Abstract
In a previous study, we reported that the OmcB protein from Chlamydia pneumoniae mediates adhesion of the infectious elementary body to human HEp-2 cells by interacting with heparin/heparan sulfate-like glycosaminoglycans (GAGs) via basic amino acids located in the first of a pair of XBBXBX heparin-binding motifs (K. Moelleken and J. H. Hegemann, Mol. Microbiol. 67:403–419, 2008). In the present study, we show that the basic amino acid at position 57 (arginine) in the first XBBXBX motif, the basic amino acid at position 61 (arginine) in the second motif, and another amino acid (lysine 69) C terminal to it play key roles in the interaction. In addition, we show that discrimination between heparin-dependent and -independent adhesion by C. trachomatis OmcBs is entirely dependent on three variable amino acids in the so-called variable domain C terminal to the conserved XBBXBX motif. Here, the predicted conformational change in the secondary structure induced by the proline at position 66 seems to be crucial for heparin recognition. Finally, we performed neutralization experiments using different anti-heparan sulfate antibodies to gain insight into the nature of the GAGs recognized by OmcB. The results suggest that C. trachomatis serovar L2 OmcB interacts with 6-O-sulfated domains of heparan sulfate, while C. pneumoniae OmcB apparently interacts with domains of heparan sulfate harboring a diverse subset of O-sulfations.
INTRODUCTION
Chlamydiae are Gram-negative bacteria that cause widespread infections in humans, other mammals, and birds. In humans, Chlamydia pneumoniae infects the respiratory tract, causing pharyngitis, sinusitis, bronchitis, and pneumonia (1–3). In addition, C. pneumoniae has been implicated in a variety of chronic diseases, including asthma, chronic obstructive pulmonary disease (COPD), atherosclerosis, multiple sclerosis (MS), and Alzheimer′s disease (AD) (2). Infections caused by C. trachomatis, on the other hand, make it the most common cause of preventable blindness (serovars A to C) and a leading cause of sexually transmitted diseases worldwide (serovars D to K and LGV serovars) (4, 5). Chlamydiae are obligate intracellular pathogens that have a unique biphasic life cycle involving alteration between two developmental forms (6).
The initial attachment of elementary bodies (EBs) to the target cell appears to be reversible (7), and numerous studies have pointed to the involvement of glycosaminoglycans (GAGs) as the host cell ligands recognized in this step (8–13). Thus, pretreatment of infectious EBs with either heparan sulfate or heparin inhibits attachment and reduces the infectivity of C. pneumoniae and the B and LGV serovars of C. trachomatis (14–22).
Heparan sulfate/heparin is one of the four major classes of GAGs (together with the chondroitin sulfate/dermatan sulfate, keratan sulfate, and hyaluronan families). GAGs are linear, acidic polysaccharides of highly complex structure, largely due to variable modification of their disaccharide subunits, which undergo N- and O-sulfation, acetylation, and carboxylation (23). With 2.7 sulfate groups per disaccharide on average, heparin is one of the most negatively charged macromolecules known (24). Heparan sulfate is similar in structure to heparin but is less highly sulfated (<1 sulfate group per disaccharide). However, it comprises highly modified domains very similar to heparin and domains with almost no modifications (25, 26). Due to the flexibility of its nonmodified regions, heparan sulfate can interact via its sulfated regions with basic domains in proteins (25, 27). Comparison of heparin-binding domains has highlighted motifs with the consensus sequences XBBXBX and XBBBXXBX, where B indicates a basic amino acid (28).
OmcB, a 60-kDa, cysteine-rich protein, was conclusively identified to interact with GAGs by heparin affinity chromatography (10). Strikingly, all the OmcB proteins of Chlamydiae possess at least one XBBXBX sequence in their N-terminal domains, and synthetic peptides corresponding to N-terminal fragments of C. trachomatis and C. pneumoniae OmcB, including the XBBXBX motif, were shown to interact with heparin (10). The OmcB protein is largely conserved among chlamydial species (70 to 80% overall identity) but can be divided into a highly conserved C-terminal portion (≥80% identity) and a species-specific N-terminal part showing pairwise identities of 19 to 93% (Fig. 1). Originally, OmcB was described as a periplasmic protein in the C. psittaci 6BC strain (29). In the same year, analysis of OmcB from the C. psittaci GPIC strain suggested that this protein is accessible at the cell surface of EBs and involved in the adhesion to the host cell (8). In accordance with the latter findings, subsequent antibody binding and proteolytic cleavage studies have shown that the N-terminal portion of OmcB in all other species and strains tested is exposed on the surface of EBs (9–13).
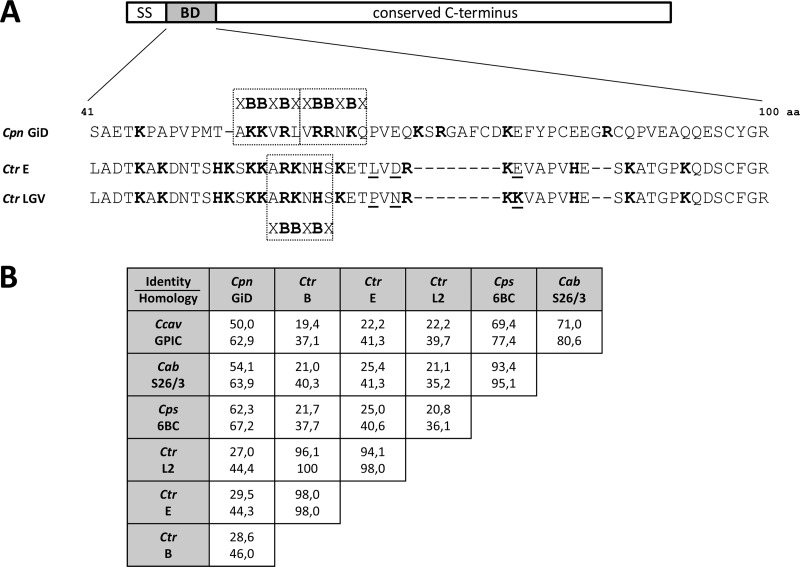
Fig 1.
Variability within the N-terminal heparin-binding domain (BD) of OmcB. (A) Amino acid sequence alignment of the binding domains (positions 41 to 100) of OmcB proteins from C. pneumoniae (Cpn) strain GiD and the C. trachomatis (Ctr) serovar E and the LGV serovars. Basic amino acids are highlighted in boldface, and heparin-binding motifs (XBBXBX) are boxed. The three amino acid differences between the binding domains of OmcB proteins from C. trachomatis E and LGV are underlined. SS, signal sequence. (B) Comparison of the levels of sequence identity (upper values) and homology (lower values) within the binding domains of OmcB proteins from the chlamydial species C. pneumoniae (Cpn) strain GiD; C. trachomatis (Ctr) serovars B (NCBI accession no. YP_002888946), E (P23603), and L2 (P0DJI2); C. psittaci (Cps) strain 6BC (AAB61619); C. abortus (Cab) strain S26/3 (YP_219610); and C. caviae (Ccav) strain GPIC (NP_829058). Alignments were performed with EMBOSS Needle.
Adhesion of C. pneumoniae and C. trachomatis LGV serovars to host cells is dependent on GAGs like heparin and heparan sulfate (14, 16, 18–20, 22, 30, 31), and it is mediated by a region in the N-terminal portion of the OmcB, which is termed the OmcB binding domain (OmcB-BD) (13). LGV serovars have a single XBBXBX motif in this domain, while C. pneumoniae OmcB is unique among chlamydial species in having two tandemly linked copies of the heparin-binding motif in this domain (Fig. 2A). All three of the basic amino acids within the first of these contribute to the adhesive capacity of C. pneumoniae OmcB (13), but the role of the second motif has remained unclear.
