Abstract
Bob1 (Obf-1 or OCA-B) is a 34-kDa transcriptional coactivator encoded by the Pou2af1 gene that is essential for normal B-cell development and immune responses in mice. During lymphocyte activation, Bob1 protein levels dramatically increase independently of mRNA levels, suggesting that the stability of Bob1 is regulated. We used a fluorescent protein-based reporter system to analyze protein stability in response to genetic and physiological perturbations and show that, while Bob1 degradation is proteasome mediated, it does not require ubiquitination of Bob1. Furthermore, degradation of Bob1 in B cells appears to be largely independent of the E3 ubiquitin ligase Siah. We propose a novel mechanism of Bob1 turnover in B cells, whereby an acidic region in the C terminus of Bob1 regulates the activity of degron signals elsewhere in the protein. Changes that make the C terminus more acidic, including tyrosine phosphorylation-mimetic mutations, stabilize the instable murine Bob1 protein, indicating that B cells may regulate Bob1 stability and activity via signaling pathways. Finally, we show that expressing a stable Bob1 mutant in B cells suppresses cell proliferation and induces changes in surface marker expression commonly seen during B-cell differentiation.
INTRODUCTION
B-lymphocyte development is regulated by an intricate network of interacting signaling pathways. In most cases, these signaling networks lead to the regulation of numerous transcription factors, thereby changing the expression of genes important for B-cell proliferation, differentiation, and function (1, 2).
We are interested in understanding the role of Bob1 (Obf-1 or OCA-B) in these signaling pathways during B-cell development and function. Originally identified as an interaction partner and transcriptional coactivator of Oct 1 and Oct 2 in B cells (3–5), Bob1 has no strong sequence similarity to other cellular proteins. Previous work has established that the N terminus of Bob1 binds to Oct 1 and/or Oct 2 and to the adenosine at position 5 of the 5′-ATGCAAAT-3′ consensus octamer motif (6). It thereby acts as a molecular clamp (7) and drives transcription via interactions between its proline-rich, C-terminal transactivation domain (8, 9) and the general transcription machinery (10–12).
Bob1 is expressed throughout B-cell development, with transcripts appearing even before B-lineage specification (13, 14). Bob1 protein abundance transiently increases in pre-B cells in the bone marrow and again in germinal center B cells (15, 16). In humans, differences in Bob1 protein levels have been correlated with the prognosis in hematopoietic malignancies (17, 18), and polymorphism in the Bob1 genetic locus (POU2AF1) has been linked to autoimmunity (19, 20).
Mice lacking Bob1 expression are immunodeficient, showing decreased B-cell production (21, 22) and defects in the activation of peripheral B cells (23). Notably, germinal centers are not formed in Bob1−/− mice (24–26), resulting in reduced titers of class-switched immunoglobulin isotypes and a poor B-cell memory response. On the other hand, overexpression of Bob1 in the B-cell compartment of transgenic mice leads to a nearly complete block in B-cell development at the pre-B-cell stage (27). These results illustrate the importance of closely regulating the level of Bob1 protein in B cells. While transcript levels modestly increase during B-cell differentiation, changes in protein levels are considerably more pronounced, indicating that Bob1 is regulated posttranscriptionally (28, 29). These studies led to the proposal that Bob1 is constitutively degraded via the proteasome during most stages of B-cell development and that this process is inhibited or exacerbated in response to certain signals, thereby modulating the amount of Bob1 protein.
Using a system for dynamically observing Bob1 protein stability, we have further characterized the mechanism and importance of this regulation. The transactivating C-terminal domain of Bob1 contains an acidic patch (AP), and we have found that these negative charges play an important role in stabilizing Bob1 by blocking an otherwise robust ubiquitin-independent, proteasome-mediated degradation process. We then show that, by increasing the negative charge at the C terminus and stabilizing Bob1, the proliferation potential of WEHI-231 B cells is reduced, and this is accompanied by a phenotypically altered surface marker pattern.
MATERIALS AND METHODS
Transgenic constructs.
All episomal expression plasmids express enhanced green fluorescent protein (EGFP; henceforth referred to as “GFP”) fusion proteins under the control of the human cytomegalovirus intermediate early (HCMV-IE) promoter and contain a β-globin intron. Tandem-dimer Tomato (tdTomato; henceforth referred to as “Tomato”) expression is initiated by the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES). Retroviral expression plasmids were constructed by cloning the GFP fusion cassette immediately 3′ of the packaging element in the vector pMItom (30). Site-directed mutageneses were performed using a QuikChange kit (Agilent). A DNA-binding mutant of murine Bob1 (mBob1) was created by mutating the following residues: V22 to D, V24 to D, and E30 to L. Random mutagenesis of mBob1 was based on the method described by Cadwell and Joyce (31). Briefly, three rounds of PCR amplification of the Bob1 open reading frame (ORF) with Titanium Taq (Clontech) were performed in the presence of 0.64 mM MnCl2 and reduced (0.2 mM) dATP before cloning into a GFP fusion vector. Individual clones were subsequently isolated, and the Bob1 ORF was sequenced. Bob1 orthologous sequences were cloned from the following sources: rabbit, rabbit splenic cDNA; chicken, cDNA isolated from the DT-40 B cell line; Xenopus, cloned from Xenopus laevis splenic cDNA; zebrafish, cloned from Danio rerio kidney cDNA; and catfish, provided by G. Warr (32). The ORFs for murine Ebf1, Hes3, Spi-B, Blimp1, Syk, E47, and human Pax5 were also cloned as GFP fusions in the episomal and retroviral expression vectors described above. A plasmid encoding human Siah1 with an N-terminal hemagglutinin (HA) tag (29) was provided by P. Matthias.
Cell culture techniques.
Unless otherwise noted, all media were supplemented with penicillin-streptomycin (Gibco), glutamine (Gibco), 10% fetal calf serum (FCS; PAN-Biotech GmbH, PAA Laboratories GmbH, or Biochrom), and 60 μM β-mercaptoethanol and cultured at 37°C in a humidified incubator with 7% CO2. Pre-B-cell lines and bone marrow cultures were maintained in Iscove's modified Dulbecco modified Eagle medium (DMEM; Biochrom) supplemented with interleukin-7 (IL-7). All other B cell lines, Ltk cells, and HEK293 cells were cultured in Iscove's modified DMEM or RPMI 1640 (PAA). Plat-E cells (33) were cultured in low-glucose DMEM (PAA) containing 10 mM HEPES, 10 μg/ml blasticidin, and 1 μg/ml puromycin. Catfish B-cell lines 1B10 and 3B11 (34) were kindly provided B. Magor (University of Alberta) and cultured at 30°C with 5% CO2 in 0.9× RPMI 1640 supplemented with 1% carp serum (G. Riegger Aquaculture, Ettenheim, Germany). B-cell transfections were performed using a Neon transfection system (Life Technologies) with 4 μg plasmid DNA/2 × 106 to 3 × 106 cells. Adherent cells were transfected with TurboFect transfection reagent (Fermentas) with a ratio of 6 μg DNA/12 μl TurboFect/6 × 105 cells. For pervanadate stimulation, B cells at a concentration of 1 × 107 cells/ml were incubated for 30 min in serum-free RPMI 1640 at 37°C. Na3VO4 and H2O2 were added to a final concentration of 220 μM and 0.039%, respectively, from a fresh 100× pervanadate premix solution (22 mM Na3VO4 and 3.9% H2O2) which had been preincubated at room temperature for 5 min. For other cellular perturbations, the following compounds were used: MG132 (5 μM; Sigma), PYR-41 (10 μM; Calbiochem), epoxomicin (250 nM; Enzo Life Sciences), 5 mM NH4Cl, 150 nM bafilomycin (Santa Cruz Biotechnology), 1 μM thapsigargin (Sigma), and cycloheximide (10 μg/ml).
Animal work and sample collection.
All animal procedures were conducted in accordance with Australian National Health and Medical Research or German regulations on the use and care of experimental animals and approved by the QIMR Berghofer Medical Research Institute Animal Experimentation Ethics Committee (P1499) or the Max Planck Institute of Immunobiology and Epigenetics Animal Ethics Committee (Re-To3). C57BL/6 Siah2−/− (35) and wild-type mice were bred and maintained at the QIMR Berghofer as previously described (36). Spleens were removed from Siah2 wild-type and knockout mice, mashed, and strained through a 40-μm mesh filter. Samples were incubated for 1 min with red blood cell lysis buffer and then washed 3 times with fluorescence-activated cell sorting (FACS) buffer (phosphate-buffered saline [PBS] containing 2% FCS). The femurs and tibias were removed from the mice and cleaned of muscle, and the bone marrow was flushed out, resuspended, and strained through 40-μm-mesh-size filters. Samples were then washed twice with FACS buffer. Half of each sample (spleen or bone marrow) was used for flow cytometry analysis, while the other half was boiled in 1× Laemmli buffer and later sonicated prior to gel electrophoresis.
Siah1 siRNA.
