Abstract
Our previous study showed that Akt phosphorylates TopBP1 at the Ser-1159 residue and induces its oligomerization. Oligomerization is required for TopBP1 to bind and repress E2F1 activity. However, the mechanism through which phosphorylation of TopBP1 by Akt leads to its oligomerization remains to be determined. Here, we demonstrate that binding between the phosphorylated Ser-1159 (pS1159) residue and the 7th and 8th BRCT domains of TopBP1 mediates TopBP1 oligomerization. Mutations within the 7th and 8th BRCT domains of TopBP1 that block binding to a pS1159-containing peptide block TopBP1 oligomerization and its ability to bind and repress E2F1 activities. The Akt-induced TopBP1 oligomerization is also directly demonstrated in vitro by size exclusion chromatography. Importantly, oligomerization perturbs the checkpoint-activating function of TopBP1 by preventing its recruitment to chromatin and ATR binding upon replicative stress. Hyperactivation of Akt inhibits Chk1 phosphorylation after hydroxyurea treatment, and this effect is dependent on TopBP1 phosphorylation at Ser-1159. Thus, Akt can switch the TopBP1 function from checkpoint activation to transcriptional regulation by regulating its quaternary structure. This pathway of regulation is clinically significant, since treatment of a specific Akt inhibitor in PTEN-mutated cancer cells inhibits TopBP1 oligomerization and causes its function to revert from promoting survival to checkpoint activation.
INTRODUCTION
Akt/protein kinase B (PKB) plays very important and diverse roles in regulating cell fate in response to external stimuli. In general, Akt stimulates cell survival signaling pathways. Akt is also known to inhibit DNA damage checkpoint response (1–3). Heightened activation of Akt is very common in cancer and has been associated with genomic instability (4). It has been a common belief that Akt exerts its diverse activities by phosphorylating many different target proteins. It remains to be determined whether one or some of its targets can participate in diverse functions and whether these different activities can be regulated by Akt at the same time.
Akt has been shown to inhibit Chk1 activation upon gamma irradiation in late G2 phase (2, 3). Akt can phosphorylate Chk1 at Ser-280 (1, 5). Loss of PTEN can inhibit Chk1 through ubiquitination and cytoplasmic sequestration of Chk1, and S280A mutant Chk1 can restore CDC25A degradation (6). These data suggest that phosphorylation of Chk1 by Akt mediates, at least in part, the checkpoint defect in PTEN loss. However, the role of Ser-280 phosphorylation by Akt in checkpoint inhibition is challenged by two other studies (2, 3). The S280A and S280D Chk1 mutants exert the same extent of G2 arrest as the wild type upon ionizing irradiation (2). Furthermore, Akt inhibition enhances irradiation-induced activation of both wild-type Chk1 and S280A-Chk1 (3). These data indicate other mechanisms are responsible for this regulation, for example, inhibition of BRCA1 foci by activated Akt (2).
TopBP1 (topoisomerase IIβ binding protein 1) contains nine BRCA1 carboxyl-terminal (BRCT) domains (7) and functions in DNA damage checkpoint activation, replication, and transcription (8). It activates ATR (ATM [ataxia-telangiectasia mutated]–Rad3-related kinase) through a conserved ATR-activating domain between the 6th and 7th BRCT domains (9). Upon replicative stress, TopBP1 is recruited to stalled replication forks either through direct binding to the stalled forks (10, 11) or via the Rad9-Hus1-Rad1 (9-1-1) clamp (12). ATR-ATRIP (ATR-interacting protein) also contributes to its activation by promoting ATR autophosphorylation at Thr-1989 and directing TopBP1 to stimulate ATR-ATRIP, which subsequently activates a checkpoint kinase, Chk1 (13).
TopBP1 also regulates several transcriptional factors, including E2F1 (14–16), p53 (17), Miz1 (16, 18), and SPBP (stromelysin 1 platelet-derived growth factor-responsive element-binding protein) (19). Regulation of E2F1 and p53 by TopBP1 is important for restricting the proapoptotic activities of both transcriptional factors during normal S phase transition. TopBP1 represses E2F1 transcriptional activities by recruiting the Brg1-Brm chromatin-remodeling complex (15), whereas TopBP1 binds to the p53 DNA-binding domain and directly inhibits its transcriptional function (17). Being an E2F target, TopBP1 is frequently overexpressed in cancer, particularly when the pRb pathway is deregulated, and inactivates p53 functions (17, 20). TopBP1 also binds to mutant p53 and mediates the gain of function of mutant p53 in cancer (20).
