Abstract
Tim23 is an essential channel-forming subunit of the presequence translocase recruiting multiple components for assembly of the core complex, thereby regulating the protein translocation process. However, understanding of the precise interaction of subunits associating with Tim23 remains largely elusive. Our findings highlight that transmembrane helix 1 (TM1) is required for homodimerization of Tim23, while, together with TM2, it is involved in preprotein binding within the channel. Based on our evidence, we predict that the TM1 and TM2 from each dimer are involved in the formation of the central translocation pore, aided by Tim17. Furthermore, TM2 is also involved in the recruitment of Tim21 and the presequence-associated motor (PAM) subcomplex to the Tim23 channel, while the matrix-exposed loop L1 generates specificity in their association with the core complex. Strikingly, our findings indicate that the C-terminal sequence of Tim23 is dispensable for growth and functions as an inhibitor for binding of Tim21. Our model conceptually explains the cooperative function between Tam41 and Pam17 subunits, while the antagonistic activity of Tim21 predominantly determines the bound and free forms of the PAM subcomplex during import.
INTRODUCTION
Mitochondria are indispensable organelles required for a wide spectrum of cellular processes, including energy metabolism. The mitochondrial genome encodes only a few proteins across the kingdoms, ranging from 7 in Saccharomyces cerevisiae to 13 in humans (1, 2). Almost all the several hundreds of proteins residing in various mitochondrial compartments are encoded by the nuclear genome and synthesized on cytosolic ribosomes. Therefore, active mitochondrial function and biogenesis require efficient protein transport destined for the different subcompartments (1–9). Greater than 60% of mitochondrial proteins are targeted to the matrix compartment and are translocated through the presequence translocase (Tim23 complex), an inner membrane-bound multisubunit machinery whose components are highly conserved. The presequence translocase consists of central channel-forming components mainly constituted by the Tim23 protein together with Tim17, which maintains the integrity of the translocation pore (10–15), while other subunits, such as Tim50 and Tim21, which contains a single transmembrane helix with a large intermembrane space (IMS)-exposed domain, assist with presorting and guided channeling of preproteins via interaction with the translocase of outer membrane (TOM) complex (16–20). Besides channel-forming components, the Tim23 complex recruits a peripherally associated import motor whose functions are essential for the translocation of proteins destined for the matrix compartment (21–23). Mitochondrial heat shock protein Hsp70 (Ssc1 in yeast) forms the core of the import motor recruited for preprotein binding in the ATP-bound state to the channel via a membrane tether, Tim44 (21–24). Tim44 provides the platform and recruits two additional terminal regulatory components, J protein Pam18 and J-like protein, Pam16 (25–29). Both Pam18 and Pam16 constitute the presequence-associated motor (PAM) subcomplex by forming a stable heterodimer that stimulates mitochondrial Hsp70s ATPase activity coupled to multiple import reaction cycles (30). Recently, the Pam17 and Tam41 proteins have been identified as additional nonessential components of the Tim23 complex that are involved in the organization and recruitment of the PAM subcomplex to the core channel (31–33).
Phylogenetically, Tim23 is a polytopic membrane protein containing conserved segments of four predicted transmembrane helices (transmembrane helix 1 [TM1] to TM4) with three connecting loops (L1 to L3) exposed to either matrix or IMS regions (34, 35). In addition to that, Tim23 also possesses a large unstructured N-terminal hydrophilic domain that consists of 100 amino acids exposed to the IMS region (36). The N terminus of Tim23 interacts with the IMS region of Tim50 and is cooperatively involved in preprotein recognition and transfer to the central channel (37–40). On the other hand, the C-terminal region of Tim23 is membrane embedded and has been shown to form a voltage-gated cation-selective channel (41, 42).
Although the multiple interacting components aid in the core complex assembly process, the precise recruitment of subunits to the Tim23 channel and their dissociation from the Tim23 channel are still enigmatic. In this report, we present a novel set of temperature-sensitive (Ts) mutants, isolated through a genetic screen, with mutations in the distinct parts of the Tim23 protein which are precisely defective in recruitment of interacting subunits. Through mutational analysis, our findings comprehensively provide evidence for the defined functions for the different segments of Tim23, such as homodimerization, preprotein binding, and the helices involved in the formation of translocation pore, thereby maintaining the inner membrane polarity. Besides, our report reveals that the C-terminal sequence of the Tim23 protein negatively regulates the association of Tim21, thus modulating the PAM-bound and -free forms of Tim23 for the import of matrix and inner membrane proteins.
MATERIALS AND METHODS
Genetic methods, plasmids, and yeast strains.
The strains used in this study are listed in Table S1 in the supplemental material. The plasmids used in this study are described in Table S2 in the supplemental material. The TIM23 gene from positions −600 to +980 was amplified by use of forward primer 5′-GGCACTAGTGACTCTGGAGGGCGTAACGG-3′ and reverse primer 5′-GGCCTCGAGGGAAATACCCGAGAGTGAGCGG-3′ using genomic DNA as the template and subsequently cloned into SpeI/XhoI-digested vector pRS314. This recombinant plasmid, designated pRS314-Tim23, was used for all further genetic modifications. Initially, a BamHI restriction site was introduced into pRS314-Tim23 by silent mutagenesis at the 531th nucleotide position using forward primer 5′-GGCAAACATGACACCGCGGGATCCATTGGCGCTGGGGCCCT-3′, reverse primer 5′-AGGGCCCCAGCGCCAATGGATCCCGCGGTGTCATGTTTGCC-3′, and Pfu Turbo DNA polymerase from Stratagene. To obtain Ts mutants, a library of random mutations in pRS314-Tim23 was created through PCR amplification of the coding region of the TIM23 gene by forward primer 5′-ATGTCGTGGCTTTTTGGAGA-3′ and reverse primer 5′ TCATTTTTCAAGTAGTCTTT-3′ using low-fidelity Taq DNA polymerase, as described previously (43, 44). After digestion with the SalI and BamHI restriction enzymes, the randomly mutagenized PCR fragment generated a fragment corresponding to nucleotide positions +157 to +534, which was subcloned into the pRS314-Tim23 vector digested with the same set of restriction enzymes. The Δtim23 strain carrying pRS316-tim23Δ1-50 (where tim23Δ1-50 represents tim23 devoid of the sequence for the first 50 amino acids of Tim23) was transformed with the library and incubated at 30°C on plates from which tryptophan was omitted. Transformants were patched onto 5-fluoroorotic acid (5-FOA) plates to select for candidates that could grow in the absence of tim23Δ1-50 and subsequently replicated to plates from which tryptophan was omitted and incubated at 37°C to score the Ts phenotype. Plasmids were rescued from conditional mutants by previously described protocols (45) and subsequently sequenced to find out the position of the mutated nucleotide.
The chromosomal deletion mutants were constructed by using homologous recombination and linear PCR-amplified DNA fragments as the substrates for transformation. Briefly, for deletion of the PAM17 gene, a PCR product containing an LEU2 auxotrophic marker cassette and short sequences homologous to the flanking regions of the PAM17 locus were amplified by primers pam17Δfor (5′-TCGCTAGTCAACCTTCTGTCACCGCTGCGGCATTGCGCTCAGATTGTACTGAGAGTGCA-3′) and pam17Δrev (5′-AATCTCTTAACCATTGCTTATATTCCTTTAAGGAACCTATCTGTGCGGTATTTCACACC-3′) using pRS415 as the template. To isolate positive clones, yeast transformants were grown on selective medium lacking leucine. The TAM41 and TIM21 genes were disrupted by homologous recombination of the corresponding PCR products containing a kanMX4 cassette and short sequences homologous to the flanking regions of the TAM41 and TIM21 loci amplified using the pFA6-kanMX4 vector. Primers corresponding to tam41Δfor (5′-AATGCCATTCAACGAACGTAAGCATGTTTAATAGGCTTCTCGTACGCTGCAGGTCG-3′) and tam41Δrev (5′-CATACTTAATTGACTTTGTGATTCCAGCTGTGAAAACACCATCGATGAATTCGAGC-3′) were used for deletion of the TAM41 gene. The primers corresponding to tim21Δfor (5′-GACATAGAAAGCCTTTGTTTCCACGATATAATACTTTCGTGAACGTACGCTGCAGGTCG-3′) and tim21Δrev (5′-AGCCCTTCGAATTAGAAACCGGATGCAATTTTGGCTTGATCAAATCGATGAATTCGAGC-3′) were used for disruption of the TIM21 gene. To isolate positive clones, yeast transformants were grown on medium containing kanamycin (400 μg/ml). The deletions were further verified by PCR and by Western blot analysis. Placing the TAM41 gene under the control of the GAL1 promoter was accomplished by transforming yeast cells with linear DNA encoding a selectable marker cassette, the GAL1 promoter flanked by sequences that are homologous to a target site in the yeast chromosome. Briefly, the 5′ untranslated region of the TAM41 gene from nucleotides −870 to −500 was amplified from the genomic DNA template using primers GAL1 for.1 (5′-GGTATAGTTTCGGTTGATCTAGATATAAAT-3′) and GAL1 rev.1 (5′-TGGTGCACTCTCAGTACAATCTTGCATACATTGCCAGAAAGAAACC-3′) having an overhang of the TRP1 marker cassette. The TRP1 cassette was amplified, using pRS314 as a template, by primers GAL1 for.2 (5′-GGTTTCTTTCTGGCAATGTATGCAAGATTGTACTGAGAGTGCACCA-3′) and GAL1 rev.2 (5′-GCCCGCTCGGCGGCTTCTAATCCGTCTGTGCGGTATTTCACACCGCAT-3′) having an overhang of the GAL1 promoter. The amplification of the GAL1 promoter was accomplished, using the pYES2.0 vector, by primers GAL1 for.3 (5′-ATGCGGTGTGAAATACCGCACAGACGGATTAGAAGCCGCCGAGCGGGC-3′) and GAL1 rev.3 (5′-GACCATTTTCAGAAACTCGTAACATGTTTTTTCTCCTTGACGTTAAAGT-3′) having an overhang of the TAM41 gene. The open reading frame (ORF) of the TAM41 gene was amplified by primers GAL1 for.4 (5′-ACTTTAACGTCAAGGAGAAAAAACATGTTACGAGTTTCTGAAAATGGTC-3′) and GAL1 rev.4 (5′-ACAAACCGGCACGGAGCTTTGAAGGATGAC-3′). These PCR fragments were subsequently annealed together by complementary overhangs using the splicing by overhang extension-PCR method. To isolate positive clones, yeast transformants were grown on selective medium lacking tryptophan. All primers used in this study were synthesized by Eurofins Inc., and sequencing reactions were performed by SciGenom labs.
Cells were grown in 1% yeast extract, 2% peptone, and 2% glucose (YPD), 1% yeast extract, 2% peptone, and 3% glycerol (YPG), 1% yeast extract, 2% peptone, and 2% galactose (YPGal), or synthetic dextrose (SD) medium having 2% glucose and 0.66% yeast nitrogen base (YNB) without amino acids and with appropriate supplements. For isolation of mitochondria, wild-type and mutant yeast cells were grown in YPG medium at a permissible temperature. However, due to the respiration-deficient behavior of mutants with a tim23 allele with a Y-to-N change at residue 105 (Y105N) and a G-to-D change at residue 120 (G120D) (tim23Y105N/G120D), mitochondria were isolated after growing the cells in SD medium. For the isolation of mitochondria of yeast strains overexpressing Tim17 or Tim21, cells were grown in SD-uracil (SD-URA) medium with appropriate supplements. For the generation of mitochondria depleted of the Tam41 protein, a yeast strain having the TAM41 gene under the control of the GAL1 promoter was grown for 48 h on galactose-containing medium, after which cells were harvested, resuspended in glucose-containing medium, and grown for a further 18 h to deplete the level of the Tam41 protein.
In vitro import assay.
For the in vitro import assay, wild-type or mutant mitochondria were subjected to heat shock at 34°C for 30 min, followed by incubation with precursor protein Cyb2(167)Δ19-DHFR(His6), comprised of the mouse dihydrofolate reductase (DHFR) fused to the N-terminal 167 amino acids of cytochrome b2 having a deletion of 19 amino acids of the inner membrane sorting signal, in import buffer (250 mM sucrose, 10 mM MOPS [morpholinepropanesulfonic acid]-KOH, pH 7.2, 80 mM KCl, 5 mM dithiothreitol, 5 mM MgCl2, 2 mM ATP, 2 mM NADH, 1% bovine serum albumin) for 5-, 10-, 15-, and 20-min intervals at 25°C. The import reaction was stopped using 10 μg/ml of valinomycin (Sigma). Protease treatment was performed by incubating the mitochondria with 0.1 mg/ml proteinase K (PK) for 20 min on ice, which was further inactivated by addition of 1 mM phenylmethylsulfonyl fluoride (PMSF; US Biological). After PK treatment, mitochondria were isolated by centrifugation and analyzed by SDS-PAGE, followed by Western blotting using specific antibodies. Image quantification was performed using ImageJ software.
