O-TMS) ethers and analyzed by gas chromatography-mass spectrometry (GC-MS) (23).
EPS (W50) was methylated according to the method of Kvernheim (24). Methylated polysaccharides were hydrolyzed with 0.5 M trifluoroacetic acid for 1.5 h at 120°C, followed by reduction with NaBD4 (22°C; 4 h), and acetylated with pyridine-acetic anhydride (1:1, by volume) at 100°C for 1 h. Methylated alditol acetates were analyzed by GC-MS.
The absolute configurations of the monosaccharides were determined as described by Gerwig et al. (25).
Acetolysis of EPS.
Acetolysis of EPS (W50) was carried out using the modified procedure of Kocourek and Ballou (26) as described in Paramonov et al. (3). De-O-acetylation and isolation of the de-O-acetylated products of acetolysis of EPS (W50) prior to matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS were performed as described in Paramonov et al. (3).
Hydrolysis of PS (BHI) by mannosidases.
PS (BHI) (3.3 mg) was incubated in sodium acetate buffer, pH 6.0, containing 4 mM Ca2+ (1.62 ml), and 0.15 ml of P. gingivalis W50 cell extract was added. A portion (0.5 ml) of the reaction mixture was withdrawn immediately (time zero), after 19 h, and after 44 h; centrifuged in an Eppendorf microcentrifuge to remove cell debris; and applied to a Bio Gel P-6 column (1.6 cm [inside diameter] by 36 cm) equilibrated in 0.05 M NH4HCO3. The column was washed with equilibration buffer at a flow rate of 60 ml/h, the column effluent was monitored using an RI detector, and fractions (1 ml) were collected. The mannan fractions eluting at the void volume were collected in each case, freeze-dried, and weighed. Aliquots (10 μl) of the reaction mixture were applied to TLC plates and chromatographed as described in “Mannosidase assays” above.
Preparation of P. gingivalis cell extracts.
P. gingivalis W50 and mutant strains were grown in BHI for 24 h, and 100 ml of culture (OD540, 3.0) was centrifuged in a Sorvall RC5C centrifuge at 10,000 × g at 4°C for 45 min. The supernatant was discarded, and the slushy pellet was centrifuged at 15,000 × g at 4°C for 30 min to pellet the cells. The supernatant was removed, and the cell pellet was washed with phosphate-buffered saline (PBS), resuspended in 4 ml of PBS containing a cocktail of protease inhibitors (50 ml of PBS plus 1 tablet of cOmplete Mini protease inhibitor cocktail), and sonicated (Soniprep 150; MSE United Kingdom Ltd., London, United Kingdom) at 10-μm amplitude 6 times for 1 min each time on ice with a cooling time of 1 min between successive sonications. The crude extract was either (i) used as such in assays for mannosidase activity or (ii) centrifuged in an Eppendorf microcentrifuge at 17,000 × g at 4°C for 10 min to separate cell debris from cytoplasmic/soluble proteins, and the supernatant was used in assays for mannosidase activity. Whole washed cells of P. gingivalis W50 and mutant strains were also used in assays directly and resuspended in PBS containing inhibitors as described above.
Fluorescent labeling of Arg-gingipains and Lys-gingipain in culture supernatants.
Culture supernatants (500 μl) from 6-day-old cultures of P. gingivalis, ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, and ΔPG1712 were treated with 1.5 volumes of ice-cold acetone and stored at −20°C for 1 h to effect complete precipitation of macromolecules. The samples were centrifuged at 13,300 rpm at 4°C for 20 min, the supernatant was discarded, and the pellet was dried in air to remove traces of acetone. Arg- and Lys-gingipains in the pellets were fluorescently labeled with DNS-EGR-CK, as described previously (27). Fluorescently labeled samples were dissolved in 60 μl of SDS sample buffer and subjected to SDS-PAGE in the Hoefer Mighty Small gel system. Fluorescent bands were visualized under UV light and photographed immediately.
Isolation of LPS.
LPS was prepared using an LPS extraction kit from Intron Biotechnology (South Korea) as described previously (4).
SDS-urea-PAGE and Western blotting.
LPS preparations (10 to 20 μg) from P. gingivalis W50 and mutant strains were subjected to SDS-urea-PAGE in polyacrylamide slab gels at 10°C for 3 to 4 h (28) using the Hoefer SE600 series gel system. The gels were stained with silver (Sigma-Aldrich Co. Ltd., Poole, Dorset, United Kingdom). Samples were transferred onto nitrocellulose membranes and probed with MAb 1B5 as described previously (4).
RESULTS
Mannosidase assays and localization of mannosidase activities.
α-Mannosidase and β-mannosidase activities of the P. gingivalis W50 parent strain were assayed using 4-nitrophenyl α-d-mannopyranoside and 4-nitrophenyl β-d-mannopyranoside as described above (see Materials and Methods). In the first instance, the assays were conducted using fresh whole cells and sonicated cell extracts in buffers containing 4 mM Ca2+, 4 mM Co2+, or 4 mM Zn2+ at pH 4.5, 5.0, 5.5, and 6.0 (0.1 M acetate buffers) and 6.5 and 7.0 (0.1 M Tris-HCl buffers) at 37°C. No activity was detectable using either 4 mM Co2+ or 4 mM Zn2+ in the buffers, whereas maximum activities were detected in the presence of 4 mM Ca2+. The rates of hydrolysis of 4-nitrophenyl α-d-mannopyranoside and 4-nitrophenyl β-d-mannopyranoside by P. gingivalis whole-cell suspensions at 37°C as a function of pH are shown in Table 3.
Table 3.
Hydrolysis of 4-nitrophenyl α-d-mannopyranoside and 4-nitrophenyl β-d-mannopyranoside by P. gingivalis whole-cell suspensions
| pH | Rate of hydrolysis (nmol 4-NP produced/h/OD600 unit of cells)a |
|
|---|---|---|
| α-Mannosidase activity | β-Mannosidase activity | |
| 4.5 | ∼0 | ∼0 |
| 5.0 | 0.0224 | 1.519 |
| 5.5 | 0.156 | 1.959 |
| 6.0 | 0.270 | 2.171 |
| 6.5 | 0.227 | 2.154 |
| 7.0 | 0.120 | 0.681 |
| 7.5 | 0.097 | 0.167 |
Rates of hydrolysis were measured in either sodium acetate buffers or Tris-HCl buffers containing 4 mM Ca2+ at 37°C.
The highest values of α-mannosidase and β-mannosidase activities were obtained between pH 6.0 and pH 6.5. The levels of β-mannosidase activity were ∼8-fold higher than α-mannosidase activity using the chromogenic substrates.