TopBP1 is phosphorylated by Akt at Ser-1159 (pS1159), which induces TopBP1 oligomerization and the ability to bind and repress E2F1 transcriptional activity (16). How phosphorylation of a single residue (Ser-1159) can induce structural change is a very intriguing question. An understanding of this mechanism may provide us novel means to therapeutically rectify the deregulation in cancer, e.g., to reactivate the proapoptotic function of E2F1 in cancer cells, which harbor high levels of TopBP1. An in vitro assay showing pS1159-dependent self-association of TopBP1 using purified proteins indicates that Ser-1159 phosphorylation and the carboxyl terminus of TopBP1 are the only required components for Akt-dependent oligomerization of TopBP1, and this process does not require other, unknown factors (16). In addition, the carboxyl terminus of TopBP1, including the 7th and 8th BRCT domains, can bind to a pS1159-containing peptide. This leads us to propose that the binding of the 7th and 8th BRCT domains and pS1159 from another TopBP1 molecule mediates Akt-dependent oligomerization of TopBP1 (16). However, this model needs to be tested experimentally.
Estrogen has been shown to inhibit ATR activation through phosphatidylinositol 3-kinase (PI3K)/Akt action, which inhibits the interaction between ATR and wild-type TopBP1 but not S1159A mutant TopBP1 (21). Estrogen also inhibits the interaction between Chk1 and claspin via phosphorylation of Chk1 by Akt (21). Therefore, the underlying mechanism by which Akt inhibits the checkpoint response may involve multiple regulators. A role for phosphorylation of TopBP1 at Ser-1159 in inhibition of Chk1 activation by Akt and the molecular details remain to be established.
While TopBP1 is involved in DNA replication, checkpoint activation, and transcriptional regulation, it is unclear how different functions of TopBP1 are regulated or coordinated. Here, we provide evidence to support the idea that the binding between the 7th and 8th BRCT domains and the Akt-phosphorylated Ser-1159 residue is the mechanism for structural regulation of TopBP1 by Akt. We also demonstrate that oligomerization hampers TopBP1 function in checkpoint activation by preventing TopBP1 recruitment to chromatin and subsequent binding to ATR, while at the same time, it induces the interaction with E2F1. Thus, Akt switches the function of TopBP1 from checkpoint activation to transcriptional regulation.
MATERIALS AND METHODS
Cell culture and transfection.
HEK293, REF52, and H1299 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (50 IU/ml), and streptomycin (50 μg/ml). All cells were grown in a humidified incubator at 37°C with 5% CO2 and 95% air. HEK293 and H1299 cells were transfected by a standard calcium phosphate method or with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After transfection, the cells were incubated for 48 h before analysis.
Molecular dynamic simulation.
The AMBER 9 simulation package (22) was used for molecular dynamic (MD) simulation. The all-atom point-charge force field of Duan et al. (AMBER ff03) (23) was applied for proteins. Solvent was represented by the TIP3P water model (24). An 8-Å-thick truncated-octahedron water box was added to the manually docked structure model. A 1,000-cycle energy minimization was first applied to remove potential collision contacts with the added water molecules. The system was then gradually heated to 300 K over the course of 50 ps, followed by a dissolving step for another 50 ps under constant temperature. Finally, the simulation was continued at 300 K for another 5-ns production run in the NVT ensemble (constant moles [N], volume [V], and temperature [T]). The simulation snapshots were saved every 2 ps for analysis.
Plasmid construction.
Construction of FLAG-TopBP1, Myc-TopBP1, FLAG-TopBP1(S1159A), GST-TopBP1, GST-TopBP1-BRCT7/8L, GST-E2F1, HA-E2F1, and HA-CA-Akt was described previously (16). To construct the different tagged TopBP1-BRCT7/8 and TopBP1-BRCT6/7/8, TopBP1 was first amplified by PCR with the following primers: TopBP1-BRCT7/8, forward, 5′-CGCCATATGGGATCCGAGACTCATGAAGAA-3′, and reverse, 5′-AGCAAAATCCATTACCTTGC-3′; TopBP1-BRCT6/7/8, forward, 5′-CGCCATATGGGATCCGAAGCCCCAAAGCCA-3′, and reverse, 5′-AGCAAAATCCATTACCTTGC-3′. The PCR products were digested with NdeI/BamHI, cloned into pET-28a, and verified by sequencing. Then, we subcloned the BamHI/EcoRI fragments of pET-28a-TopBP1-BRCT7/8 and -6/7/8 to a BamHI/EcoRI-digested pGEX6P1 vector to obtain pGEX6P1-TopBP1-BRCT7/8 and -6/7/8, respectively. The K1317M mutations of pcDNA3-TopBP1 and GST-TopBP1-BRCT7/8 and -6/7/8 were generated using the GeneEditor in vitro site-directed mutagenesis system (Promega) with primers 5′-CTTCGAAACGAGATGTATTTAGCCTCA-3′ and 5′-TGAGGCTAAATACATCTCGTTTCGAAG-3′, respectively. The S1273A mutations in pcDNA3-TopBP1 and GST-TopBP1-BRCT7/8 and -6/7/8 were also generated with primers 5′-CATATTTCAGTTAGCATCTCTGAATCC-3′ and 5′-GGATTCAGAGATGCTAACTGAAATATG-3′, respectively. The sequences of mutants were verified by DNA sequencing. FLAG-TopBP1-BRCT7/8 K1317M and S1273A mutants were created by digesting the respective pGEX6P1-TopBP1-BRCT7/8 mutants with BamHI/EcoRI. These fragments were then subcloned into a similarly digested Tag2B empty vector. FLAG- and Myc-tagged TopBP1 K1317M and S1273A mutants were further constructed by swapping the mutant TopBP1 cDNAs from pcDNA3 vectors to pCMV-Tag vectors. FLAG-ATR was described previously (25).