WEHI-231 cells were cotransfected with 4 μg GFP-mBob1 plasmid DNA and 100 pmol TriFECTa dicer-substrate RNA duplexes (Integrated DNA Technologies) against either Siah1a (catalog no. 95300460) or Siah1b (catalog no. 65587509) or 50 pmol of each. Knockdown efficiency was measured by small interfering RNA (siRNA) against HPRT (70 to 95% decrease), and all values were normalized to the value for a nonspecific negative-control siRNA. FACS and quantitative PCR (qPCR) analysis were performed at 16 to 20 h posttransfection. RNA was isolated using Direct-zol columns (Zymo Research) after phase separation with Tri reagent (Sigma) and on-column DNase treatment. Oligo(dT)-primed cDNA was synthesized using a First Strand cDNA synthesis kit (Thermo Scientific), and reverse transcriptase-negative controls were used to measure genomic DNA contamination. Primer sequences for qPCR were as follows: Siah1a forward, 5′-GAAGTGAGTGCAGGATGCTCTTGTC-3′; Siah1a reverse, 5′-ATTAAAACATGTGATTTGCCCCAGAC-3′; Siah1b forward, 5′-TCAGGGTACGCTCTCCAT-3′; Siah1b reverse, 5′-ATAATGCTGTAGCAGCCTGA-3′; HPRT forward, 5′-GCTGGTGAAAAGGACCTCT-3′; and HPRT reverse, 5′-CACAGGACTAGAACACCTGC-3′.
Bob1 immunoprecipitation.
K46 cells transfected with an 8× HA-ubiquitin expression plasmid (37) were incubated overnight before treatment with MG132. Cells were washed in PBS containing 20 mM N-ethylmaleimide and lysed in isotonic buffer containing 1% NP-40, 20 mM N-ethylmaleimide, 2 mM EDTA, and 1× HALT protease inhibitor cocktail (Thermo Scientific). The lysate from 8 × 106 cells was diluted to 0.5% NP-40 before addition of 15 μl protein G-Sepharose (Amersham Biosciences) and 4 μg anti-Bob1 (mouse IgG2A monoclonal antibody recognizing an epitope between amino acids 133 and 145 of mBob1; provided by T. Wirth, University of Ulm, Ulm, Germany). After overnight incubation at 4°C, immobilized proteins were washed, eluted with 1× Laemmli buffer, and analyzed via SDS-PAGE and immunoblotting.
Protein electrophoresis and immunoblotting.
Cell lysates were typically extracted with isotonic lysis buffer containing 0.1 to 1.0% NP-40 and protease inhibitors (without EDTA for PhosTag applications). SDS-urea-polyacrylamide (12.5% acrylamide) gels were cast by adding urea to 6 M in the gel solution prior to polymerization. Lysate samples were boiled in 1× Laemmli buffer prior to electrophoresis. SDS-urea samples were boiled in 2× Laemmli buffer and cooled to room temperature before adding an equal volume of 9.6 M urea. PhosTag gels were cast by adding equimolar amounts of PhosTag (Manac Inc.) and MnCl2 prior to polymerization. Following electrophoresis, PhosTag gels were soaked in transfer buffer containing 1 mM EDTA, followed by subsequent washing to remove EDTA and manganese ions. Proteins were wet blotted onto nitrocellulose membranes, which were then blocked with 5% milk before immunoblotting. The following antibodies were used for immunoblotting: rat anti-Bob1 (6F10; Santa Cruz Biotechnology), mouse anti-CD79A (24C2.5; eBioscience), rabbit anti-GFP polyclonal serum (provided by P. Heun, Max Planck Institute, Freiburg, Germany), 5D3 mouse monoclonal anti-eukaryotic initiation factor 4A (anti-eIF4A) (38), and horseradish peroxidase-conjugated goat anti-mouse, anti-rabbit, and anti-rat IgG (Cell Signaling Technology).
Flow cytometry.
Measurements were performed on a FACSCalibur II, FACS Canto II, or LSR II flow cytometer (BD Biosciences). Stability assays using fluorescent proteins were performed a minimum of two times. Antibody staining and proliferation assays were performed at least three times. For cellular proliferation assays, cells were loaded with 10 μM eFluor 670 (eBioscience) at 37°C before washing away excess dye. Cellular apoptosis staining was performed with Alexa Fluor 647-conjugated annexin V in annexin V binding buffer (BioLegend). For DNA content measurement, cells were cultured at 37°C with 10 μM Hoechst 33342 (AAT Bioquest) for 1.5 h before analysis. The following antibodies were used for surface marker analysis: anti-CD2-biotin (RM2-5; BD Pharmingen), anti-IgD-Alexa Fluor 647 (11-26c.2a; BioLegend and eBioscience), anti-IgM-phycoerythrin Cy7 (II/41; eBioscience), anti-CD45.2-fluorescein isothiocyanate (104; eBioscience), anti-CD81-biotin (Eat-2; BioLegend), and anti-CD268-Alexa Fluor 647 (BAFFR; 7H22-E16; BioLegend). Biotinylated antibodies were incubated with streptavidin-DyLight 649 (BioLegend) before analysis.
Luciferase activity assay.
Cells of the Ltk (murine) or HEK293 (human) cell line were seeded at 105 cells/ml and incubated overnight to adhere. Cells were then transfected at a ratio of 1 μg total DNA and 2 μl TurboFect (Fermentas) per ml culture volume and incubated for 24 h before harvesting and lysis. The Photinus reporter construct and a Bob1-, GFP-Bob1-, mutant-, or GFP-only-containing test construct were transfected in a 1:1 ratio. Photinus luciferase abundance was normalized to that for a cotransfected reporter plasmid for Renilla luciferase under the control of the eIF4A promoter (25 ng/μg Photinus reporter). Luciferase measurements were obtained with a dual assay system (Dual-Glo; Promega), and technical duplicates of biological triplicate values are given with error bars, representing 1 standard deviation (SD).
Bioinformatic analyses and statistics.
Phosphorylation site prediction was performed with the GPS, version 2.1, software (https://gps.biocuckoo.org) (39). For proteome-wide analyses, protein sequences were extracted from the Ensembl Genes 70 database (human, build GRCh37.p10; mouse, build GRCm38.p1). Transcription factors were identified using the Gene Ontology (GO) terms 0003712 (transcription cofactor activity) and 0001071 (nucleic acid binding transcription factor activity). PatchScan analysis written in the perl programming language was used to search protein sequences for 20-amino-acid windows containing 8 or more aspartic or glutamic acid residues. For correlations to stability, analysis was restricted to the subset of genes from the human ORFeome library for which a protein stability index (PSI) (40) was available. Significance was calculated using a Fisher exact test with gamma function approximation for large numbers or testing for deviation from a slope of 0 by linear regression. Mann-Whitney U tests for pairwise comparisons and Spearman's rank correlation tests were used in place of Student's t and Pearson tests due to the non-Gaussian distribution of protein charge, acidic content, and stability.
RESULTS
Murine Bob1 contains destabilizing motifs.
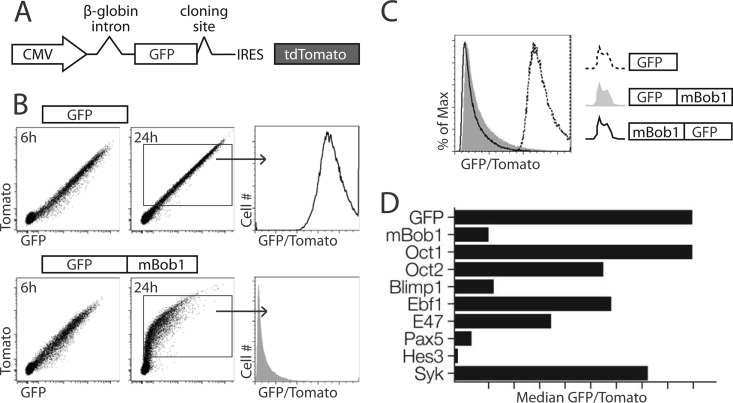
To measure protein turnover rates, we designed a dual-fluorescent-protein expression system (Fig. 1A). Fusing an instable protein of interest to GFP destabilizes the fluorescent molecule, leading to a lower abundance of cellular GFP relative to that of an IRES-driven transcript-internal Tomato control. The relative turnover rates of different GFP fusion partners in transiently transfected or retrovirally transduced cells can be determined by calculating the ratio of the mean fluorescent intensities of GFP and Tomato for each individual cell. Full-length murine Bob1 (mBob1) strongly destabilizes GFP in all transiently transfected B-cell lines tested, including the murine lymphoma-derived K46 (41) or WEHI-231 (42) cell line, the IL-7-dependent Bob1−/− pre-B-cell line BB2, or the human Burkitt lymphoma-derived BJAB cell line. Fusion proteins were robustly expressed at early time points (e.g., 6 h), but most of the protein was eliminated from the cell after 24 h (shown for WEHI-231 in Fig. 1B). This instability was also observed following retroviral transduction, although in this case, the GFP signal was not completely lost over time due to constitutive new synthesis of GFP-Bob1. Fusing Bob1 to either the C or N terminus of GFP resulted in its destabilization (Fig. 1C). Relative to several B-cell-specific proteins and transcription factors, Bob1 was quite instable (Fig. 1D). Notably, Oct 1 and Oct 2, cofactors of Bob1 on chromatin, were much more stable than Bob1. In non-B cells, the kinetics of degradation differed (B cells degraded Bob1 much faster than fibroblasts), but mBob1 was always less stable than GFP alone.
Fig 1.
Murine Bob1 is an instable protein. (A) Schematic representation of the expression construct used to determine relative protein stability. (B) Transient expression of GFP-IRES-tdTomato compared to that of GFP-mBob1-IRES-tdTomato in WEHI-231 cells analyzed at 6 and 24 h after transfection. Histograms represent the ratio of GFP to Tomato for each cell. (C) Fusions of mBob1 to the C terminus and N terminus of GFP relative to GFP alone. (D) Comparison of mBob1 stability with that of selected proteins cloned into the GFP fusion vector. Flow cytometric analysis of WEHI-231 cells evaluated the stability of the following proteins at 16 h posttransfection: murine octamer-binding proteins 1 (Oct 1) and 2 (Oct 2), murine B lymphocyte-induced maturation protein 1/Prdm1 (Blimp 1), murine early B cell factor 1 (Ebf1), murine immunoglobulin enhancer-binding factor E2a/Tcf3 (E47), human paired box protein Pax5 (Pax5), murine hairy and enhancer of split 3 (Hes3), and murine spleen tyrosine kinase (Syk).
