Abstract
A 94-kb integrative conjugative element (ICESluvan) transferable to Enterococcus faecium and Enterococcus faecalis from an animal isolate of Streptococcus lutetiensis consists of a mosaic of genetic fragments from different Gram-positive bacteria. A variant of ICESluvan was confirmed in S. lutetiensis from a patient. A complete Tn5382/Tn1549 with a vanB2 operon is integrated into a streptococcal ICESde3396-like region harboring a putative bacteriophage exclusion system, a putative agglutinin receptor precursor, and key components of a type IV secretion system. Moreover, ICESluvan encodes a putative MobC family mobilization protein and a relaxase and, thus, in total has all genetic components essential for conjugative transfer. A 9-kb element within Tn5382/Tn1549 encodes, among others, putative proteins similar to the TnpX site-specific recombinase in Faecalibacterium and VanZ in Paenibacillus, which may contribute to the detected low-level teicoplanin resistance. Furthermore, ICESluvan encodes a novel bacitracin resistance locus that is associated with reduced susceptibility to bacitracin when transferred to E. faecium. The expression of a streptococcal pezAT toxin-antitoxin-encoding operon of ICESluvan in S. lutetiensis, E. faecium, and E. faecalis was confirmed by reverse transcription (RT)-PCR, indicating an active toxin-antitoxin system which may contribute to stabilizing ICESluvan within new hosts. Junction PCR and DNA sequencing confirmed that ICESluvan excised to form a circular intermediate in S. lutetiensis, E. faecalis, and E. faecium. Transfer between E. faecalis cells was observed in the presence of helper plasmid pIP964. Sequence analysis of the original S. lutetiensis donor and enterococcal transconjugants showed that ICESluvan integrates in a site-specific manner into the C-terminal end of the chromosomal tRNA methyltransferase gene rumA.
INTRODUCTION
Horizontal gene transfer is a key factor in bacterial evolution, and mobile genetic elements (MGEs) play an important role in the dissemination and persistence of antimicrobial resistance in enterococci. Genome sequence analysis and comparative genome hybridizations of seven Enterococcus faecium isolates from various sources have revealed large differences in genome size, mostly due to the variable presence of mobile genetic elements. The E. faecium pan-genome is considered to be unrestricted in size (1), implying that excess genes, such as those involved in environmental persistence, colonization, and virulence, can easily be incorporated into the E. faecium gene pool. Up to 38% of the E. faecium genome may be noncore, and differences in gene content indicate that gain and loss of genes as important in the evolution of E. faecium (1, 2, 3). These findings are consistent with the presence of more than 25% mobile or foreign DNA in Enterococcus faecalis V583 (3, 4), suggesting extensive genome plasticity.
The first high-level vancomycin-resistant enterococci (VRE) were described in 1988 (5, 6). VRE have since become an increasing nosocomial problem both in Europe and the United States (7, 8). The origin of vancomycin resistance determinants is not known, although soil bacteria seem to represent a rich and assorted reservoir of genes closely related to vanA (9). Moreover, analyses of genomes from invasive vanB-positive E. faecium isolates and vanB-positive anaerobic gut commensals demonstrate that vanB resistance in enterococci commonly arises through gene transfer from members of other bacterial genera in the human gastrointestinal tract (10). The vanB cluster is mainly found in E. faecium and E. faecalis, though it has also been described in isolates of vanB-resistant Enterococcus gallinarum (11, 12, 13, 14), Enterococcus hirae (15), Enterococcus durans (16), Enterococcus casseliflavus (17), Staphylococcus (18), Streptococcus (19, 20, 21, 22), Eggerthella, Clostridium, Ruminococcus (23, 24, 25, 26, 27), and Atopobium (27).
The vanB gene cluster has a conserved gene order and can be divided into three genetic subtypes, vanB1, vanB2, and vanB3 (28, 29, 30, 31, 32). As an integral part of the conjugative transposon Tn5382/Tn1549, vanB2 is the most-widespread subtype in clinically important enterococci (30, 33, 34, 35, 36, 37, 38). Tn5382/Tn1549 is able to support transfer of the vanB2 operon from Clostridium to enterococci in the intestinal environment (39), but the transposon is more often transferred as a part of larger chromosomal elements or plasmids (30, 35, 37, 40, 41).
An approximately 100-kb transferable chromosomal element containing vanB2 has been described in a patient isolate of Streptococcus lutetiensis previously designated Streptococcus bovis biotype II with vanB3 (22, 42) and in the plasmid-free S. lutetiensis strain 5-F9 from animal feces (20). The vanB2 cluster of the 5-F9 strain has previously been shown to be an integral part of a Tn5382/Tn1549 element (20) and was transferred to E. faecium and E. faecalis at relatively high frequencies (10−7 to 10−5). In contrast, the element was unable to transfer between E. faecium and E. faecalis strains (20, 42), which suggests a coresident transfer system in Streptococcus (42). Retransfer of the ∼100-kb element from the patient isolate between E. faecalis strains, including to a recombination-deficient recipient, was obtained in the presence of a conjugative helper plasmid (42), and the ∼100-kb element appeared to integrate in a site-specific manner (20, 42). Transfer of this ∼100-kb chromosomal element from the S. lutetiensis strain that contains no visible plasmids into enterococci suggests self-encoded transfer functions consistent with an integrative and conjugative element (ICE) (20). The transfer mechanism has not been resolved.
Here, we describe the sequencing, annotation, and functional analysis of the 94-kb element designated ICESluvan, originally detected in S. lutetiensis from animal feces in the Netherlands. A variant of this element was confirmed in the S. lutetiensis patient isolate from France. The element encodes VanB-type vancomycin resistance on conjugative transposon Tn5382/Tn1549, a VanZ homologue which may contribute to the detected low-level resistance to teicoplanin, a novel bacitracin locus which seems to contribute to bacitracin resistance in S. lutetiensis and E. faecium, and a streptococcal toxin-antitoxin (TA) pezAT locus, as well as all essential components for conjugative transfer. ICESluvan excises to form a circular intermediate in S. lutetiensis, E. faecalis, and E. faecium and integrates in a site-specific manner. However, transfer between enterococci required a helper plasmid. The expression of the pezAT operon indicates a functional toxin-antitoxin system.
MATERIALS AND METHODS
Bacterial strains and construction of BAC clones.
The bacterial strains used in this study and their relevant characteristics are given in Table 1. Briefly, genomic DNA from the E. faecium transconjugant MM5-F9a was used to generate a bacterial artificial chromosome (BAC) library with an average insert size of 90 kb (MWG Biotech AG). MM5-F9a was obtained by transfer of the ICESluvan element from S. lutetiensis 5-F9, isolated from the feces of a veal calf in the Netherlands, to recipient E. faecium BM4105-RF by filter mating (20). The vanB2-containing strains S. lutetiensis NEM760, Streptococcus gallolyticus 4-C11, and S. gallolyticus 4-G10 were included to search for ICESluvan in streptococci. E. faecalis transconjugants OG5-F9a, JH5-F9a, and UV5-F9d and laboratory strains OG1-RF, JH2-2, BM4110, and UV202 were used for various studies of ICESluvan as indicated below. Total DNA from E. faecium C68 (40) was used as the template for probe synthesis.
Table 1.