CoIP and BN-PAGE.
For coimmunoprecipitation (CoIP) analysis, protein A Sepharose CL-4B (GE Healthcare) beads were used after they were thoroughly washed with water, followed by Tris-buffered saline (TBS) buffer. The washed beads were incubated with purified Tim23 antibodies raised against an N-terminal domain (amino acids 1 to 98) of the protein. The beads were washed with TBS to remove any unbound antibodies. To eliminate the contamination of the light and heavy chains of IgGs, antibodies were covalently coupled to protein A beads using a dimethyl pimelimidate (DMP) cross-linker. The wild-type and mutant mitochondria were first subjected to heat shock to induce the Ts phenotype. The lysates were than prepared by solubilizing 2 mg of mitochondria in a lysis buffer (20 mM MOPS-KOH, pH 7.4, 250 mM sucrose, 80 mM KCl, 5 mM EDTA, 1 mM PMSF) containing 1% digitonin for 45 min at 4°C (46). After a clarifying spin, the supernatant was incubated with antibody-coated beads for 2 h at 4°C. Subsequently, the beads were washed with the same lysis buffer. Samples were separated on SDS-polyacrylamide gels, followed by immunoblotting using specific antibodies.
In order to test for the homodimerization of Tim23 in vivo, wild-type or mutant Tim23 plasmids carrying 1× FLAG tag at the C terminus were transformed into a yeast strain harboring a copy of pRS316-tim23Δ1-50, in which tim23 is devoid of the sequence for the first 50 amino acids of Tim23. Mitochondria were isolated and solubilized with 1% digitonin containing buffer and subsequently incubated with protein G-Sepharose beads coupled with FLAG antibody. After several washes, samples were analyzed by SDS-PAGE, followed by immunoblotting.
In order to test for the interaction with the substrate preprotein, approximately 2 mg of wild-type and mutant mitochondria was solubilized in a lysis buffer (20 mM MOPS-KOH, pH 7.4, 250 mM sucrose, 80 mM KCl, 5 mM EDTA, 1 mM PMSF) containing 1% Triton X-100 for 30 min at 4°C. After centrifugation, the supernatant was incubated with beads cross-linked to Tim23 antibodies for 30 min at 4°C, followed by several washes with lysis buffer. Subsequently, beads were incubated with 5 μg of purified Cyb2(167)Δ19-DHFR(His6) protein for 1 h at 4°C and analyzed by SDS-PAGE, followed by Western blotting.
Blue native PAGE (BN-PAGE) analysis was performed according to previously published protocols (47). Briefly, mitochondria (200 μg) were first subjected to heat shock, followed by lysis in 90 μl of 1% digitonin buffer for 45 min at 4°C, as described above. The unsolubilized material was removed by centrifugation for 20 min at 50,000 × g. After addition of 10 μl of sample buffer (5% [wt/vol] Coomassie brilliant blue G-250, 100 mM bis-Tris, pH 7.0, 500 mM 6-aminocaproic acid), the supernatant was analyzed directly by BN-PAGE, followed by immunoblotting with specific antibodies.
Quantification of protein half-lives.
Quantification of protein half-lives was performed as described previously (48). Wild-type and mutant yeast cells were grown in YPD medium to log phase (A600, ∼0.5) at a permissible temperature. Cycloheximide (US Biological) was used as a translational inhibitor (50 μg/ml). Following treatment, equal amounts of cells were collected at 0, 15, 30, and 60 min and analyzed by SDS-PAGE, followed by immunoblotting using Tim23 antibody. Data were quantified using ImageJ and analyzed using GraphPad Prism, version 5.0, software.
Fluorescence and flow cytometric analysis.
Analysis of mitochondrial membrane potential and mitochondrial mass was accomplished by previously described protocols (49). Briefly, to test for the loss in membrane potential, roughly 50 μg of mitochondria was first subjected to heat shock for 15 min, followed by incubation with JC-1 dye for 10 min at room temperature (25°C) in the dark. Wild-type mitochondria incubated in 0.1 M valinomycin for 10 min prior to dye staining were used as a control for complete depolarization. The fluorescence measurements were performed using a Jasco spectrofluorometer (model 6300) at a fixed excitation wavelength of 490 nm, and emission spectra were recorded from 500 to 620 nm. The ratios of the peaks at 590 nm (aggregate form of the JC-1 dye) and 530 nm (monomeric form) were used as an indicator of membrane polarization.
The functional mitochondrial masses of various yeast strains were determined using MitoTracker Deep Red-A dye (Invitrogen). Briefly, yeast cells were harvested during early log phase and, after washing with PBS, subjected to heat shock for 2 h, followed by incubation with 10 mM dye for 15 min in the dark. Fluorescence-activated cell sorter (FACS) analysis was performed using an excitation wavelength of 488 nm and an emission wavelength of 520 nm in a Becton, Dickinson (BD) FACSCanto II flow cytometer. The data were analyzed using WinMDI, version 2.9, software.
Fluorescence anisotropy peptide binding assays.
Fluorescence anisotropy analysis was performed with fluorescein-labeled peptide corresponding to the presequence of cytochrome c oxidase 4 (Cox4) (MLSLRQSIRFFKPTRRLC), as described previously, with minor modifications (49). Briefly, increasing concentrations of Tim23 were incubated with 25 nM fluorescein-labeled Cox4 (F-Cox4) in a buffer containing 25 mM Tris-Cl, pH 7.5, 100 mM KCl, and 10% glycerol for 1 h at room temperature. After binding reached equilibrium, the anisotropy measurements were recorded on a Beacon-2000 fluorescence polarization system (Panvera, Madison, WI) using excitation at 490 nm and emission at 535 nm. The data were fitted to a one-site binding hyperbola equation using GraphPad Prism, version 5.0, to calculate the equilibrium dissociation constant (Kd).
Miscellaneous.
Mitochondrion isolation and Hsp60 precursor accumulation analysis were performed as described previously (50). The analysis of the loss of mitochondrial DNA (mtDNA) was performed according to previously published protocols (49). The purification of Tim23 was performed using the RIL expression strain according to previously published protocols (42). The antisera used for immunodecoration against yeast-specific proteins, such as Hsp60, Mge1, Ydj1, Tim23, Tim50, Tim44, Pam18, and Pam16, were raised in rabbits, as previously reported (50). Research involving animal experimentation were carried out after approval from the Institutional Ethics Committee and from the Institutional Biosafety Committee. For preparation of antibody to Tim21, a fragment corresponding to amino acids 103 to 229 was cloned into the pET-3a vector and purified from the supernatant fraction using Escherichia coli. BL21(DE3) cells, as previously described (51). For preparation of antibody to Pam17, the ORF was cloned in the pET-3a vector and protein was reconstituted from the pellet fraction using the C41 expression strain according to previously published protocols (51). To raise antibodies against Tim17, a C-terminal peptide (CEAPSSQPLQA) was used for immunization. The Tim23 protein tagged with FLAG was detected using anti-FLAG antibody purchased from Sigma. All immunoblot analysis was carried out by using an enhanced chemiluminescence system (PerkinElmer) according to the manufacturer's instructions.
RESULTS
Isolation of conditional mutants in different segments of Tim23 protein involved in the subunit organization of the translocation channel.
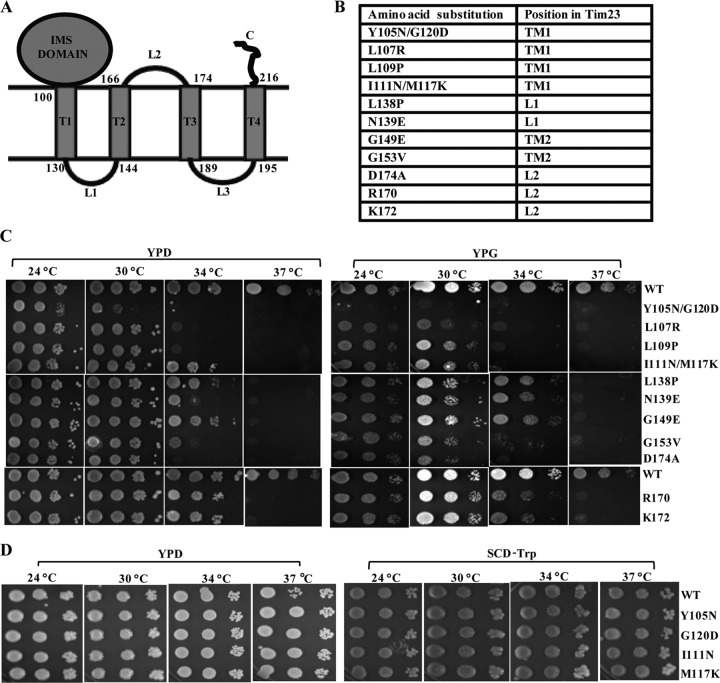
Tim23 is a multispanning inner membrane protein anchored via four transmembrane helices (TM1, TM2, TM3, and TM4) with both its amino and carboxyl termini facing toward the IMS region (Fig. 1A). The topological organization orients two loops, namely, loops L1 and L3, into the matrix, while loop L2 faces toward the IMS region. The functional significance of each of the defined segments of the Tim23 protein in the recruitment of import motor and channel-forming components is poorly understood due to the lack of conditional mutants which are selectively impaired in subunit organization of the core complex. The present study aimed to identify the critical amino acids in the different regions of the Tim23 protein which are involved in the dynamic association and dissociation of translocase components during the import process. As a step toward obtaining the conditional mutants, we constructed a mutagenic library of the region spanning from amino acids 101 to 176 of Tim23 protein by an error-prone PCR method. The mutant library was transformed into a Δtim23 haploid strain harboring a functional copy of the TIM23 gene on a URA3-based plasmid. The presence of the URA3 gene allowed selection of cells that had lost the plasmid carrying the wild-type TIM23 gene on 5-fluoroorotic acid (5-FOA) medium incubated under different permissive and nonpermissive temperature conditions. Mutant copies of the plasmid were rescued from selected Ts candidates, and the phenotype was confirmed by retransformation into a Δtim23 haploid strain. Figure 1B indicates the Ts mutants with mutations that were identified in the different segments of the Tim23 protein, including the TM1, TM2, L1, and L2 regions. In addition to that, C-terminal truncated proteins that lacked the TM3, L3, and TM4 segments together were isolated due to insertion of stop codons at positions 170 and 172, suggesting that the amino acid sequence between positions 170 and 222 is dispensable in vivo for function under permissive conditions. In order to confirm the growth phenotype, the mutant cells were subjected to drop test analysis by serially diluting and spotting them on YPD and YPG media, followed by incubation at different temperatures (Fig. 1C). As indicated in Fig. 1C (left and right), all Tim23 mutants were unable to grow at 37°C in YPD and YPG media. However, the mutants with mutations in the TM1 region displayed severe growth defects at 34°C compared to the growth of mutants with mutations in other segments. In particular, in the case of the double mutants, including the tim23Y105N/G120D and tim23I111N/M117K mutants, those with the respective single point mutations, such as the tim23Y105N, tim23G120D, and tim23I111N, tim23M117K mutants, failed to show any growth defect under our experimental conditions (Fig. 1D).
Fig 1.
Isolation of Ts mutants with mutations in different segments of the Tim23 protein. (A) Modular representation of the Tim23 protein. The topology of different helices and loops and the positions of the amino acids spanning each of the segments of the Tim23 protein are highlighted. (B) Amino acid positions of mutations in the different segments of the Tim23 protein of Ts mutants. (C and D) Growth phenotype analysis of the mutants. Serially diluted wild-type (WT) and mutant strains were spotted on the indicated medium and incubated at different temperatures. SCD, synthetic complete dextrose.
To show that the observed growth phenotypes were due to compromised import function, the Tim23 mutants were subjected to in vivo precursor accumulation analysis using Hsp60 as a model protein by monitoring the removal of the N-terminal signal sequence, which is cleaved off by the matrix-processing peptidase. After inducing the phenotype by briefly exposing the cells to nonpermissive temperature conditions, the mutants showed a robust accumulation of the Hsp60 precursor form at 34°C, indicating impaired protein import in the mutant cells (Fig. 2A). Notably, the tim23I111N/M117K, tim23L138P, tim23G149E, tim23R170, and tim23K172 mutants did not show the accumulation of detectable levels of the Hsp60 precursor form, consistent with their growth phenotypes (Fig. 2A).