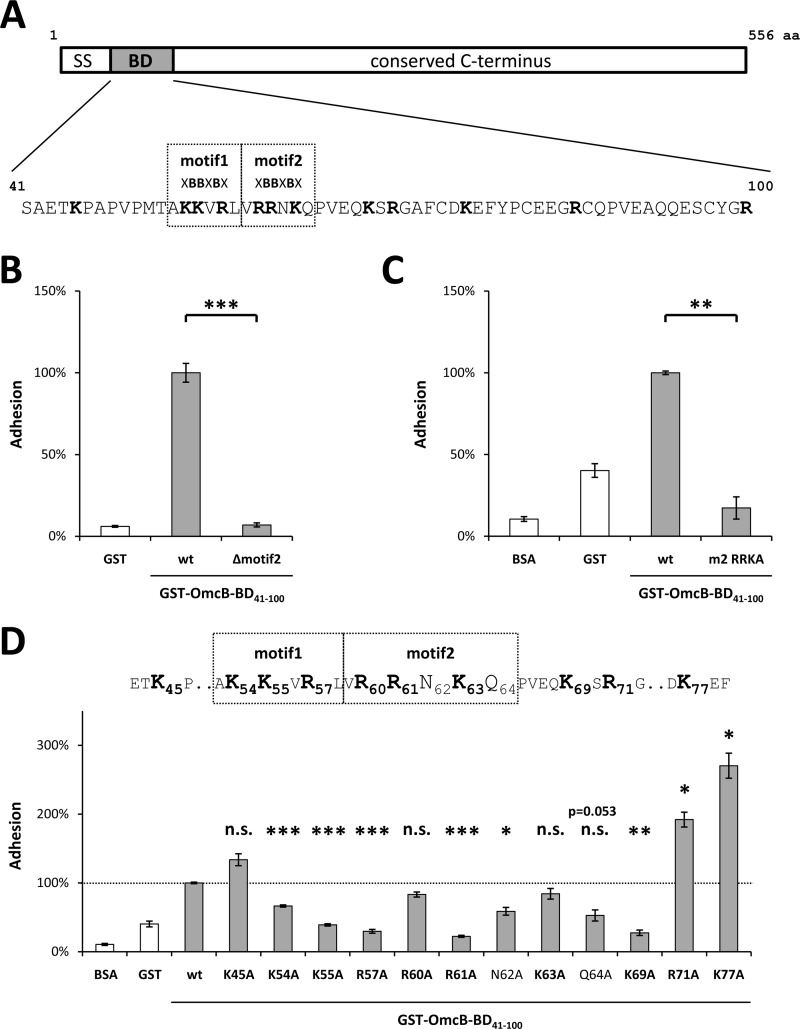
Fig 2.
Single amino acids within the binding domain (BD) differ in their impact on the binding properties of C. pneumoniae OmcB. (A) Schematic depiction of C. pneumoniae OmcB, together with the sequence of the binding domain (amino acids 41 to 100). Heparin-binding motifs (XBBXBX) are boxed, basic amino acids are in boldface. SS, signal sequence. (B) Adhesion of protein-coated latex beads to human HEp-2 cells. A total of 1 × 106 HEp-2 cells were incubated with 1 × 107 beads coated with either recombinant GST (rGST) or rGST-tagged wild-type OmcB-BD (wt) or rGST-tagged OmcB-BD lacking the second heparin-binding motif (Δmotif2). The number of beads bound by 1,000 HEp-2 cells was determined microscopically. Binding of wt OmcB-BD was set to 100% adhesion (n = 4). (C and D) Adhesion of protein-coated, fluorescent latex beads to human HEp-2 cells. A total of 1 × 106 HEp-2 cells were incubated with 1 × 107 fluorescent beads. Adhesion was quantified by measuring the overall fluorescence of attached beads per 1 × 105 HEp-2 cells by flow cytometry. Binding of wt OmcB-BD was set to 100% adhesion. (C) Beads were coated with either rGST, BSA, rGST-tagged wild-type OmcB-BD (wt), or an rGST-tagged variant of OmcB-BD in which all three basic amino acids in the second heparin-binding motif were replaced by alanines (m2 RRKA). (n = 3). (D) Beads were coated with either rGST, BSA, or rGST-tagged wild-type OmcB-BD (wt) or with rGST-tagged OmcB-BD variants in which single amino acids were replaced by an alanine residue as indicated. Heparin-binding motifs (XBBXBX) are boxed, and basic amino acids are in boldface (n = 3). Statistically significant differences are denoted by ***, **, and *, which indicate P < 0.001, P < 0.01, and P < 0.05, respectively. n.s., not significant.
As the entire OmcB GAG binding domain is enriched for basic amino acids, residues outside the XBBXBX motifs could also play a role in ligand recognition. Interestingly, attachment of C. trachomatis serovar E still occurs at wild-type (wt) levels after heparan sulfate treatment or in the absence of heparan sulfate-like GAGs on the human cell (18, 19, 31). Indeed, while the OmcB proteins of C. trachomatis serovars E and LGV differ from each other in just 11 out of 548 positions, none of the variable sites is located within the conserved heparin-binding motif (Fig. 3A). However, three of them lie immediately C terminal to the single heparin-binding motif (amino acid [aa] 66 to 71; here referred to as the variable domain), and the residues at these positions very likely determine the serovar-specific difference in the heparin dependence of adhesion seen between OmcB proteins from the LGV (OmcB-L) and E (OmcB-E) serovars (12, 13, 32). Indeed, it has previously been shown that exchange of the variable amino acid at position 66 abrogates the heparin dependency of adhesion by serovar L1 OmcB and, conversely, induces partial heparin dependency of cell binding by serovar E OmcB (13). The contribution of the other two variable amino acids within the variable region to GAG binding was not tested.
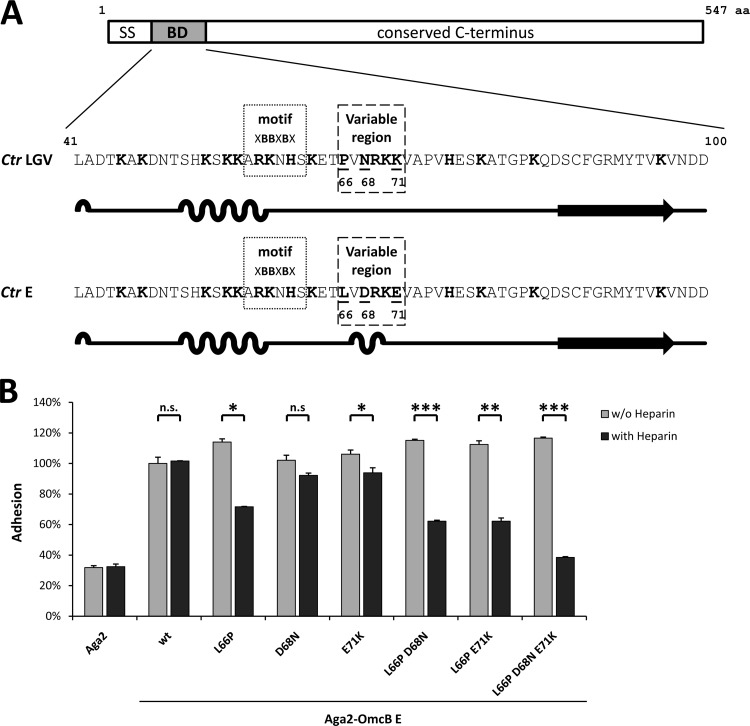
Fig 3.