Bacterial strains used in this study and their relevant characteristics
| Strain | Species | Propertiesa | MIC mg liter−1 |
Reference(s) | ||
|---|---|---|---|---|---|---|
| Vancomycin | Teicoplanin | Bacitracin | ||||
| 5-F9 | S. lutetiensis | Vanr veal calf strain with vanB2 Tn5382 chromosomally located | ≥256 | 3 | ≥256 | 19, 20 |
| NEM760 | S. lutetiensis | Vanr human patient strain with vanB2 chromosomally located | ≥256 | 4 | ≥256 | 22, 42 |
| 4-C11 | S. gallolyticus | Vanr veal calf strain with nontransferable vanB2 Tn5382 and vanA | ≥256 | 32 | 24 | 19, 20 |
| 4-G10 | S. gallolyticus | Vanr veal calf strain with nontransferable vanB2 Tn5382 and vanA | ≥256 | 64 | ≥256 | 19, 20 |
| C68 | E. faecium | vanB2 Tn5382 type strain | 40 | |||
| BM4105-RF | E. faecium | Rifr Fusr plasmid-free recipient | 1,5 | 0,5 | 48 | 46 |
| MM5-F9a | E. faecium | Transconjugant resulting from mating of 5-F9 and BM4105-RF | 32 | 3 | ≥256 | 20 |
| OG1-RF | E. faecalis | Rifr Fusr recipient derived from OG1 | 4 | 0,25 | 24 | 73 |
| OG5-F9a | E. faecalis | Transconjugant resulting from mating of 5-F9 and OG1-RF | ≥256 | 3 | 32 | This study |
| JH2-2 | E. faecalis | Rifr Fusr plasmid-free recipient derived from JH2 | 3 | 1 | 16 | 74 |
| JH5-F9a | E. faecalis | Transconjugant resulting from mating of 5-F9 and JH2-2 | ≥256 | 3 | 8 | 20 |
| UV202 | E. faecalis | Rifr Fusr plasmid-free recipient, recombination-deficient derivative of JH2-2 | 1,5 | 0,5 | 128 | 75 |
| UV5-F9d | E. faecalis | Transconjugant resulting from mating of 5-F9 and UV202 | ≥256 | 3 | 192 | 20 |
| BM4110 | E. faecalis | Strr plasmid-free recipient strain derived from JH2 | 1,5 | 0,25 | 96 | 76 |
| BM4110 pIP964 | E. faecalis | Strr recipient strain derived from JH2 containing plasmid pIP964 (pCF10 rep agg+ Tra+, 65 kb) | 1,5 | 0,38 | ≥256 | 76 |
| ICESluvan BM4110 pIP964 | E. faecalis | 1st-generation transconjugant resulting from mating of donor 5-F9 and BM4110 pIP964 | ≥256 | 3 | ≥256 | This study |
| ICESluvan pIP964 JH2-2 | E. faecalis | 2nd-generation transconjugant resulting from mating of ICESluvan BM4110 pIP964 and JH2-2 | ≥256 | 3 | ≥256 | This study |
| ICESluvan pIP964 UV202 | E. faecalis | 2nd-generation transconjugant resulting from mating of ICESluvan BM4110 pIP964 and UV202 | ≥256 | 3 | 192 | This study |
Vanr, vancomycin resistant; Rifr, rifampin resistant; Fusr, fusidic acid resistant; Strr, streptomycin resistant.
Selection and sequencing of BAC clones.
Sequential dot blot and Southern hybridizations were used to identify BAC clones positive for both vanB2 and Tn5382/Tn1549. PCR-based probes were made (PCR digoxigenin [DIG] probe synthesis kit; Boehringer Mannheim) for vanB using the consensus primers 5′-CAAAGCTCCGCAGCTTGCATG-3′ and 5′-TGCATCCAAGCACCCGATAATAC-3′ (29) and for Tn5382 using primers 5′-GTTCTTATTCCGCAGGTGGTGATT-3′ and 5′-ACGCCATGCTATTTACTTCCGGC-3′ (40). Hybridizations were detected using the DIG luminescent detection kit (Boehringer Mannheim). Plasmid DNA was isolated from BAC clones that scored positive for one or both probes (n = 24). Clones were grown overnight in yeast extract-tryptone (YT) medium containing 12.5 μg ml−1 chloramphenicol, plasmid replication was induced (Epicentre induction solution; Epicentre), and BAC DNA was purified using the E.Z.N.A BAC/PAC kit (Omega Bio-Tek). NotI (New England BioLabs)-digested plasmid DNA was separated on a 1.2% agarose gel, 5 to 15 s, 6 V/cm, 16 h, at 15°C on a CHEF-DR III (Bio-Rad) and analyzed by Southern hybridization. Five clones positive for both probes and with an average insert size of 90 kb were selected for sequencing.
Libraries of the BAC inserts of each selected clone were constructed with the use of multiplex identifiers (MIDs) and sequenced to an average depth of 91.5-times coverage using 454-GS-FLX technology. The BAC inserts were de novo assembled individually with Newbler (Roche).
End sequencing of the BAC inserts provided additional geographical information for contig ordering. End sequencing and extension of contigs were set up using extracted BAC DNA and BigDye 3.1 for cycle sequencing and an ABI prism 377 genetic analyzer (Applied Biosystems). The cycle sequencing program was as follows: initial denaturation at 96°C for 5 min followed by 99 cycles of 96°C for 30 s, 52°C for 10 s, and 60°C for 4 min (primers available on request).
DNA extraction and sequencing of transconjugants.
Genomic DNA from transconjugants was obtained using a bacterial DNA kit (E.Z.N.A) with the following modifications: lysis was performed at 30°C for 40 min using 1.5 mg lysozyme and 100 U mutanolysin in a total volume of 220 μl. Direct genome sequencing of the transconjugants was performed as for BAC end sequencing.
Gap closure and annotation.
Abacas (http://abacas.sourceforge.net/index.html) was used to map contigs from the de novo assembly against Streptococcus suis BM407 (FM252032) and Tn1549 (AF192329). The sequence was annotated using Artemis software (43). Homology comparisons to nonredundant protein databases were performed with BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and FASTA (http://www.ebi.ac.uk/Tools/fasta33/index.html) software. Protein motifs were identified using Pfam (http://pfam.sanger.ac.uk/search) and Prosite (http://au.expasy.org/prosite/). Transmembrane domains were identified with TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), and signal sequences were identified with SignalP version 3.0 (44). Comparison of the large mobile elements was facilitated using the Artemis Comparison Tool (ACT) (45), which enabled the visualization of BLASTN and TBLASTX comparisons between these sequences.
Verification of sequence assembly by PCRs.
To verify the sequence assembly, over 40 PCRs with an average product size of 2.5 kb were performed in ICESluvan (see Table S1 in the supplemental material), using genomic DNA of transconjugants E. faecium MM5-F9a and OG5-F9a and E. faecalis JH5-F9a as templates. Some of these primers were used to confirm the presence of ICESluvan regions (see Table S1) in streptococci (S. lutetiensis 5-F9 and NEM760 and S. gallolyticus 4-C11 and 4-G10) and to sequence the bacitracin locus in the E. faecalis transconjugants OG5-F9a and JH5-F9a.
RNA extraction for expression analyses.
The expression of the pezAT operon was analyzed by semiquantitative reverse transcription (RT)-PCR. Total RNA was extracted from stationary-phase cultures using an RNeasy minikit (Qiagen) with an extended lysis step of 1 h with 10 mg lysozyme and 10 U mutanolysin in a total volume of 200 μl. DNA contamination was removed by DNase I treatment, and RNA quality and quantity examined by gel analyses and spectrophotometric measurements prior to cDNA synthesis. Reverse transcription of 100 ng total RNA was performed using SuperScript II (Invitrogen). For each sample, a control reaction (without reverse transcriptase) with no enzyme was included. PCR was performed using primer sets specific for each of the genes (pezAF [5′-TGACCTGGAATCGTCTTTCC-3′] and pezAR [5′-TGGCCGAATTGATTAACACA-3′] and pezTF [5′-TTTTTCCTGCAAATCCCTTG-3′] and pezTR [5′-GACAAAGCGGAGCAGGTAAG-3′]). In addition, PCR using primers pezTF and pezAR was performed. Amplicons were visualized on an agarose gel and confirmed by sequencing using BigDye 3.1 technology (Applied Biosystems).
Verification of ICESluvan circularization.
Circularization of the ICESluvan element was examined by PCR using primers located on the very end of the element (see Fig. 2), as follows: 5′-AAGGGAAAATTAGCGATACC-3′ (nucleotides [nt] 93779 to 93798), 5′-CTGCTGCAAATGTAAATGGC-3′ (nt 93383 to 93402), and 5′-TCCAAACAGACATTCACATCAA-3′ (nt 130 to 151). PCR products were confirmed by sequencing.