Fig 2.
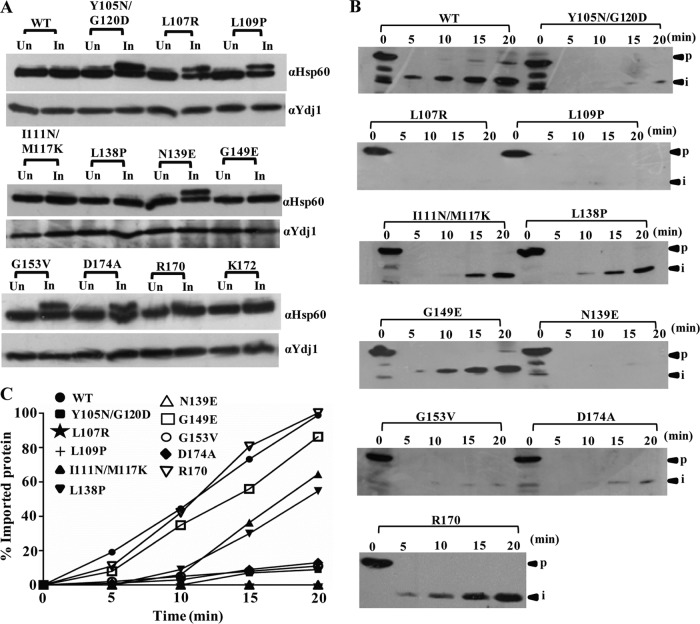
Analysis of protein translocation defects in Tim23 mutants. (A) In vivo precursor accumulation analysis. Wild-type and mutant yeast strains were grown at 30°C to early log phase and subjected to heat shock at 34°C for 2 h to induce the phenotype. The whole-cell lysates were analyzed by Western blotting using Hsp60-specific antibodies. Un, uninduced; In, induced. (B) In vitro import kinetic analysis using isolated mitochondria. Wild-type and mutant mitochondria were exposed to 34°C for 30 min to induce the Ts phenotype. Import was performed at 25°C using purified Cyb2(167)Δ19-DHFR precursor. Aliquots were taken at different time intervals, treated with proteinase K, and subsequently analyzed by Western blotting. p, precursor; i, intermediate. (C) Quantification of imported proteins on the Western blots. The signal of the imported intermediate form of Cyb2(167)Δ19-DHFR was quantified using ImageJ and is represented in the graph as a percentage of the total precursor imported in the wild type at 20 min.
In order to quantitate and kinetically monitor the translocation defects in mutants, we performed the import of chemical amounts of recombinant precursor protein Cyb2(167)Δ19-DHFR, comprised of the mouse DHFR fused to the N-terminal 167 amino acids of cytochrome b2 having a deletion of 19 amino acids of the inner membrane sorting signal to ensure localization to the matrix (52). Mitochondria were isolated from yeast cells grown at permissive temperatures, preincubated at 34°C to induce the mutant phenotype, and subjected to the import reaction using saturating amounts of the Cyb2(167)Δ19-DHFR precursor protein. However, the tim23R170 mutant did not show any import defect at 34°C, supporting the wild-type growth phenotype, while the tim23G149E, tim23I111N/M117K, and tim23L138P mutants showed an intermediate import defect consistent with their mild growth phenotype (Fig. 2B and C). Importantly, all other mutants exhibited a significant defect in the import efficiency of the mitochondrial preprotein, thus highlighting the amino acids of the different segments of Tim23 critical for in vivo function (Fig. 2B and C). The observed relative differences in the import of Hsp60 and Cyb2(167)Δ19-DHFR in the tim23G149E, tim23I111N/M117K, and tim23L138P mutants can be attributed to the length of the presequence, where a shorter presequence-containing preprotein such as Hsp60 was imported efficiently inside the mutant mitochondria, while a longer presequence-containing preprotein such as Cyb2(167)Δ19-DHFR exhibited an intermediate import defect.
The TM1 segment of Tim23 is critical for homodimerization and substrate capture in the channel during import.
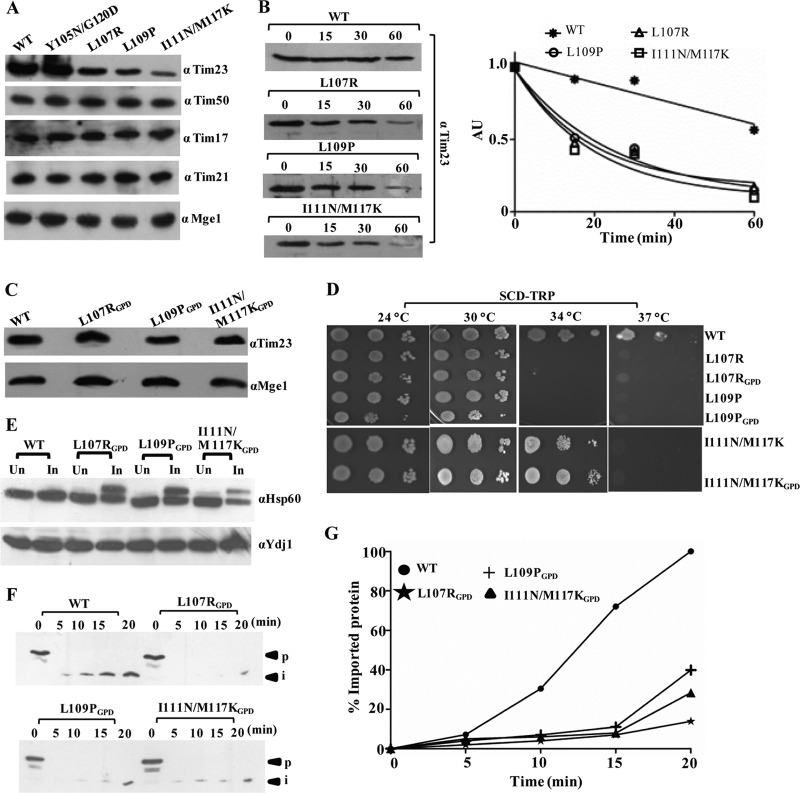
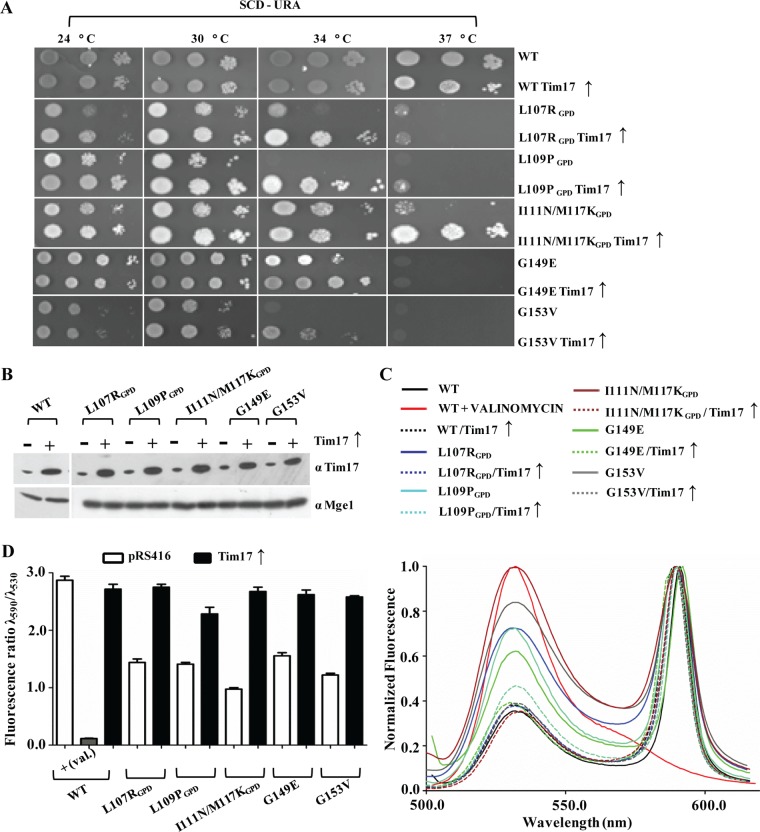
Four Ts mutants with single and double amino acid substitutions corresponding to L107R, L109P, Y105N/G120D, and I111N/M117K within the core of the TM1 segment were isolated. To test the stability of the mutant proteins, we first tested for their expression levels in the mitochondrial lysates using Western blotting. As indicated in Fig. 3A, the tim23L107R, tim23L109P, and tim23I111N/M117K mutants showed a significant reduction in protein levels. However, the levels of other channel-interacting components remained unaffected (Fig. 3A). We hypothesized that the relative decrease in the TM1 mutant level was presumably due to a reduction in the half-life of the protein. To determine the half-lives of the mutant proteins, we monitored the abundance of each mutant protein by immunoblot analysis of whole-cell extracts after inhibiting protein synthesis with cycloheximide. Importantly, the tim23L107R, tim23L109P, and tim23I111N/M117K mutants showed an enhancement in degradation as a function of time that followed first-order kinetics (Fig. 3B). To rule out the possibility that the observed growth phenotype of the TM1 mutants was due to reduced protein expression levels, we overexpressed them using the GPD promoter on a centromeric plasmid. Even though they had higher expression levels (Fig. 3C), the mutants were unable to complement the growth sensitivity under nonpermissible temperature conditions (Fig. 3D). Furthermore, the overexpressed mutant protein under the control of a stronger GPD promoter also failed to rescue the import defect under nonpermissible temperature conditions, as analyzed by in vivo precursor accumulation analysis (Fig. 3E) and determination of the in vitro import of recombinant Cyb2(167)Δ19-DHFR protein (Fig. 3F and G). These results clearly highlight that the Ts phenotype of TM1 mutants is a consequence of functional defects at the biochemical level.
Fig 3.
TM1 mutants have low expression levels caused by a reduced half-life. (A) Protein expression analysis. Wild-type and mutant yeast mitochondria were analyzed by immunoblotting using specific antibodies. (B) Determination of half-lives of mutant proteins. Wild-type and mutant yeast cells were grown to log phase, followed by addition of the translational inhibitor cycloheximide (50 μg/ml). Cells corresponding to an A600 of 0.1 were collected at different time intervals and analyzed by immunoblotting. The numbers above the lanes are times (in minutes). AU, arbitrary units. (C) Overexpression analysis of mutant proteins. Wild-type and mutant yeast cells expressing tim23 under the control of the centromeric GPD promoter were grown overnight, and cells corresponding to an A600 of 0.1 were loaded on SDS-polyacrylamide gels, followed by Western blotting. (D) Growth phenotype of mutant strains upon overexpression of Tim23. Serially diluted mutant strains overexpressing tim23 under the control of the centromeric GPD promoter were spotted on medium from which tryptophan was omitted. (E) In vivo precursor accumulation analysis upon overexpression of the TM1 mutant. Wild-type and TM1 mutant yeast strains overexpressing tim23 under the control of the CEN GPD promoter were subjected to Hsp60 precursor accumulation analysis, as described in the legend to Fig. 2A, except that the heat shock was given at 37°C. (F and G) In vitro import kinetic analysis. (F) Wild-type and mutant mitochondria were subjected to in vitro import using purified Cyb2(167)Δ19-DHFR precursor, as described in the legend to Fig. 2B, except that the heat shock was given at 37°C. (G) The signals were quantified using ImageJ software, as described in the legend to Fig. 2C.