Therefore, all assays were henceforth performed at pH 6.0 in 0.1 M sodium acetate buffer containing 4 mM Ca2+ at 37°C. α-Mannosidase and β-mannosidase activities in whole cells, sonicated cell suspensions, and cell supernatants were measured at pH 6.0, which indicated that there was no detectable α-mannosidase or β-mannosidase activity in cell supernatants and that all the activity was associated with cell membranes. No α-mannosidase or β-mannosidase activity was detected in cells that had been stored at −20°C or in freeze-dried cells. P. gingivalis W50 whole cells and cell sonicates gave identical rates of hydrolysis of 4-nitrophenyl α-d-mannopyranoside and 4-nitrophenyl β-d-mannopyranoside (data not shown).
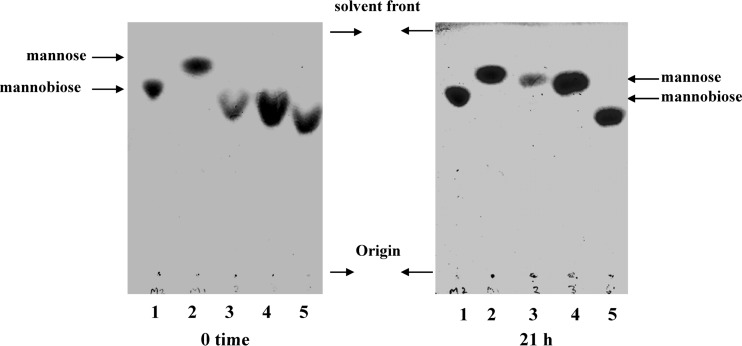
The α-mannosidase activities of P. gingivalis cell sonicates were measured using α-1→2, α-1→3, and α-1→6 mannobioses at pH 6.0 in 0.1 M acetate buffer containing 4 mM Ca2+ and detection of the products of hydrolysis by TLC. The results are shown in Fig. 1. α-1→2 mannobiose and α-1→3 mannobiose were both hydrolyzed by P. gingivalis W50 cell sonicates under the conditions of the experiments, whereas α-1→6 mannobiose was not hydrolyzed to mannose under the experimental conditions used. The hydrolysis of α-1→6 mannobiose by P. gingvalis cell sonicates was not measured at any other pH or using any other cofactors/additives.
Fig 1.
Hydrolysis of α-mannobioses by sonicated cell extracts of P. gingivalis detected by TLC on Keiselgel F254 HPTLC plates in n-butanol–ethanol–water (5:5:3, by volume). α-Mannosidase assays using α-1→2-, α-1→3-, and α-1→6-linked mannobioses were performed as described in Materials and Methods, and the assay mixtures contained 125 μg of disaccharide in 30 μl of 0.125 M sodium acetate buffer, pH 6.0, with 1.25 mM Ca2+. Ten-microliter aliquots were withdrawn and stored at 4°C, and this served as the zero time point. Ten microliters of sonicated cell extracts of P. gingivalis was added to the reaction mixture and incubated at 37°C for 20 h. Aliquots (10 μl) were withdrawn and centrifuged in an Eppendorf microcentrifuge. The supernatant was spotted onto Keiselgel F254 HPTLC glass-coated plates, and TLC was performed in a tank equilibrated for 1 h in n-butanol–ethanol–water (50:50:30, by volume). The plates were dried in air, rechromatographed in the same solvent system, dried in air, sprayed with 3% sulfuric acid in methanol, air dried, and placed in an oven at 85°C to develop the spots. Lanes: 1, α-1→2 mannobiose; 2, mannose; 3, α-1→2 mannobiose plus P. gingivalis cell extract; 4, α-1→3 mannobiose plus P. gingivalis extract; 5, α-1→6 mannobiose plus P. gingivalis extract.
Properties of mannosidase mutant strains.
Single isogenic mutants were isolated as described in Materials and Methods in the genes PG0032, PG0902, PG0973, PG1711, and PG1712, and a single isolate from each mutant strain was used in all the subsequent experiments. The growth rates and stabilities of culture turbidity of the parent P. gingivalis W50 strain and the ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, and ΔPG1712 isogenic mutants in BHI broth were similar over 6 days (not shown).
α-Mannosidase and β-mannosidase activities.
Whole-cell suspensions and cell extracts were prepared from 24-h cultures of P. gingivalis W50 and the ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, and ΔPG1712 isogenic mutant strains and used in assays to measure α-d-mannosidase activities using 4-nitrophenyl α-d-mannopyranoside and α-1→2 mannobiose, α-1→3 mannobiose, and α-1→6 mannobiose as substrates. The results obtained are shown in Table 4 and Fig. 2. β-Mannosidase activity was measured using 4-nitrophenyl β-d-mannopyranoside as the substrate.
Table 4.
Hydrolysis of 4-nitrophenyl α-d-mannopyranoside, α-1→2 mannobiose, and α-1→3 mannobiose by cell extracts of P. gingivalis W50 and isogenic mutant strains
| P. gingivalis strain | Rate of hydrolysis of 4-nitrophenyl α-d-mannopyranoside (nmol 4-NP produced/h/ml)a | % Activity relative to the parent W50 strain | Activity toward α-1→2 mannobioseb | Activity toward α-1→3 mannobioseb |
|---|---|---|---|---|
| W50 | 1.564 | 100 | Active | Active |
| ΔPG0032 | 1.585 | 102 | Active | Active |
| ΔPG0902 | 0.293 | 19 | Active | Active |
| ΔPG0973 | 1.628 | 101 | Active | Active |
| ΔPG1711 | 1.229 | 101 | Active | Inactive |
| ΔPG1712 | 1.596 | 76 | Inactive | Active |
Rates of hydrolysis were measured at pH 6.0 in 0.1 M sodium acetate buffer containing 4 mM Ca2+ at 37°C.
Hydrolysis of α-1→2 and α-1→3 mannobiose, followed by TLC on Keisel gel plates in n-butanol–ethanol–water (50:50:30, by volume).
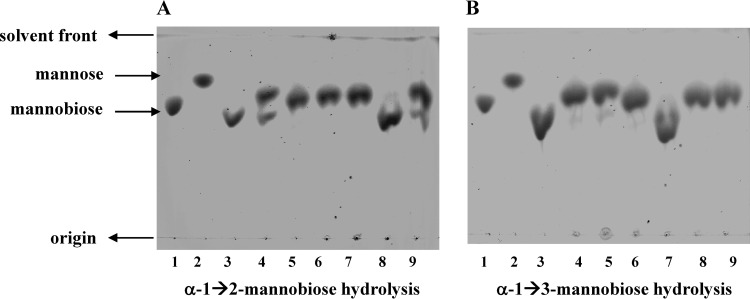
Fig 2.
Hydrolysis of α-1→2 and α-1→3 mannobioses by P. gingivalis W50 and single-isogenic mutant strains analyzed by TLC on Keiselgel F254 HPTLC plates in n-butanol–ethanol–water (5:5:3, by volume). The experiments were performed with P. gingivalis W50, ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, and ΔPG1712 as described in the legend to Fig. 1. (A) α-1→2 mannobiose. (B) α-1→3 mannobiose. Lanes: 1, mannobiose standard; 2, mannose standard; 3, W50, time 0; 4, W50, 21 h; 5, ΔPG0032, 21 h; 6, ΔPG0902, 21 h; 7, ΔPG1711, 21 h; 8, ΔPG1712, 21 h; 9, ΔPG0973, 21 h.