All three divergent residues in the variable region of the binding domain of C. trachomatis OmcB contribute to serovar-specific differences in heparin dependency of adhesion. (A) Schematic illustration of the OmcB proteins of C. trachomatis LGV serovars and serovar E showing the sequences and predicted secondary structures of their binding domains (BD; amino acids 41 to 100). The heparin-binding motif is highlighted (XBBXBX; dotted box), and basic amino acids are depicted in boldface. The variable region is indicated by a dashed box, and residues that differentiate the sequences are underlined. Secondary structure was predicted using GOR IV. Loops indicate alpha-helical segments, lines indicate random-coil regions, and arrows represent extended strands. SS, signal sequence. (B) Adhesion of protein-presenting yeast cells to human HEp-2 cells. A total of 5 × 105 HEp-2 cells were incubated with a 2-fold excess of yeast cells displaying either Aga2, Aga2-OmcB serovar E wild type (wt), or Aga2-OmcB-E variants in which the native amino acids at positions 66, 68, and 71 were replaced by the corresponding amino acids found in OmcB from LGV serovars (see panel A). Thus, the OmcB mutant L66P D68N E71K is identical to the OmcB-BD from LGV serovars. Yeast cells were pretreated either with PBS (without heparin) or with 500 μg/ml soluble heparin (with heparin). Adhesion was quantified by counting the numbers of yeast cells attached to HEp-2 cells in 10 fields viewed under the microscope. Binding of Aga2-OmcB-E was set to 100% adhesion (n = 2). Statistically significant differences are denoted by ***, **, and *, which indicate P < 0.001, P < 0.01, and P < 0.05, respectively. n.s., not significant.
In this study we show that, in C. pneumoniae, three basic amino acids within OmcB-BD are the key players in GAG binding. Interestingly, one of these is located C terminal to the two heparin-binding motifs. Second, we ruled out the possibility that all of the variant positions within the variable domain of C. trachomatis OmcB contribute to the heparin-dependent/-independent nature of the adhesion process. Finally, using various antibodies that recognize heparan sulfates with different modifications, we confirmed that the presence of heparan sulfate on the host cell surface is a prerequisite for infection by C. pneumoniae and C. trachomatis serovar L2.
MATERIALS AND METHODS
Bacterial strains, yeast strains, and cell culture.
The Escherichia coli strain XL1-Blue (Stratagene) was used for plasmid amplification, while the Rosetta strain (Novagen) was employed for protein expression. C. pneumoniae GiD and the C. trachomatis serovars L2 (L2/434/Bu) and E (DK-20) were propagated in HEp-2 cells in the presence of 1.2 μg/ml cycloheximide. Chlamydial EBs were purified using 30% gastrographin (Schering).
The Saccharomyces cerevisiae strain YKM2 [MATa ura3-52 trp1 leu2Δ1::pCM149 (LEU2) his3Δ200 pep4::HIS3::loxP-kanMX-loxP met25p-GFP prb1Δ1.6R can1 tetO7 AGA1::pIU211 (URA3)], a green fluorescent protein (GFP)-expressing version of EBY100 (Invitrogen), was grown in synthetic medium containing 2% raffinose (SR) and then in synthetic medium plus 2% galactose (SG) to induce protein expression as described before (13).
HEp-2 cells (epithelial larynx carcinoma cell line) were cultured in Dulbecco's modified Eagle medium (DMEM) GlutaMax (Invitrogen) supplemented with 10% fetal calf serum (FCS), vitamins, nonessential amino acids, amphotericin B (2.5 μg/ml), and gentamicin (50 μg/ml).
DNA manipulation and plasmid construction.
Site-specific mutagenesis was performed by homologous recombination in S. cerevisiae using PCR products generated with oligonucleotides containing the specific substitutions and either the yeast adhesion plasmid expressing C. trachomatis serovar E OmcB (13) cleaved with Bpu1102I and SpeI or the E. coli expression vector glutathione S-transferase (GST)-OmcB-BD6×His (aa 41 to 100) cleaved with Bpu1102I or SmaI. The GST-OmcB-BD6×His plasmid was generated by cloning OmcB-BD into the E. coli expression vector pFT8. pFT8 is a derivative of the expression plasmid pGEX-3X (Pharmacia), which was modified by integration of a C-terminal 6×His tag and a precision cleavage site C terminal to the GST tag. All constructs were verified by sequencing.
Affinity purification of GST-tagged proteins.
GST-tagged fusion proteins were purified under native conditions by following the protocol supplied by Sigma-Aldrich. Proteins (1-ml elution fractions) were dialyzed overnight against 1 liter of phosphate-buffered saline (PBS) at 4°C to remove the reduced glutathione (Sigma) using dialysis tubing with a 3.5-kDa cutoff (Serva). Proteins were separated by SDS-PAGE and detected with an anti-His antibody (Roche) after immunoblotting.
Yeast adhesion assay.
Yeast adhesion assays were performed on 50% confluent layers of HEp-2 cells cultured on glass coverslips. Aliquots (1 × 106) of yeast cells with the surface expression of a specific protein were washed with PBS, suspended in 500 μl PBS with or without 500 μg/ml soluble heparin (Sigma), and incubated for 30 min at 4°C. The yeast cells were then washed with PBS and suspended in 500 μl PBS, and the whole aliquot was added to coverslips bearing 5 × 105 HEp-2 cells (2-fold excess). Coverslips were incubated for 1 h at 37°C in the presence of 6% CO2, washed with PBS, and fixed with 3% paraformaldehyde. Adhesion was quantified by counting the HEp-2 cells and attached yeast cells in 10 visual fields. The average number of wt OmcB-displaying yeast cells bound to human cells varied between 15 and 20% of the input.
Bead assay.
Latex beads were coated with recombinant protein as described previously (33). Coating efficiency was analyzed by immunoblotting. Assays using fluorescence-labeled beads (diameter, 1 μm; Polysciences) were performed on 100% confluent layers of HEp-2 cells (1 × 106), and binding was quantified by flow cytometry (FACSAria; BD Biosciences). Unlabeled beads (diameter, 1.1 μm; Sigma) were incubated with HEp-2 cells growing on glass coverslips and quantified by microscopy. A 10-fold excess of beads (1 × 107) was added to the cell sample, which was incubated for 1 h at 37°C in the presence of 6% CO2. Unattached beads were then removed by washing with PBS, cells were fixed with 3% paraformaldehyde, and the numbers of beads attached to 103 HEp-2 cells were counted under the microscope. For fluorescence-based assays, the cells were detached with cell dissociation solution (Sigma) and fixed with 3% paraformaldehyde, and the numbers of beads attached to aliquots of 104 HEp-2 cells were determined by flow cytometry, measuring the fluorescence intensity of every single cell. The average number of wt OmcB-coated beads bound to human cells varied between 15 and 20% of the input.
Neutralization assay.
Fully confluent HEp-2 cell layers (1 × 106) grown on glass coverslips were preincubated with the indicated antibodies for 1 h at 37°C in the presence of 6% CO2 and infected with C. pneumoniae or C. trachomatis serovar E or L2 at a multiplicity of infection (MOI) of 10 for 2 h. Cells were then incubated for 48 h (Cpn) or 24 h (Ctr) in medium containing cycloheximide before being fixed with methanol and stained with a fluorescein isothiocyanate (FITC)-conjugated anti-chlamydial lipopolysaccharide (LPS) antibody (Bio-Rad). The number of inclusions found in 20 visual fields per coverslip was determined under the microscope. IgG concentration was analyzed by immunoblotting prior to the experiment. Antibodies were diluted 1:5 before use (34–37). The GAG-specific single-chain antibodies were obtained from T. van Kuppevelt.
Bioinformatic analysis.
EMBOSS Needle (matrix, EBLOSSUM62; gap penalty, 10.0; extend penalty, 0.5) was used for sequence alignments; GOR IV (38) was used for secondary structure prediction.
RESULTS
The second heparin-binding motif in C. pneumoniae OmcB is essential for its adhesive function.
The heparin-binding domain (aa 41 to 100) of OmcB from C. pneumoniae is rich in basic amino acids and harbors a tandem duplication of the XBBXBX motif (Fig. 2A). As we had found the first copy to be essential for binding of OmcB to host cells (13), we wished to probe the role of the second. Therefore, we constructed a deletion variant of the binding domain of OmcB (OmcB-BD) lacking the second XBBXBX motif (GST-OmcB-BDΔmotif2) and performed a bead adhesion assay with the purified recombinant protein. Latex beads coated with recombinant GST-OmcB-BDΔmotif2 failed to bind to HEp-2 cells, indicating that motif 2 is essential for adhesion (Fig. 2B).