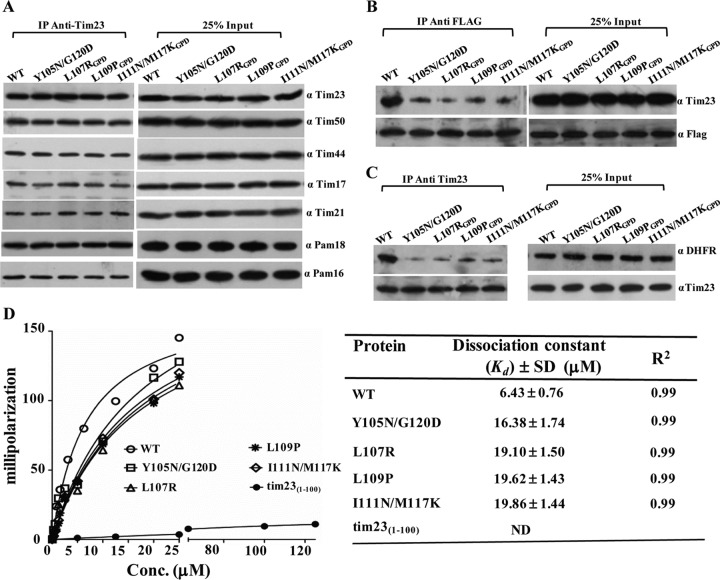
To determine the functional defects of the TM1 mutants, we tested their ability to interact with other core channel components by CoIP. For the mutants, including the tim23L107R, tim23L109P, and tim23I111N/M117K mutants, experimental analyses were carried out using a stronger GPD promoter, where the expression levels of mutant Tim23 were found to be comparable to those of the wild type (Fig. 3C). The digitonin-solubilized mitochondrial lysates prepared from overexpressed mutant strains were subjected to immunoprecipitation (IP) using cross-linked Tim23-specific antibodies. The samples were resolved on SDS-polyacrylamide gels and subjected to immunodecoration against all the components of the Tim23 translocase. Interestingly, as indicated in Fig. 4A, none of the TM1 mutants showed any significant defects in terms of their association with the other import motor components. Our results suggest that the TM1 segment is probably involved in orchestrating the other events of the import function. Hence, we tested the TM1 mutants for their self-dimerization and preprotein interaction property. Previously, it has been proposed that the gating of the channel requires dimerization of the Tim23 protein (12). The deletion of the first 50 amino acids of Tim23 was reported not to impair the function of the protein (12). Therefore, we first monitored the homodimerization propensity by coexpressing the truncated version of Tim23 (pRS316-tim23Δ1-50), followed by its affinity copurification using C-terminally FLAG-tagged full-length Tim23 and variants. The use of the N-terminally truncated version of Tim23 allowed us to achieve better separation between tim23Δ1-50 and full-length Tim23 on SDS-polyacrylamide gels. In the wild type, the FLAG-tagged Tim23 efficiently immunoprecipitated along with tim23Δ1-50 in digitonin-solubilized mitochondria (Fig. 4B). On the other hand, a significant reduction in the immunoprecipitation of tim23Δ1-50 protein was observed with FLAG-tagged TM1 mutants, thus highlighting the importance of TM1 in homodimerization (Fig. 4B). The truncation of the N terminus did not show any synthetic lethal effect with any of the TM1 mutations; therefore, the defect in homotypic interaction is solely attributed to the point mutations in the TM1 region (data not shown). Second, we checked for the interaction of Tim23 with substrate protein using Cyb2(167)Δ19-DHFR as a model protein. To demonstrate this, we first dissociated the import motor components from the translocase by addition of Triton X-100 and subsequently incubated them with Tim23-cross-linked antibodies. The Tim23-bound beads were then incubated with equal amounts of substrate protein and analyzed on SDS-polyacrylamide gels, followed by immunoblotting using substrate-specific antibodies. As shown in Fig. 4C, all TM1 mutants displayed poor interaction with substrate proteins compared to the wild type, which thus indicates that the TM1 segment is required for the interaction with the precursor polypeptides. To quantitatively measure the substrate-binding properties of the TM1 mutants, we utilized a fluorescence anisotropy-based assay using cytochrome oxidase 4 (Cox4) peptide (MLSLRQSIRFFKPTRRLC) derived from the presequence of yeast Cox4 labeled with a fluorescein fluorophore covalently attached to the cysteine residue. This assay measures the relative changes in the tumbling rates of fluorescence-labeled peptides upon binding of Tim23 in solution. The full-length wild-type and mutant proteins were purified to homogeneity (data not shown) and subjected to anisotropy analysis. The wild-type Tim23 showed a strong affinity for the peptide and yielded a Kd of 6.43 ± 0.76 μM (mean ± standard deviation [SD]), while the IMS domain of Tim23 (tim231-100) failed to show any significant binding to the peptide (Fig. 4D), which is in agreement with previously published results (40). However, all TM1 mutants exhibited a 2.5- to 3-fold reduced affinity for the peptide compared to the wild type (Fig. 4D), therefore further highlighting the significance of the TM1 segment in stabilizing the interaction with the precursor polypeptides within the core channel.
Fig 4.
Essentiality of the TM1 helix for homodimerization and substrate capture in the channel. (A) CoIP analysis of the Tim23 complex in mutants. Wild-type and mutant mitochondria were solubilized in 1% digitonin buffer and subjected to immunoprecipitation using Tim23 antibody-cross-linked beads, followed by Western blotting. (B) Tim23 homodimerization analysis. Mitochondria expressing a C-terminal FLAG-tagged form of wild-type and mutant Tim23 were solubilized in 1% digitonin buffer and subjected to CoIP using FLAG-specific antibodies, followed by immunodecoration with Tim23 antibodies. (C) Analysis of interaction of substrate proteins with Tim23. Wild-type and mutant mitochondria were solubilized in 1% Triton X-100 and incubated with Tim23 antibody-cross-linked beads, followed by incubation with purified Cyb2(167)Δ19-DHFR protein. The samples were analyzed by immunoblotting. (D) Fluorescence anisotropy assay for wild-type and mutant Tim23 proteins. F-Cox4 peptide (25 nM) was incubated in the presence of the indicated concentrations of wild-type and mutant Tim23 proteins. Anisotropy measurements were recorded and plotted against increasing concentrations of different proteins using GraphPad Prism, version 5.0, software. The table shows the Kd and R2 value of different proteins with F-Cox4 peptide. ND, not determined. Twenty-five percent of the input was used as a loading control.
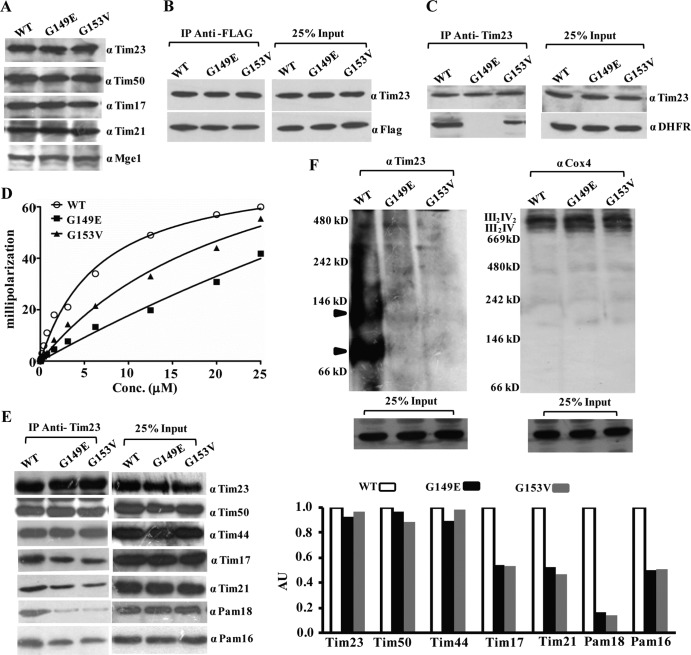
The TM2 segment of Tim23 is essential for Tim17 association and substrate interaction within the core channel.
TM2 mutants with a single amino acid substitution, namely, tim23G149E and tim23G153V, expressed Tim23 protein up to wild-type levels, as observed upon immunoblotting of the lysates, unlike the TM1 mutants (Fig. 5A and 3A). Together, the levels of the other components of the channel also remained unaltered (Fig. 5A). To determine the biochemical defects, we first asked whether the TM2 segment plays any role in the homodimerization of Tim23, a property similar to that of TM1. To monitor this, we copurified the tim23Δ1-50 truncation with C-terminally FLAG-tagged TM2 mutations, which are expressed on a CEN plasmid under the control of the TEF promoter. As indicated in Fig. 5B, none of the mutations in the TM2 segment exerted any direct impairment of the Tim23 homodimerization property, whereas mutations in the TM1 segment did (Fig. 4B). To determine the involvement of the TM2 segment in the substrate interaction, the Tim23-cross-linked beads were incubated with wild-type and mutant mitochondria, followed by incubation with Cyb2(167)Δ19-DHFR substrate. Importantly, the tim23G149E mutant completely failed to interact with the substrate (Fig. 5C). On the other hand, the tim23G153V mutant protein showed a significant reduction in substrate interaction compared to that of the wild-type Tim23 protein (Fig. 5C). These results were further validated by anisotropy analysis of the TM2 mutants using the F-Cox4 peptide. Both mutant proteins (the tim23G149E and tim23G153V proteins) were first purified to homogeneity (data not shown) and subjected to anisotropy analysis. As shown in Fig. 5D, tim23G153V yielded a Kd of 20.22 ± 1.66 μM (mean ± SD), indicating an ∼3-fold reduced affinity compared to that of the wild type (Kd, 6.50 ± 0.68 μM [mean ± SD]), while the tim23G149E mutant exhibited a very poor interaction with the F-Cox4 peptide, yielding a Kd of 39.84 ± 3.92 μM (mean ± SD), suggesting that the TM2 segment closely cooperates with the TM1 region for preprotein capture within the channel during import (Fig. 5D).
Fig 5.
TM2 plays a critical role in substrate recognition and Tim17 interaction. (A) Protein expression analysis of TM2 mutants. Wild-type and mutant yeast mitochondria were subjected to immunoblotting using the indicated antibodies. (B) Analysis of Tim23 homodimerization in TM2 mutants. Wild-type and mutant mitochondria expressing a C-terminal FLAG-tagged form of Tim23 were solubilized in digitonin buffer, subjected to immunoprecipitation using anti-FLAG antibodies, and analyzed by Western blotting. (C) Substrate binding analysis of TM2 mutants. After solubilization in 1% Triton X-100-containing buffer, wild-type and mutant mitochondria were analyzed for substrate interaction, as described in the legend to Fig. 4C (D) Fluorescence anisotropy analysis for wild-type and mutant Tim23 proteins. F-Cox4 peptide (25 nM) was incubated in the presence of the indicated concentrations of wild-type and mutant proteins, and anisotropy values were recorded, as described in the legend to Fig. 4D. (E) CoIP analysis of the Tim23 complex in TM2 mutants. (Left) For CoIP analyses, digitonin-solubilized mitochondria were incubated with Tim23 antibody-cross-linked beads and subsequently analyzed by immunoblotting; (right) the amount of different immunoprecipitated proteins was quantified using ImageJ software. (F) Determination of stability of the core-Tim23 complex and respiratory chain complexes in TM2 mutants by BN-PAGE. Digitonin-solubilized mitochondria were subjected to BN-PAGE analysis, followed by Western blotting using Tim23 antibodies (left) and Cox4 antibodies (right). The locations of the complexes of the wild type are indicated by arrowheads. Twenty-five percent of the input was used as a loading control.
To determine the influence of mutations within the TM2 segment in the organization of the presequence translocase, we performed CoIP using cross-linked Tim23 antibodies and analyzed samples on SDS-polyacrylamide gels, followed by immunodecoration for different subunits of the Tim23 complex. As indicated in Fig. 5E, both the tim23G149E and tim23G153V mutants showed impairment in their association with the core channel subunit, Tim17, and import motor components, including Pam18 and Pam16. Additionally, both TM2 mutants showed a reduction in the association of the Tim21 subunit with the core Tim23 component (Fig. 5E). However, the association of Tim44 and Tim50 with the core Tim23 channel remained unaltered. To confirm the findings presented above, the stabilities of the core complexes were further analyzed by BN-PAGE using the digitonin-solubilized mitochondria. As anticipated, both TM2 mutants failed to assemble a stable core complex of 90 kDa with the Tim23 subunit, thus additionally confirming the immunoprecipitation results (Fig. 5F). However, as a control, we checked for the stability of the respiratory chain complexes (III2IV2 and III2IV), which remained unaltered in both TM2 mutants, as analyzed with anti-Cox4 antibodies (Fig. 5F) (53). These observations highlight the fact that the TM2 helix plays a critical role in recruitment of Tim17 to the Tim23 core subunit and regulates the association of Tim21 and the PAM subcomplex.