The hydrolysis of 4-nitrophenyl α-d-mannopyranoside by cell extracts of P. gingivalis W50 and isogenic single-mutant strains showed that the ΔPG0902 and ΔPG1712 strains had reduced activities of ∼20% and ∼80%, respectively, toward the chromogenic substrate compared to the parent W50 strain, whereas the ΔPG0032, ΔPG0973, and ΔPG1711 mutant strains had ∼100% activity compared to the parent strain (Table 4). This indicates that there are only two gene products from PG0902 and PG1712 that contribute ∼80% and ∼20% activity, respectively, to the hydrolysis of the chromogenic substrate 4-nitrophenyl α-d-mannopyranoside, and inactivation of PG0032, PG0973, and PG1711 has no influence on this activity. In the case of β-mannosidase activity, only ΔPG0032 showed complete loss of activity toward the aryl chromogenic substrate 4-nitrophenyl β-d-mannopyranoside, whereas all other single-isogenic mutant strains, ΔPG0902, ΔPG0973, ΔPG1711, and ΔPG1712, showed β-mannosidase activity identical to that of the parent W50 strain (data not shown). Hence, PG0032 is the principal β-mannosidase of this organism.
α-1→2-, α-1→3-, and α-1→6-d-mannobioses were used as substrates for the assay of cell extracts from P. gingivalis W50 and single-isogenic mutant strains, and the hydrolysis products were detected by TLC (Fig. 2). The ΔPG1712 mutant strain was not able to hydrolyze α-1→2-d-mannobiose, whereas all the other strains tested showed conversion of α-1→2-d-mannobiose to mannose (Fig. 2A). Hence, PG1712 is an α-1→2 mannosidase. Similarly, when α-1→3-d-mannobiose was used as the substrate in the assays, only ΔPG1711 was unable to hydrolyze the substrate to mannose (Fig. 2B). Therefore, PG1711 is an α-1→3 mannosidase. None of the P. gingivalis strains was able to convert α-1→6 mannobiose to mannose under the experimental conditions used (data not shown). Thus, it appears that the three genes PG0902, PG1711, and PG1712 account for all the α-d-mannosidase activity measured in P. gingivalis W50. PG0973 does not appear to be involved in the hydrolysis of any of the substrates tested. All these results are summarized in Table 4.
Arg-gingipain and Lys-gingipain activities.
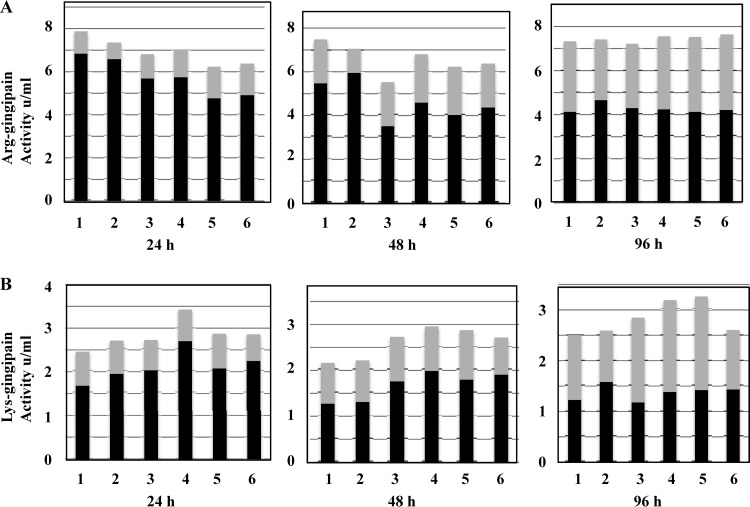
Arg-gingipan activities in whole cultures and culture supernatants of P. gingivalis W50 and ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, and ΔPG1712 single-isogenic mutant strains were measured over 4 days, and the results obtained were similar to those of the parent W50 strain after 4 days of growth (Fig. 3A). The Arg-gingpain activity in whole cultures varied between 6.2 U/ml (80%) and 7.4 U/ml (94%) for the mutant strains compared to 7.9 U/ml (100%) for the parent W50 strain after 24 h of growth. However, after 4 days of growth, the Arg-gingipain activities in all the strains were comparable to that of the parent W50 strain (Fig. 3A). Arg-gingipain activities in culture supernatants showed a range of ∼90% to ∼100% of the levels present in the parent W50 strain after 4 days (Fig. 3A). The levels and distribution of Arg gingipain activity between cell-bound and culture supernatants in P. gingivalis W50 are a very sensitive indicator of changes in the stability of the cell surface glycosylation status of the Arg gingipains and the biosynthesis of A-LPS in the organism (29). The data obtained here indicate that mutations in the mannosidase genes appear to have a minimal effect(s) on the growth and Arg-X protease activities of P. gingivalis.
Fig 3.
Arg-gingipain and Lys-gingipain activities in whole cultures and culture supernatants of P. gingivalis W50 and mutant strains. (A) Arg-gingipain activities were measured as described in Materials and Methods. The cultures were adjusted to the same OD600 with BHI broth prior to the assays. The results are expressed as activity (absorbance at 405 nm) units/ml of whole cultures or culture supernatants. (B) Lys-gingipain activities were measured as described in Materials and Methods. The cultures were adjusted to the same OD600 with BHI broth prior to the assays. The data are expressed as activity (absorbance at 405 nm) units/ml of whole cultures or culture supernatants. Bars: 1, W50; 2, ΔPG0032; 3, ΔPG0902; 4, ΔPG0973; 5, ΔPG1711; 6, ΔPG1712. The filled bars represent cell-bound activity, and the gray bars represent activity in culture supernatants. The sum of the two activities represents the Arg-gingipain/Lys-gingipain activity in whole cultures.
Lys-gingipain activities were measured in whole cultures and culture supernatants of P. gingivalis W50 and single-isogenic mutant strains (Fig. 3B) throughout the duration of the experiment (24 h to 96 h) and were approximately equal to (ΔPG0032 and ΔPG1712) or higher than (ΔPG0902, ΔPG0973, and ΔPG1711) the activities present in the parent W50 strain (Fig. 3B). Similarly, the Lys-gingipain activities in the culture supernatants were variable, with ΔPG0032 and ΔPG1712 showing between ∼80 and 90% of the levels present in the parent W50 strain and ΔPG0902, ΔPG0973, and ΔPG1711 showing higher levels (between ∼130% and 142%) of the levels in the parent strain after 96 h (Fig. 3B).