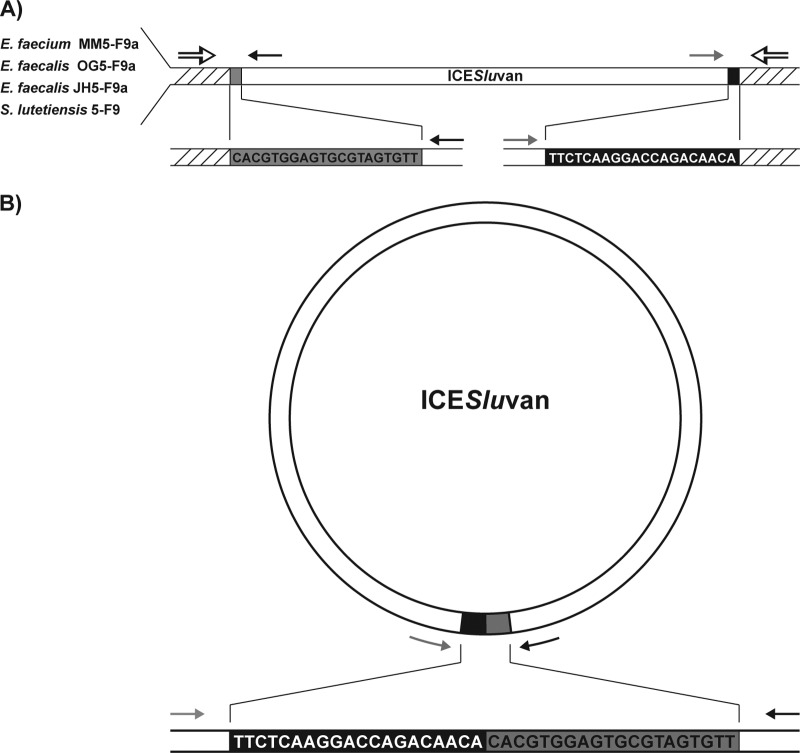
Fig 2.
Integrated (A) and circular (B) forms of ICESluvan in transconjugants E. faecium MM5-F9a, E. faecalis OG5-F9a, and E. faecalis JH5-F9a, as well as donor S. lutetiensis 5-F9. The left and right ends of ICESluvan are colored gray and black, respectively, while the native chromosomal DNA is shown by hatching. The sequences of the 20-bp ends of ICESluvan are also shown. Black and gray arrows indicate the positions and directions of primers for the left and right end of ICESluvan, respectively. The positions of primers designed from native E. faecium MM5-F9a regions are indicated with black-and-white arrows. PCR and sequencing confirmed both the integrated and the circular form of ICESluvan in E. faecium MM5-F9a, E. faecalis OG5-F9a, E. faecalis JH5-F9a, and S. lutetiensis 5-F9.
Transfer of ICESluvan.
Transfer of the ICESluvan element to E. faecalis strains BM4110 and JH2-2 with pIP964 and retransfer from these first-generation transconjugants to recipients (E. faecalis strains JH2-2, UV202, and BM4110) without this helper plasmid were performed by filter mating as previously described (33), with some modifications. Briefly, donor and recipient strains were grown to an A600 of 0.6 and mixed in a ratio of 1:1 in 2 ml of culture. The suspension was pelleted, resuspended in 150 μl brain heart infusion (BHI), and used for filter mating before application on selective agar plates using 8 mg liter−1 vancomycin, 1,000 mg liter−1 streptomycin, 20 mg liter−1 rifampin, and 10 mg liter−1 fusidic acid. A control experiment (E. faecalis JH5-F9a × BM4110) with recipients containing no helper plasmid was also carried out. The presence of the vanB gene of ICESluvan was confirmed by PCR (29), and ICESluvan insertion in the transconjugant genome was confirmed by visual inspection of the SmaI pulsed-field gel electrophoresis (PFGE) pattern. The presence of the pheromone-responsive helper plasmid pIP964 (46) was determined by PCRs that detect the pCF10 replication initiation gene reppCF10 as described by Jensen et al. (47), and the presence of the pheromone plasmid's conserved aggregation substance (agg) was detected using primers 5′-GCTCGTGGTGATGTTCTTTC-3′ and 5′-CTTTTCTACCACTAATGGCTCTAC-3′.
PFGE analyses.
Total DNA was digested with SmaI (New England BioLabs) and analyzed by PFGE as previously described (48), with some modifications. Briefly, 5 μl lysozyme (100 mg ml−1) and 2 μl mutanolysin (10 U μl−1) were added to the agarose plugs before molding, as well as to the lysis buffer.
Antimicrobial susceptibility testing.
Susceptibility testing was performed using MIC test strips for vancomycin, teicoplanin (bioMérieux), and bacitracin (Liofilchem) on S. lutetiensis 5-F9, E. faecalis JH5-F9a, JH2-2, OG5-F9a, and OG1-RF, and E. faecium MM5-F9a and BM4105-RF according to the manufacturers' recommendations.
Nucleotide sequence accession number.
The whole sequence of ICESluvan is available in GenBank under the accession number HE963029.
RESULTS AND DISCUSSION
Gap closure and ICESluvan mosaic structure.
The five selected BAC clones were sequenced to an average depth of 91.5-times coverage and assembled individually into an average of 28 contigs (>500 bp). Mapping of contigs against reference sequences and end sequencing of BAC inserts provided scaffold information for the gap closure that resulted in one large, 94-kb contig (GenBank accession number HE963029) containing 86 open reading frames (ORFs) (Fig. 1; see also Table S2 in the supplemental material). Nine smaller contigs of >1 kb with BLAST similarities to unculturable organisms and transposases were discarded from further analysis. Extensive control PCRs (see Table S1) verified the assembly for E. faecium transconjugant MM5-F9a and confirmed the presence of the ICESluvan element in E. faecalis transconjugants OG5-F9a and JH5-F9a (Table 1). Some primer combinations used for MM5-F9a did not give PCR products with OG5-F9a and JH5-F9a. However, the presence of these regions was confirmed by altering the combination of the primers (denoted by plus signs in Table S1). Some of the PCRs worked at one time point and not at another, and when we sequenced the bacitracin locus of the E. faecalis transconjugants, we found that some of the primer combinations did not give a PCR product even though both primers matched the sequence 100%. Thus, we think there might be other reasons than nucleotide differences, possibly related to the primary or secondary DNA sequence, why PCR products were not achievable for all transconjugants in some specific regions.
Fig 1.
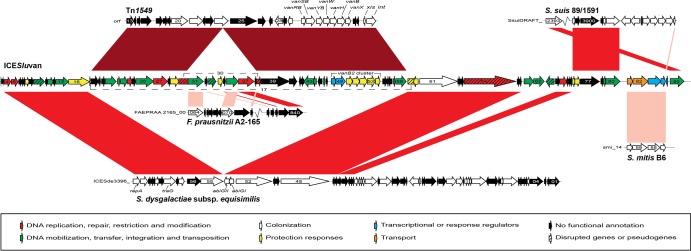
Genetic organization of the 94,189-bp ICESluvan element and linear DNA comparison against Tn1549, the 64-kb ICESde3396 element from S. dysgalactiae subsp. equisimilis, and regions from S. suis 89/1591, S. mitis B6, and Faecalibacterium prausnitzii A2-165. In ICESluvan, coding sequences (CDSs) are colored according to predicted functions. In the other genetic elements, black arrows indicate CDSs with no assigned functions and white arrows indicate CDSs with putative assigned functions. Gene identifiers for Tn1549, S. dysgalactiae subsp. equisimilis, S. suis 89/1591, S. mitis B6, and F. prausnitzii A2-165 are captured from records with GenBank accession numbers AF192329, EU142041, NZ_AAFA03000001, FN568063, and NZ_ACOP00000000, respectively. For simplicity, the CDS prefixes are omitted in the figure. Gene names are indicated if available from annotation. Disrupted genes are linked via dashed lines, and both disrupted and pseudogenes are indicated as crosshatched arrows. For the comparison, red shading between the genetic elements represents regions with similarity: light red indicates regions with >65% amino acid sequence identity; red indicates regions with >90% amino acid sequence identity; and dark red indicates regions with >99% amino acid sequence identity.