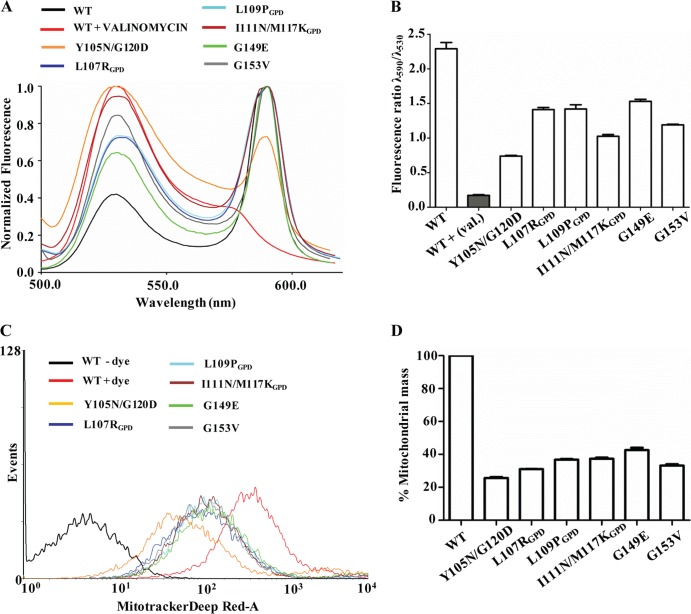
The TM1 and TM2 helices play a critical role in the maintenance of membrane potential and polarity across the inner membrane.
Mitochondrial inner membrane integrity largely depends upon the accurate assembly of multiple transporters and translocation channels. In order to test the effect of mutations in the TM1 and TM2 helices of Tim23 on inner membrane integrity and polarity, we tested for the retention of membrane potential by JC-1 dye staining. The uptake of polarity-sensitive dye JC-1 is dependent on the presence of an intact inner membrane potential, resulting in an aggregated form of dye yielding red fluorescence. On the other hand, the loss of membrane potential leads to the retention of dye outside the mitochondrial compartment, leading to green fluorescence. Therefore, the ratio of red to green fluorescence is used as an indicator of functionality and retention of the inner membrane polarity of the mitochondria. A complete disruption of the polarity was obtained by treating mitochondria with valinomycin, which served as a positive control (P < 0.001) (Fig. 6A). A greater than 30% reduction in the ratio of red (590 nm) to green (530 nm) fluorescence intensity was observed with both the TM1 and TM2 mutants, indicating a significant loss of polarity across the inner membrane (P < 0.001) (Fig. 6A and B). To test whether alteration of the membrane polarity influences the functionality of the mitochondria, mutant yeast cells were stained with MitoTracker Deep Red-A dye, followed by fluorescence measurement using FACS-based analysis. Importantly, both the TM1 and TM2 mutants showed a significant reduction in the functional mitochondrial mass due to decreased polarity across the mitochondrial membrane, thus highlighting the critical role played by the TM1 and TM2 helices in the maintenance of architecture and the permeability barrier of the Tim23 channel (P < 0.001) (Fig. 6C and D).
Fig 6.
TM1 and TM2 are required for maintenance of membrane polarity of the Tim23 channel. (A and B) Measurements of inner membrane polarity. The mitochondrial membrane potential of wild-type and TM1 and TM2 mutant mitochondria were measured by staining with JC-1 dye with excitation at 490 nm and recording of the emission from 500 to 620 nm (A), and the relative distribution of dye was quantified and is represented in a bar chart (B). val., valinomycin. (C and D) Estimation of functional mitochondrial mass. (C) Wild-type and TM1 and TM2 mutant strains were grown at the permissive temperature, followed by heat shock for 2 h, and were subjected to functional mitochondrial mass determination using MitoTracker Deep Red-A dye and a FACS-based method. (D) The total masses estimated for the wild type and mutants are represented in a bar chart.
To probe whether other channel subunits are able to cooperate in the assembly of the Tim23 core channel, we overexpressed all the components of the presequence translocase in a mutant background using the CEN plasmid under the control of the TEF promoter (data not shown). Strikingly, overexpression of only the wild-type copy of the Tim17 subunit could rescue the growth sensitivity of the TM1 and TM2 mutants (Fig. 7A and B). To assess the reversal of membrane polarity by the Tim17 protein, mutants were subjected to JC-1 staining in the presence or absence of overexpressed Tim17. As depicted in Fig. 7C and D, a significant suppression of membrane polarity was achieved in the TM helix mutants, highlighting that Tim17 is critical for the structural integrity of the Tim23 core channel (P < 0.001).
Fig 7.
Overexpression of Tim17 rescues the membrane potential defect of TM1 and TM2 mutants. (A) Growth phenotype analysis of TM1 and TM2 mutants upon overexpression of Tim17. Mutant yeast cells overexpressing Tim17 under the control of the centromeric TEF promoter were subjected to serial dilution analysis on plates from which uracil was omitted. (B) Protein expression analysis. The overexpression of Tim17 in mutant strains was analyzed by immunoblotting, and Mge1 was used as a loading control. (C and D) Measurements of inner membrane polarity upon overexpression of Tim17. The intact mitochondrial membrane potential of mutant strains upon overexpression of Tim17 (broken lines) was measured using JC-1 dye (C) and was quantified (D) as described in the legends to Fig. 6A and B.
Loop L1 is critical for PAM subcomplex organization.
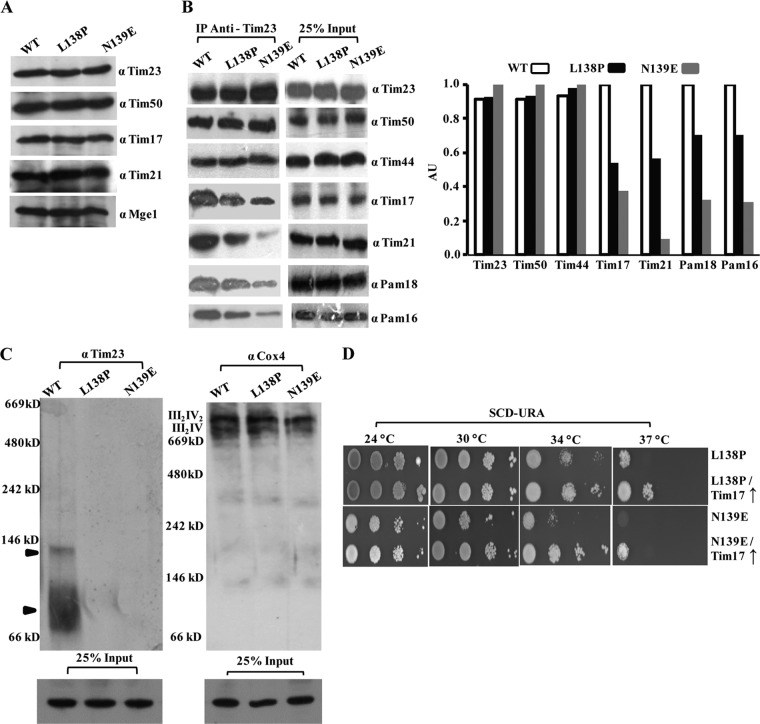
Loop L1, which is exposed to the matrix side, connects the TM1 and TM2 helices, and its functional significance is still unclear. Our genetic screen led to the isolation of two novel Ts mutants with a single amino acid substitution in loop L1: the tim23L138P and tim23N139E mutants. Both mutants showed comparable levels of expression of all the channel subunits, including Tim23, suggesting that the targeting remained unaltered (Fig. 8A).
Fig 8.
Loop L1 of Tim23 is indispensable for recruitment of the PAM subcomplex. (A) Protein expression profile of loop L1 mutants. Wild-type and loop L1 mutant mitochondria were analyzed by immunoblotting using specific antibodies. (B) CoIP analysis of loop L1 mutants. The CoIP analysis of loop L1 mutants was performed by solubilizing mitochondria in 1% digitonin buffer, followed by incubation with Tim23 antibody-cross-linked beads. (Left) The samples were separated on SDS-polyacrylamide gels and immunoblotted for Tim23 channel subunits; (right) the amounts of the different immunoprecipitated proteins were quantified using ImageJ software. (C) Determination of stability of the Tim23 complex by BN-PAGE. Digitonin-solubilized wild-type and mutant mitochondria were subjected to BN-PAGE analysis, followed by Western blotting using Tim23 (left) and Cox4 (right) antibodies. The locations of the complexes are indicated by arrowheads. Twenty-five percent of the input was used as a loading control. (D) Growth phenotype analysis of loop L1 mutants upon overexpression of Tim17. Mutant yeast cells overexpressing Tim17 using plasmid pRS316 were subjected to serial dilution analysis on plates from which uracil was omitted.
To uncover the compromised import defect of loop L1 mutants, we analyzed the complex organization by CoIP of mitochondrial lysates using Tim23-specific antibodies. Importantly, both loop L1 mutants showed impairment in recruitment of the PAM subcomplex and Tim21, in association with the diminished interaction of Tim17 with Tim23 protein. (Fig. 8B). However, compared to the tim23L138P mutant, the tim23N139E mutant exhibited a drastic reduction in the recruitment of the Pam18 and Pam16 subcomplex together with the Tim17 and Tim21 component (Fig. 8B). To confirm the findings presented above, we performed BN-PAGE analysis to test the integrity of higher-molecular-mass core complexes in the mutants. Compared to the wild type, a complete lack of a distinct core complex with a molecular mass of 90 kDa in the mutants suggested the critical importance of loop L1 in the recruitment of Tim17 subunits to the Tim23 channel (Fig. 8C). On the other hand, as a control, the stability of the respiratory chain complexes (III2IV2 and III2IV) remained unaffected in both the loop L1 mutants, as analyzed with anti-Cox4 antibodies (Fig. 8C).
Previous studies have demonstrated that Tim17 functions as a docking point for the PAM machinery (19). To investigate a possible connection, we overexpressed Tim17 in a mutant background and scored for the growth defect by serial dilution analysis. Notably, overexpression of Tim17 partially rescued the growth defect of both loop L1 mutants, thus further confirming that the motor recruitment defect observed in the loop L1 mutants is a consequence of the diminished interaction between Tim23 and Tim17 (Fig. 8D).
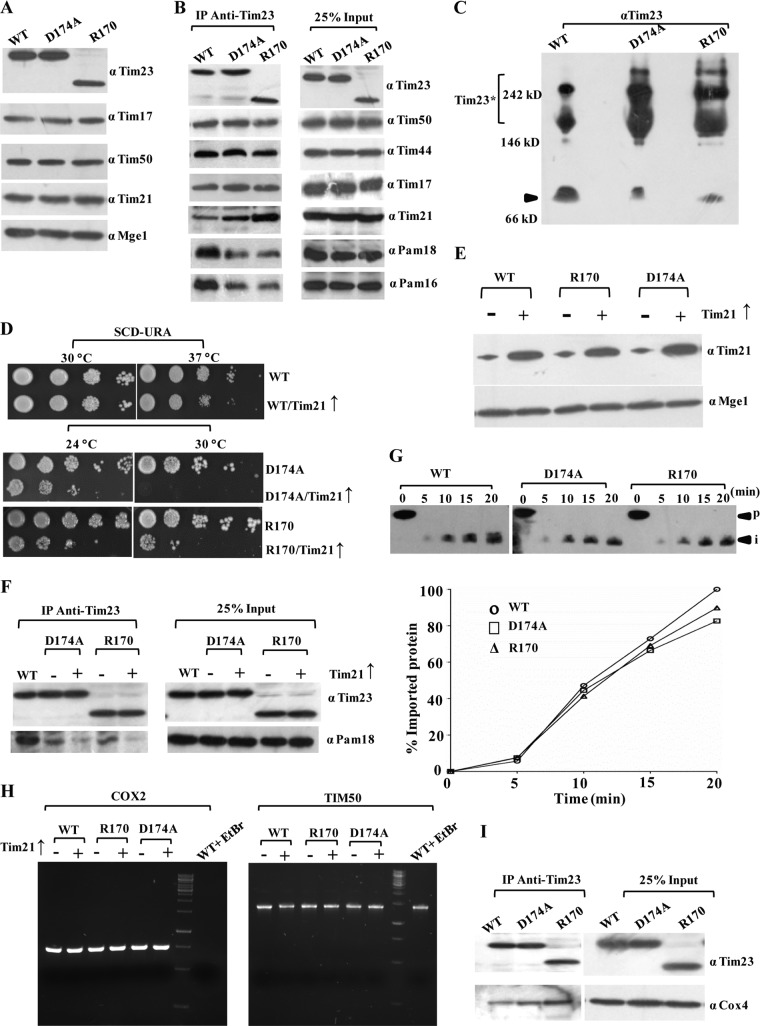
Loop L2 and the C-terminal fragment of Tim23 (amino acids 170 to 222) determine the association of Tim21 with the core complex.