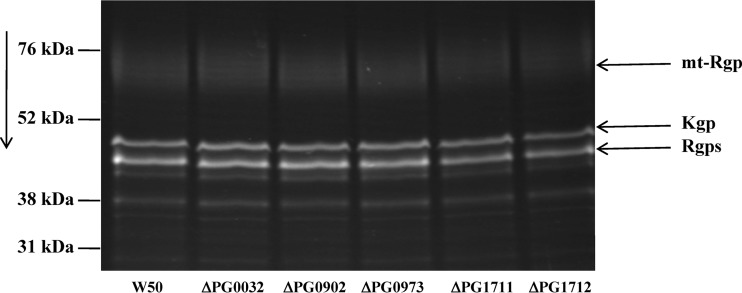
Any differences in the Arg- and Lys-gingipains present in the culture supernatants with respect to size or glycosylation status can be distinguished by SDS-PAGE of the enzymes in culture supernatants fluorescently labeled with the protease inhibitor DNS-EGR-chloromethylketone (Fig. 4) and SDS-PAGE of proteins in culture supernatants, followed by Western blotting versus MAb 1B5 (data not shown). Figure 4 shows the profiles of Arg-gingipains and Lys-gingipain present in the culture supernatants of 6-day cultures of P. gingivalis W50, ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, and ΔPG1712 strains labeled with DNS-EGR-chloromethylketone.
Fig 4.
SDS-PAGE of fluorescently labeled Arg- and Lys-gingipains in the culture supernatants of P. gingivalis W50, ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, and ΔPG1712 strains. Culture supernatants (500 μl) from 6-day-old cultures of P. gingivalis W50 and mutant strains were treated with 750 μl of ice-cold acetone and left at −20°C for 1 h to effect complete precipitation of proteins. The suspension was centrifuged in an Eppendorf centrifuge at 13,300 rpm at 4°C for 20 min, the supernatant was discarded, and the protein pellet was air dried. Arg-gingipains and Lys-gingipains in the pellets were labeled with DNS-EGR-chloromethylketone as described previously (27). The samples were subjected to SDS-PAGE in 12.5% acrylamide gels, viewed under fluorescent light, and photographed immediately. The direction of migration is indicated by the arrow. The positions of mt-Rgps, Kgp, and Rgps are indicated.
Culture supernatants from all the strains described above showed identical profiles of fluorescent bands on SDS-PAGE. The mt-Rgps, which appear as a fuzzy, diffuse band on SDS-PAGE due to the presence of 30 to 40% (by weight) carbohydrate, did not appear to differ in any of the mutant strains compared to the wild-type W50 strain. Similarly, no differences were found in Kgp and in the soluble Rgps (representing HRgpA, RgpAcat, and RgpB), which migrate as a single band under these conditions.
Characterization of EPS.
Spent culture medium (1.7 liters) from 24-h cultures of P. gingivalis W50 grown in BHI yielded 46 mg of EPS (27 mg/liter), which was neutral at pH 6.5 and did not show any cross-reactivity with MAb 1B5 [referred to here as EPS (W50)]. The structure of EPS (W50) was determined by NMR spectroscopy and methylation analysis.
Analysis of EPS (W50).
Monosaccharide analysis of EPS (W50) by methanolysis, followed by GC-MS of O-TMS ethers of methyl glycosides, showed Man to be the main component in both preparations, with very low levels of Gal and Glc, which may be contaminants.
Methylation analysis of EPS (W50) revealed the presence of terminal Man and 2-substituted, 3-substituted, 6-substituted, and 2,6-disubstituted Man in the ratio 8.4:3.13:1:1:4.56. The expected ratios are 4:2:1:1:4 for terminal Man and 2-substituted, 3-substituted, 6-substituted, and 2,6-disubstituted Man based on the integration of areas under the signals of anomeric protons in the 1H NMR spectra, which suggests that the lower-than-expected values for 2,6-disubstituted Man were due to the acid lability of 3,4-di-O-methylmannose (30) and the low values for 6-, 2-, and 3-substituted Man could be due to steric reasons.
NMR spectroscopy of EPS (W50).
The residues of the repeating units for EPS (W50) were labeled A to G in decreasing order of chemical shifts of their H-1 resonances. As judged from 1D 31P NMR analysis data, there were no phosphorylated glycosidic residues in EPS (W50). 1H and 13C NMR spectra for EPS (W50) showed anomeric signals for H-1 of α-linked residues at δH 5.30, 5.18, 5.13, 5.10, 5.08, 5.05, and 4.91 and seven anomeric signals at δC 101.76, 103.63, 99.59, 99.48, 103.35, 103.60, and 100.69, respectively (Table 5). Their assignment was achieved using 2D homonuclear or heteronuclear TOCSY, HMQC, and HSQC-TOCSY experiments (Table 5). The position of the resonances of the anomeric protons, together with the vicinal coupling constant (3JH,H) values, indicated α configurations of Man units.
Table 5.
1H NMRa and 13C NMR data for EPS (W50)
| Residue | δ (ppm) |
|||||
|---|---|---|---|---|---|---|
| H-1/C-1 | H-2/C-2 | H-3/C-3 | H-4/C-4 | H-5/C-5 | H-6, H-6′/C-6 | |
| Unit A | 5.30 | 4.14 | 3.95 | 3.74 | 3.74 | 3.91; 3.78 |
| →2)-Man | 101.76 | 79.54b | 71.72 | 68.13 | 74.48 | 62.30 |
| Unit B | 5.18 | 4.08 | 3.88 | 3.71 | 3.81 | 3.91; 3.78 |
| α-Man→3) | 103.63 | 70.96 | 72.20 | 68.12 | 74.60 | 62.30 |
| Unit C | 5.13 | 4.06 | 3.92 | 3.86 | 3.72 | 4.03; 3.80 |
| →2,6) α-Man | 99.59 | 79.82 | 71.63 | 67.78 | 74.51 | 66.99 |
| Unit D | 5.10 | 4.05 | 3.92 | 3.82 | 3.70 | 4.02; 3.80 |
| →2,6) α-Man | 99.48 | 79.82 | 71.68 | 67.75 | 72.58 | 66.99 |
| Units E and E1c | 5.08 | 4.10 | 3.86 | 3.69 | 3.76 | 3.91; 3.78 |
| α-Man→2) | 103.35 | 72.28 | 71.88 | 68.22 | 74.48 | 62.30 |
| Unit F | 5.05 | 4.23 | 3.98 | 3.71 | 3.81 | 3.91; 3.78 |
| →3)-α-Man | 103.60 | 70.89 | 79.25 | 68.12 | 74.60 | 62.30 |
| Unit G | 4.91 | 4.00 | 3.8–3.9 | 3.8–3.9 | 3.82 | 3.95; 3.75 |
| →6)-Man | 100.69 | 71.32 | 71–72 | 71–72 | 68.5 | 66.86 |
The coupling constants are not reported, but when measured were in agreement with the expected values.
The positions of substituted carbons are shown in boldface.
The chemical shifts for units E and E1 are given in the same row, as they are identical.