As basic amino acids within XBBXBX motifs mediate the interaction with heparin (24, 27), we tested a variant of OmcB in which the basic amino acids at positions 60 (arginine), 61 (arginine), and 63 (lysine) were all replaced by alanines (GST-OmcB-BDR60R61K63A). This triple mutant was completely unable to bind to human cells (Fig. 2C). Hence, the basic amino acids within the second heparin-binding motif are essential for C. pneumoniae OmcB interaction with GAGs on host cells.
The GAG-binding domain of C. pneumoniae OmcB extends beyond the two XBBXBX motifs.
Since a contribution of additional basic amino acids surrounding the two XBBXBX motifs to GAG recognition had not been ruled out, we constructed a series of recombinant OmcB proteins in which each of the basic amino acids in the two heparin-binding motifs and the regions flanking them was replaced in turn (Fig. 2D). In addition, we analyzed the contribution of the polar amino acids in the second motif (asparagine 62 and glutamine 64), as it is known that the polar residues in heparin-binding motifs stabilize the interaction of the basic amino acids with GAGs (24, 27).
Relative to beads bearing wild-type GST-OmcB-BD, beads coated with recombinant GST-OmcB-BDK54A, GST-OmcB-BDK55A, and GST-OmcB-BDR57A exhibited significantly reduced attachment efficiencies to HEp-2 cells of 66, 39, and 30%, respectively (Fig. 2D). Of the corresponding mutations within motif 2, R61A had the greatest impact on binding, significantly reducing adhesion to 22% of the control value. In contrast, replacement of the arginine at position 60, or the lysine at position 63, by alanine (GST-OmcB-BDR60A and GST-OmcB-BDK63A, respectively) resulted in only a moderate decrease in adhesion to 83 and 84%, respectively. Interestingly, substitution of the lysine at position 69 (OmcB-BDK69A), which is the first basic amino acid C terminal to the heparin-binding motifs, had a much greater effect, significantly reducing adhesion to 27%. Thus, the contribution to adhesion of this amino acid, which lies beyond motif 2, is comparable to that of each of the basic residues at positions 57 and 61. Therefore, we investigated the contribution of the next two basic amino acids in the sequence. To our surprise, the alanine substitutions at positions 71 (arginine) and 77 (lysine) strongly increased binding, to 192 and 270% of the control value, respectively, in the bead assay. In addition, replacement of the first basic amino acid N terminal to the heparin-binding motifs, the lysine at position 45 (GST-OmcB-BDK45A), also increased adhesion, albeit to a lesser and not statistically significant extent (134%). In contrast, substitution of the two polar amino acids within the second heparin-binding motif (GST-OmcB-BDN62A and GST-OmcB-BDQ64A) had a negative effect on binding, reducing bead adhesion to 59% (P = 0.0346) and 53% (P = 0.0535), respectively (Fig. 2D).
These results demonstrate that several basic and polar amino acids within the binding domain of C. pneumoniae OmcB contribute significantly to the protein's capacity to adhere to human cells. Furthermore, basic amino acids located on either side of the heparin-binding motifs are also relevant for adhesion, as their substitution actually results in an increase in adhesion.
Three amino acids within the N-terminal variable domain of C. trachomatis OmcB determine the glycosaminoglycan dependency of adhesion.
Recently, we showed that the amino acid at position 66 within the variable domain of the C. trachomatis OmcB is critical for its interaction with heparin-like GAGs on human cells (13). The sequence differences observed between the OmcB proteins of serovars E and LGV at these positions are predicted to correlate with changes in secondary structure (Fig. 3A). Thus, we employed the yeast adhesion assay to analyze the contribution of each of the three variable amino acids in this region by substituting those found in LGV serovars for their counterparts in the heparin-independent OmcB-E (Fig. 3).
Exchange of the amino acid at position 66 (OmcB-EL66P; leucine to proline) increased overall adhesion to 113% (P = 0.0559) and induced a partial dependence on heparin, as preincubation of yeast cells bearing this OmcB variant with soluble heparin significantly reduced attachment to 70% of the control value (Fig. 3B). In contrast, replacements of the amino acids at positions 68 and 71 (OmcB-ED68N, aspartic acid to asparagine; OmcB-EE71K, glutamic acid to lysine) did not alter overall adhesion and induced a slight reduction in heparin-dependent adhesion of 9 and 6%, respectively. We then tested double substitutions of amino acids 66 and 68 or 71. Again, adhesion of doubly substituted OmcB-EL66P, D68N (leucine and aspartic acid to proline and asparagine) and OmcB-EL66P, E71K (leucine and glutamic acid to proline and lysine) expressed on yeast cells was increased slightly, to 113 and 111%, respectively. Moreover, pretreatment with soluble heparin significantly decreased adhesion to 61 and 62%, respectively, reflecting the additive effects of the single substitutions at positions 66, 68, and 71 (Fig. 3B). Since neither of the double substitutions in OmcB-E conferred the completely heparin-dependent adhesion described for OmcB-L (13), we constructed a triply substituted OmcB-E mutant in which all three variant amino acids in the variable domain were exchanged, making the sequence of the binding domain identical to that of OmcB-L. Like all other L66P-harboring variants, the OmcB-EL66P, D68N, E71K (leucine, aspartic acid, and glutamic acid to proline, asparagine, and lysine, respectively) variant showed a slight increase in overall adhesion to 114%, while preincubation of yeast cells expressing the triple mutant with soluble heparin significantly reduced adhesion to 38%, which is comparable to the background level observed for the negative-control Aga2; thus, it is compatible with a complete dependence of adhesion on heparin (Fig. 3B).
These findings show that heparin dependence of C. trachomatis OmcB adhesion is entirely determined by the identity of the amino acids found at only three positions within the variable region. Furthermore, the proline at position 66 and the resulting change in the secondary structure is critical for the induction of heparin dependency (see Fig. S1 in the supplemental material). Interestingly, the presence of proline at position 66 also enhances overall adhesion relative to OmcB variants harboring a leucine at this position.
Infection by C. pneumoniae or C. trachomatis serovar L2 can be blocked by treating human HEp-2 cells with specific anti-heparan sulfate antibodies.
The exact chemical nature of the heparan sulfate-like GAGs recognized by Chlamydiae is unknown, although at least a decasaccharide (dodecasaccharide) with 6-O-sulfation is necessary to neutralize infection by C. trachomatis L2, while 2-O-sulfation and N-sulfation do not seem to be required (15, 21). In order to gain insight into the heparan sulfate structure recognized by OmcB, we performed antibody neutralization experiments using single-chain antibodies produced in E. coli, which were selected by phage display for the ability to recognize different natural heparan sulfates and require different specific modifications for binding (Fig. 4A) (35). All antibodies recognized epitopes on the HEp-2 cells used in this experiment (Fig. 4A).