Loop L2 is exposed to the IMS region and links the TM2 and TM3 helices. A single amino acid substitution in the loop L2 region, D174A, led to a Ts growth phenotype (Fig. 1C). In addition to that, insertion of a stop codon at positions 170 and 172 in the L2 region resulted in truncated proteins with growth sensitivity only under high-temperature (37°C) conditions (Fig. 1C). Importantly, these findings suggest that the TM3 and TM4 helices and connecting loop L3 are completely dispensable for growth at the permissive temperature. This is in contrast to the previous observation where the tim23 mutant with deletion of the sequence for TM3 and TM4 failed to support the growth of yeast cells due to the possible involvement of this sequence in insertion of the protein in the inner membrane (34, 35). However, both the point and truncation mutants showed comparable levels of expression of Tim23 and other subunits of the presequence translocase (Fig. 9A).
Fig 9.
The C-terminal segment of Tim23 is dispensable for growth and negatively regulates the interaction with Tim21. (A) Protein expression profile for loop L2 and truncation mutants. Mitochondrial lysates prepared from wild-type and loop L2 and truncation mutants were subjected to immunoblotting. (B) CoIP analysis for interaction of the subunits with the Tim23 channel. CoIP analysis of the loop L2 and truncation mutants was performed by solubilizing wild-type and mutant mitochondria in 1% digitonin, followed by incubation with Tim23 antibody-cross-linked beads. The samples were separated on SDS-polyacrylamide gels and immunoblotted for Tim23 channel subunits. (C) BN-PAGE analysis of loop L2 mutants. Digitonin-solubilized mitochondria were subjected to BN-PAGE analysis, followed by Western blotting using Tim23 antibodies. The locations of the core complex (arrowhead) and Tim23* are indicated. (D) Testing of the synthetic interaction between C-terminal Tim23 mutants and overexpressed Tim21. Yeast strains overexpressing Tim21 under the control of the centromeric TEF promoter were subjected to drop test analysis on plates from which uracil was omitted. (E) Protein expression analysis. The overexpression of Tim21 was tested by Western blotting using Mge1 as a loading control. (F) CoIP analysis for the motor recruitment upon overexpression of Tim21. The motor recruitment in loop L2 mutants upon Tim21 overexpression was analyzed by solubilizing mitochondria in 1% digitonin buffer, followed by immunoprecipitation using Tim23 antibody-cross-linked beads. (G) In vitro import kinetic analysis of wild-type and mutant strains. In vitro import of the Cyb2(167)-DHFR precursor was performed in wild-type and mutant mitochondria as described in the legends to Fig. 2B and C. (H) Analysis of mtDNA loss. The abundance of mitochondrial DNA was analyzed upon Tim21 overexpression using a PCR-mediated approach. Genomic DNA was isolated from wild-type and mutant strains overexpressing Tim21, followed by PCR amplification of the COX2 gene to assess mtDNA loss and the TIM50 gene to serve as a control for nuclear DNA. Yeast cells treated with ethidium bromide (EtBr) to induce mtDNA loss were used as a positive control. (I) Analysis of respiratory chain coupling with presequence translocase. The digitonin-solubilized wild-type and mutant mitochondria were subjected to immunoprecipitation using Tim23 antibody-cross-linked beads, followed by immunoblotting using Cox4 and Tim23 antibodies. Twenty-five percent of the input was used as a loading control.
To determine the role of loop L2 and the C-terminal fragment of Tim23 in the recruitment of other components in the channel, we performed CoIP with digitonin-solubilized mitochondrial lysates using Tim23-cross-linked beads. Strikingly, as illustrated in Fig. 9B, a significant enhancement of the interaction of the Tim21 subunit was observed with the mutant Tim23 protein, and this was more pronounced in the truncation mutant. On the other hand, a concomitant reduction in the association of Pam16 and Pam18 was observed (Fig. 9B). The enhanced association of Tim21 was further verified by BN-PAGE analysis, where both loop L2 mutants showed an enhancement in Tim21-containing Tim23* complexes and a concomitant reduction in the amount of the 90-kDa core complex (Fig. 9C). These results further suggest that the amino acid sequence beyond the terminal part of the loop L2 region negatively modulates the recruitment of Tim21 protein to the import-competent form of the Tim23-core complex during the translocation of presequence-containing proteins. To further validate this observation, we performed a genetic analysis by overexpressing Tim21 under the control of the TEF promoter in a wild-type or mutant background and analyzed the strain for growth sensitivity at different temperatures (Fig. 9D and E). The viability of the wild-type strain remained unaffected when Tim21 was overexpressed using the TEF promoter. In contrast, overexpression of Tim21 in a mutant background using the TEF promoter led to a stronger growth defect at all temperatures tested (Fig. 9D). Furthermore, the motor coupling in mutant strains upon Tim21 overexpression was directly assessed by IP using Tim23-cross-linked beads. Interestingly, overexpression of Tim21 in the mutant strain led to a further decrease in the recruitment of motor components, such as Pam18 (Fig. 9F), thus additionally confirming the negative role played by Tim21 at the in vivo level and that the interaction is regulated through the C-terminal region of Tim23. How does the enhancement of Tim21 binding in loop L2 mutants influence the sorting of preproteins into the inner membrane compartment? To address this, we performed the import of an inner membrane-sorting protein comprised of a shorter presequence, such as Cyb2(167)-DHFR, whose translocation is independent of the PAM complex (54). Strikingly, the import of Cyb2(167)-DHFR in both loop L2 mutants was found to be almost as efficient as that of the wild type (Fig. 9G). These findings confirm that the Ts phenotype of loop mutants is attributed to the import defect of matrix-targeting proteins as a consequence of the enhanced interaction of Tim21 with the core complex. However, the massive overproduction of inner mitochondrial membrane proteins is often associated with a number of pleiotropic effects, ranging from morphological rearrangements to a loss of mtDNA. Therefore, to check the pleiotropic effects due to overexpression of Tim21 in wild-type or mutant strains, we monitored the loss of mtDNA by checking the amplicon amount of a mitochondrial gene, COX2, as described previously (49). Notably, analysis of the mutant strains, including tim23D174A and tim23R170 mutant strains, by PCR showed levels of COX2 equivalent to those of the wild type upon Tim21 overexpression, thus demonstrating that the increased levels of Tim21 did not cause any loss of mtDNA under our experimental conditions (Fig. 9H). Furthermore, the mitochondrial morphology was found to be indistinguishable upon Tim21 overexpression and exhibited a complex reticulated network of mitochondria in all strains, as analyzed using confocal laser microscopy (data not shown).
Earlier reports have indicated that the sorting form of the presequence translocase interacts with the respiratory chain in a Tim21-dependent manner (55–57). Therefore, we checked for Tim23-respiratory chain coupling in loop L2 mutants by immunoprecipitation using Tim23 antibody-cross-linked beads, followed by immunoblotting using Cox4 antibody. Strikingly, in both truncation mutants and the point mutant, an enhancement in the binding of Cox4 was observed, which reveals the role of the C terminus in modulating the dynamics of the presequence translocase with Tim21 (Fig. 9I).
Recruitment of the PAM subcomplex to the Tim23-core channel is regulated by the Tim21 protein.
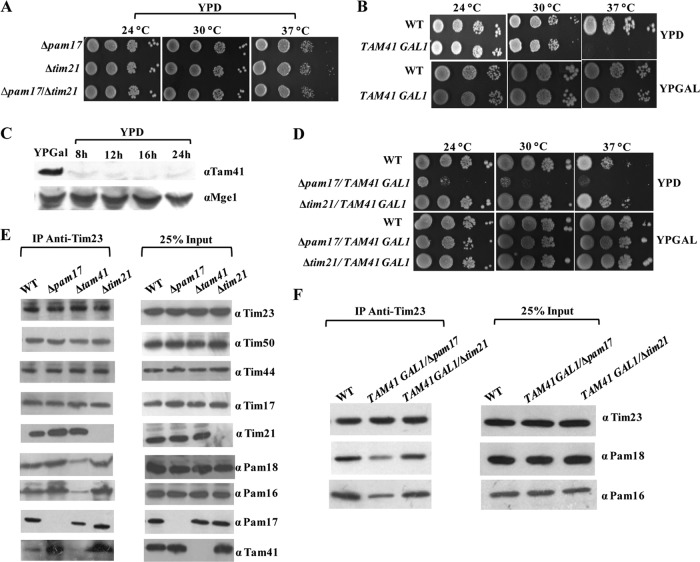
Previous studies have indicated that the Pam17 and Tam41 subunits mediate the association of the Pam18 and Pam16 heterodimer to the Tim23 channel (31–33). However, both subunits are dispensable for growth in yeast cells. In addition to that, Tim21, another nonessential component, plays an antagonistic role in the recruitment of the Pam16 and Pam18 heterodimer (19, 58). How these three components dynamically regulate the association of the terminal PAM subcomplex to the Tim23 channel and dissociation from the Tim23 channel is largely elusive. To understand the contribution of individual subunits in PAM subcomplex recruitment, we employed a systematic genetic approach using combinations of deletions. As shown in Fig. 10A, a strain with a deletion of PAM17 with TIM21 did not show a defect at all temperatures tested. In order to test for the genetic interaction of the Tim21 and Pam17 subunits with Tam41, we constructed a yeast strain in which the TAM41 gene was inserted under the control of the GAL1 promoter to deplete the protein levels by allowing it to grow on YPD medium. Pam17 or Tim21 was subsequently disrupted in the strain using appropriate selection cassettes. The strain harboring the TAM41 gene under the control of the GAL1 promoter showed a Ts phenotype on YPD medium, further confirming previously reported findings as well as excluding a possibility of leaky basal expression from the promoter (Fig. 10B and C). Strikingly, the Δpam17/TAM41 GAL1 strain showed a robust growth defect on YPD medium in comparison to the growth of a strain with a tam41 deletion alone, suggesting a strong genetic interaction between these two components (Fig. 10D). Most importantly, the temperature sensitivity of the Δtam41 strain was completely relieved upon deletion of Tim21 (Δtim21/TAM41 GAL1), as tested on YPD medium, highlighting an antagonistic in vivo function of Tim21 with Tam41 (Fig. 10D).
Fig 10.
Functional antagonism between Tam41 and Tim21 in recruitment of the PAM subcomplex to the Tim23 channel. (A) Serial dilution analysis of different knockout yeast strains. Δpam17 and Δtim21 single-knockout and Δpam17/Δtim21 double-knockout strains were spotted on rich medium and incubated at different temperatures. (B) Growth phenotype analysis of yeast cells under Tam41-depleted conditions. Serially diluted yeast cells from the wild type and cells expressing Tam41 under the control of the GAL1 promoter were spotted on YPD and YPGal media. (C) Analysis of Tam41 expression in galactose-rich and depleted media. The yeast cells expressing Tam41 under the control of the GAL1 promoter were grown overnight on galactose-containing medium. The cells were harvested and resuspended in glucose-containing medium for the indicated times. The cells equivalent to 0.1 optical density unit were collected and analyzed by immunoblotting using Tam41 antibodies. (D) Interaction network among Pam17, Tim21, and Tam41 subunits of presequence translocase. Serially diluted yeast cells from Δpam17 and Δtim21 yeast strains expressing Tam41 under the control of the GAL1 promoter were spotted on YPD and YPGal media. (E and F) CoIP analysis of various knockout yeast strains. CoIP analysis of mitochondrial lysates prepared from either a Δpam17, Δtim21, or Δtam41 yeast strain (E) or from a Δpam17 or Δtim21 strain expressing Tam41 under the control of the GAL1 promoter (F) was performed using Tim23 antibody-cross-linked beads. The samples were analyzed by immunoblotting against the indicated antibodies. Twenty-five percent of the input was used as a loading control.
To provide direct evidence in favor of this hypothesis, we performed CoIP in digitonin-solubilized mitochondrial lysates prepared from various knockout strains using Tim23 antibody-cross-linked antibodies. As depicted in Fig. 10E, deletion of Pam17 led to increased binding of Tim21, consistent with the previous observation highlighting the antagonistic function between these two subunits (58). A previous study has indicated that Tam41 is not a subunit of the presequence translocase and its association with the Tim23 channel is transient in nature (32). However, we observed a weak basal level of interaction of Tam41 with Tim23 in the wild-type background which was further significantly enhanced upon deletion of either Pam17 or Tim21 (Fig. 10E). Strikingly, the binding of Pam17 was also reduced upon deletion of Tam41, indicating that Tam41 plays a significant role in the recruitment of the Pam17 subunit, other than mediating the association of the Pam18/Pam16 complex with the presequence translocase (Fig. 10E). In addition to that, upon CoIP of digitonin-solubilized mitochondria from a TAM41 GAL1/Δpam17 strain), the amount of Pam18/Pam16 immunoprecipitated with the Tim23 complex was significantly decreased compared to that for the wild-type control (Fig. 10F). At the same time, the levels of Pam18/Pam16 were restored back to the normal levels in mitochondria isolated from the TAM41 GAL1/Δtim21 strain (Fig. 10F). Therefore, our combined genetic and biochemical analysis provides the first compelling evidence demonstrating that Pam17 and Tam41 act synergistically to recruit the PAM subcomplex to the Tim23 channel, while Tim21 acts in a manner antagonistic to these two components.