The linkage positions and sequences of the glycosidic residues in the repeating units of EPS (W50) were established on the basis of the analysis of 2D NMR experimental data, and the linkage sites of the residues in the repeating unit of EPS (W50) were identified by comparing the 13C NMR chemical shift data with reference values for methyl α-mannopyranosides (31). The significant deshielding of the resonances for C-2 of residue A at 79.54 ppm, C-2 and C-6 of residues C and D at 79.82 ppm and 66.99 ppm, and C-6 of residue G at 66.86 ppm identify these as linkage carbons. The signals for anomeric carbon residues at 103.63 ppm (B) and 103.35 ppm (E) could be assigned to two different types of terminal mannose residues, while those at 101.76 ppm, 100.69 ppm, and 99.48 to 99.59 ppm belonged to 2-substituted, 6-substituted, and 2,6-disubstituted mannose, respectively (32). The sequence of the residues in the repeating unit of EPS was established from 1H-1H NOESY and 1H-13C HSQC-NOESY NMR experiments (data not shown).
On the basis of the NMR spectral-analysis data, the repeating unit of EPS (W50) is built up from a dodecasaccharide and has at least a pentasaccharide backbone consisting of α-1→6-linked d-mannopyranose residues in such a manner that every fifth α-1→6-linked Man is not branched at C-2 and the side chains of EPS (W50) are made up of 2- or 3-substituted or unsubstituted mannose residues attached to the backbone at position 2. The results of methylation analysis and integration of areas under the signals of anomeric protons were in accordance with the proposed structure of EPS (W50). The lengths of the α-mannose-containing side chains were confirmed by MALDI-TOF MS of the products of acetolysis of EPS (W50) (3), which showed the presence of Man-disaccharide-, Man-trisaccharide-, and Man-tetrasaccharide-containing fragments (not shown).
The structure of the EPS (W50) isolated from spent culture medium is a highly branched PS constructed around an α-1,6-linked Man backbone, with side chains averaging 2 or 3 sugar residues in length containing α-1,2 and α-1,3 linkages attached to the backbone by α-1,2 linkages, and is almost identical to that of yeast mannan (33, 34).
The mannan component makes up ∼30 to 45% of the dry weight of the yeast cell wall, and since BHI broth contains yeast extract, it was important to establish whether the mannans isolated from spent culture medium and from BHI broth were similar. Therefore, we isolated the PS present in BHI broth using the same procedures used for isolating the EPS (W50) from spent culture medium. The yield (26 mg/liter), 1H NMR and 13C NMR data (Table 5), and methylation analysis and acetolysis data were identical to those obtained for EPS (W50) from spent culture medium (data not shown), indicating that the EPS described by Farquharson et al. (9) is likely to be derived from the yeast extract present in the culture medium.
The fact that the structure of the A-PS repeating units of A-LPS from P. gingivalis W50 bears a strong resemblance to the structure of the yeast mannan and that there is a preponderance of α-d-Man-linked residues in A-PS and in the outer-core OS of O-LPS raised the possibility that P. gingivalis could harness the yeast mannan present in the medium by using a combination of α-d-mannosidases to hydrolyze the mannan into Man or Man-containing oligosaccharides for incorporation into the repeating unit of A-PS and the outer-core OS.
The EPS (W50) from spent culture medium and PS (BHI) broth were subjected to digestion by P. gingivalis mannosidase as described below.
Digestion of EPS (W50) and PS (BHI) by P. gingivalis mannosidases.
EPS (W50) and PS (BHI) were incubated with P. gingvalis W50 whole-cell suspensions/cell extracts to determine whether they were digested to oligosaccharides. The weights of undigested EPS (W50)/PS (BHI) recovered from the reaction mixture (by gel filtration) after 0 h, 19 h, and 44 h of incubation were identical, suggesting that P. gingvalis W50 α-d-mannosidase(s) was unable to hydrolyze/release mannose and/or mannose-oligosaccharides from EPS (W50)/PS (BHI) under the experimental conditions tested. The reaction mixture was also subjected to TLC, and no mannose or short oligosaccharides were detected (data not shown).
Expression of α- and β-mannosidase activities in P. gingivalis W50.
In order to determine whether the α- and β-mannosidase activities were constitutively expressed in P. gingivalis W50, the organism was grown in a chemically defined medium (CDM) that contained mineral salts and bovine serum albumin (BSA) as the sole source of carbon and nitrogen (35) and also in CDM supplemented with 0.03% yeast mannan. Since BSA is not a glycoprotein, the levels of α- and β-mannosidase activities in P. gingivalis W50 grown in a medium devoid of added carbohydrate give a clear indication of the nature of expression of these enzyme activities. The Arg- and Lys-gingipain activities in whole cultures of P. gingivalis W50 grown in CDM were approximately 3% and 6%, respectively, of values obtained when P. gingivalis W50 was grown in BHI (data not shown). Since the concentration of BSA in the culture supernatant is very high, it is not possible to detect/label the low levels of Arg- and Lys-gingipains by labeling with DNS-EGR-chloromethylketone. Total LPS isolated from P. gingivalis W50 grown in CDM and subjected to SDS-urea-PAGE followed by silver staining shows a laddering pattern similar to that of W50 grown in BHI, indicating the presence of O-LPS, and shows cross-reactivity with MAb 1B5 on Western blotting (data not shown).
P. gingivalis W50 grown in CDM contains only ∼25% as much α-mannosidase activity (using 4-nitrophenyl α-d-mannopyranoside as the substrate) as W50 grown in BHI broth and higher β-mannosidase activity (138%, using 4-nitrophenyl β-d-mannopyranoside as the substrate). P. gingivalis W50 grown in CDM supplemented with yeast mannan contains the same low level of α-mannosidase activity (∼25%) as the W50 strain grown in BHI medium, and the β-mannosidase activity is slightly higher (127%).
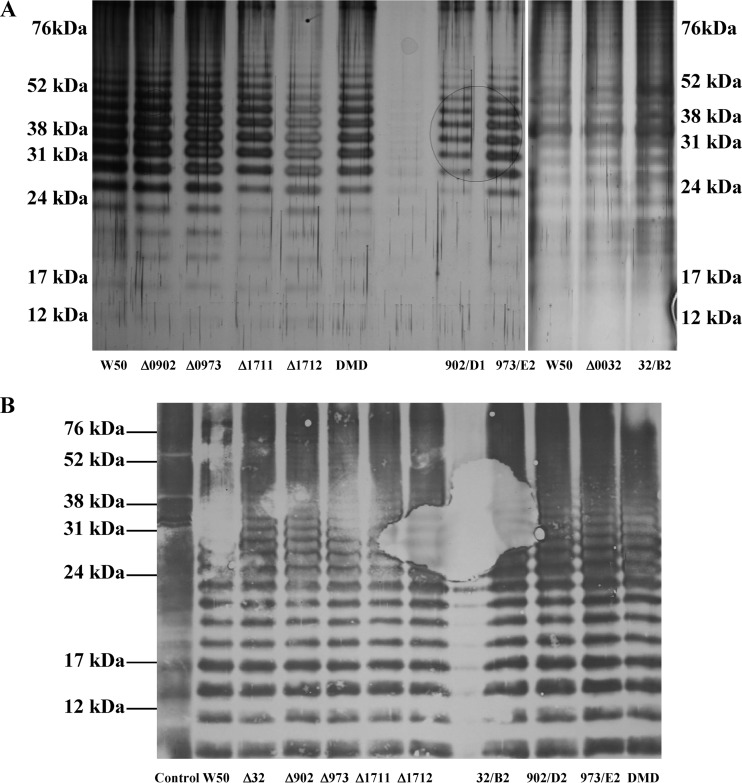
LPS from P. gingivalis W50 and isogenic mutant strains.