The degradation of Bob1 is proteasome mediated but not dependent on lysine ubiquitination.
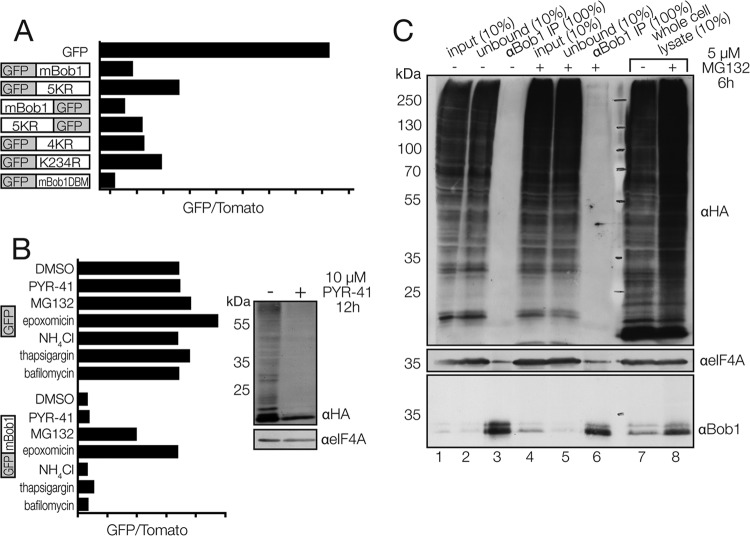
Many cellular proteins are targeted for proteasomal degradation via lysine ubiquitination. We sequentially mutated the five lysines in mBob1 to arginine in an effort to identify which residues might be ubiquitinated prior to degradation. In the fully mutated mBob1 (5KR), some stabilization did occur (Fig. 2A), but this stabilizing effect was incomplete (relative to that for a GFP-only control) and depended on the location of Bob1 relative to that of GFP in the fusion protein. Moreover, the compound mutation of the first four lysines (4KR; mBob1 K5, K25, K29, and K35) had a small stabilizing effect, as did the single mutation of the fifth lysine (K234). These results suggest either that Bob1 is not directly ubiquitinated or that ubiquitination and subsequent proteasomal degradation are not the major pathway for Bob1 turnover in vivo.
Fig 2.
Bob1 degradation is proteasome mediated but independent of Bob1 lysine ubiquitination. (A) Lysine mutagenesis of Bob1 and analysis in WEHI-231 cells. 5KR, all five lysines in mBob1 (K5, K25, K29, K35, and K234) mutated to arginine; 4KR, the first four lysines in mBob1 mutated to arginine; mBob1DBM, mBob1 DNA-binding mutant. (B) (Left) WEHI-231 cells were transfected with either GFP or GFP-mBob1, incubated for 4 h, and treated with dimethyl sulfoxide (DMSO), 10 μM PYR-41 (an E1 inhibitor), 5 μM MG132, or 250 nM epoxomicin (proteasome inhibitors) or 5 mM NH4Cl, 1 μM thapsigargin, or 150 nM bafilomycin (autophagy inhibitors) for 12 h before flow cytometric analysis; (right) to demonstrate the E1-inhibitory effect of PYR-41 on WEHI-231 cells, cells were transfected with HA-ubiquitin, incubated for 4 h, and treated with PYR-41 for 12 h before whole-cell lysis and Western blotting. (C) K46 murine B cells were transfected with HA-ubiquitin and incubated overnight before treatment with dimethyl sulfoxide (lanes 1 to 3 and 7) or MG132 (lanes 4 to 6 and 8) for 6 h. Cells were extracted with 1% NP-40 for Bob1 immunoprecipitation (lanes 1 to 6). Ubiquitinated Bob1 was probed for by incubating the Western blot with anti-HA (anti-Bob1 immunoprecipitation [αBob1 IP]; lanes 3 and 6).
To further test the possibility that ubiquitination is not required for Bob1 degradation, we inhibited the E1 ubiquitin-activating enzyme using PYR-41. WEHI-231 cells treated with PYR-41 showed strongly reduced ubiquitination of cellular substrates (Fig. 2B, right); however, there was very little effect on the stability of mBob1 (Fig. 2B, left). To test whether proteasomal activity is required for Bob1 degradation, we inhibited its activity with the proteasome inhibitor MG132 or epoxomicin. In contrast to the result with PYR-41, this led to a strong stabilization of GFP-mBob1 in transfected B cells (Fig. 2B, left). Additionally, Bob1 stability was not increased following inhibition of the autophagocytic pathway using NH4Cl, thapsigargin, or bafilomycin (Fig. 2B), suggesting that the proteasome is the major contributor to Bob1 degradation but that this process does not require ubiquitin-substrate ligation.
Ubiquitin-dependent proteasomal degradation is typically initiated by conjugation of a K48-linked polyubiquitin chain to lysine residues on a target protein and subsequent recognition by the proteasomal 19S regulatory subunit. In an attempt to directly detect ubiquitination of Bob1, we expressed HA-tagged ubiquitin in the B-cell line K46 before immunoprecipitating endogenous Bob1. No ubiquitinated forms of Bob1 were detected, even after proteasome inhibition and lysis in the presence of deubiquitinase inhibitors (Fig. 2C).
We also tested whether Bob1 transcription factor activity is necessary for degradation, a model that has been posited for several other transcription factors (43, 44). A Bob1 mutant that was unable to stimulate transcription was also instable, suggesting that Bob1 transcriptional activity is not essential for degradation (Fig. 2A). We conclude that ubiquitination of Bob1 does not play a major role in regulating Bob1 stability.
A Siah-independent mechanism for Bob1 degradation in B cells.
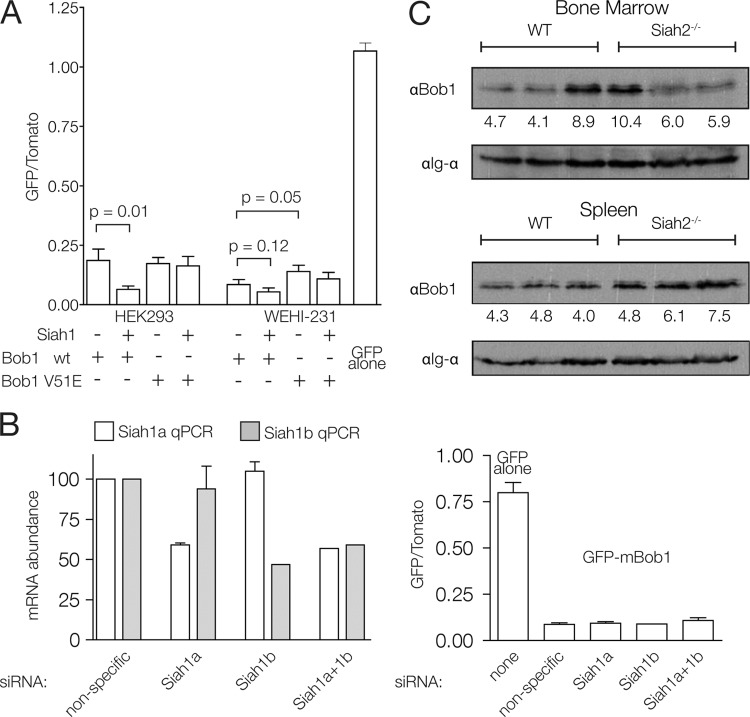
The E3 ubiquitin ligase Siah1 was previously shown to bind Bob1 and facilitate its degradation (28, 29). Bob1 stability was lower in non-B cells cotransfected with Bob1 and Siah1, and this effect was dependent on valine 51 in Bob1. We reproduced this result with our GFP fusion system in non-B cells (Fig. 3A). Interestingly, in the B-cell line WEHI-231, the stability of Bob1 was not significantly affected by Siah1 cotransfection (Fig. 3A), suggesting that a Siah-independent mechanism is largely responsible for Bob1 degradation in B cells. A mutation in the Siah interaction motif (V51E) (45) of Bob1 did cause a small but statistically significant increase in Bob1 stability. This could indicate a contribution of endogenous Siah to Bob1 turnover which cannot be augmented by Siah1 cotransfection. Nevertheless, this increase was small relative to the stability of GFP alone or to that of other Bob1-stabilizing mutants, as presented below.
Fig 3.
Effect of Siah on Bob1 stability in B cells. (A) FACS analysis of HEK293 or WEHI-231 cells at 16 h posttransfection. Error bars represent 1 SD from three independent experiments. wt, wild type. (B) siRNA-mediated Siah1 knockdown. (Left) qPCR analysis of siRNA efficiency in WEHI-231 cells; (right) GFP-mBob1 stability analysis at 16 h posttransfection with siRNA specific for Siah1a and/or Siah1b. Error bars represent 1 SD from two independent experiments. (C) Western blot analysis of Bob1 protein levels in wild-type (WT) and Siah2−/− mice. Each lane corresponds to a cell lysate from each of three mice tested per genotype. The numbers below the lanes are a quantitation of the anti-Bob1 signal normalized to the anti-Igα signal.