Interestingly, homology searches of the 86 predicted ORFs showed that the ICESluvan element contains genes similar to 24 ORFs (ORF4 to -13, ORF15 to -17, ORF60 to -66, and ORF71 to -74) of ICESde3396 found in the beta-hemolytic Streptococcus dysgalactiae subsp. equisimilis NS3396 (49). Furthermore, ICESluvan ORF1 to -3, ORF14, ORF69, and ORF75 to -86 showed similarities to ORFs from three non-beta-hemolytic streptococci, Streptococcus pneumoniae, S. suis, and Streptococcus mitis.
A complete vanB2 Tn5382/Tn1549 (ORF18 to -30 and ORF39 to -59) putative conjugative transposon was integrated into the partial ICESde3396-like sequence in ORF17, similar to ICESde3396_54. The vanB2 Tn5382/Tn1549 sequence of ICESluvan was 99% identical at the nucleotide level to the fully sequenced Tn1549 (Fig. 1; see also Table S2 in the supplemental material) (30). Five more ORFs were identified in the Tn5382/Tn1549 element inserted in ICESluvan compared to those from previous reports (see Table S2). Identical sequences were also present in Tn1549 (30) but have not been annotated previously. In addition, a 9-kb contig located within ORF30 in Tn5382/Tn1549 was flanked by directly repetitive sequences with 291 of 292 nucleotides identical. Annotation of the 9-kb contig revealed a TnpX site-specific recombinase (ORF31), a pseudotransposase of the IS30 family (ORF33), a putative VanZ family protein (ORF34), a putative mobilization protein (ORF36), and a DNA primase (ORF37), as well as hypothetical proteins (ORF32, -35, and -38) with 40 to 96% amino acid identity to putative proteins in Faecalibacterium, Lactobacillus, and Paenibacillus species, respectively (see Table S2).
The mosaic pattern of the ICESluvan element, which consists of genes from several different streptococcal species, Faecalibacterium, enterococci, and possibly other intestinal bacteria (see Table S2 in the supplemental material), indicate that mobilization and recombination events have been key factors in the assembly of this element. A mosaic pattern was also observed for ICESde3396, which is a montage of genes derived from group A, B, and G Streptococcus organisms, in addition to genes acquired from nonstreptococcal Gram-positive bacteria, such as Listeria innocua and E. faecalis (49).
ICESluvan variant confirmed in an S. lutetiensis isolate from a patient in France.
During examination of mixed fecal samples from 556 veal calf herds in the Netherlands, four vancomycin-resistant Streptococcus isolates were found. In addition to S. lutetiensis 5-F9, S. gallolyticus isolates 4-C11 and 4-G10 were positive for the vanB gene and found to have a vanB2 cluster integrated in Tn5382/Tn1549 (20). Furthermore, S. lutetiensis NEM760, isolated from a stool swab of a patient in France, showed transfer of an approximately 100-kb element containing vanB2 (22, 42) that inserted into the same SmaI PFGE fragment as ICESluvan in both E. faecium BM4105-RF- and E. faecalis JH2-derived recipients (20, 22, 42). Since 4-C11, 4-G10, and NEM760 are likely candidates to contain ICESluvan or variants of this element, we have examined these isolates by using primers selected to confirm the ICESluvan sequence assembly (see Table S1 in the supplemental material). The circa-100-kb element found in NEM760 was indeed very similar to ICESluvan, with some differences in the right end of the element, while 4-C11 and 4-G10 showed the presence of Tn5382/Tn1549 and adjacent regions but not the left or right end of ICESluvan, which is not surprising since transfer of vanB was not achieved for these isolates (19, 20). The presence of highly similar variants of ICESluvan capable of interspecies transfer in two S. lutetiensis isolates from completely different environments implies that ICESluvan is a successful element that may contribute to resistance in enterococci.
ICESluvan type IV secretion system components.
A putative transfer system (ORF11 to -16 and ORF79 to -80) (see Table S2 in the supplemental material) was identified in the ICESluvan element, in addition to those encoded by Tn5382/Tn1549 (ORF21 to -23, ORF26, and ORF43 and -44) (see Table S2) (30). ORF11 to -15 were 94 to 99% identical to ICESde3396_59 to ICESde3396_55 (49). The ORFs display the same genetic order in the ICESluvan element and belong to a type IV secretion system. Three factors have been recognized as important for DNA transfer by type IV secretion system in Gram-positive bacteria. These are murein hydrolase (50), which is involved in controlled local degradation of the peptidoglycan, making space for the formation of a mating channel, the VirB4 ATPase, which provides energy for the translocation, and the VirD4 coupling protein, which links the DNA transfer intermediate to the mating channel (50, 51). In the ICESluvan element, we identified two putative virB4 genes (ORF15 and OFR26) encoding ATPases, a putative virB6 gene (ORF13) encoding a putative membrane protein that has previously been shown to interact with several other Vir proteins mediating DNA substrate transfer through the cytoplasmic membrane channel (50), and two putative virD4 genes (ORF11 and ORF21) encoding coupling proteins of the TraG/TraD family. ORF11 is truncated by a frameshift after codon 465, but ORF21 encodes a putative coupling protein which belongs to the same coupling-protein family. Moreover, ICESluvan ORF16 encodes a putative N-acetylmuramoyl l-alanine amidase that may aid in local degradation of the peptidoglycan by hydrolyzing the amide bond between the N-acetylmuramic acid side chain and l-alanine of the short peptide (52). Genes encoding putative mobilization proteins of the MobC family (ORF79 and ORF44) and relaxases (ORF80 and ORF43) were also identified in ICESluvan. Taken together, this indicates that the ICESluvan element contains most genes necessary for mobilization and conjugative transfer. Furthermore, ICESluvan is transferable by conjugation from S. lutetiensis organisms with no visible plasmids, strongly suggesting that its transfer system is intact (20).
ICESluvan circularization, transfer, and integration.
A putative site-specific serine recombinase (ORF86) (see Table S2 in the supplemental material) that can be involved in the genomic exit and integration of mobile genetic elements was found at the right flank of the ICESluvan. Although it lacks 1 or 20 amino acids in the C-terminal end compared to the sequences of its closest homologues, SsuiDRAFT_2393, identified in S. suis (98% amino acid identity), and SP70585_1107, identified in S. pneumoniae (95% amino acid identity), we hypothesize that the putative site-specific recombinase functions as normal in a transfer situation. This hypothesis was supported by circularization of the ICESluvan element both in the S. lutetiensis donor 5-F9 and the Enterococcus transconjugants, as shown by PCR and confirmed by sequence analyses (Fig. 2). Furthermore, we have transferred ICESluvan from S. lutetiensis 5-F9 to both E. faecium and E. faecalis (20). Retransfer was only possible when ICESluvan was transferred into recipients that contain the helper plasmid pIP964 (Tra+), which is restricted to E. faecalis. We did not test broad-host-range plasmids to achieve retransfer between E. faecium strains. Retransfer from E. faecalis BM4110 ICESluvan pIP964 to E. faecalis JH2-2 or UV202 (Fig. 3, lanes 4 to 6) and from JH2-2 ICESluvan pIP964 to BM4110 (data not shown) was confirmed by growth on selective plates, ICESluvan- and pIP964-specific PCRs, SmaI PFGE analysis (Fig. 3), and hybridization with vanB probe (data not shown) showing transconjugant patterns with an approximately 100-kb enlargement of one of the two 240-kb fragments compared to the recipient patterns (Fig. 3, lanes 4 to 6). However, transconjugant BM4110 ICESluvan pIP964 had gained an extra copy of ICESluvan, as shown by the presence of a fragment around 220 kb in Fig. 3, lane 4, which hybridized with vanB (data not shown). The presence of more than one copy of ICESluvan has been shown before in other JH-derived transconjugants (20). Both SmaI PFGE patterns and DNA sequencing show that the ICESluvan element is integrated in a site-specific manner into the recipient chromosome, although the E. faecium and E. faecalis integration sites within the C-terminal end of tRNA methyltransferase gene rumA showed some sequence differences (Fig. 4). The same integration site was identified within each species for all transconjugants tested (data not shown). The ICESluvan integration site in S. lutetiensis is also within the rumA gene. However, the right flank of ICESluvan in S. lutetiensis showed an extended 5.7-kb region identical to part of the E. faecalis G1-01247 vanG operon which was not transferred together with ICESluvan into the enterococci (see Fig. S1 in the supplemental material). Interestingly, ICESluvan ORF1 is a truncated homolog identical to the 13-amino-acid C-terminal part of S. pneumoniae rumA (see Table S2). This C-terminal part is in the same translational frame as the N-terminal part of the enterococcal rumA, and together, they form a recombined full-length rumA gene that is likely to be functional.