LPSs isolated from P. gingivalis W50 and mannosidase mutant strains were subjected to SDS-urea-PAGE and stained with silver, and also subjected to Western blotting and probed with MAb 1B5. In all the strains tested, i.e., single-, double-, and triple-isogenic mutant strains, ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, ΔPG1712, DMD, PG0032B2, PG0902D1, and PG0973E2, the LPS showed a laddering pattern identical to that of the parent W50 strain (Fig. 5A). SDS-urea-PAGE and Western blotting versus MAb 1B5 of LPS isolated from P. gingivalis W50, ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, ΔPG1712, DMD, PG0032B2, PG0902D1, and PG0973E2 demonstrated that all the LPSs showed cross-reactivity, indicating that A-LPS was being synthesized by all the isogenic mutant strains of P. gingivalis (Fig. 5B).
Fig 5.
SDS-urea-PAGE, followed by silver staining and Western blotting versus MAb 1B5 of LPSs isolated from P. gingivalis W50 and single-, double-, and triple-isogenic mannosidase mutant strains. LPSs from P. gingivalis W50 and single-isogenic mutant strains ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, and ΔPG1712; double-isogenic mutant strain DMD; and triple-isogenic mutant strains 32/B2, 902/D2, and 973/E2 were isolated using the LPS isolation kit from Intron Biotechnology (South Korea). Aliquots containing 10 μg of LPS were subjected to SDS-urea-PAGE (28). (A) Silver staining was performed using the kit from Sigma Chemical Company, Poole, Dorset, United Kingdom. (B) Western blotting versus MAb 1B5 was performed as described previously (4). Δ32, ΔPG0032; Δ902, ΔPG0902; Δ973, ΔPG0973; Δ1711, ΔPG1711; Δ1712, ΔPG1712.
DISCUSSION
Mannose is a major constituent of the surface polysaccharides and glycoproteins of P. gingivalis (3, 4, 8, 27). The EPS described by Farquharson et al. (9), which is rich in mannose residues, has been established in this study to be identical to the yeast cell wall mannan (33, 34) present in brain heart infusion broth used for culturing P. gingivalis.
Five mannosidase genes (PG0032, PG0902, PG0973, PG1711, and PG1712) are present in P. gingivalis, and none of the enzymes has been characterized. This led us to hypothesize that the mannosidases are involved in Man acquisition for biosynthetic purposes. PG0032 is a hydrophilic protein with a putative signal peptide (25 amino acids long) and, based on conserved domains, belongs to β-glycoside hydrolase family 2 (10), typical members of which are LacZ (β-galactosidase)/β-glucuronidase enzymes, and has 22% identity (39% similarity) to PG0665 (β-galactosidase) of P. gingivalis (residues 196 to 477). The highest amino acid sequence homology is to proteins belonging to the phylum Bacteroidetes, including Bacteroides fragilis, Bacteroides ovatus, and Bacteroides thetaiotaomicron, as well as proteins from Xanthomonas spp. PG0902 (CSL Ltd.; accession number AAD51075) and PG0973 (CSL Ltd.; accession number AAD51077) are classified as putative α-1,2-mannosidases based on conserved domain architecture, are similar in hydrophobicity profile and organization to PG0032, and belong to glycoside hydrolase family 92. Primary structure homology is highest in Bacteroides spp. Unlike PG0902, the SIGNALP algorithm acknowledges a signal peptide (1 to 22 amino acids) in PG0973.
PG1711 is also annotated as an α-1,2-mannosidase (glycoside hydrolase family 92) and has counterparts in other bacteria, including members of the Bacteroides and Xanthomonas groups (10). PG1712 (CSL Ltd.; accession number AAD51077), which is relatively hydrophilic in spite of the presence of a transmembrane domain at the N terminus, is annotated as an immunoreactive 89-kDa antigen PG87 and exhibits extensive sequence similarity to proteins from the Bacteroides group (74% similarity to B. fragilis over 569 amino acids), and it is also classed as an α-1,2-mannosidase belonging to glycosidic hydrolase family 92. At the nucleotide level, PG1711and PG1712 are separated by a 69-bp intergenic region but could still form an operon. The overall identity between PG1711 and PG1712 is 25.7%. In addition, PG0902 shows amino acid similarity of 29% to PG0973 (residues 11 to 725), 26% similarity to PG1711 (residues 14 to 740), and 32% similarity to PG1712 (residues 30 to 738).
Homer et al. (36) reported the presence of a β-mannosidase in P. gingivalis, which was inhibited by plant extracts used for the control of oral hygiene in Kenya. Other β-mannosidases from Gram-negative bacteria include B. thetaiotaomicron exo-β-mannosidase (BtMan2A), described by Tailford et al. (37), which is a classical exoenzyme and is very active against aryl-mannosides, namely, 2,4-dinitrophenyl β-d-mannopyranoside and 4-nitrophenyl β-d-mannopyranoside. Mannose-OSs, namely, biose, triose, tetraose, and pentaose, were hydrolyzed to mannose and oligosacchariden−1, and Man-β1,4-GlcNAc was hydrolyzed to Man at a rate similar to that of mannobiose.
We have established in this work that PG0032 is the principal β-mannosidase in P. gingivalis W50. Although we have not tested the activity of P. gingivalis W50 β-mannosidase against β-mannose–OS, it is possible that it is an exo-β-mannosidase.
There are only a few reports in the literature on the occurrence of α-mannosidases in Gram-negative bacteria. Thompson et al. (38) described the 3D structures and mechanism of N-glycan processing by endo-α-mannosidases from B. thetaiotaomicron and Bacteroides xylanisolvens. These endomannosidases are classified in the Carbohydrate-Active Enzymes Database family GH99 (www.cazy.org) and hydrolyze the α-1,2-mannosidic bond between the glucose-substituted mannose and the remainder of the N-glycan and act on structures of Glc1-3Man9GlcNAc2, as well as structures that have been trimmed by ER-mannosidases ERM1 and ERM2 in the 6′-pentamannosyl branch, releasing Glc1-3-1,3-α-Man oligosaccharides (38). Endo-α-mannosidases are membrane-associated proteins, whereas the endo-α-mannosidases from B. thetaiotaomicron and B. xylanisolvens are soluble proteins and may have been acquired by horizontal gene transfer, because these organisms are common and beneficial components of the human gut (39). The role of endomannosidases in B. thetaiotaomicron and B. xylanisolvens is unclear but may include mannose foraging for basic metabolic needs. Since incomplete deglucosylation of the N-glycan Glc1-3Man9GlcNAc2 in the mammalian cell prevents the action of exo-α-mannosidases, a similar problem with the bacterial exo-α-mannosidases might have led to the beneficial acquisition of the enzyme by the bacteria.