The mouse genome contains two functional Siah1 genes, Siah1a and Siah1b. Based on mass spectrometric analysis, we estimate that the Siah1b protein is almost 10-fold more abundant than Siah1a in WEHI-231 cells (data not shown). To further determine the extent to which a Siah-independent mechanism contributes to Bob1 destabilization in B cells, we performed an siRNA-mediated knockdown of both Siah1 isoforms in B cells transfected with GFP-mBob1. A 2-fold knockdown of Siah1a and Siah1b, either separately or simultaneously, did not significantly affect Bob1 stability (Fig. 3B).
In addition to Siah1a and Siah1b, an additional Siah homolog, known as Siah2, is present in the murine and human genomes and can also interact with Bob1 (28). Mice deficient for Siah1a (46) and Siah2 (35) have been generated and show no overt defect in B-cell development or response to immunization. Furthermore, mice hematopoietically reconstituted with Siah1a−/− Siah2−/− fetal liver precursors also produce normal peripheral B-cell populations (35). Bob1 protein levels in B cells from these mice have not been investigated. We confirmed that B-cell development proceeds normally in Siah2−/− mice (data not shown) and show here that Bob1 protein abundance is not significantly affected in the bone marrow or spleen of these mice (Fig. 3C). We conclude that Bob1 stability is not changed in Siah2-deficient B cells.
Bob1 contains several degrons and a C-terminal degradation switch.
To identify degron sequences within mBob1, we cloned a series of C-terminal truncations and partial fragments of the full-length protein. Removal of sequences in the central part of Bob1 (amino acids 87 to 159) indicated the presence of a degron both necessary and sufficient to induce Bob1 instability. The N-terminal 45-amino-acid fragment was also very instable, suggesting the presence of additional destabilizing elements (Fig. 4A). Additional constructs spanning the central region (Fig. 4B) revealed that amino acids 117 to 145 contain sequences that are sufficient to destabilize GFP almost to the same extent as full-length mBob1. However, removing this fragment did not fully stabilize the protein, consistent with the presence of multiple degrons in mBob1.
Fig 4.
Bob1 contains degron sequences and a C-terminal stability-regulating domain. (A, B) The stability of successive C-terminal truncations (A) and mBob1 fragments fused to GFP (B) is shown. (C) Bob1 ortholog stability. GFP was combined with Bob1 from (top to bottom) mouse (Mus musculus), human (Homo sapiens), rabbit (Oryctolagus cuniculus), chicken (Gallus gallus), frog (Xenopus laevis), zebrafish (Dario rerio), and American channel catfish (Ictalurus punctatus). For panels A to C, analysis was performed in WEHI-231 cells at 16 h posttransfection. (D) Identification of the stabilizing domain in cfBob1. (Left) Amino acids 85 to 150 from either mouse (upper) or catfish (lower) Bob1 fused to GFP (black lines); (middle) the C-terminal 45 amino acids from cfBob1 in place of those from mBob1 (mcBob1; top) and the converse chimeric Bob1 (bottom) fused to GFP (black lines); (right), the C-terminal domains fused directly to GFP (mouse, top; catfish, bottom). (E) From top to bottom, stability of mBob1, catfish Bob1 (cfBob1), and the chimeric mcBob1 in American channel catfish-derived B cells (1B10; the analysis was performed at 72 h posttransfection) fused to GFP (black lines) relative to that of GFP-mBob1 (shaded regions) and GFP (dashed lines). (F) Cycloheximide (CHX) chase of Bob1−/− pre-B cells transduced with either mBob1 (left) or mcBob1 (right) IRES-GFP-containing constructs.
Phylogenetically, Bob1 appears concomitantly with V(D)J recombination-driven adaptive immunity. To date, no Bob1 ortholog has been identified in jawless vertebrates or invertebrates. To determine whether the instability of Bob1 is a conserved feature, we cloned Bob1 orthologs from several vertebrates and analyzed protein stability in the GFP/Tomato system. All Bob1 orthologs were functional as transcriptional activators in a luciferase reporter assay (Fig. 5A), but they showed different stabilities in the cellular fluorescence assay (Fig. 4C). Interestingly, Bob1 from the American channel catfish, Ictalurus punctatus (cfBob1), was almost as stable as the GFP-only control.
Fig 5.
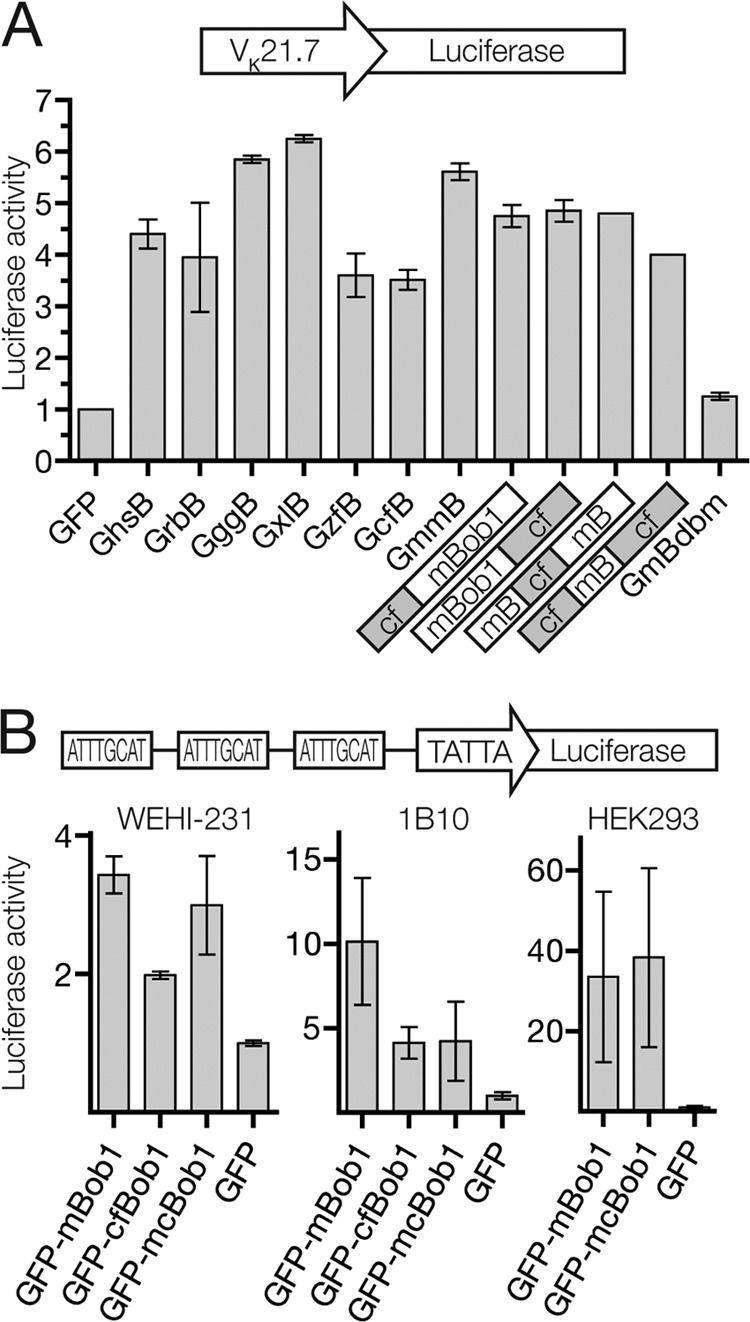
Bob1 ortholog and chimera transcriptional activity. (A) Renilla-normalized luciferase activity of Bob1-containing fusion constructs in the murine fibroblast line Ltk. Photinus luciferase production is driven by the murine immunoglobulin light-chain VK21.7 promoter. From left to right: GFP only (GFP), GFP-human Bob1 (GhsB), GFP-rabbit Bob1 (GrbB), GFP-chicken Bob1 (GggB), GFP-Xenopus Bob1 (GxlB), GFP-zebrafish Bob1 (GzfB), GFP-catfish Bob1 (GcfB), GFP-mBob1 (GmmB), GFP-cf1-83m86-256Bob1 (where cf1-83 is amino acids 1 to 83 of cfBob1 and m86-256 is amino acids 86 to 256 of mBob1) (cf mBob1), GFP-m1-151cf152-245Bob1 (mBob1 cf), GFP-m1-85cf84-151m152-256Bob1 (mB cf mB), GFP-cf1-83m86-151cf152-245Bob1 (cf mB cf), and a GFP-mBob1 DNA-binding mutant (GmBdbm). (B) Renilla-normalized luciferase activity of Bob1 fusion constructs in (from left to right) WEHI-231, 1B10, and HEK293 cells. Photinus luciferase production is driven by an artificial promoter containing 3 octamer motifs and a TATA box.
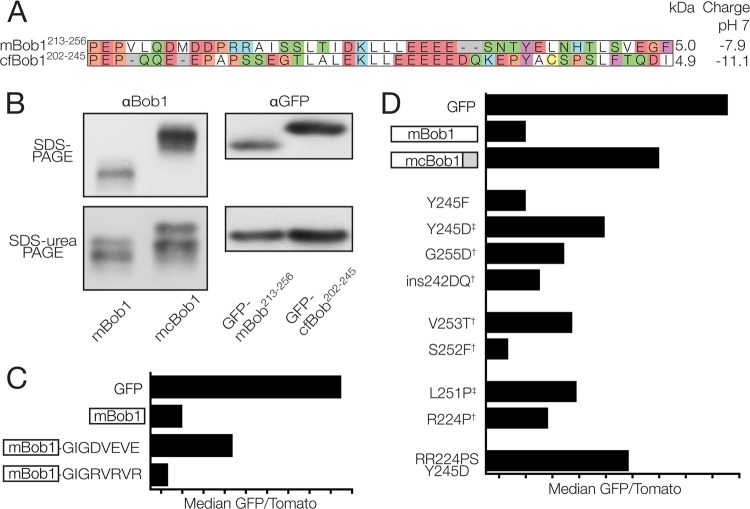
To identify sequences in cfBob1 that contribute to the stability of the protein, we fused N-terminal, internal, and C-terminal fragments from either murine or catfish Bob1 to GFP. Although the relative stability of the two full-length proteins was very different, the central 65 amino acids of both orthologs had a strongly destabilizing effect when isolated from surrounding sequences (Fig. 4D, left), suggesting that the degron that we described in this region of mBob1 is conserved in cfBob1. However, in the context of the full-length protein, these degradation-inducing segments of the catfish Bob1 protein appeared to be protected from recognition by the cellular degradation machinery, thus creating a more stable protein.