Fig 3.
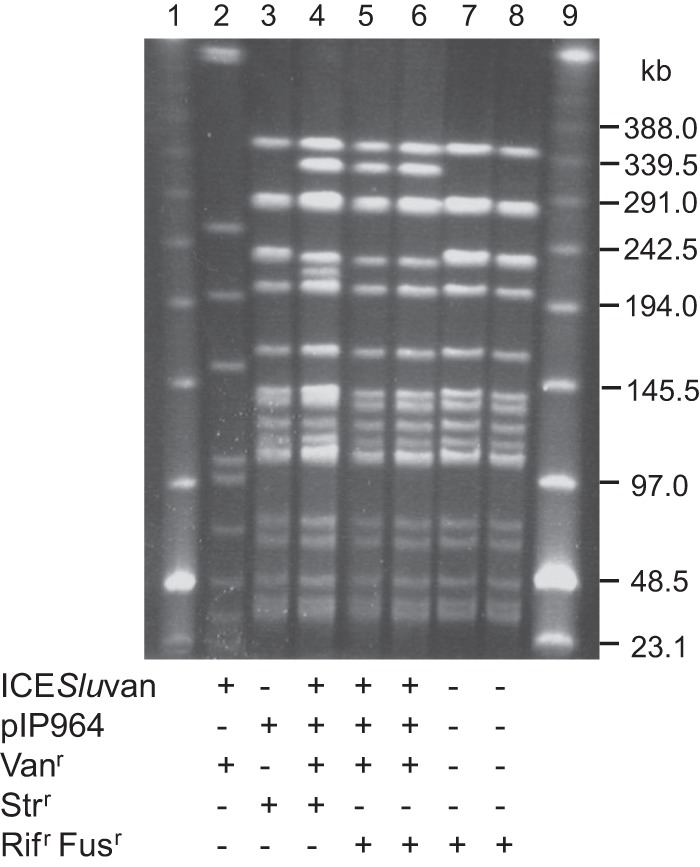
PFGE of SmaI-digested total DNA of recipient, donor, and transconjugants from filter matings. Lanes 1 and 9, low-range PFGE marker; lane 2, ICESluvan donor S. lutetiensis 5-F9; lane 3, recipient BM4110 containing pIP964; lane 4, 1st-generation transconjugant/2nd-generation donor BM4110 ICESluvan pIP964; lane 5, 2nd-generation transconjugant JH2-2 ICESluvan pIP964; lane 6, 2nd-generation transconjugant UV202 ICESluvan pIP964; lane 7, recipient JH2-2; lane 8, recipient UV202. Vancomycin, streptomycin, and rifampin and fusidic acid resistance are indicated by plus signs opposite Vanr, Strr, and Rifr Fusr, respectively. The presence of ICESluvan was verified by vanB PCR and visual inspection of the SmaI PFGE pattern, where ICESluvan insertion in a 240-kb band results in replacement of one of the 240-kb double bands with a 340-kb band, as shown previously for JH2-derived recipients (20). The presence of the pheromone-responsive plasmid pIP964 was verified by reppCF10 and agg PCRs.
Fig 4.
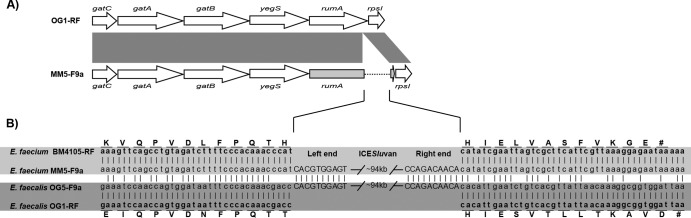
(A) Operon into which ICESluvan has been integrated in E. faecium MM5-F9a compared to the corresponding region in the fully sequenced E. faecalis OG1-RF (accession number CP002621). Dark gray shading indicates regions of similarity between compared CDSs. The tRNA (uracil-5)-methyltransferase rumA disrupted by the insertion is colored light gray, and the insertion is indicated with a dotted line. (B) Sequence comparison of the insertion regions of ICESluvan in E. faecium MM5-F9a and E. faecalis OG5-F9a and the corresponding regions in their recipient strains, E. faecium BM4105-RF and E. faecalis OG1-RF, respectively. Vertical lines indicate identical nucleotides. Left- and right-end sequences of ICESluvan are shown in uppercase. Amino acid residues of CDSs that have been disrupted upon insertion of ICESluvan are marked (capital and boldface), together with their respective codons. #, stop codon.
Since ICESluvan was shown to transfer from Streptococcus to Enterococcus but retransfer between enterococci required a helper plasmid, we hypothesize that a host factor is necessary for transfer. This has been shown for SXT elements in Vibrio cholerae, which require the host factors IHF and Fis for excision, recombination, and conjugation (53). Host factors may be chromosomally encoded and not on the MGE itself, leading to the inability of the MGE to conjugate in their absence. Transfer of the E. faecalis V583 pathogenicity island (PAI), containing the features of an ICE, by an ICE-independent mechanism has been reported. Characterization of V583 PAI transconjugants showed cotransfer of selectable markers representing virtually all regions of the chromosome, including a vancomycin resistance transposon, capsule genes, and alleles which are used for multilocus sequence typing. PAI transfer was dependent upon helper plasmid function in the donors (54).
Putative bacteriophage exclusion system.
The ICESluvan ORF17 located upstream and ORF60 to -64 located downstream from Tn5382/Tn1549 are 81 to 97% identical to ICESde3396_54 to ICESde3396_48. ORF60 and ORF17 are homologous to ICESde3396_54 and ICESde3396_53, encoding the abortive infection proteins AbiGI and AbiGII, respectively. The Lactococcus lactis abiG resistance genes confer complete resistance to ϕ712 phages (936 phage species) and partial resistance to ϕc2 phages (2 species) (55). Most Abi systems are plasmid encoded and are widespread in bacteria (56). The abi genes are also known to be involved in increased stress tolerance of the host. These genes are commonly found in Streptococcus (57). Transposon Tn5382/Tn1549 was inserted in the C-terminal end after codon 251 of abiGII in ICESluvan (Fig. 1), thereby disrupting this gene. Since this leads to an AbiGII protein lacking 30 amino acids compared to its closest homologue, it is not known whether these abi genes encode a functional system.
Putative virulence determinants.
The ICESluvan ORF61 was 95% identical at the nucleotide level to ICESde3396_52, which encodes a putative agglutinin receptor precursor protein that harbors an LPXTG motif known to be common among several virulence factors in enterococci (58). Human salivary agglutinin has previously been shown to interact with streptococci in a calcium-dependent reaction for oral bacterial aggregation (59, 60). The ICESluvan putative agglutinin receptor precursor is located next to a putative Ca2+ binding protein (ORF62). Studies have shown that transformation of nonaggregating E. faecalis with the streptococcal surface antigen SSP-5 confers an aggregation-positive phenotype in the presence of saliva agglutinin (61).
Resistance to glycopeptides and bacitracin.