We have established in this study that the α-mannosidase activity toward the aryl-chromogenic substrate 4-nitrophenyl α-d-mannopyranoside in P. gingivalis is due to PG0902 (80% of the total) and PG1712 (20% of total). We have shown that PG1711 is an α-1→3 mannosidase and that PG1712 also possesses α-1→2 mannosidase activity (Table 4 and Fig. 2), in addition to 20% of the activity toward 4-nitrophenyl α-d-mannopyranoside (see above). Inactivation of PG0973 did not affect the mannosidase activity of any of the substrates tested, and thus, no mannosidase activity could be ascribed to PG0973.
Based on the analysis of total LPS from P. gingivalis W50 and the ΔPG0032, ΔPG0902, ΔPG0973, ΔPG1711, and ΔPG1712 isogenic mutant strains, there is no direct link between the biosynthesis of O-LPS and A-LPS and the α- and β-mannosidases of P. gingivalis. Since the yields of EPS (yeast mannan) isolated from spent culture medium and BHI broth were identical and the mannosidases were unable to digest this PS into Man or Man-oligosaccharides, these enzymes play no role in the acquisition of Man from the medium for the biosynthesis of A-LPS and the core region of the LPS of P. gingivalis.
In several bacteria, glycosidases appear to be induced when the bacteria are grown on specific carbohydrates as the sole carbon source. In B. ovatus, higher levels of β-mannanases and α-galactosidases are produced when the organism is grown on guar gum (40). The marine organism Vibrio sp. strain MA-138 secretes multiple β-mannanases into the growth medium in the presence of an inducer, such as β-mannan, konjac powder, or mannose (41). Sampaio et al. (42) reported the presence of an α-mannosidase encoded by mngB in Escherichia coli K-12 grown on 2-O-α-mannosyl-d-glycerate as the sole carbon source. It was thought that mngB was part of a gene cluster comprising mngRAB, encoding a 2-O-α-mannosyl-d-glycerate-utilizing system.
The level of α-mannosidase activity in P. gingivalis W50 grown in CDM containing BSA (which is nonglycosylated) as the sole carbon and nitrogen source, before and after being supplemented with 0.03% yeast mannan, was ∼25%, and β-mannosidase activity was slightly higher than the values in P. gingivalis W50 grown in BHI broth. Thus, the α-mannosidases of P. gingivalis W50 are not induced by the presence of α-mannan in the growth medium. The β-mannosidase activity of P. gingivalis W50 grown in CDM was slightly higher than the values present in BHI-grown cells, suggesting that the enzyme is expressed constitutively.
The study of the mannosidases of P. gingivalis W50 allows us to conclude that they are not able to forage for mannose from the yeast mannan present in the growth medium, and they do not appear to have an effect on the biosynthesis of O-LPS and A-LPS or the Arg-gingipains of the organism. Thus, the roles of the one β-mannosidase and three α-mannosidases identified in this study in the biology of P. gingivalis remain to be determined.
ACKNOWLEDGMENTS
This investigation was supported by Medical Research Council (United Kingdom) grant G0501478, by Research Advisory Board of the Barts and The London Charity grant RAB06/PJ/14, and by the Wellcome Trust Value in People (VIP) Scheme.
Footnotes
Published ahead of print 20 September 2013
REFERENCES
- 1.Curtis MA, Aduse-Opoku J, Rangarajan M. 2001. Cysteine proteases of Porphyromonas gingivalis. Crit. Rev. Oral Biol. Med. 12:192–216 [DOI] [PubMed] [Google Scholar]
- 2.Paramonov N, Bailey D, Rangarajan M, Hashim A, Kelly G, Curtis MA, Hounsell EF. 2001. Structural analysis of the polysaccharide from the lipopolysaccharide of Porphyromonas gingivalis strain W50. Eur. J. Biochem. 268:4698–4707 [DOI] [PubMed] [Google Scholar]
- 3.Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, Hounsell E, Curtis MA. 2005. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol. Microbiol. 58:847–863 [DOI] [PubMed] [Google Scholar]
- 4.Rangarajan M, Aduse-Opoku J, Paramonov N, Hashim A, Bostanci N, Fraser OP, Tarelli E, Curtis MA. 2008. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 190:2920–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aduse-Opoku J, Muir J, Slaney JM, Rangarajan M, Curtis MA. 1995. Characterization, genetic analysis, and expression of a protease antigen (PrpRI) of Porphyromonas gingivalis W50. Infect. Immun. 63:4744–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher A, Aduse-Opoku J, Rangarajan M, Slaney JM, Curtis MA. 2003. Glycosylation of the Arg-gingipains of Porphyromonas gingivalis and comparison with glycoconjugate structure and synthesis in other bacteria. Curr. Protein Pept. Sci. 4:427–441 [DOI] [PubMed] [Google Scholar]
- 7.Curtis MA, Thickett A, Slaney JM, Rangarajan M, Aduse-Opoku J, Shepherd P, Paramonov N, Hounsell EF. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paramonov NA, Aduse-Opoku J, Hashim A, Rangarajan M, Curtis MA. 2009. Structural analysis of the core region of O-lipopolysaccharide of Porphyromonas gingivalis from mutants defective in O-antigen ligase and O-antigen polymerase. J. Bacteriol. 191:5272–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farquharson SI, Germaine GR, Gray GR. 2000. Isolation and characterization of the cell-surface polysaccharides of Porphyromonas gingivalis ATCC 53978. Oral Microbiol. Immunol. 15:151–157 [DOI] [PubMed] [Google Scholar]
- 10.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher HM, Schenkein HA, Morgan RM, Bailey KA, Berry CR, Macrina FL. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aduse-Opoku J, Slaney JM, Hashim A, Gallagher A, Gallagher RP, Rangarajan M, Boutaga K, Laine ML, Van Winkelhoff AJ, Curtis MA. 2006. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect. Immun. 74:449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, Feldman MF. 2011. Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 286:1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haake SK, Yoder SC, Attarian G, Podkaminer K. 2000. Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J. Bacteriol. 182:1176–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aduse-Opoku J, Davies NN, Gallagher A, Hashim A, Evans HE, Rangarajan M, Slaney JM, Curtis MA. 2000. Generation of Lys-gingipain protease activity in Porphyromonas gingivalis W50 is independent of Arg-gingipain protease activities. Microbiology 146:1933–1940 [DOI] [PubMed] [Google Scholar]