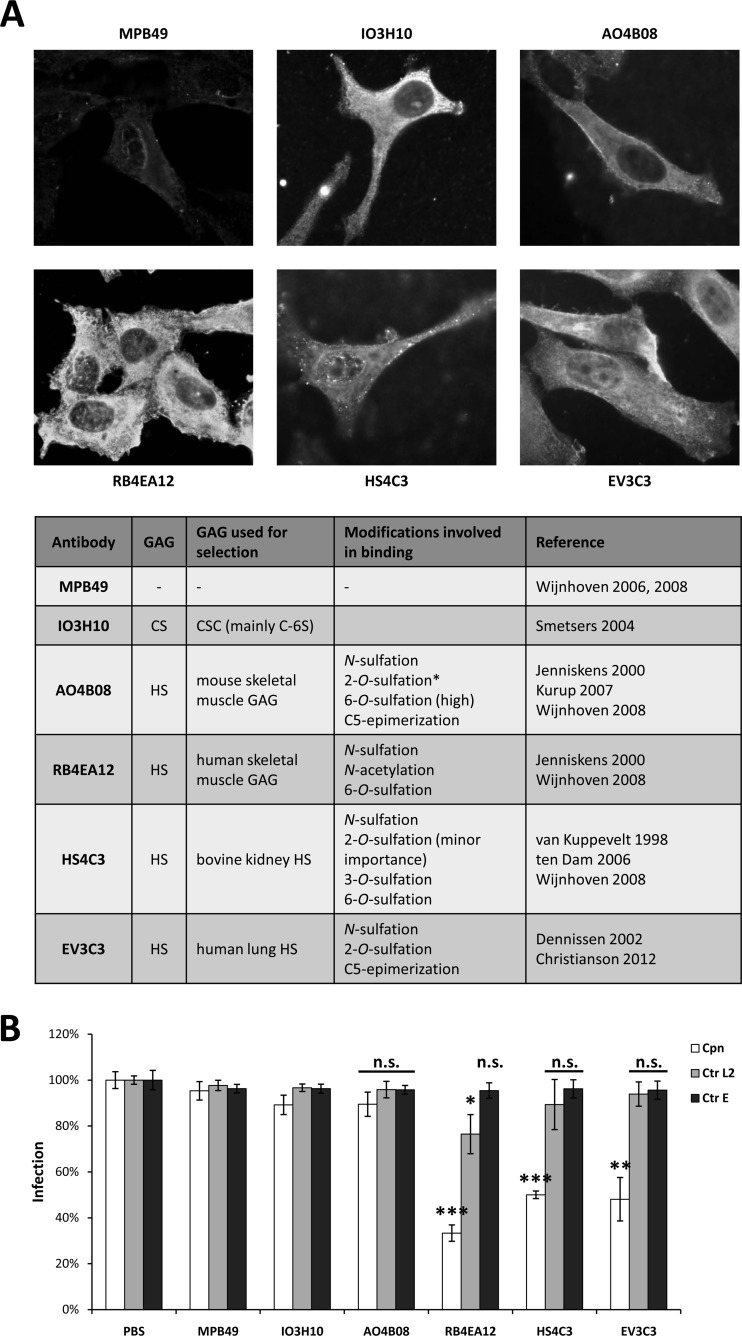
Fig 4.
Antibodies directed against endogenous heparan sulfate neutralize infection by C. pneumoniae and C. trachomatis serovar L2. (A) Human HEp-2 cells were stained with single-chain anti-GAG antibodies provided by T. van Kuppevelt. The specificity of each antibody is shown in the table below the panel (34–37, 40–43, 50, 51). Cells were fixed with methanol. The asterisk indicates that this antibody requires an internal 2-O-sulfated iduronic acid residue for binding. CS, chondroitin sulfate; CSC, chondroitin sulfate C; C-6S, chondroitin-6 sulfate; HS, heparan sulfate; GAG, glycosaminoglycan. (B) Neutralization of chlamydial infection by pretreatment of HEp-2 cells with the indicated antibodies. Cells were pretreated with each antibody prior to chlamydial infection (MOI, 10). Infectivity was determined by microscopy 24 (Ctr) or 48 (Cpn) h postinfection. Infectivity following pretreatment with PBS was set to 100%. Cpn, C. pneumoniae; Ctr L2, C. trachomatis serovar L2; Ctr E, C. trachomatis serovar E (n = 4). Statistically significant differences are denoted by ***, **, and *, which indicate P < 0.001, P < 0.01, and P < 0.05, respectively. n.s., not significant. P values state significant changes relative to the corresponding PBS control.
Pretreatment of human cells with anti-heparan sulfate (HS) antibodies that bind specific epitopes found in human skeletal muscle GAGs (RB4EA12), bovine kidney HS (HS4C3), or human lung HS (EV3C3) significantly reduced the incidence of infection by C. pneumoniae to 35, 52, and 50% of the control, respectively (Fig. 4B). All three antibodies require N-sulfation of the antigen for efficient binding. RB4EA12 and HS4C3 also require 6-O- and 6-O-/3-O-sulfation, respectively, whereas EV3C3 requires 2-O-sulfation and C5-epimerization of d-glucoronic acid (GlcA) to l-iduronic acid (IdoA) (Fig. 4A). On the other hand, antibodies recognizing chondroitin sulfate (IO3H10) or heparan sulfate obtained by selection against mouse skeletal muscle GAGs (AO4B08) were unable to block infection by C. pneumoniae (Fig. 4B). As the latter antibody requires a specific internal 2-O-sulfated IdoA residue in the antigen for recognition, this specific modification may inhibit heparan sulfate targeting by C. pneumoniae (Fig. 4A).
Infection by C. trachomatis L2 was reduced significantly (to 78%) only by preincubation of human cells with RB4EA12, which also inhibited infection by C. pneumoniae (Fig. 4B). None of the other antibodies had any significant effect on infection by C. trachomatis L2.
None of the antibodies tested neutralized infection by C. trachomatis E when preincubated with human cells prior to infection, confirming that attachment mediated by OmcB-E is indeed independent of the presence of highly modified heparan sulfate on the host cell (Fig. 4B).
Thus, our findings indicate that C. pneumoniae OmcB mediates attachment by interacting with heparan sulfate GAGs that display N-sulfation and a specific subset of O-sulfations. In contrast, the OmcB on C. trachomatis L2 seems to interact specifically with simply 6-O-sulfated heparan sulfate domains. Thus, the OmcB-GAG interaction seems to be highly adapted to the tissues infected by the different species and serovars.
DISCUSSION
All OmcB proteins possess an XBBXBX heparin-binding motif; however, uniquely, C. pneumoniae OmcB has tandem copies. Interestingly, the heparin-binding motifs of all chlamydial OmcB proteins are predicted to fold into an α-helix. Recently, using the yeast display system, we showed that replacement of all three basic residues present in the first copy eliminates OmcB's ability to attach to human cells. Of the individual residues, the arginine at position 57 has the strongest impact on binding ability (13). In this study, we show that the second motif is equally essential for binding. As with the first motif, an arginine residue at position 61 in the second motif makes the largest contribution to adhesion, in agreement with the observation that arginine binds more tightly to heparin than lysine does (24, 27). As in the case of the first motif (13), simultaneous replacement of all basic residues in the second motif causes complete loss of adhesion capacity (Fig. 2C), implying that both motifs are essential for adhesion. Interestingly, the lysine at position 69 C terminal to motif 2 has a function similar in importance to that of the arginines at positions 57 and 61 within the duplicated motifs. Thus, the domain of C. pneumoniae OmcB that directly interacts with heparin-like GAGs is not restricted to the XBBXBX motifs but extends further C terminally. Indeed, it has been demonstrated that heparin-binding domains need not consist of an unbroken linear sequence of amino acids (24). On the contrary, heparin can be efficiently recognized by basic residues separated by a distance of approximately 20 Å in the native conformation of the binding protein (39). Furthermore, we identified the polar amino acids asparagine (at position 62) and glutamine (at position 64) as important for C. pneumoniae OmcB-mediated attachment. Asparagine and glutamine are capable of hydrogen bonding; thus, they stabilize the interaction of the basic amino acids with the negatively charged heparin (27). Surprisingly, replacement of the lysines at positions 71 and 77 by alanines resulted in a strong increase in the binding capacity of OmcB but may lead to a loss of specificity. In fact, enhancement of adhesion may compromise the function of OmcB, as OmcB-mediated adhesion is assumed to be reversible (7), allowing the infectious EB to surf the host cell surface to find the specific domain for invasion.
Taken together, these findings show that the GAG-binding domain of C. pneumoniae OmcB encompasses at least 32 aa, including the two XBBXBX motifs; thus, it is one of the largest linearly contiguous GAG binding sites described so far (27).