By creating chimeric full-length proteins with fragments from both murine and catfish Bob1, we found that replacing the C-terminal 44 amino acids from mouse Bob1 with those from the catfish sequence (mcBob1) (Fig. 4D, middle) was sufficient to stabilize the protein. The converse was also true; the C terminus of mouse Bob1, when placed into the catfish sequence, destabilized the protein (Fig. 4D, bottom middle). The stable mcBob1 chimera was transcriptionally active (Fig. 5B) and located in the nucleus (data not shown), similar to mBob1. Exchanging smaller C-terminal peptide fragments between mouse and catfish did not produce chimeric proteins with the same stability as the catfish full-length or mcBob1 chimeric protein, although both halves of the catfish Bob1 C-terminal 44 amino acids could partially stabilize the mouse sequence (data not shown). Similarly, single mutations replacing conserved mBob1 amino acids with those from cfBob1 could somewhat, but not completely, stabilize the protein (Fig. 6D), indicating that the stability-regulating domain encompasses a longer stretch of amino acids in the C terminus. The isolated 44-amino-acid C-terminal fragments from mouse and catfish could destabilize or stabilize GFP, but not as strongly as full-length mBob1 (Fig. 4D, right). These observations were not dependent on the host cell species: the same relative stability was observed in the catfish B-cell line 1B10 (Fig. 4E). Importantly, in a cycloheximide chase assay with constructs not fused to GFP, the chimeric mcBob1 was also more stable than mBob1 (Fig. 4F), thus excluding the possibility that the differences in stability are specific for the GFP fusion.
Fig 6.
Charged residues in the Bob1 C terminus regulate protein stability. (A) Protein alignment of the C-terminal 44 amino acids from mouse (top row) and catfish (bottom row). (B) (Top left) Full-length mBob1 (instable) and mcBob1 (stable) migration in an SDS-polyacrylamide gel (expression in HEK293 cells); (top right) fusions of the C termini of mBob1 or cfBob1 (see panel A) to the C terminus of GFP after expression in K46 cells; (bottom) electrophoresis of the same lysates from the top panels in SDS-urea gels. (C) Median GFP/Tomato ratios for C-terminal acidic and basic tags fused to GFP-mBob1 relative to those for GFP only (GFP) and GFP-mBob1 (mBob1). WEHI-231 murine B cells were analyzed at 24 h posttransfection. (D) Point mutations in the C terminus of mBob1 affecting stability. WEHI-231 cells were analyzed at 16 h posttransfection. †, mutations of mBob1 to the corresponding residue in cfBob1; ‡, stabilizing mutations identified by mBob1 random mutagenesis.
The net charge of the Bob1 C terminus is important for regulating stability.
The predicted molecular masses of mouse and catfish Bob1 are 27.7 and 26.1 kDa, respectively, and the C-terminal 44-amino-acid domains from murine and catfish Bob1 also have similar predicted molecular masses (5.0 and 4.9 kDa, respectively) (Fig. 6A). Surprisingly, both the stable full-length mcBob1 protein and the catfish C-terminal fragment (fused to GFP) migrated more slowly than the respective full-length mBob1 protein and C-terminal fragment in an SDS-polyacrylamide gel (Fig. 6B, top). This migration difference was reduced in an SDS-urea-polyacrylamide gel (Fig. 6B, bottom), suggesting that conformational or SDS-binding/charge differences could be responsible.
Bob1 frequently migrated as at least two major bands (Fig. 2C, 6B, and 7), commonly referred to as p34 and p35 (47). The p35 isoform of Bob1 has been described to be myristoylated and cytoplasmic. Our constructs expressing mBob1 or the GFP-mBob1 fusion protein both lacked the N-terminal sequences necessary for myristoylation, yet they also showed two major bands (Fig. 6B and 7B). We conclude that there are probably additional, as yet uncharacterized, species of Bob1 which could comigrate with the myristoylated isoform.
Fig 7.
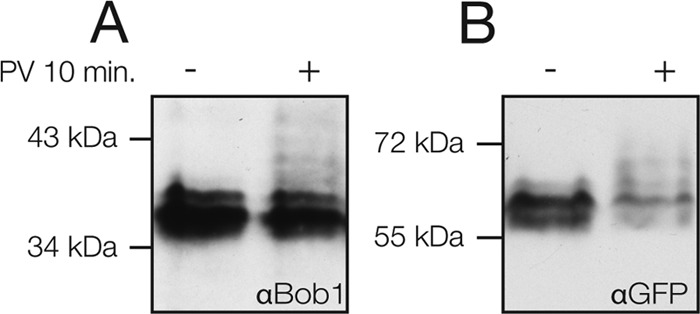
Phosphorylated mBob1 accumulates in pervanadate-treated B cells. (A) K46 cells were treated with pervanadate (PV) for 10 min prior to lysis and electrophoresis on an SDS-PhosTag gel. Phosphate groups bind to the PhosTag compound in the gel, decreasing mobility and resulting in the appearance of additional bands in an immunoblot. Endogenous mBob1 was detected with an anti-Bob1 antibody. (B) WEHI-231 cells were transiently transfected with GFP-mBob1. At 16 h, the cells were pervanadate treated and lysates were subjected to SDS-PhosTag electrophoresis. The fusion protein and phosphorylated isoforms were detected with an anti-GFP antibody.
Given the electrophoretic migration differences between the mouse and catfish C termini, we wanted to know if charge differences could explain the increased stability. Interestingly, the catfish sequence was more acidic, having a predicted net −3 charge relative to the mouse sequence at physiological pH. We added additional charged residues to the C terminus of mBob1 to artificially raise or lower the net charge. An acidic tag (GIGDVEVE) clearly increased the stability relative to that of the wild-type mBob1 sequence, while a basic tag (GIGRVRVR) decreased the stability (Fig. 6C). By themselves, the acidic and basic tags did not appreciably influence the stability of GFP, indicating that a positively charged C-terminal tag does not directly serve as a degradation signal but, rather, regulates the recognition of existing instability motifs elsewhere in the Bob1 protein.
In a separate series of experiments aimed at identifying individual amino acids that affect the stability of Bob1, we randomly mutagenized the mouse Bob1 sequence or replaced specific amino acids in the C terminus of mouse Bob1 with the catfish equivalent. Several mutations of mBob1 amino acids to their orthologous catfish counterparts resulted in a modified charge (R224P, G255D, and ins242DQ) or potentially modified charge (by the addition [V253T] or removal [S252F] of predicted phosphorylation sites) and altered stability (Fig. 6D).
For the random mutagenesis, we analyzed a total of 74 clones containing 187 amino acid mutations and observed several clones with increased or reduced stability. Because many of the clones contained multiple mutations, we individually mutated selected positions in a second round of screening and examined their effect on stability. Two point mutations identified by our screen increased mBob1 stability (L251P and Y245D) (Fig. 6D), further supporting the importance of the C terminus. Although the L251P mutation does not alter the charge of mBob1, proline is a known helix breaker, and its introduction could interrupt secondary structure necessary for degradation (this may also explain the stabilization seen in the R224P mutant). Combining several mutations (RR224PS and Y245D) could have an additive effect. Thus, it is likely that several factors, such as structure and charge, are responsible for the activity of this stability-regulating domain. We observed that the phosphomimetic Y245D mutation stabilized Bob1, while mutation to phenylalanine (Y245F) did not affect Bob1 stability (Fig. 6D). Interestingly, the tyrosine residue at position 245 is predicted to be a target of Src family kinases and was recently identified as a Syk phosphorylation substrate in human B cells (48). This opens up the possibility that B cells use kinase-mediated signaling pathways to regulate the stability of Bob1. Phosphorylation of other Bob1 residues has previously been described to be inducible in T cells and constitutive in B cells (49). We found that phosphorylated forms of both endogenous Bob1 and GFP-mBob1 accumulate in response to pervanadate treatment of B cells (Fig. 7).
Acidic patches are enriched and associated with stability in transcription factors.
Tagging the C terminus of Bob1 with acidic residues stabilized the protein, while basic residues made it even less stable (Fig. 6C). To investigate whether the regulation of protein stability by the C terminus, as seen for Bob1, may be a common characteristic among transcription factors, we analyzed protein charge and stability in silico. Since the acidic amino acids in the C terminus of Bob1 are clustered, we scanned the proteome for patches of acidic amino acids. Defining an acidic patch (AP) as a minimum of eight aspartic or glutamic acid residues within a 20-amino-acid window, the abundance of APs was significantly higher in transcription factors than in the rest of both the human (37% versus 25%) and mouse (36% versus 25%) proteomes (Fig. 8A). However, transcription factors, on average, were not particularly more acidic than nontranscription factors. In fact, the average net charge at pH 7.0 was slightly higher for transcription factors (Fig. 8B), indicating that the significance of finding an AP in a transcription factor is not merely because it is more acidic on the whole. Interestingly, basic patches were also found more frequently in transcription factors than nontranscription factors (54% versus 32% for human, P = 10−50).