- 16.Maley J, Shoemaker NB, Roberts IS. 1992. The introduction of colonic-Bacteroides shuttle plasmids into Porphyromonas gingivalis: identification of a putative P. gingivalis insertion-sequence element. FEMS Microbiol. Lett. 72:75–81 [DOI] [PubMed] [Google Scholar]
- 17.Rangarajan M, Smith SJ, U S, Curtis MA. 1997. Biochemical characterization of the arginine-specific proteases of Porphyromonas gingivalis W50 suggests a common precursor. Biochem. J. 323:701–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bax A, Davis DG. 1985. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65:355–360 [Google Scholar]
- 19.Wagner G, Wüthrich K. 1982. Sequential resonance assignments in protein 1H nuclear magnetic resonance spectra. Basic pancreatic trypsin inhibitor. J. Mol. Biol. 155:347–366 [DOI] [PubMed] [Google Scholar]
- 20.Bax A, Griffey RH, Hawkins BL. 1983. Correlation of proton and N-15 chemical-shifts by multiple quantum NMR. J. Magn. Reson. 55:301–315 [Google Scholar]
- 21.Bax A, Subramanian S. 1986. Sensitivity-enhanced two-dimentional heteronuclear shift correlation NMR-spectroscopy. J. Magn. Reson. 67:565–569 [Google Scholar]
- 22.Altman E, Perry MB, Brisson JR. 1989. Structure of the lipopolysaccharide antigenic O-chain produced by Actinobacillus pleuropneumoniae serotype 4 (ATCC 33 378). Carbohydr. Res. 191:295–303 [DOI] [PubMed] [Google Scholar]
- 23.Kakehi K, Honda S. 1989. Analysis of carbohydrates by GLC and MS. CRC Press, Boca Raton, FL [Google Scholar]
- 24.Kvernheim A. 1987. Methylation analysis of polysaccharides with butyllithium in dimethylsulphoxide. Acta Chem. Scand. B 41:150–152 [Google Scholar]
- 25.Gerwig G, Kamerling J, Vliegnthart J. 1978. Determination of D and L configuration of neutral monosaccharides by high-resolution capillary GLC. Carbohydr. Res. 62:349–357 [Google Scholar]
- 26.Kocourek J, Ballou CE. 1969. Method for fingerprinting yeast cell wall mannans. J. Bacteriol. 100:1175–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rangarajan M, Hashim A, Aduse-Opoku J, Paramonov N, Hounsell EF, Curtis MA. 2005. Expression of Arg-gingipain RgpB is required for correct glycosylation and stability of monomeric Arg-gingipain RgpA from Porphyromonas gingivalis W50. Infect. Immun. 73:4864–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inzana T, Apicella M. 1999. Use of a bilayer stacking gel to improve resolution of lipopolysaccharides and lipooligosaccharides in polyacrylamide gels. Electrophoresis 20:462–465 [DOI] [PubMed] [Google Scholar]
- 29.Shoji M, Ratnayake DB, Shi Y, Kadowaki T, Yamamoto K, Yoshimura F, Akamine A, Curtis MA, Nakayama K. 2002. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148:1183–1191 [DOI] [PubMed] [Google Scholar]
- 30.Barreto-Berter ME, Travassos RL, Gorin PAJ. 1980. Chemical structure of the d-galacto-d-mannan component from hyphae of Aspergillus niger and other Aspergillus spp. Carbohydr. Res. 86:273–285 [Google Scholar]
- 31.Bock K, Thøgersen H. 1983. Nuclear magnetic resonance spectroscopy in the study of mono- and oligosaccharides, p 1–57 In Webb GA. (ed), Annual reports on NMR spectroscopy, vol 13 Academic Press, London, United Kingdom [Google Scholar]
- 32.Kogan G, Pavliak V, Masler L. 1988. Structural studies of mannans from the cell walls of the pathogenic yeasts Candida albicans serotype A and serotype B and Candidia parapsilosis. Carbohydr. Res. 172:243–253 [DOI] [PubMed] [Google Scholar]
- 33.Jones GH, Ballou CE. 1968. Isolation of an alpha-mannosidase which hydrolyzes yeast mannan. Structure of the backbone of yeast mannan. J. Biol. Chem. 243:2442–2446 [PubMed] [Google Scholar]
- 34.Vinogradov E, Petersen B, Bock K. 1998. Structural analysis of the intact polysaccharide mannan from Saccharomyces cerevisiae yeast using H-1 and C-13 NMR spectroscopy at 750 MHz. Carbohydr. Res. 307:177–183 [DOI] [PubMed] [Google Scholar]
- 35.Milner P, Batten JE, Curtis MA. 1996. Development of a simple chemically defined medium for Porphyromonas gingivalis: requirement for alpha-ketoglutarate. FEMS Microbiol. Lett. 140:125–130 [DOI] [PubMed] [Google Scholar]
- 36.Homer KA, Manji F, Beighton D. 1992. Inhibition of peptidase and glycosidase activities of Porphyromonas gingivalis, Bacteroides intermedius and Treponema denticola by plant extracts. J. Clin. Periodontol. 19:305–310 [DOI] [PubMed] [Google Scholar]
- 37.Tailford LE, Money VA, Smith NL, Dumon C, Davies GJ, Gilbert HJ. 2007. Mannose foraging by Bacteroides thetaiotaomicron: structure and specificity of the beta-mannosidase, BtMan2A. J. Biol. Chem. 282:11291–11299 [DOI] [PubMed] [Google Scholar]
- 38.Thompson AJ, Williams RJ, Hakki Z, Alonzi DS, Wennekes T, Gloster TM, Songsrirote K, Thomas-Oates JE, Wrodnigg TM, Spreitz J, Stütz AE, Butters TD, Williams SJ, Davies GJ. 2012. Structural and mechanistic insight into N-glycan processing by endo-α-mannosidase. Proc. Natl. Acad. Sci. U. S. A. 109:781–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD, MetaHIT Consortium 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gherardini FC, Salyers AA. 1987. Characterization of an outer membrane mannanase from Bacteroides ovatus. J. Bacteriol. 169:2031–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamaru Y, Araki T, Amagoi H, Mori H, Morishita T. 1995. Purification and characterization of an extracellular beta-1,4-mannanase from a marine bacterium, Vibrio sp. strain MA-138. Appl. Environ. Microbiol. 61:4454–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sampaio MM, Chevance F, Dippel R, Eppler T, Schlegel A, Boos W, Lu YJ, Rock CO. 2004. Phosphotransferase-mediated transport of the osmolyte 2-O-alpha-mannosyl-d-glycerate in Escherichia coli occurs by the product of the mngA (hrsA) gene and is regulated by the mngR (farR) gene product acting as repressor. J. Biol. Chem. 279:5537–5548 [DOI] [PubMed] [Google Scholar]