The binding specificity of the chlamydial OmcB proteins for particular subdomains of heparan sulfate proteoglycans was elucidated on the basis of antibody neutralization experiments (Fig. 4). Various anti-HS antibodies recognizing different HS modifications were able to partially inhibit infection by C. pneumoniae (Fig. 4B). All of the antibodies tested require N-sulfation for binding but recognize different (combinations of) O-sulfated positions: 2-O-sulfation (HS4C3 and EV3C3), 3-O-sulfation (HS4C3), and 6-O-sulfation (RB4EA12 and HS4C3) (Fig. 4A) (34–36, 40–42). Therefore, C. pneumoniae OmcB may interact with a domain of heparan sulfate that harbors N-sulfated subunits and a specific subset of O-sulfated subunits, which can be masked by any of several antibodies, thereby inhibiting the infection. Strikingly, although it also recognizes N-sulfated and 6-O-sulfated domains (40, 41, 43), the antibody AO4B08 had no effect on infection by C. pneumoniae (Fig. 4A). This could be due to the fact that AO4B08 requires an internal 2-O-sulfated iduronic acid residue for binding (43), which may not be needed for adhesion mediated by C. pneumoniae OmcB.
Although the OmcB proteins of the 19 known C. trachomatis serovars differ at just 13 out of 548 positions (94 to 100% identity) (Fig. 1B), they show remarkable differences in their dependence on heparin/heparan sulfate for adhesion. It was previously demonstrated that C. pneumoniae OmcB adheres to heparin-like GAGs via the XBBXBX heparin-binding motif, which is also present in all C. trachomatis OmcB proteins (this study and 10, 13). Strikingly, however, in the C. trachomatis OmcB proteins a variable region occurs immediately C terminal to the XBBXBX motif (amino acids 66 to 71). The residues at three positions (66, 68, and 71) within this segment differ between serovars (Fig. 3A). In this study, we analyzed the contribution of each of the three variable amino acids to the heparin dependency of OmcB-mediated adhesion of serovar E. We found the amino acid at position 66 to be the major determinant of heparin-dependent and heparin-independent attachment. Binding of OmcB-E to human host cells is heparin independent. However, it becomes heparin dependent to a significant degree when the leucine at position 66 is replaced by the proline found at the corresponding site in OmcB of the LGV serovars (Fig. 3B). The converse is also true: substitution of leucine for proline in the normally heparin-dependent OmcB-L makes adhesion to human cells completely independent of heparin, as in the case of OmcB-E (13). Interestingly, the exchange at position 66 in the OmcB-L protein results in a complete mimic of the OmcB-E phenotype, while the reverse exchange results in only partial heparin dependency (Fig. 3B) (13).
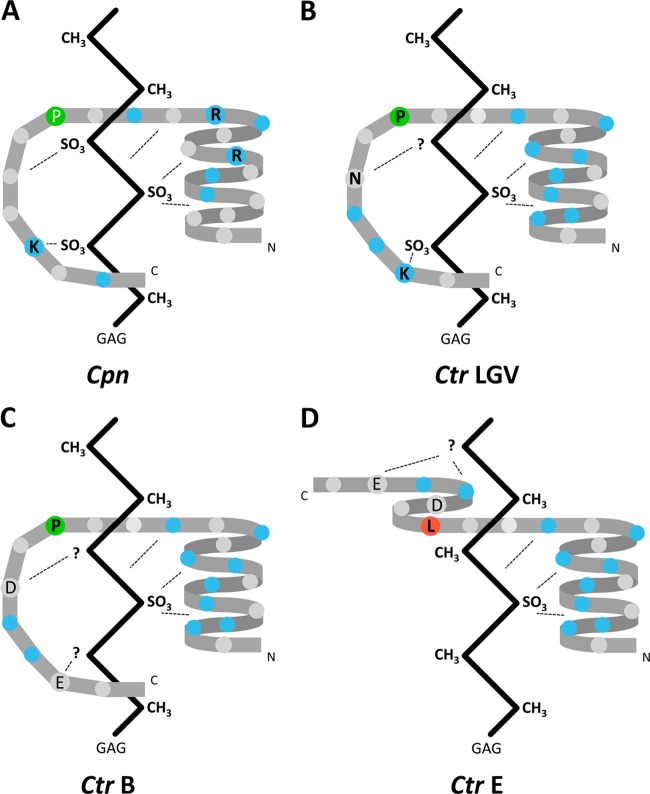
The exchange of either of the amino acids at the other two variable positions, 68 and 71, in the variable domain of OmcB-E had only a slight influence on heparin-independent adhesion (Fig. 3B). The phenotypes observed are in agreement with the predicted effects of the replacements on secondary structure. In OmcB-E, the leucine at position 66 is compatible with an alpha-helical structure, whereas proline disrupts alpha-helices, inducing the adoption of an extended random-coil structure. Conversely, the replacement of proline by leucine in OmcB-L allows an alpha-helical structure to form. Exchanges at positions 68 and 71 are not expected to disrupt the alpha-helical structure in OmcB-E (see Fig. S1 in the supplemental material). Therefore, the secondary structure defined by the amino acid at position 66 is likely to be crucial for determining whether OmcB-mediated adhesion is dependent on or independent of heparin. Interestingly, the heparin-dependent OmcB adhesins from C. trachomatis LGV serovars and C. pneumoniae have a proline at a similar position C terminal to the heparin-binding motifs and are predicted to have similar secondary structures in this area of the GAG-binding domain (Fig. 5). However, to mimic the OmcB-L phenotype completely, all three variable amino acids in OmcB-E had to be changed to those found at the corresponding positions in LGV serovars. This indicates that both the positively charged lysine at position 71 and the polar, hydrogen-bonding asparagine at position 68 act to stabilize the OmcB-heparin interaction (27). Again, the comparison to the OmcB sequence from C. pneumoniae reveals that a lysine C terminal to the heparin-binding motif(s) also contributes significantly to heparin-dependent adhesion (Fig. 2D). Thus, we conclude that while a random-coil structure C terminal to the XBBXBX motifs is necessary for efficient interaction of OmcB with heparin-like GAGs, basic residues C terminal to the heparin-binding motif(s) also contribute appreciably to the binding characteristics of OmcB (Fig. 5).
Fig 5.
Model of the interaction between the chlamydial adhesin OmcB and human glycosaminoglycans. The schematic diagrams of protein conformation are based on sequence-based prediction of secondary structures. (A) C. pneumoniae; (B) C. trachomatis LGV serovars; (C) C. trachomatis serovar B; (D) C. trachomatis serovar E. Critically important amino acids are labeled by the single-letter code in black. The proline (P) in panel A has not been experimentally analyzed but has an influence on the secondary structure prediction similar to that of the corresponding proline residues shown in panels B and C. Basic amino acids are highlighted in blue, and important proline and leucine residues are in green and red, respectively. A hypothetical GAG chain is represented by a vertical zigzag pattern. Based on the results from Fig. 4, the GAG chain recognized by the C. pneumoniae OmcB carries more sulfate groups.
The partially heparin-dependent adhesion of the L66P variant of C. trachomatis OmcB-E has previously been observed for the attachment of EBs of serovars B and C to host cells (14, 16, 30). Indeed, the binding domain of the OmcB proteins of serovars B and C show sequence identity to the OmcB-EL66P mutant. In fact, among all serovars, only three amino acid combinations are found in the short stretch C terminal to the heparin binding motif. The sequence of this variable domain in the three LGV serovars is PVNRKK, in serovars E and F it is LVDRKE, and all other serovars have the sequence PVDRKE (underlined letters represent the variable amino acids). This naturally occurring smooth transition from heparin-dependent to heparin-independent OmcB-mediated adhesion of EBs makes it very likely that all chlamydial OmcB adhesins recognize and bind to GAGs. However, the precise nature of the GAG recognized, including the modifications it bears, seems to be serovar specific. Indeed, none of the antibodies selected for binding to heparan sulfate bearing specific modifications was able to reduce the C. trachomatis serovar E infection (Fig. 4B), indicating that OmcB-E interacts with less or nonsulfated regions of heparan sulfate. That would also explain why prior exposure to soluble heparin, which is highly sulfated, does not reduce adhesion of C. trachomatis serovar E EBs (13, 14, 18, 19, 30, 31). Moreover, even heparan sulfate with its heterogeneous and reduced sulfate pattern is unable to block adhesion of C. trachomatis OmcB-E-presenting E. coli cells (32). In contrast, infection by C. trachomatis serovar L2 was inhibited only by the antibody RB4EA12, which recognizes N-sulfated, N-acetylated, and 6-O-sulfated regions (Fig. 4A) (40, 41). In contrast, the antibodies HS4C3 and EV3C3, which require N-sulfation and 6-O-/3-O-/2-O-sulfation (HS4C3) or 2-O-sulfation (EV3C3) of the antigen for binding, did not block C. trachomatis LGV infection significantly (34–36, 41, 42). Thus, it is likely that the OmcB-L protein interacts with N-acetylated, N-sulfated, and 6-O-sulfated domains of heparan sulfate, while 2-O-sulfation and/or 3-O-sulfation might be inhibitory (Fig. 4B) (42). The observation that AO4B08, which also recognizes 6-O-sulfated domains (Fig. 4A) (40, 41, 43), has no effect on infection by C. trachomatis serovar L2 could be explained by the fact that AO4B08 requires an internal 2-O-sulfated iduronic acid residue for binding (43) which may not be needed for OmcB-L-mediated adhesion. This is in agreement with previous work on the impact of differentially modified heparan sulfate oligosaccharides and CHO cells deficient in special GAG modifications on infection by C. trachomatis serovar L2. In summary, these studies showed that 6-O-sulfation, but not 2-O-sulfation or N-sulfation, is necessary for adhesion and infection by L2 (15, 21).