DISCUSSION
The multispanning transmembrane protein Tim23 forms the central core of the presequence translocase and recruits interacting subunits to facilitate the import and sorting of proteins into different subcompartments. Therefore, understanding the organization of the Tim23 protein is the central important element that needs to be known in order to obtain mechanistic insights into the early events of precursor protein capture coupled to the translocation process. The present study comprehensively addresses the key unresolved questions and highlights the contribution of each of the segments of Tim23, which regulates the association and dissociation of partner subunits during import.
The transmembrane helices TM1 and TM2 play the most critical functions in terms of the organization of the channel through homodimerization (which maintains the inner membrane polarity), preprotein capture, and recruitment of terminal interacting subunits (Fig. 11A). The N-terminal segment spanning from amino acids 51 to 100 of Tim23 has previously been implicated in the homodimerization process (12). However, a recent nuclear magnetic resonance structural analysis of the N-terminal 100 amino acids indicates that it exists in a monomeric form (59). Interestingly, our findings highlight that the TM1 helix plays a critical role in aiding the homodimerization of the Tim23 protein, thus clarifying the previous contrasting observations. The TM1 Ts mutants showed a great reduction in affinity for homodimerization (Fig. 11B). The homotypic and heterotypic association between TM helices is facilitated by a small set of core sequence motifs. Out of these, the GXXXG motif is one of the motifs overrepresented in TM segments, which are known to act as a universal scaffold for the assembly of TM helices by providing a site of close contact between them (60–62). The GXXXG motifs are common in several TM proteins, including glycophorin A, the subunits of the mitochondrial ATP synthase complex, and the amyloid precursor protein (60–63). Interestingly, three tandem GXXXG motifs are present in TM1 of Tim23, defined by glycines at positions 108, 112, 116, and 120, respectively, which are highly conserved across the phylogenetic species (see Fig. S1 in the supplemental material). Furthermore, the TM1 mutations isolated in our library reside in the GXXXG motif, therefore showing that such motifs are instrumental for the homodimerization of the protein. Equally, TM1 mutants also displayed reduced steady-state levels due to lower stability, suggesting that dimer formation is essential in vivo for Tim23 organization in the inner membrane.
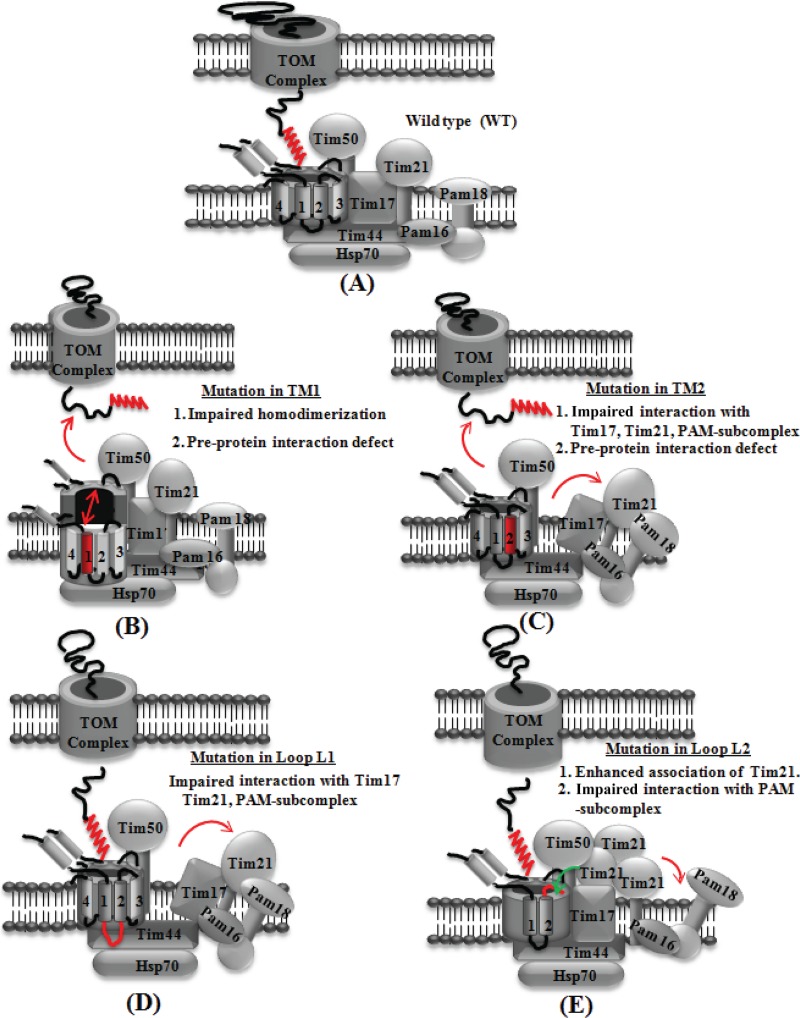
Fig 11.
Proposed model highlighting the functional significance of different segments of Tim23 protein identified using conditional mutant analysis. (A) Organization of the partner subunits with the wild-type Tim23 protein; (B) TM1 mutation leads to impairment in homodimerization and association with substrate preprotein; (C) TM2 mutation impairs the Tim17 interaction, thereby causing the dissociation of the PAM subcomplex and Tim21; (D) loop L1 mutation destabilizes the Tim17, Tim21, and PAM subcomplex association; (E) a point mutation or truncation in the loop L2 region results in the enhanced interaction with Tim21 and the concomitant release of the PAM subcomplex.
Previously, it has been reported that the TM1 helix of Tim23 plays a critical role in the interaction with Tim17, while TM2 is required for substrate protein recognition (64, 65). Interestingly, our genetic and biochemical analysis highlights the fact that both the TM1 and TM2 helices possess preprotein recognition and binding properties during import. The mutants of both helices displayed a significantly compromised ability to bind to signal sequence-containing precursor proteins, indicating that TM1 and TM2 are essential for early substrate capture in the channel (Fig. 11B and C). Furthermore, the TM2 helix also plays a vital role in the recruitment of partner subunits of translocase via interaction with Tim17. The TM2 mutants with reduced Tim17 interaction showed a substantial decrease in the recruitment of the PAM subcomplex as well as Tim21 subunits, thus establishing a link between channel and import motor association (Fig. 11C). Most notably, the G149E and G153V mutations isolated in our library are within the tandem GXXXG motifs of TM2 and are highly conserved among orthologs (see Fig. S1 in the supplemental material). Our data provide evidence that a mutation in any one of the glycine residues of this motif impairs the interaction of Tim23 with Tim17, thereby causing the destabilization of the core complex. Based on our observations, we propose a model depicting an organization in which the Tim23 dimer forms a single translocation pore having two TM1 and two TM2 helices arranged in a tetrahelical bundle (Fig. 11A). In this regard, the TM2 helices can independently recruit two Tim17 and terminal import motor subunits on either side of the channel for efficient translocation. By considering the dispensability of the C-terminal TM3 and TM4 helices for permissive growth, assembling a single protein-conducting pore consisting of two TM1 and two TM2 helices from the Tim23 dimer is the most distinct possibility in a normal physiological state (Fig. 11A).
The integrity of the central translocation channel is critical for the maintenance of membrane polarity. Any structural perturbation in the pore leads to inner membrane depolarization, as observed in both TM1 and TM2 conditional mutants. Most of the amino acid substitutions identified in the TM1 and TM2 segments that lead to growth sensitivity are drastic in nature and generate the imbalance in the overall charge of the hydrophobicity ratio of the helices. Therefore, it is reasonable to believe that such perturbations could influence the helical structure and, ultimately, pore integrity, leading to membrane depolarization. This observation is further supported by the reduction in the overall functional mitochondrial mass associated with TM1 and TM2 mutations, thereby affecting import and cellular viability. Previously, it has been proposed that the Tim17 protein might cooperatively function in the organization of the Tim23 channel (66, 67). Intriguingly, the defects associated with TM1 and TM2 mutations are suppressed by overexpression of only the Tim17 subunit. Importantly, Tim17 also restores the inner membrane polarity, suggesting that it directly aids in the organization of the translocation pore.
One of the important aspects of the organization of the Tim23 protein is to generate an interacting surface on which partner components may assemble as a multisubunit complex to maximize the efficiency of the import reaction. In this regard, loop L1 of Tim23 plays a critical role in the recruitment of interacting proteins, including the PAM subcomplex, together with the Tim21 subunit (Fig. 11D). Intriguingly, loop L1 mutants also exhibit a reduction in their association with Tim17, similar to the findings for the TM2 segment, suggesting that for these components, the dynamic association with and dissociation from the Tim23 channel are mediated through the Tim17 protein (Fig. 11D). Based on our results, we hypothesize that the interaction of Tim17 with the TM2 helix of Tim23 is perhaps required for the modulation of central translocation channel activity, while loop L1 provides the specificity for the association of partner subunits.
One of the noteworthy observations obtained from the conditional mutants was that the C-terminal region of the Tim23 protein beyond loop L2 negatively regulates the association of the Tim21 subunit with the core channel (Fig. 11E). Due to the presence of an inhibitory sequence within the Tim23 protein, the binding of Tim21 is self-limited and is associated nonstoichiometrically with the core channel. Therefore, we predict that the majority of Tim21 is redistributed to distinct inner membrane machinery, such as respiratory chain complexes, as previously reported (55–57). Retrospectively, this would promote the translocation of the matrix-driven proteins through the PAM-bound Tim23 complex. Based on the results, we envisioned that the sequestration of proteins containing a hydrophobic transmembrane-targeting sequence might aid with a unique conformational switch in the core, thereby releasing the negative effect of the C-terminal region of Tim23. This would favor the association of Tim21, leading to the organization of a PAM-free Tim23 complex, thus promoting the import of proteins destined for the inner membrane. Enhanced binding of Tim21 to the Tim23 channel is deleterious in nature due to changes in the equilibrium between the bound and free forms of PAM with the Tim23 complex. This is evident from the truncation and loop L2 mutants, which exhibited an increased association of Tim21 with the Tim23 channel concomitantly with a dissociation of PAM components. Our findings highlight that the binding of Tim21 to the Tim23 channel is a critical regulatory event required to maintain the balance between the import of matrix and inner membrane proteins, which is largely governed by the C-terminal sequence of Tim23.
The organization of the PAM subcomplex is mediated through the Tam41 and Pam17 subunits. So, how does the association of Tam41 and Pam17 influence Tim21 recruitment to the Tim23 complex? Previously, it has been indicated that Pam17 and Tim21 function antagonistically at the translocation complex (58). Strikingly, our observations indicate that Tim21 and Tam41 exhibit a strong antagonistic function in vivo. The yeast cells in which Tam41 was deleted showed a significant enhancement in the association of Tim21 with the Tim23-core complex, even though the presence of Pam17 does not provide any growth advantage for the yeast cells by suppressing the negative impact of the Tim21 subunit. Remarkably, the growth sensitivity of yeast cells in which Tam41 is downregulated was completely suppressed by deleting Tim21 in the same strain background, suggesting a strong functional antagonism between these two subunits. Based on this result, we propose that Tam41 functions as a predominant subunit in the recruitment of the PAM subcomplex to the Tim23 channel by suppressing the antagonistic functions of Tim21, while a subunit such as Pam17 which is less conserved in higher eukaryotes plays a partial supporting role in the recruitment of the PAM subcomplex.
In summary, our combined genetic and biochemical analyses provide valuable insights into the functions of different segments of Tim23, which is highly conserved across phylogenetic boundaries; hence, we envision similarly defined functional parameters attributed to the distinct segments of the protein. The specificity associated with the recruitment of subunits and the dynamic organization of the Tim23 complex in response to protein import is still poorly understood. Therefore, having conditional mutants of Tim23 with predefined functional defects in the different segments would be an asset in dissecting the regulatory events leading to the import of a large variety precursor proteins across the mitochondrial inner membrane.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Toshiya Endo for the Δtim23 haploid yeast strain and Tam41 antibody and Carla Koehler for Cox4 antibody. We thank Prasenjit Prasad Saha for the preparation of the model and Ajeet Singh for technical assistance during the work. We thank the FACS Facility and Confocal Facility of the Indian Institute of Science, Bangalore, India.
P.D. acknowledges funding from the SwarnaJayanthi fellowship scheme (DST) and grant-in-aid (DBT). G.P. acknowledges a research fellowship from the Council of Scientific and Industrial Research.