The ICESluvan element encodes vancomycin resistance mediated by a vanB2 cluster (ORF48 to -54) located in Tn5382/Tn1549. S. lutetiensis 5-F9 and the transconjugants, as well as S. lutetiensis NEM760 and S. gallolyticus 4-C11 and 4-G10, accordingly expressed resistance to vancomycin, whereas the recipient strains E. faecium BM4105-RF and E. faecalis OG1-RF, JH2-2, UV202, BM4110, and BM4110 pIP964 were susceptible to vancomycin according to EUCAST clinical breakpoints for enterococci (MIC, ≤4 mg liter−1) (Table 1). In addition, ORF34 on the 9-kb element inserted into Tn5382/Tn1549 encoded a protein with 40% amino acid similarity to a putative VanZ family protein from Paenibacillus (see Table S2 in the supplemental material). The vanZ gene, contributing to teicoplanin resistance, is normally located within the vanA gene cluster, whereas vanB usually only confers resistance to vancomycin. The functionality of the VanZ homologue was supported by Etest results showing MICs of 3 or 4 mg liter−1 to teicoplanin (low-level resistance according to EUCAST clinical breakpoints for enterococci) for S. lutetiensis NEM760 and 5-F9 and the 5-F9 transconjugants, which was at least 3-fold higher than the MICs of the corresponding susceptible recipient strains (BM4105-RF and UV202 MIC, 0.5 mg liter−1; JH2-2 MIC, 1 mg liter−1; BM4110 pIP964 MIC, 0.38 mg liter−1; and OG1-RF MIC, 0.25 mg liter−1) (Table 1). The finding of this vanZ gene illustrates the dynamics and the potential of the Streptococcus and Enterococcus genomes to acquire and collect genes which may increase their ability for environmental adaptation.
The streptococcal ICESde3396 encodes resistance toward cadmium and arsenic (49). These resistance genes were not identified in ICESluvan. Rather, ORF81 to -84 encode an ABC transporter (ATPase and permease) and a putative two-component system (histidine kinase and response regulator) resembling proteins BceA, -B, -R, and -S (previously designated MbrABCD), which are involved in bacitracin resistance in S. mutans through active efflux of bacitracin (GenBank accession number AB078507) (62, 63). ORF81 encodes 249 amino acids with 48% identity to the original S. mutans BceA (250 amino acids) and 82% identity to a putative BceA of S. mitis (GenBank accession number EFN94736) (245 amino acids), ORF82 encodes 672 amino acids with 29% identity to the original S. mutans BceB (667 amino acids) and 67% identity to a putative ABC transporter permease of S. mitis (GenBank accession number EFN94737) (671 amino acids), ORF83 encodes 524 amino acids with >20% identity to BceS (249 amino acids) and 65% identity to a putative membrane protein with a histidine kinase domain of S. mitis (GenBank accession number EFN94738) (524 amino acids), and ORF84 encodes 198 amino acids with 24% identity to BceR (223 amino acids) and 75% identity to a putative response regulator of S. mitis (GenBank accession number EFN94739) (198 amino acids). Compared to the bceABRS locus of S. mutans, ICESluvan holds a different gene order of the putative response regulator and histidine kinase. The putative bacitracin locus of ICESluvan shows about 20% amino acid identity to components encoded by a bcrRABD locus conferring high-level bacitracin resistance in E. faecalis, which show yet another synteny with the regulator gene upstream from the ATPase, permease, and kinase genes (64). Phenotypic testing indeed shows at least a 5-fold-increased MIC for bacitracin associated with transfer of ICESluvan from S. lutetiensis 5-F9 to E. faecium. The E. faecium transconjugant MM5-F9a expressed a bacitracin MIC of ≥256 mg liter−1, while the MIC of the corresponding E. faecium recipient BM4105-RF was 48 mg liter−1, implying functionality of the putative bacitracin locus of ICESluvan as a novel bacitracin resistance locus. On the other hand, the E. faecalis transconjugants and their corresponding recipients showed similar low or high MICs to bacitracin, except for the second-generation transconjugant JH2-2 ICESluvan pIP964, which after transfer from the high-bacitracin-MIC background in E. faecalis BM4110 pIP964 showed at least a 16-fold-increased MIC compared to that of JH2-2 (Table 1). Undecaprenyl pyrophosphate phosphatase has recently been shown to account for the low-level resistance to bacitracin (MICs of 32 to 48 mg liter−1) in both laboratory (JH2-2) and clinical (V583) strains of E. faecalis (65), while variants of the transferable bcrRABD locus may be responsible for the high-level bacitracin resistance (MIC ≥128 mg liter−1) in enterococci (64, 66). Alternative primer combinations had to be used to confirm the bacitracin locus region of ICESluvan (see Q38 in Table S1 in the supplemental material) in E. faecalis JH5-F9a and OG5-F9a compared to that in E. faecium MM5-F9a, indicating some sequence differences. However, sequencing E. faecalis JH5-F9a and OG5-F9a revealed identical nucleotides of the bacitracin locus (nt 85669 to 91794) compared to the sequence of MM5-F9a (GenBank accession number HE963029). Thus, host-related factors of E. faecalis and E. faecium may be a possible explanation for the lack of increased bacitracin MICs when ICESluvan was introduced directly into E. faecalis strains with low-level resistance to bacitracin.
ICESluvan expresses a pezAT TA system.
ICESluvan ORF75 and ORF76 are 93 and 89% identical at the nucleotide level to the pezAT plasmid maintenance system previously described in S. pneumoniae (67), S. agalacticae (57, 67), and S. suis (57). The pezAT gene cassette consists of two genes, encoding an epsilon antitoxin and a zeta toxin, respectively. Toxin-antitoxin systems are traditionally known as plasmid addiction systems that ensure stable maintenance of plasmids in a bacterial population (68). However, during recent years, these gene loci have been found on MGEs and shown to promote the maintenance of ICE elements in V. cholerae (69, 70). Experimental evidence also indicated that TA systems are involved in the stress response, enabling the cell to survive hostile growth conditions (71, 72). Analyses of the DNA sequence upstream from the pezAT locus showed the presence of a putative −10 promoter sequence (TATAAT) 34 bp upstream from an indicated start codon and a −35 promoter sequence (GTGCGTT) 19 bp from the putative Pribnow box. In addition, a putative ribosome-binding site (AGGAG) was shown 12 bp upstream from the putative start codon. All except the −35 sequence were identical to the S. pneumoniae sequences (67). RT-PCR analyses revealed the expression of both pezAT genes (data not shown) in all of the donor and transconjugant strains tested. Figure 5 shows the PCR products of both genes. Moreover, both genes were detected in a single transcript (Fig. 5), confirming that pezA and pezT constitute an operon, as in S. pneumoniae (67). PCR products were not detected in the absence of reverse transcriptase or in the recipient strain (BM4105-RF). All sequences were identical to the sequence found in our E. faecium transconjugant. These observations suggest that this is a functional toxin-antitoxin system that may contribute to the stabilization of ICESluvan after integration into new hosts.
Fig 5.
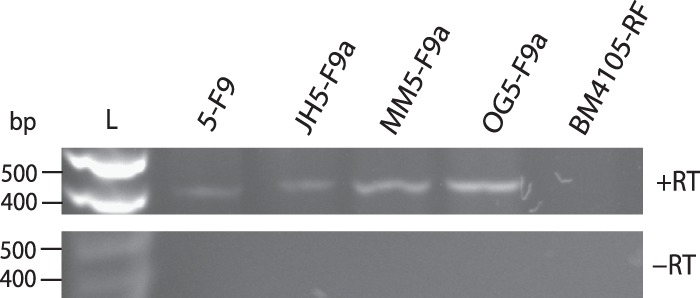
Expression of pezAT toxin-antitoxin loci in original donor S. lutetiensis 5-F9, transconjugants E. faecalis JH5-F9a, E. faecium MM5-F9a, and E. faecalis OG5-F9a, and recipient strain E. faecium BM4105-RF tested with RT-PCR (+RT). As DNA contamination controls, reaction mixtures with no reverse transcriptase added were included (-RT). L, kb ladder.
In conclusion, our report describes the complete genetic structure of a 94-kb element, named ICESluvan, originally detected in S. lutetiensis from veal calf feces in the Netherlands. A variant of ICESluvan is confirmed for an S. lutetiensis isolate from a patient in France. ICESluvan encodes glycopeptide and bacitracin resistance loci, a putative bacteriophage exclusion system, a putative virulence gene, and components necessary for conjugative transfer, as well as a toxin-antitoxin stabilization system. This element is a novel ICE, since it forms a circular intermediate, is self-transferable from streptococci to enterococci, and integrates into the chromosome in a site-specific manner.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by research grants from the Norwegian Research Council (projects no. 165997 and 183653/S19), Northern Norway Regional Health Authority Medical Research Program, and the European Commission (LSHE-CT-2007-03410 ACE).