Our data indicate that OmcB accounts for the tissue specificity and contributes to the success of infection. Most C. trachomatis infections of the urogenital tract are attributable to serovars E and F (44–47), whose OmcB adhesins share the same variable region. In contrast, all LGV serovars harbor the OmcB-L variable region and are capable of penetrating deeper into tissues and entering the lymphatic system, and all of this may be related. Interestingly, a recent study showed that 6-O-sulfation is required for adhesion of and, as a result, infection by C. muridarum (48). While the XBBXBX heparin binding motif of C. muridarum OmcB is very similar to that of C. trachomatis OmcB, the variable regions of the two proteins differ completely. Interestingly, however, like all heparin-dependent C. trachomatis OmcB proteins, C. muridarum OmcB is not predicted to exhibit an alpha-helical secondary structure in the variable region. Thus, in conclusion, the OmcB proteins from C. pneumoniae, C. trachomatis LGV serovars, and C. muridarum all seem to recognize 6-O-sulfated heparan sulfate domains, suggesting that this modification is a key player in their interactions with GAGs. Tissue and host specificity mediated by different versions of a single adhesin has previously been reported for the PapG adhesin of uropathogenic E. coli (UPEC) strains that recognize different but related α-d-galactopyranosyl-(1-4)-β-d-galactopyranoside moieties of glycolipids (49).
To elucidate how OmcB interacts with heparin sulfate-like GAGs and identify the kinds of modification required, it will be necessary to determine the three-dimensional structure of the OmcB binding domain. The identification of the precise chemical nature of the GAG structures it recognizes will be an important step toward an understanding of the host and tissue specificity of OmcB-mediated attachment by different chlamydial species and serovars.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to Toin van Kuppevelt for providing the GAG-specific single-chain antibodies and to Wolfgang Kaisers for his help performing the statistical analysis.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Priority Program 590, Project C5) and the Federal Ministry for Education and Research (BMBF, Project CHI) to J.H.H. T.F. was a scholarship holder of the Graduate School Molecules of Infection (MOI), funded by the Jürgen Manchot Foundation.
Footnotes
Published ahead of print 20 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00780-13.
REFERENCES
- 1.Kuo CC, Jackson LA, Campbell LA, Grayston JT. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8:451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasi F, Tarsia P, Aliberti S. 2009. Chlamydophila pneumoniae. Clin. Microbiol. Infect. 15:29–35 [DOI] [PubMed] [Google Scholar]
- 3.Grayston JT, Campbell LA, Kuo CC, Mordhorst CH, Saikku P, Thom DH, Wang SP. 1990. A new respiratory tract pathogen: Chlamydia pneumoniae strain TWAR. J. Infect. Dis. 161:618–625 [DOI] [PubMed] [Google Scholar]
- 4.Whitcher JP, Srinivasan M, Upadhyay MP. 2001. Corneal blindness: a global perspective. Bull. World Health Organ. 79:214–221 [PMC free article] [PubMed] [Google Scholar]
- 5.Bebear C, de Barbeyrac B. 2009. Genital Chlamydia trachomatis infections. Clin. Microbiol. Infect. 15:4–10 [DOI] [PubMed] [Google Scholar]
- 6.Abdelrahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949–959 [DOI] [PubMed] [Google Scholar]
- 7.Carabeo RA, Hackstadt T. 2001. Isolation and characterization of a mutant Chinese hamster ovary cell line that is resistant to Chlamydia trachomatis infection at a novel step in the attachment process. Infect. Immun. 69:5899–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ting LM, Hsia RC, Haidaris CG, Bavoil PM. 1995. Interaction of outer envelope proteins of Chlamydia psittaci GPIC with the HeLa cell surface. Infect. Immun. 63:3600–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mygind P, Christiansen G, Birkelund S. 1998. Topological analysis of Chlamydia trachomatis L2 outer membrane protein 2. J. Bacteriol. 180:5784–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens RS, Koshiyama K, Lewis E, Kubo A. 2001. Heparin-binding outer membrane protein of chlamydiae. Mol. Microbiol. 40:691–699 [DOI] [PubMed] [Google Scholar]
- 11.Montigiani S, Falugi F, Scarselli M, Finco O, Petracca R, Galli G, Mariani M, Manetti R, Agnusdei M, Cevenini R, Donati M, Nogarotto R, Norais N, Garaguso I, Nuti S, Saletti G, Rosa D, Ratti G, Grandi G. 2002. Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect. Immun. 70:368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fadel S, Eley A. 2007. Chlamydia trachomatis OmcB protein is a surface-exposed glycosaminoglycan-dependent adhesin. J. Med. Microbiol. 56:15–22 [DOI] [PubMed] [Google Scholar]
- 13.Moelleken K, Hegemann JH. 2008. The Chlamydia outer membrane protein OmcB is required for adhesion and exhibits biovar-specific differences in glycosaminoglycan binding. Mol. Microbiol. 67:403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JC, Stephens RS. 1994. Trachoma and LGV biovars of Chlamydia trachomatis share the same glycosaminoglycan-dependent mechanism for infection of eukaryotic cells. Mol. Microbiol. 11:501–507 [DOI] [PubMed] [Google Scholar]
- 15.Chen JC, Zhang JP, Stephens RS. 1996. Structural requirements of heparin binding to Chlamydia trachomatis. J. Biol. Chem. 271:11134–11140 [PubMed] [Google Scholar]
- 16.Chen JCR, Stephens RS. 1997. Chlamydia trachomatis glycosaminoglycan dependent and independent attachment to eukaryotic cells. Microb. Pathog. 22(1):23–30 [DOI] [PubMed] [Google Scholar]
- 17.Zaretzky FR, Pearce-Pratt R, Phillips DM. 1995. Sulfated polyanions block Chlamydia trachomatis infection of cervix-derived human epithelia. Infect. Immun. 63:3520–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis CH, Wyrick PB. 1997. Differences in the association of Chlamydia trachomatis serovar E and serovar L2 with epithelial cells in vitro may reflect biological differences in vivo. Infect. Immun. 65:2914–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taraktchoglou M, Pacey AA, Turnbull JE, Eley A. 2001. Infectivity of Chlamydia trachomatis serovar LGV but not E is dependent on host cell heparan sulfate. Infect. Immun. 69:968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wuppermann FN, Hegemann JH, Jantos CA. 2001. Heparan sulfate-like glycosaminoglycan is a cellular receptor for Chlamydia pneumoniae. J. Infect. Dis. 184:181–187 [DOI] [PubMed] [Google Scholar]
- 21.Yabushita H, Noguchi Y, Habuchi H, Ashikari S, Nakabe K, Fujita M, Noguchi M, Esko JD, Kimata K. 2002. Effects of chemically modified heparin on Chlamydia trachomatis serovar L2 infection of eukaryotic cells in culture. Glycobiology 12:345–351 [DOI] [PubMed] [Google Scholar]