Fig 8.
Acidic patches are enriched in and affect the stability of transcription factors. (A) Human and mouse whole-proteome analysis of transcription factor (TF) APs (defined as ≥8 D/E residues in a 20-amino-acid window) compared to nontranscription factors. Significance is determined by Fisher's exact test with a gamma function approximation for large numbers. (B) Total acidic amino acid composition and net predicted charge of transcription factors versus nontranscription factors. (C) Stability versus acidic amino acid composition plots for transcription factors (linear regression [black line] was not significantly nonzero; Spearman correlation test, P = 0.82) and nontranscription factors (black line, P < 0.0001 by linear regression analysis and P < 0.0001 by the Spearman correlation test). (D) Average stability and distribution of the stability of transcription factors containing APs in the C terminus (C-terminal 10% of the protein), outside the C terminus, or without an AP relative to those of nontranscription factors containing a C-terminal AP, non-C-terminal AP, or no AP.
To see whether charge correlates with stability, we analyzed data available from a dual-fluorescent-protein approach similar to ours (40), performed in HEK293T cells using the human ORFeome library. For nontranscription factors, there was a clear positive correlation between stability and acidic amino acid content which did not exist for transcription factors (Fig. 8C). The location of APs was also different between transcription factors and nontranscription factors. Transcription factors tended to be more stable if they had an AP in the C terminus (defined as the last 10% of the protein) and less stable if an AP occurred elsewhere in the protein (Fig. 8D). For nontranscription factors and consistent with the results in Fig. 8C, AP-containing proteins tended to be more stable, regardless of the location of the AP. Among these groups, the presence of a basic patch was not correlated with stability.
Increased levels of Bob1 prolong the cell cycle in B cells.
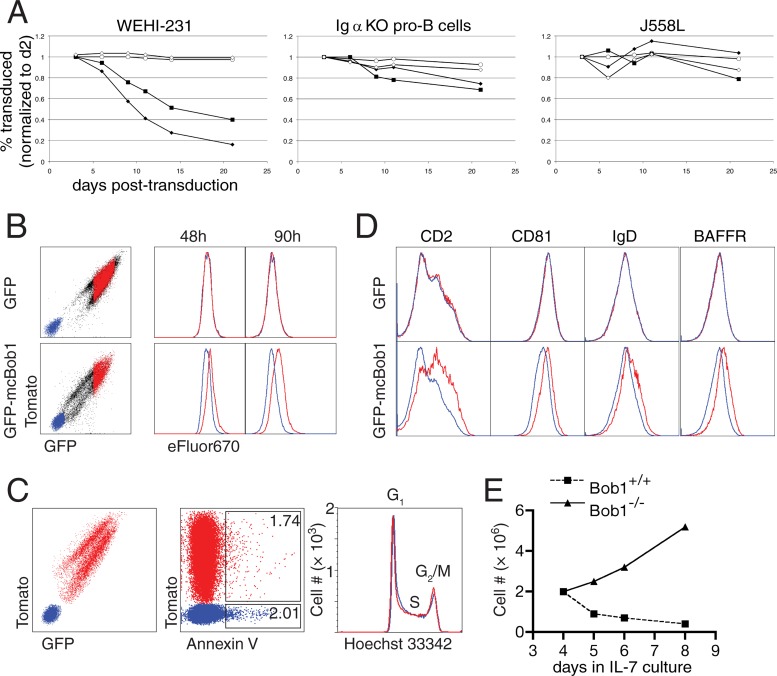
Retroviral expression of Bob1 in WEHI-231 murine B cells led to a significant growth disadvantage of transgenic cells, resulting in their loss in a mixed culture with untransduced cells over time (Fig. 9A). Overexpressing the stable mouse-catfish chimeric protein had a more drastic effect, while the DNA-binding mutant (even in the context of the stable chimeric protein) had no influence on the cells. Expression levels varied among transduced cells, and those cells with the strongest transgene expression were lost first, suggesting that this phenotype is dependent on the amount of Bob1 protein present.
Fig 9.
Increased Bob1 protein levels inhibit cellular proliferation. (A) Transgene-positive cells in B-cell lines [WEHI-231 cells, Igα−/− pro-B cells, and J558L myeloma-derived cells] retrovirally transduced with GFP-Bob1 fusion constructs (as a percentage of the total culture on day 2 [d2] postransduction) over time. Circles, GFP alone; squares, GFP-mBob1; black diamonds, GFP-mcBob1; open diamonds, GFP-mcBob1 DNA-binding mutant. KO, knockout. (B) WEHI-231 cells were labeled with the proliferation dye eFluor 670 and gated for transduced (red) and nontransduced cells (blue) at subsequent time points. Top row, GFP only; bottom row, GFP-mcBob1. (C) Annexin V staining (middle) and Hoechst 33342 DNA content staining (right) of GFP-mcBob1-transduced cells (red) and nontransduced cells (blue, gated as shown in the left panel). (D) Surface expression of CD2, CD81, IgD, and the BAFF receptor (CD268) on transduced (red) versus untransduced (blue) WEHI-231 cells in mixed culture. Top row, GFP only; bottom row, GFP-mcBob1. (E) IL-7-supported bone marrow culture growth curves from wild-type and Bob1−/− mice.
Transduction of additional B-cell lines showed that WEHI-231 cells are the most sensitive to overexpression of Bob1. Cells of an IL-7-dependent, Igα−/− pro-B-cell line were also sensitive to Bob1 overexpression, though to a lesser degree than WEHI-231 cells (Fig. 9A, middle), while expression in the plasma cell-derived cell line J558L had no effect on growth (Fig. 9A, right).
WEHI-231 cells expressing exogenous Bob1 showed altered expression of several cell surface receptors associated with B-cell development. IgD, CD81, BAFFR, and CD2 surface protein levels were markedly higher (Fig. 9D). These changes were stronger upon expression of stable mcBob1 and were not present in cells transduced with the DNA-binding mutant. Surface markers which did not change upon expression of GFP-mcBob1 included IgM, CD5, CD21, CD22, CD23, CD24, CD38, CD40, CD62L, and CD72 (not shown). Taken together, these observations are reminiscent of changes in surface proteins seen in vivo as splenic B cells progress from a transitional stage (from which WEHI-231 cells are suggested to have been derived) to a more mature stage.
We found no evidence for increased apoptosis in GFP-mBob1-transduced WEHI-231 cells (Fig. 9C, middle) but observed a prolonged division time, indicating a partial block in progression through the cell cycle (Fig. 9B). There did not appear to be a distinct block at any particular point during the cell cycle in the transduced cells (Fig. 9C, right). These results could be corroborated in primary cells: pre-B cells from the bone marrow of Bob1−/− mice showed increased proliferation ex vivo in IL-7 culture relative to cells isolated from Bob1-sufficient littermates (Fig. 9E), further supporting a role for Bob1 as a suppressor of the cell cycle.
DISCUSSION
Protein abundance can be regulated by changes in transcription, translation, or protein stability. Wide-scale approaches to characterize protein stability have been performed using a dual-fluorescent-protein approach (40) or by mass spectrometry (50). The former study was performed with a cDNA library of the human ORFs exogenously expressed in HEK293T cells, while the latter used stable isotope labeling to measure proteome-wide turnover in NIH 3T3 murine fibroblasts. Since neither experiment was performed in B cells, the experiments provide no information on the stability of Bob1 compared to that of the rest of the proteome. The proteins identified by mass spectrometry are strongly enriched for acidic patch-containing sequences (36% compared to 25% predicted by the genome). Since we calculated that acidic proteins tend to be more stable (Fig. 8C), we attribute this enrichment to a methodological detection bias.
Using an IRES-driven secondary fluorescent protein (in this case, Tomato) for transcript internal normalization, we could observe the effect of any peptide on the stability of GFP. Fusions to the C terminus of GFP minimize possible differences in translation initiation efficiency, and the highly structured GFP molecule may serve as a barrier against the destruction of otherwise un- or misfolded proteins. To ensure that the differences in stability that we observed were not due to the possible influence of upstream sequences on IRES translation efficiency, we observed the same stability ratios with a third fluorescent protein (super cyan fluorescent protein) (51) carried by a separate, cotransfected plasmid.
Previous work has established that Bob1 protein levels increase in pre-B cells and germinal center B cells without equivalent increases in mRNA abundance and suggests that the E3 ubiquitin ligase Siah1 is responsible for the degradation of Bob1 (28, 29). However, in our system, mutations abolishing the ability of Siah1 to bind Bob1 had only a small effect on Bob1 stability in B cells (Fig. 3A). Notably, cotransfection with Siah1 in B cells caused a much smaller decrease in Bob1 stability than that in non-B cells, and this decrease could also be seen in the Bob1 V51E mutant (Fig. 3A). It is interesting to note that Siah1 cotransfection in B cells (but not in non-B cells) led to a decrease in the stability of every protein that we investigated, including GFP alone, mcBob1, Blimp 1, and Ebf1 (data not shown), suggesting that Siah1 globally contributes to B-cell protein degradation and thus nonspecifically affects the stability of Bob1. Furthermore, siRNA depletion of Siah1 and genetic deletion of Siah2 did not appreciably affect Bob1 protein levels. While we do not exclude the possibility of a contribution of the Siah proteins to Bob1 turnover, these data suggest that a novel, Siah1-independent mechanism is likely more relevant for Bob1 degradation in B cells. Although we provide evidence that this mechanism does not require direct ubiquitination of Bob1 for degradation (Fig. 2), the publicly available PhosphoSitePlus database (52) describes lysine 29 ubiquitination of Bob1. However, this result was observed in only one of several B-cell lines analyzed and has not been verified by additional methods. We also showed that mutation of K29 does not stabilize mBob1 any more than mutation of K234 and that mutation of all lysine residues in Bob1 has only a modest stabilizing effect on the protein (Fig. 2A). Finally, inhibiting the ubiquitination of substrate proteins by depleting ubiquitin-activated E1 from the cell did not affect Bob1 stability (Fig. 2B). The drug used to inhibit E1 activation (PYR-41) has been shown to block the E1 enzyme Ube1. Its effect on the recently identified ubiquitin E1 Uba6 (53) has not yet been described, so we cannot rule out the possibility that Bob1 is still ubiquitinated in a Uba6-dependent manner. However, our results involving lysine mutants and ubiquitin tagging support the hypothesis that Bob1 degradation is primarily ubiquitination independent. As has been demonstrated for other labile proteins, it is possible that Bob1 binds to another protein, which then ferries it to the proteasome (54, 55), interacts noncovalently with ubiquitin (56), or binds the proteasome directly (57).