We thank Tracy Munthali Lunde, Christina Borch-Pedersen, and Trine Tessem for their excellent technical assistance We also thank Eirik W. Lundblad and Johanna E. Sollid for critical reading of the manuscript.
Footnotes
Published ahead of print 27 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02165-12.
REFERENCES
- 1.Van Schaik W, Top Riley JD, Boekhorst J, Vrijenhoek J, Schapendonk C, Hendrickx A, Nijman I, Bonten M, Tettelin H, Willems R. 2010. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239. 10.1186/1471-2164-11-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam MM, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PD, Howden BP, Stinear TP. 2012. Comparative analysis of the first complete Enterococcus faecium genome. J. Bacteriol. 194:2334–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Schaik W, Willems RJ. 2010. Genome-based insights into the evolution of enterococci. Clin. Microbiol. Infect. 16:527–532 [DOI] [PubMed] [Google Scholar]
- 4.Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 5.Uttley AH, Collins CH, Naidoo J, George RC. 1988. Vancomycin-resistant enterococci. Lancet i:57–58 [DOI] [PubMed] [Google Scholar]
- 6.Leclercq R, Derlot E, Duval J, Courvalin P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157–161 [DOI] [PubMed] [Google Scholar]
- 7.Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinsson KG, Leclercq R, Lester CH, Lillie M, Novais C, Olsson-Liljequist B, Peixe LV, Sadowy E, Simonsen GS, Top Vuopio-Varkila JJ, Willems RJ, Witte W, Woodford N. 2008. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 13:1–11 [PubMed] [Google Scholar]
- 8.Galloway-Pena JR, Nallapareddy SR, Arias CA, Eliopoulos GM, Murray BE. 2009. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J. Infect. Dis. 200:1566–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guardabassi L, Agersø Y. 2006. Genes homologous to glycopeptide resistance vanA are widespread in soil microbial communities. FEMS Microbiol. Lett. 259:221–225 [DOI] [PubMed] [Google Scholar]
- 10.Howden BP, Holt KE, Lam MM, Seemann T, Ballard S, Coombs GW, Tong SY, Grayson ML, Johnson PD, Stinear TP. 2013. Genomic insights to control the emergence of vancomycin-resistant enterococci. mBio 4(4):e00412–13. 10.1128/mBio.00412-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii Y, Ohno A, Kashitani S, Iwata M, Yamaguchi K. 1996. Identification of VanB-type vancomycin resistance in Enterococcus gallinarum from Japan. J. Infect. Chemother. 2:102–105 [DOI] [PubMed] [Google Scholar]
- 12.Simonsen GS, Andersen BM, Digranes A, Harthug S, Jacobsen T, Lingaas E, Natas OB, Olsvik Ø Ringertz SH, Skulberg A, Syversen G, Sundsfjord A. 1998. Low faecal carrier rate of vancomycin resistant enterococci in Norwegian hospital patients. Scand. J. Infect. Dis. 30:465–468 [DOI] [PubMed] [Google Scholar]
- 13.Liassine N, Frei R, Jan I, Auckenthaler R. 1998. Characterization of glycopeptide-resistant enterococci from a Swiss hospital. J. Clin. Microbiol. 36:1853–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schooneveldt JM, Marriott RK, Nimmo GR. 2000. Detection of a vanB determinant in Enterococcus gallinarum in Australia. J. Clin. Microbiol. 38:3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres C, Tenorio C, Portillo A, Garcia M, Martinez C, Del CR, Ruiz-Larrea F, Zarazaga M. 2003. Intestinal colonization by vanA- or vanB2-containing enterococcal isolates of healthy animals in Spain. Microb. Drug Resist. 9(Suppl 1):S47–S52 [DOI] [PubMed] [Google Scholar]
- 16.Jenney A, Franklin C, Liolios L, Spelman D. 2000. Enterococcus durans vanB. J. Antimicrob. Chemother. 46:515. [DOI] [PubMed] [Google Scholar]
- 17.Ameri S, Talebi M, Rahimi F, Pourshafie MR, Ebrahimipour G. 2009. The homogeneity of vanB gene cluster among enterococcal isolates in Iran. Lett. Appl. Microbiol. 48:157–161 [DOI] [PubMed] [Google Scholar]
- 18.Roberts MC, Soge OO, Giardino MA, Mazengia E, Ma G, Meschke JS. 2009. Vancomycin-resistant Enterococcus spp. in marine environments from the West Coast of the U. S. A. J. Appl. Microbiol. 107:300–307 [DOI] [PubMed] [Google Scholar]
- 19.Mevius D, Devriese L, Butaye P, Vandamme P, Verschure M, Veldman K. 1998. Isolation of glycopeptide resistant Streptococcus gallolyticus strains with vanA, vanB, and both vanA and vanB genotypes from faecal samples of veal calves in The Netherlands. J. Antimicrob. Chemother. 42:275–276 [DOI] [PubMed] [Google Scholar]
- 20.Dahl KH, Sundsfjord A. 2003. Transferable vanB2 Tn5382-containing elements in fecal streptococcal strains from veal calves. Antimicrob. Agents Chemother. 47:2579–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poyart C, Quesne G, Trieu-Cuot P. 2002. Taxonomic dissection of the Streptococcus bovis group by analysis of manganese-dependent superoxide dismutase gene (sodA) sequences: reclassification of ‘Streptococcus infantarius subsp. coli' as Streptococcus lutetiensis sp. nov. and of Streptococcus bovis biotype 11.2 as Streptococcus pasteurianus sp. nov. Int. J. Syst. Evol. Microbiol. 52:1247–1255 [DOI] [PubMed] [Google Scholar]
- 23.Stinear TP, Olden DC, Johnson PD, Davies JK, Grayson ML. 2001. Enterococcal vanB resistance locus in anaerobic bacteria in human faeces. Lancet 357:855–856 [DOI] [PubMed] [Google Scholar]
- 24.Ballard SA, Pertile KK, Lim M, Johnson PDR, Grayson ML. 2005. Molecular characterization of vanB elements in naturally occurring gut anaerobes. Antimicrob. Agents Chemother. 49:1688–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ballard SA, Grabsch EA, Johnson PDR, Grayson ML. 2005. Comparison of three PCR primer sets for identification of vanB gene carriage in feces and correlation with carriage of vancomycin-resistant enterococci: interference by vanB-containing anaerobic bacilli. Antimicrob. Agents Chemother. 49:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domingo MC, Huletsky A, Giroux R, Boissinot K, Picard FJ, Lebel P, Ferraro MJ, Bergeron MG. 2005. High prevalence of glycopeptide resistance genes vanB, vanD, and vanG not associated with enterococci in human fecal flora. Antimicrob. Agents Chemother. 49:4784–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marvaud JC, Mory F, Lambert T. 2011. Clostridium clostridioforme and Atopobium minutum clinical isolates with vanB-type resistance in France. J. Clin. Microbiol. 49:3436–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evers S, Courvalin P. 1996. Regulation of VanB-type vancomycin resistance gene expression by the VanS(B)-VanR(B) two-component regulatory system in Enterococcus faecalis V583. J. Bacteriol. 178:1302–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahl KH, Simonsen GS, Olsvik Ø Sundsfjord A. 1999. Heterogeneity in the vanB gene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 43:1105–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garnier F, Taourit S, Glaser P, Courvalin P, Galimand M. 2000. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146:1481–1489 [DOI] [PubMed] [Google Scholar]
- 31.Gold HS, Unal S, Cercenado E, Thauvin-Eliopoulos C, Eliopoulos GM, Wennersten CB, Moellering RC., Jr 1993. A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes of enterococci. Antimicrob. Agents Chemother. 37:1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel R, Uhl JR, Kohner P, Hopkins MK, Steckelberg JM, Kline B, Cockerill FR., III 1998. DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcus isolates. Antimicrob. Agents Chemother. 42:202–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahl KH, Lundblad EW, Røkenes TP, Olsvik Ø Sundsfjord A. 2000. Genetic linkage of the vanB2 gene cluster to Tn5382 in vancomycin-resistant enterococci and characterization of two novel insertion sequences. Microbiology 146:1469–1479 [DOI] [PubMed] [Google Scholar]