- 22.Yan Y, Silvennoinen-Kassinen S, Leinonen M, Saikku P. 2006. Inhibitory effect of heparan sulfate-like glycosaminoglycans on the infectivity of Chlamydia pneumoniae in HL cells varies between strains. Microbes Infect. 8:866–872 [DOI] [PubMed] [Google Scholar]
- 23.Kjellen L, Lindahl U. 1991. Proteoglycans: structures and interactions. Annu. Rev. Biochem. 60:443–475 [DOI] [PubMed] [Google Scholar]
- 24.Capila I, Linhardt RJ. 2002. Heparin-protein interactions. Angew. Chem. Int. Ed. Engl. 41:391–412 [DOI] [PubMed] [Google Scholar]
- 25.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. 1999. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68:729–777 [DOI] [PubMed] [Google Scholar]
- 26.Smits NC, Shworak NW, Dekhuijzen PN, van Kuppevelt TH. 2010. Heparan sulfates in the lung: structure, diversity, and role in pulmonary emphysema. Anat. Rec. (Hoboken) 293:955–967 [DOI] [PubMed] [Google Scholar]
- 27.Hileman RE, Fromm JR, Weiler JM, Linhardt RJ. 1998. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays 20:156–167 [DOI] [PubMed] [Google Scholar]
- 28.Cardin AD, Weintraub HJ. 1989. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 9:21–32 [DOI] [PubMed] [Google Scholar]
- 29.Everett KD, Hatch TP. 1995. Architecture of the cell envelope of Chlamydia psittaci 6BC. J. Bacteriol. 177:877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JP, Stephens RS. 1992. Mechanism of C. trachomatis attachment to eukaryotic host cells. Cell 69:861–869 [DOI] [PubMed] [Google Scholar]
- 31.Beswick EJ, Travelstead A, Cooper MD. 2003. Comparative studies of glycosaminoglycan involvement in Chlamydia pneumoniae and C. trachomatis invasion of host cells. J. Infect. Dis. 187:1291–1300 [DOI] [PubMed] [Google Scholar]
- 32.Fadel S, Eley A. 2008. Differential glycosaminoglycan binding of Chlamydia trachomatis OmcB protein from serovars E and LGV. J. Med. Microbiol. 57:1058–1061 [DOI] [PubMed] [Google Scholar]
- 33.Dersch P, Isberg RR. 1999. A region of the Yersinia pseudotuberculosis invasin protein enhances integrin-mediated uptake into mammalian cells and promotes self-association. EMBO J. 18:1199–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennissen MA, Jenniskens GJ, Pieffers M, Versteeg EM, Petitou M, Veerkamp JH, van Kuppevelt TH. 2002. Large, tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J. Biol. Chem. 277:10982–10986 [DOI] [PubMed] [Google Scholar]
- 35.van Kuppevelt TH, Dennissen MA, van Venrooij WJ, Hoet RM, Veerkamp JH. 1998. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J. Biol. Chem. 273:12960–12966 [DOI] [PubMed] [Google Scholar]
- 36.Ten Dam GB, Kurup S, van de Westerlo EM, Versteeg EM, Lindahl U, Spillmann D, van Kuppevelt TH. 2006. 3-O-sulfated oligosaccharide structures are recognized by anti-heparan sulfate antibody HS4C3. J. Biol. Chem. 281:4654–4662 [DOI] [PubMed] [Google Scholar]
- 37.Smits NC, Robbesom AA, Versteeg EM, van de Westerlo EM, Dekhuijzen PN, van Kuppevelt TH. 2004. Heterogeneity of heparan sulfates in human lung. Am. J. Respir. Cell Mol. Biol. 30:166–173 [DOI] [PubMed] [Google Scholar]
- 38.Combet C, Blanchet C, Geourjon C, Deleage G. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147–150 [DOI] [PubMed] [Google Scholar]
- 39.Margalit H, Fischer N, Ben-Sasson SA. 1993. Comparative analysis of structurally defined heparin binding sequences reveals a distinct spatial distribution of basic residues. J. Biol. Chem. 268:19228–19231 [PubMed] [Google Scholar]
- 40.Jenniskens GJ, Oosterhof A, Brandwijk R, Veerkamp JH, van Kuppevelt TH. 2000. Heparan sulfate heterogeneity in skeletal muscle basal lamina: demonstration by phage display-derived antibodies. J. Neurosci. 20:4099–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wijnhoven TJ, van de Westerlo EM, Smits NC, Lensen JF, Rops AL, van der Vlag J, Berden JH, van den Heuvel LP, van Kuppevelt TH. 2008. Characterization of anticoagulant heparinoids by immunoprofiling. Glycoconj. J. 25:177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christianson HC, van Kuppevelt TH, Belting M. 2012. ScFv anti-heparan sulfate antibodies unexpectedly activate endothelial and cancer cells through p38 MAPK: implications for antibody-based targeting of heparan sulfate proteoglycans in cancer. PLoS One 7:e49092. 10.1371/journal.pone.0049092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurup S, Wijnhoven TJ, Jenniskens GJ, Kimata K, Habuchi H, Li JP, Lindahl U, van Kuppevelt TH, Spillmann D. 2007. Characterization of anti-heparan sulfate phage display antibodies AO4B08 and HS4E4. J. Biol. Chem. 282:21032–21042 [DOI] [PubMed] [Google Scholar]
- 44.Suchland RJ, Eckert LO, Hawes SE, Stamm WE. 2003. Longitudinal assessment of infecting serovars of Chlamydia trachomatis in Seattle public health clinics: 1988–1996. Sex Transm. Dis. 30:357–361 [DOI] [PubMed] [Google Scholar]
- 45.Morre SA, Rozendaal L, van Valkengoed IG, Boeke AJ, van Voorst Vader PC, Schirm J, de Blok S, van Den Hoek JA, van Doornum GJ, Meijer CJ, van Den Brule AJ. 2000. Urogenital Chlamydia trachomatis serovars in men and women with a symptomatic or asymptomatic infection: an association with clinical manifestations? J. Clin. Microbiol. 38:2292–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lysen M, Osterlund A, Rubin CJ, Persson T, Persson I, Herrmann B. 2004. Characterization of ompA genotypes by sequence analysis of DNA from all detected cases of Chlamydia trachomatis infections during 1 year of contact tracing in a Swedish county. J. Clin. Microbiol. 42:1641–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang B, Zheng HP, Feng ZQ, Xue YH, Wu XZ, Huang JM, Xue XJ, Jiang HN. 2010. The prevalence and distribution of Chlamydia trachomatis genotypes among sexually transmitted disease clinic patients in Guangzhou, China, 2005–2008. Jpn. J. Infect. Dis. 63:342–345 [PubMed] [Google Scholar]
- 48.Kim JH, Chan C, Elwell C, Singer MS, Dierks T, Lemjabbar-Alaoui H, Rosen SD, Engel JN. 2013. Endosulfatases SULF1 and SULF2 limit Chlamydia muridarum infection. Cell Microbiol. 15:1560–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hultgren SJ, Normark S, Abraham SN. 1991. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45:383–415 [DOI] [PubMed] [Google Scholar]
- 50.Wijnhoven TJ, Lensen JF, Rops AL, van der Vlag J, Kolset SO, Bangstad HJ, Pfeffer P, van den Hoven MJ, Berden JH, van den Heuvel LP, van Kuppevelt TH. 2006. Aberrant heparan sulfate profile in the human diabetic kidney offers new clues for therapeutic glycomimetics. Am. J. Kidney Dis. 48:250–261 [DOI] [PubMed] [Google Scholar]
- 51.Smetsers TF, van de Westerlo EM, ten Dam GB, Overes IM, Schalkwijk J, van Muijen GN, van Kuppevelt TH. 2004. Human single-chain antibodies reactive with native chondroitin sulfate detect chondroitin sulfate alterations in melanoma and psoriasis. J. Investig. Dermatol. 122:707–716 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.