Protein turnover requires protein-intrinsic signals that target labile proteins to cellular degradation complexes. Since adding additional proline or glutamic acid residues to the C terminus of mBob1 actually increases stability, classical PEST motifs (58) do not appear to explain Bob1 instability. Several other known degradation signals (59), such as the N-end rule, can also be excluded because most constructs described here have the same N terminus, namely, GFP.
Full-length mBob1 was less stable than the isolated C terminus (Fig. 4D, upper right), suggesting that the C terminus of mBob1 is a stability-regulating domain rather than a motif that directly signals to the cellular degradation machinery. If this is the case, a direct or functional interaction between this domain and other cellular components may exist. Another speculative possibility is that the C terminus interacts intramolecularly with Bob1 degron sequences (such as those shown in Fig. 4C) to regulate their function. Interaction of negatively charged acidic C-terminal amino acids with degron-associated basic domains seems unlikely because the minimal degron sequence in Bob1 (amino acids 117 to 145) does not contain any clear positively charged regions. However, the N-terminal DNA-binding domain (which, in isolation, is very destabilizing) (Fig. 4A) contains a stretch of basic amino acids and could interact with the C terminus, as has been described for other transcription factors, such as interferon response factor (IRF) family members (60).
We showed (Fig. 6) that when a tyrosine in the C terminus (that was reported to be phosphorylated in B cells following anti-B-cell receptor stimulation [48]) is mutated to aspartic acid to mimic phosphorylation, Bob1 is stabilized. This suggests that phosphorylation could stabilize Bob1. Our observation that phosphorylated Bob1 isoforms accumulate following inhibition of phosphotyrosine protein phosphatases with pervanadate, together with published results of phosphothreonine and phosphoserine residues in Bob1, is compatible with such a model.
Like the C terminus of Bob1, transactivation domains tend to be enriched for acidic amino acids (61). This is consistent with our observation that transcription factors are more likely to contain an acidic patch without being generally more acidic (Fig. 8A and B). While several studies have associated acidic transactivation domains with instability (62), acidic patches in the C terminus may have a counteracting, stabilizing effect on transcription factors, as we saw for Bob1. Indeed, transcription factors with C-terminal acidic patches are significantly more stable than those with patches elsewhere (Fig. 8D). Strikingly, the C terminus of the instable mBob1 does not meet our criterion that it contain an acidic patch (it contains only 7 acidic amino acids in a 20-residue window), but the C termini of the stable cfBob1 and mcBob1 do, and the C terminus of a phosphorylated form of mBob1 also could. Our findings could illustrate a more general scheme for the regulation of transcription factor stability, in which acidic amino acid clusters play a position-specific role in regulating protein turnover. In any case, the negative charges in the C terminus of Bob1 have been functionally implicated in B-cell processes (10). The authors concluded that a Bob1 mutant in which acidic residues in the C terminus were replaced with basic amino acids had a lower transcriptional output than wild-type Bob1. Our findings suggest that this reduced transcriptional capacity may be due to decreased stability.
What are the cellular consequences if Bob1 cannot be degraded? We show that the cell cycle is inhibited by stable Bob1 and, conversely, that Bob1−/− B cells show increased proliferation ex vivo. We propose a new role for Bob1 as a brake on the cell cycle. A destabilization of Bob1 would enhance proliferation in developing and activated B cells, and an increase in Bob1 stability would support exit from the cell cycle and permit further differentiation. Supporting this model, Bob1 expression at constitutively high levels in transgenic mice using an IgH promoter/enhancer combination was shown to block development of mature B cells (27). Interestingly, the authors used a C-terminal HA-tagged Bob1 construct, which could influence the stability of the Bob1 protein. During B-cell development, pre-B cells undergo a phase of limited proliferation, followed by rearrangement of the immunoglobulin light-chain locus. A similar process also takes place during the germinal center reaction, where B cells proliferate before maturing to memory B or plasma cells. Thus, Bob1 may promote differentiation in proliferating cells, partly by slowing the cell cycle.
Since Bob1 has been implicated in autoimmunity and malignancy in humans, these findings may help shed light on its role in such processes, especially since Bob1 protein levels have been suggested to be prognostic indicators of clinical outcome. The specific expression of Bob1 in B lymphocytes means that the ability to specifically modulate Bob1 stability, if developed, could provide a useful pharmacological tool in B cells without affecting other cell lineages.
ACKNOWLEDGMENTS
We thank Elias Hobeika and Patrick Heun for cells and antibodies, Michael Mitterer, Sabine Sané, Luzia Ballmer, and Osric Forrest for technical assistance, Marinus Lamers for critical reading of the manuscript, Patrick Matthias for providing the human Siah1 construct and comments regarding the manuscript, Gregory Warr for the catfish Bob1 DNA construct, Brad Magor for the catfish B-cell lines, Georg Riegger for fish serum, members of the group of Wolfgang Schamel for help with the PhosTag system, Verdon Taylor for the Hes3 cDNA, Thomas Wirth for the anti-Bob1 hybridoma, the lab of Rudolf Grosschedl for the Ebf1 and Pax5 cDNA clones, and the lab of Robert Schneider for qPCR reagents.
J.M.L. is supported by an IMPRS-MCB fellowship from the Max Planck Society, and A.M. is supported by a fellowship and grants from the National Breast Cancer Foundation and Cancer Council Queensland.
Footnotes
Published ahead of print 23 September 2013
REFERENCES
- 1.Kurosaki T, Shinohara H, Baba Y. 2010. B cell signaling and fate decision. Annu. Rev. Immunol. 28:21–55 [DOI] [PubMed] [Google Scholar]
- 2.Matthias P, Rolink AG. 2005. Transcriptional networks in developing and mature B cells. Nat. Rev. Immunol. 5:497–508 [DOI] [PubMed] [Google Scholar]
- 3.Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens CM. 1995. A B-cell coactivator of octamer-binding transcription factors. Nature 373:360–362 [DOI] [PubMed] [Google Scholar]
- 4.Strubin M, Newell JW, Matthias P. 1995. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell 80:497–506 [DOI] [PubMed] [Google Scholar]
- 5.Luo Y, Roeder RG. 1995. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol. Cell. Biol. 15:4115–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chasman D, Cepek K, Sharp PA, Pabo CO. 1999. Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein-DNA interface. Genes Dev. 13:2650–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauter P, Matthias P. 1998. Coactivator OBF-1 makes selective contacts with both the POU-specific domain and the POU homeodomain and acts as a molecular clamp on DNA. Mol. Cell. Biol. 18:7397–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gstaiger M, Georgiev O, van Leeuwen H, van der Vliet P, Schaffner W. 1996. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 15:2781–2790 [PMC free article] [PubMed] [Google Scholar]
- 9.Krapp A, Strubin M. 1999. B-cell coactivator OBF-1 exhibits unusual transcriptional properties and functions in a DNA-bound Oct-1-dependent fashion. Mol. Cell. Biol. 19:4247–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo Y, Ge H, Stevens S, Xiao H, Roeder R. 1998. Coactivation by OCA-B: definition of critical regions and synergism with general cofactors. Mol. Cell. Biol. 18:3803–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolstein O, Silkov A, Revach M, Dikstein R. 2000. Specific interaction of TAFII105 with OCA-B is involved in activation of octamer-dependent transcription. J. Biol. Chem. 275:16459–16465 [DOI] [PubMed] [Google Scholar]
- 12.Ren X, Siegel R, Kim U, Roeder RG. 2011. Direct interactions of OCA-B and TFII-I regulate immunoglobulin heavy-chain gene transcription by facilitating enhancer-promoter communication. Mol. Cell 42:342–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121:1109–1121 [DOI] [PubMed] [Google Scholar]
- 14.Mansson R, Zandi S, Welinder E, Tsapogas P, Sakaguchi N, Bryder D, Sigvardsson M. 2010. Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood 115:2601–2609 [DOI] [PubMed] [Google Scholar]
- 15.Greiner A, Müller KB, Hess J, Pfeffer K, Müller-Hermelink HK, Wirth T. 2000. Up-regulation of BOB.1/OBF.1 expression in normal germinal center B cells and germinal center-derived lymphomas. Am. J. Pathol. 156:501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin XF, Reichlin A, Luo Y, Roeder RG, Nussenzweig MC. 1998. OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J. 17:5066–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toman I, Loree J, Klimowicz AC, Bahlis N, Lai R, Belch A, Pilarski L, Reiman T. 2011. Expression and prognostic significance of Oct2 and Bob1 in multiple myeloma: implications for targeted therapeutics. Leuk. Lymphoma 52:659–667 [DOI] [PubMed] [Google Scholar]