- 34.McGregor KF, Nolan C, Young HK, Palepou MF, Tysall L, Woodford N. 2001. Prevalence of the vanB2 gene cluster in vanB glycopeptide-resistant enterococci in the United Kingdom and the Republic of Ireland and its association with a Tn5382-like element. Antimicrob. Agents Chemother. 45:367–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng B, Tomita H, Inoue T, Ike Y. 2009. Isolation of VanB-type Enterococcus faecalis strains from nosocomial infections: first report of the isolation and identification of the pheromone-responsive plasmids pMG2200, encoding VanB-type vancomycin resistance and a Bac41-type bacteriocin, and pMG2201, encoding erythromycin resistance and cytolysin (Hly/Bac). Antimicrob. Agents Chemother. 53:735–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demertzi E, Palepou MF, Kaufmann ME, Avlamis A, Woodford N. 2001. Characterisation of vanA and vanB elements from glycopeptide-resistant Enterococcus faecium from Greece. J. Med. Microbiol. 50:682–687 [DOI] [PubMed] [Google Scholar]
- 37.Hanrahan J, Hoyen C, Rice LB. 2000. Geographic distribution of a large mobile element that transfers ampicillin and vancomycin resistance between Enterococcus faecium strains. Antimicrob. Agents Chemother. 44:1349–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdezate S, Labayru C, Navarro A, Mantecon MA, Ortega M, Coque TM, Garcia M, Saez-Nieto JA. 2009. Large clonal outbreak of multidrug-resistant CC17 ST17 Enterococcus faecium containing Tn5382 in a Spanish hospital. J. Antimicrob. Chemother. 63:17–20 [DOI] [PubMed] [Google Scholar]
- 39.Launay A, Ballard SA, Johnson P, Grayson M, Lambert T. 2006. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob. Agents Chemother. 50:1054–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carias LL, Rudin SD, Donskey CJ, Rice LB. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dahl KH, Røkenes TP, Lundblad EW, Sundsfjord A. 2003. Nonconjugative transposition of the vanB-containing Tn5382-like element in Enterococcus faecium. Antimicrob. Agents Chemother. 47:786–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poyart C, Pierre C, Quesne G, Pron B, Berche P, Trieu-Cuot P. 1997. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob. Agents Chemother. 41:24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 44.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 45.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 46.Poyart C, Trieu-Cuot P. 1994. Heterogeneric conjugal transfer of the pheromone-responsive plasmid pIP964 (IncHlyI) of Enterococcus faecalis in the apparent absence of pheromone induction. FEMS Microbiol. Lett. 122:173–179 [DOI] [PubMed] [Google Scholar]
- 47.Jensen LB, Garcia-Migura L, Valenzuela AJ, Lohr M, Hasman H, Aarestrup FM. 2010. A classification system for plasmids from enterococci and other Gram-positive bacteria. J. Microbiol. Methods 80:25–43 [DOI] [PubMed] [Google Scholar]
- 48.Saeedi B, Hallgren A, Jonasson J, Nilsson LE, Hanberger H, Isaksson B. 2002. Modified pulsed-field gel electrophoresis protocol for typing of enterococci. APMIS 110:869–874 [DOI] [PubMed] [Google Scholar]
- 49.Davies MR, Shera J, Van Domselaar GH, Sriprakash KS, McMillan DJ. 2009. A novel integrative conjugative element mediates genetic transfer from group G Streptococcus to other beta-hemolytic streptococci. J. Bacteriol. 191:2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grohmann E, Muth G, Espinosa M. 2003. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Layec S, Decaris B, Leblond-Bourget N. 2008. Diversity of Firmicutes peptidoglycan hydrolases and specificities of those involved in daughter cell separation. Res. Microbiol. 159:507–515 [DOI] [PubMed] [Google Scholar]
- 53.McLeod SM, Burrus V, Waldor MK. 2006. Requirement for Vibrio cholerae integration host factor in conjugative DNA transfer. J. Bacteriol. 188:5704–5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manson JM, Hancock LE, Gilmore MS. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc. Natl. Acad. Sci. U. S. A. 107:12269–12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Connor L, Coffey A, Daly C, Fitzgerald GF. 1996. AbiG, a genotypically novel abortive infection mechanism encoded by plasmid pCI750 of Lactococcus lactis subsp. cremoris UC653. Appl. Environ. Microbiol. 62:3075–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chopin MC, Chopin A, Bidnenko E. 2005. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 8:473–479 [DOI] [PubMed] [Google Scholar]
- 57.Holden MT, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, Goodhead I, Mungall K, Quail MA, Price C, Rabbinowitsch E, Sharp S, Croucher NJ, Chieu TB, Mai NT, Diep TS, Chinh NT, Kehoe M, Leigh JA, Ward PN, Dowson CG, Whatmore AM, Chanter N, Iversen P, Gottschalk M, Slater JD, Smith HE, Spratt BG, Xu J, Ye C, Bentley S, Barrell BG, Schultsz C, Maskell DJ, Parkhill J. 2009. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One 4:e6072. 10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hendrickx APA, Willems RJL, Bonten MJM, van Schaik W. 2009. LPxTG surface proteins of enterococci. Trends Microbiol. 17:423–430 [DOI] [PubMed] [Google Scholar]
- 59.Malamud D, Appelbaum B, Kline R, Golub EE. 1981. Bacterial aggregating activity in human saliva: comparisons of bacterial species and strains. Infect. Immun. 31:1003–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rundegren J. 1986. Calcium-dependent salivary agglutinin with reactivity to various oral bacterial species. Infect. Immun. 53:173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demuth DR, Berthold P, Leboy PS, Golub EE, Davis CA, Malamud D. 1989. Saliva-mediated aggregation of Enterococcus faecalis transformed with a Streptococcus sanguis gene encoding the SSP-5 surface antigen. Infect. Immun. 57:1470–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsuda H, Yamashita Y, Shibata Y, Nakano Y, Koga T. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob. Agents Chemother. 46:3756–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ouyang J, Tian XL, Versey J, Wishart A, Li YH. 2010. The BceABRS four-component system regulates the bacitracin-induced cell envelope stress response in Streptococcus mutans. Antimicrob. Agents Chemother. 54:3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manson JM, Keis S, Smith JM, Cook GM. 2004. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob. Agents Chemother. 48:3743–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shaaly A, Kalamorz F, Gebhard S, Cook GM. 2013. Undecaprenyl pyrophosphate phosphatase confers low-level resistance to bacitracin in Enterococcus faecalis. J. Antimicrob. Chemother. 68:1583–1593 [DOI] [PubMed] [Google Scholar]
- 66.Matos R, Pinto VV, Ruivo M, Lopes MF. 2009. Study on the dissemination of the bcrABDR cluster in Enterococcus spp. reveals that the BcrAB transporter is sufficient to confer high-level bacitracin resistance. Int. J. Antimicrob. Agents 34:142–147 [DOI] [PubMed] [Google Scholar]
- 67.Khoo SK, Loll B, Chan WT, Shoeman RL, Ngoo L, Yeo CC, Meinhart A. 2007. Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J. Biol. Chem. 282:19606–19618 [DOI] [PubMed] [Google Scholar]
- 68.Gerdes K, Christensen SK, Lobner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371–382 [DOI] [PubMed] [Google Scholar]
- 69.Wozniak RA, Waldor MK. 2009. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 5:e1000439. 10.1371/journal.pgen.1000439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szekeres S, Dauti M, Wilde C, Mazel D, Rowe-Magnus DA. 2007. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 63:1588–1605 [DOI] [PubMed] [Google Scholar]
- 71.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U. S. A. 98:14328–14333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christensen SK, Pedersen K, Hansen FG, Gerdes K. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809–819 [DOI] [PubMed] [Google Scholar]
- 73.Dunny GM, Brown BL, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. U. S. A. 75:3479–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jacob AE, Hobbs SJ. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yagi Y, Clewell DB. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143:966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Courvalin P, Carlier C. 1986. Transposable multiple antibiotic resistance in Streptococcus pneumoniae. Mol. Gen. Genet. 205:291–297 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.