Footnotes
Published ahead of print 23 Setember 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00876-13.
REFERENCES
- 1.Neupert W, Herrmann JM. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749 [DOI] [PubMed] [Google Scholar]
- 2.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker MJ, Frazier AE, Gulbis JM, Ryan MT. 2007. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 17:456–464 [DOI] [PubMed] [Google Scholar]
- 4.Neupert W, Brunner M. 2002. The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 3:555–565 [DOI] [PubMed] [Google Scholar]
- 5.Bauer MF, Hofmann S, Neupert W, Brunner M. 2000. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 10:25–31 [DOI] [PubMed] [Google Scholar]
- 6.Truscott KN, Brandner K, Pfanner N. 2003. Mechanisms of protein import into mitochondria. Curr. Biol. 13:326–337 [DOI] [PubMed] [Google Scholar]
- 7.Schmidt O, Pfanner N, Meisinger C. 2010. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11:655–667 [DOI] [PubMed] [Google Scholar]
- 8.van der Laan M, Hutu DP, Rehling P. 2010. On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim. Biophys. Acta 1803:732–739 [DOI] [PubMed] [Google Scholar]
- 9.Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. 2008. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 9:42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emtage JL, Jensen RE. 1993. MAS6 encodes an essential inner membrane component of the yeast mitochondrial protein import pathway. J. Cell Biol. 122:1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dekker PJ, Keil P, Rassow J, Maarse AC, Pfanner N, Meijer M. 1993. Identification of MIM23, a putative component of the protein import machinery of the mitochondrial inner membrane. FEBS Lett. 330:66–70 [DOI] [PubMed] [Google Scholar]
- 12.Bauer MF, Sirrenberg C, Neupert W, Brunner M. 1996. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87:33–41 [DOI] [PubMed] [Google Scholar]
- 13.Ryan KR, Menold MM, Garrett S, Jensen RE. 1994. SMS1, a high-copy suppressor of the yeast mas6 mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol. Biol. Cell 5:529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maarse AC, Blom J, Keil P, Pfanner N, Meijer M. 1994. Identification of the essential yeast protein MIM17, an integral mitochondrial inner membrane protein involved in protein import. FEBS Lett. 349:215–221 [DOI] [PubMed] [Google Scholar]
- 15.Kübrich M, Keil P, Rassow J, Dekker PJ, Blom J, Meijer M, Pfanner N. 1994. The polytopic mitochondrial inner membrane proteins MIM17 and MIM23 operate at the same preprotein import site. FEBS Lett. 349:222–228 [DOI] [PubMed] [Google Scholar]
- 16.Geissler A, Chacinska A, Truscott KN, Wiedemann N, Brandner K, Sickmann A, Meyer HE, Meisinger C, Pfanner N, Rehling P. 2002. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111:507–518 [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto H, Esaki M, Kanamori T, Tamura Y, Nishikawa S-i, Endo T. 2002. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 111:519–528 [DOI] [PubMed] [Google Scholar]
- 18.Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE, Neupert W, Hell K. 2003. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 22:816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P. 2005. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120:817–829 [DOI] [PubMed] [Google Scholar]
- 20.Mokranjac D, Popov-Celeketić D, Hell K, Neupert W. 2005. Role of Tim21 in mitochondrial translocation contact sites. J. Biol. Chem. 280:23437–23440 [DOI] [PubMed] [Google Scholar]
- 21.Schneider HC, Berthold J, Bauer MF, Dietmeier K, Guiard B, Brunner M, Neupert W. 1994. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371:768–774 [DOI] [PubMed] [Google Scholar]
- 22.Rassow J, Maarse AC, Krainer E, Kübrich M, Müller H, Meijer M, Craig EA, Pfanner N. 1994. Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol. 127(Pt 1):1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berthold J, Bauer MF, Schneider HC, Klaus C, Dietmeier K, Neupert W, Brunner M. 1995. The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ATP driving system. Cell 81:1085–1093 [DOI] [PubMed] [Google Scholar]
- 24.Liu Q, D'Silva P, Walter W, Marszalek J, Craig EA. 2003. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300:139–141 [DOI] [PubMed] [Google Scholar]
- 25.Mokranjac D, Sichting M, Neupert W, Hell K. 2003. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 22:4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truscott KN, Voos W, Frazier AE, Lind M, Li Y, Geissler A, Dudek J, Müller H, Sickmann A, Meyer HE, Meisinger C, Guiard B, Rehling P, Pfanner N. 2003. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163:707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Silva PD, Schilke B, Walter W, Andrew A, Craig EA. 2003. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. U. S. A. 100:13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frazier AE, Dudek J, Guiard B, Voos W, Li Y, Lind M, Meisinger C, Geissler A, Sickmann A, Meyer HE, Bilanchone V, Cumsky MG, Truscott KN, Pfanner N, Rehling P. 2004. Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 11:226–233 [DOI] [PubMed] [Google Scholar]
- 29.Kozany C, Mokranjac D, Sichting M, Neupert W, Hell K. 2004. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 11:234–241 [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Dudek J, Guiard B, Pfanner N, Rehling P, Voos W. 2004. The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J. Biol. Chem. 279:38047–38054 [DOI] [PubMed] [Google Scholar]
- 31.van der Laan M, Chacinska A, Lind M, Perschil I, Sickmann A, Meyer HE, Guiard B, Meisinger C, Pfanner N, Rehling P. 2005. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol. Cell. Biol. 25:7449–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura Y, Harada Y, Yamano K, Watanabe K, Ishikawa D, Ohshima C, Nishikawa S, Yamamoto H, Endo T. 2006. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J. Cell Biol. 174:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallas MR, Dienhart MK, Stuart RA, Long RM. 2006. Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Mol. Biol. Cell 17:4051–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis AJ, Ryan KR, Jensen RE. 1998. Tim23p contains separate and distinct signals for targeting to mitochondria and insertion into the inner membrane. Mol. Biol. Cell 9:2577–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Káldi K, Bauer MF, Sirrenberg C, Neupert W, Brunner M. 1998. Biogenesis of Tim23 and Tim17, integral components of the TIM machinery for matrix-targeted preproteins. EMBO J. 17:1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donzeau M, Káldi K, Adam A, Paschen S, Wanner G, Guiard B, Bauer MF, Neupert W, Brunner M. 2000. Tim23 links the inner and outer mitochondrial membranes. Cell 101:401–412 [DOI] [PubMed] [Google Scholar]
- 37.Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T. 2009. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184:129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gevorkyan-Airapetov L, Zohary K, Popov-Celeketic D, Mapa K, Hell K, Neupert W, Azem A, Mokranjac D. 2009. Interaction of Tim23 with Tim50 is essential for protein translocation by the mitochondrial TIM23 complex. J. Biol. Chem. 284:4865–4872 [DOI] [PubMed] [Google Scholar]
- 39.Schulz C, Lytovchenko O, Melin J, Chacinska A, Guiard B, Neumann P, Ficner R, Jahn O, Schmidt B, Rehling P. 2011. Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex. J. Cell Biol. 195:643–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marom M, Dayan D, Demishtein-Zohary K, Mokranjac D, Neupert W, Azem A. 2011. Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 protein complex. J. Biol. Chem. 286:43809–43815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohret TA, Jensen RE, Kinnally KW. 1997. Tim23, a protein import component of the mitochondrial inner membrane, is required for normal activity of the multiple conductance channel, MCC. J. Cell Biol. 137:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Truscott KN, Kovermann P, Geissler A, Merlin A, Meijer M, Driessen AJ, Rassow J, Pfanner N, Wagner R. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8:1074–1082 [DOI] [PubMed] [Google Scholar]
- 43.Cirino PC, Mayer KM, Umeno D. 2003. Generating mutant libraries using error-prone PCR. Methods Mol. Biol. 231:3–9 [DOI] [PubMed] [Google Scholar]
- 44.McCullum EO, Williams BA, Zhang J, Chaput JC. 2010. Random mutagenesis by error-prone PCR. Methods Mol. Biol. 634:103–109 [DOI] [PubMed] [Google Scholar]
- 45.Robzyk K, Kassir Y. 1992. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 20:3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D'Silva P, Schilke BA, Hayashi M, Craig EA. 2008. Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol. Biol. Cell 19:424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chacinska A, van der Laan M, Mehnert CS, Guiard B, Mick DU, Hutu DP, Truscott KN, Wiedemann N, Meisinger C, Pfanner N, Rehling P. 2010. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol. Cell. Biol. 30:307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. 2006. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. U. S. A. 103:13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goswami AV, Samaddar M, Sinha D, Purushotham J, D'Silva P. 2012. Enhanced J-protein interaction and compromised protein stability of mtHsp70 variants lead to mitochondrial dysfunction in Parkinson's disease. Hum. Mol. Genet. 21:3317–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinha D, Joshi N, Chittoor B, Samji P, D'Silva P. 2010. Role of magmas in protein transport and human mitochondria biogenesis. Hum. Mol. Genet. 19:1248–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pareek G, Samaddar M, D'Silva P. 2011. Primary sequence that determines the functional overlap between mitochondrial heat shock protein 70 Ssc1 and Ssc3 of Saccharomyces cerevisiae. J. Biol. Chem. 286:19001–19013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geissler A, Krimmer T, Bömer U, Guiard B, Rassow J, Pfanner N. 2000. Membrane potential-driven protein import into mitochondria; the sorting sequence of cytochrome b2 modulates the Δψ-dependence of translocation of the matrix-targeting sequence. Mol. Biol. Cell 11:3977–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kutik S, Rissler M, Guan XL, Guiard B, Shui G, Gebert N, Heacock PN, Rehling P, Dowhan W, Wenk MR, Pfanner N, Wiedemann N. 2008. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J. Cell Biol. 183:1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voos W, Gambill BD, Guiard B, Pfanner N, Craig EA. 1993. Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J. Cell Biol. 123:119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N. 2006. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr. Biol. 16:2271–2276 [DOI] [PubMed] [Google Scholar]
- 56.Wiedemann N, van der Laan M, Hutu DP, Rehling P, Pfanner N. 2007. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J. Cell Biol. 179:1115–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mick DU, Dennerlein S, Wiese H, Reinhold R, Pacheu-Grau D, Lorenzi I, Sasarman F, Weraarpachai W, Shoubridge EA, Warscheid B, Rehling P. 2012. MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell 151:1528–1541 [DOI] [PubMed] [Google Scholar]
- 58.Popov-Čeleketić D, Mapa K, Neupert W, Mokranjac D. 2008. Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 27:1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de la Cruz L, Bajaj R, Becker S, Zweckstetter M. 2010. The intermembrane space domain of Tim23 is intrinsically disordered with a distinct binding region for presequences. Protein Sci. 19:2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melnyk RA, Kim S, Curran AR, Engelman DM, Bowie JU, Deber CM. 2004. The affinity of GXXXG motifs in transmembrane helix-helix interactions is modulated by long-range communication. J. Biol. Chem. 279:16591–16597 [DOI] [PubMed] [Google Scholar]
- 61.Doura AK, Kobus FJ, Dubrovsky L, Hibbard E, Fleming KG. 2004. Sequence context modulates the stability of a GXXXG-mediated transmembrane helix-helix dimer. J. Mol. Biol. 341:991–998 [DOI] [PubMed] [Google Scholar]
- 62.Walters RF, DeGrado WF. 2006. Helix-packing motifs in membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 103:13658–13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McClain MS, Iwamoto H, Cao P, Vinion-Dubiel AD, Li Y, Szabo G, Shao Z, Cover TL. 2003. Essential role of a GXXXG motif for membrane channel formation by Helicobacter pylori vacuolating toxin. J. Biol. Chem. 278:12101–12108 [DOI] [PubMed] [Google Scholar]
- 64.Alder NN, Sutherland J, Buhring AI, Jensen RE, Johnson AE. 2008. Quaternary structure of the mitochondrial TIM23 complex reveals dynamic association between Tim23p and other subunits. Mol. Biol. Cell 19:159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alder NN, Jensen RE, Johnson AE. 2008. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell 134:439–450 [DOI] [PubMed] [Google Scholar]
- 66.Martinez-Caballero S, Grigoriev SM, Herrmann JM, Campo ML, Kinnally KW. 2007. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J. Biol. Chem. 282:3584–3593 [DOI] [PubMed] [Google Scholar]
- 67.Meier S, Neupert W, Herrmann JM. 2005. Conserved N-terminal negative charges in the Tim17 subunit of the TIM23 translocase play a critical role in the import of preproteins into mitochondria. J. Biol. Chem. 280:7777–7785 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.