- 18.Advani AS, Lim K, Gibson S, Shadman M, Jin T, Copelan E, Kalaycio M, Sekeres MA, Sobecks R, Hsi E. 2010. OCT-2 expression and OCT-2/BOB.1 co-expression predict prognosis in patients with newly diagnosed acute myeloid leukemia. Leuk. Lymphoma 51:606–612 [DOI] [PubMed] [Google Scholar]
- 19.The Collaborative Group GAMES 2006. Linkage disequilibrium screening for multiple sclerosis implicates JAG1 and POU2AF1 as susceptibility genes in Europeans. J. Neuroimmunol. 179:108–116 [DOI] [PubMed] [Google Scholar]
- 20.Nakamura M, Nishida N, Kawashima M, Aiba Y, Tanaka A, Yasunami M, Nakamura H, Komori A, Nakamuta M, Zeniya M, Hashimoto E, Ohira H, Yamamoto K, Onji M, Kaneko S, Honda M, Yamagiwa S, Nakao K, Ichida T, Takikawa H, Seike M, Umemura T, Ueno Y, Sakisaka S, Kikuchi K, Ebinuma H, Yamashiki N, Tamura S, Sugawara Y, Mori A, Yagi S, Shirabe K, Taketomi A, Arai K, Monoe K, Ichikawa T, Taniai M, Miyake Y, Kumagi T, Abe M, Yoshizawa K, Joshita S, Shimoda S, Honda K, Takahashi H, Hirano K, Takeyama Y, Harada K, Migita K, Ito M, et al. 2012. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am. J. Hum. Genet. 91:721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hess J, Nielsen PJ, Fischer KD, Bujard H, Wirth T. 2001. The B lymphocyte-specific coactivator BOB.1/OBF.1 is required at multiple stages of B-cell development. Mol. Cell. Biol. 21:1531–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubart DB, Rolink A, Schubart K, Matthias P. 2000. Cutting edge: lack of peripheral B cells and severe agammaglobulinemia in mice simultaneously lacking Bruton's tyrosine kinase and the B cell-specific transcriptional coactivator OBF-1. J. Immunol. 164:18–22 [DOI] [PubMed] [Google Scholar]
- 23.Corcoran LM, Hasbold J, Dietrich W, Hawkins E, Kallies A, Nutt SL, Tarlinton DM, Matthias P, Hodgkin PD. 2005. Differential requirement for OBF-1 during antibody-secreting cell differentiation. J. Exp. Med. 201:1385–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. 1996. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature 383:538–542 [DOI] [PubMed] [Google Scholar]
- 25.Nielsen PJ, Georgiev O, Lorenz B, Schaffner W. 1996. B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur. J. Immunol. 26:3214–3218 [DOI] [PubMed] [Google Scholar]
- 26.Kim U, Qin XF, Gong S, Stevens S, Luo Y, Nussenzweig M, Roeder RG. 1996. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature 383:542–547 [DOI] [PubMed] [Google Scholar]
- 27.Bordon A, Bosco N, Du Roure C, Bartholdy B, Kohler H, Matthias G, Rolink AG, Matthias P. 2008. Enforced expression of the transcriptional coactivator OBF1 impairs B cell differentiation at the earliest stage of development. PLoS One 3:e4007. 10.1371/journal.pone.0004007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boehm J, He Y, Greiner A, Staudt L, Wirth T. 2001. Regulation of BOB.1/OBF.1 stability by SIAH. EMBO J. 20:4153–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiedt R, Bartholdy BA, Matthias G, Newell JW, Matthias P. 2001. The RING finger protein Siah-1 regulates the level of the transcriptional coactivator OBF-1. EMBO J. 20:4143–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herzog S, Hug E, Meixlsperger S, Paik J-H, Depinho RA, Reth M, Jumaa H. 2008. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat. Immunol. 9:623–631 [DOI] [PubMed] [Google Scholar]
- 31.Cadwell RC, Joyce GF. 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2:28–33 [DOI] [PubMed] [Google Scholar]
- 32.Richard M, Hikima J, Wilson M, Miller N, Cunningham C, Warr G. 2009. BOB.1 of the channel catfish, Ictalurus punctatus: not a transcriptional coactivator? Mol. Immunol. 46:481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita S, Kojima T, Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066 [DOI] [PubMed] [Google Scholar]
- 34.Miller NW, Rycyzyn MA, Wilson MR, Warr GW, Naftel JP, Clem LW. 1994. Development and characterization of channel catfish long term B cell lines. J. Immunol. 152:2180–2189 [PubMed] [Google Scholar]
- 35.Frew IJ, Hammond VE, Dickins RA, Quinn JM, Walkley CR, Sims NA, Schnall R, Della NG, Holloway AJ, Digby MR, Janes PW, Tarlinton DM, Purton LE, Gillespie MT, Bowtell DD. 2003. Generation and analysis of Siah2 mutant mice. Mol. Cell. Biol. 23:9150–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong CS, Sceneay J, House CM, Halse HM, Liu MC, George J, Hunnam TC, Parker BS, Haviv I, Ronai Z, Cullinane C, Bowtell DD, Moller A. 2012. Vascular normalization by loss of Siah2 results in increased chemotherapeutic efficacy. Cancer Res. 72:1694–1704 [DOI] [PubMed] [Google Scholar]
- 37.Treier M, Staszewski LM, Bohmann D. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787–798 [DOI] [PubMed] [Google Scholar]
- 38.Edery I, Hümbelin M, Darveau A, Lee KA, Milburn S, Hershey JW, Trachsel H, Sonenberg N. 1983. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J. Biol. Chem. 258:11398–11403 [PubMed] [Google Scholar]
- 39.Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. 2008. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics 7:1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen H-CS, Xu Q, Chou DM, Zhao Z, Elledge SJ. 2008. Global protein stability profiling in mammalian cells. Science 322:918–923 [DOI] [PubMed] [Google Scholar]
- 41.Kim KJ, Kanellopoulos-Langevin C, Merwin RM, Sachs DH, Asofsky R. 1979. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J. Immunol. 122:549–554 [PubMed] [Google Scholar]
- 42.Gutman GA, Warner NL, Harris AW. 1981. Immunoglobulin production by murine B-lymphoma cells. Clin. Immunol. Immunopathol. 18:230–244 [DOI] [PubMed] [Google Scholar]
- 43.Molinari E, Gilman M, Natesan S. 1999. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 18:6439–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernandez-Hernandez A, Ray P, Litos G, Ciro M, Ottolenghi S, Beug H, Boyes J. 2006. Acetylation and MAPK phosphorylation cooperate to regulate the degradation of active GATA-1. EMBO J. 25:3264–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.House CM, Frew IJ, Huang HL, Wiche G, Traficante N, Nice E, Catimel B, Bowtell DD. 2003. A binding motif for Siah ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 100:3101–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dickins RA, Frew IJ, House CM, O'Bryan MK, Holloway AJ, Haviv I, Traficante N, de Kretser DM, Bowtell DDL. 2002. The ubiquitin ligase component Siah1a is required for completion of meiosis I in male mice. Mol. Cell. Biol. 22:2294–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu X, Wang L, Luo Y, Roeder RG. 2001. Identification and characterization of a novel OCA-B isoform. Implications for a role in B cell signaling pathways. Immunity 14:157–167 [PubMed] [Google Scholar]
- 48.Xue L, Wang W-H, Iliuk A, Hu L, Galan JA, Yu S, Hans M, Geahlen RL, Tao WA. 2012. Sensitive kinase assay linked with phosphoproteomics for identifying direct kinase substrates. Proc. Natl. Acad. Sci. U. S. A. 109:5615–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zwilling S, Dieckmann A, Pfisterer P, Angel P, Wirth T. 1997. Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science 277:221–225 [DOI] [PubMed] [Google Scholar]
- 50.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342 [DOI] [PubMed] [Google Scholar]
- 51.Kremers GJ, Goedhart J, van Munster EB, Gadella TW., Jr 2006. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius. Biochemistry 45:6570–6580 [DOI] [PubMed] [Google Scholar]
- 52.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. 2012. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40:D261–D270. 10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin J, Li X, Gygi SP, Harper JW. 2007. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447:1135–1138 [DOI] [PubMed] [Google Scholar]
- 54.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. 1992. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360:597–599 [DOI] [PubMed] [Google Scholar]
- 55.Dang Y, Siew LM, Zheng Y-H. 2008. APOBEC3G is degraded by the proteasomal pathway in a Vif-dependent manner without being polyubiquitylated. J. Biol. Chem. 283:13124–13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Y, Zhu H, Zou M-H. 2012. Non-covalent interaction between polyubiquitin and GTP cyclohydrolase 1 dictates its degradation. PLoS One 7:e43306. 10.1371/journal.pone.0043306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Touitou R, Richardson J, Bose S, Nakanishi M, Rivett J, Allday MJ. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 20:2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers S, Wells R, Rechsteiner M. 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234:364–368 [DOI] [PubMed] [Google Scholar]
- 59.Ravid T, Hochstrasser M. 2008. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9:679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brass AL, Kehrli E, Eisenbeis CF, Storb U, Singh H. 1996. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 10:2335–2347 [DOI] [PubMed] [Google Scholar]
- 61.Ptashne M. 1988. How eukaryotic transcriptional activators work. Nature 335:683–689 [DOI] [PubMed] [Google Scholar]
- 62.Thomas D, Tyers M. 2000. Transcriptional regulation: kamikaze activators. Curr. Biol. 10:R341–R343. 10.1016/S0960-9822(00)00462-0 [DOI] [PubMed] [Google Scholar]