In vitro peptide binding assay.
GST-TopBP1-BRCT7/8 and -6/7/8 (wild type [WT], K1317M, and S1273A) were produced in Escherichia coli and purified. The glutathione S-transferase (GST) portion was removed with PreScission protease. A biotin-labeled nonphosphorylated peptide (biotin-REERARLAS1159NLQWPS) or phosphorylated peptide (biotin-REERARLApSer1159NLQWPS) was synthesized by Sigma-Genosys. The biotin-labeled peptides (1 μg) were incubated with 1 μg of TopBP1-BRCT7/8 or TopBP1-BRCT6/7/8 protein in 1 ml of NETN-A buffer (20 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA, pH 8.0, and 0.5% NP-40) at 4°C overnight. The mixtures were then incubated with streptavidin-Sepharose (Amersham) at 4°C for 3 h and washed with NETN-B buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, pH 8.0, and 0.5% NP-40) six times. The bound TopBP1-BRCT7/8 or -BRCT6/7/8 was subjected to 10% SDS-PAGE and analyzed by Western blotting with TopBP1 antibody and Sypro Ruby staining for peptides.
GST pulldown assay.
GST, GST-TopBP1-BRCT7/8L, GST-TopBP1-BRCT7/8 (WT, K1317M, and S1273A), and GST-TopBP1-BRCT6/7/8 (WT, K1317M, and S1273A) proteins were induced by 0.1 mM IPTG in E. coli strain BL21 and purified as described previously (14). Akt kinase (Akt1/PKB, active; Upstate; 1 μg) was used for incubation with 2 μg of purified TopBP1-BRCT6/7/8 protein or its K1317M or S1273A mutant at 30°C for 30 min, and then 2 μg of GST-TopBP1-BRCT7/8 or its K1317M or S1273A mutant was added and incubation was continued for another 3 h. GST-tagged proteins were pulled down with glutathione-Sepharose, and the beads were washed six times with Tris-buffered saline (TBS) buffer and then subjected to 10% SDS-PAGE, and Western blot analysis was carried out.
His pulldown assay.
Two micrograms of purified GST-TopBP1-BRCT7/8 or GST-TopBP1-BRCT7/8L protein was incubated with 1 μg Akt kinase at 30°C for 30 min, and then 2 μg of purified His-TopBP1-BRCT7/8 protein (a gift from Rongbao Li) was added to continue incubation for another 3 h. His-TopBP1-BRCT7/8 was pulled down with nickel-Sepharose, and the beads were washed six times with TBS buffer and then subjected to 10% SDS-PAGE and analyzed by Western blotting.
Immunoprecipitation and Western blot analysis.
The transfected cells were harvested 48 h later with TNN buffer as described previously (14). An aliquot of cell lysates was lysed with SDS lysis buffer, and the rest of the cell lysates were incubated with appropriate antibodies or beads for 3 to 12 h at 4°C. Anti-FLAG (M2) beads were purchased from Sigma. The beads were washed three times with TNN buffer. Immunoprecipitates were fractionated by SDS-PAGE and electrotransferred to an Immobilon-P membrane (Millipore). Equal protein loading was confirmed by Ponceau S staining. The specific signals were detected with appropriate antibodies. E2F1 (C-20 and KH-95), GST (B-14), c-Myc (A14), hemagglutinin (HA) (Y11), Chk1 (G4), cyclin-dependent kinase 1 (CDK1)/Cdc2 p34 (17), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (6C5) antibodies were purchased from Santa Cruz. TopBP1 (monoclonal antibody), PARP, and Akt antibodies were purchased from BD Transduction Laboratories. TopBP1 (BL893; rabbit polyclonal) antibody was purchased from Bethyl. Anti-phospho-TopBP1 (Ser-1159) antibody was purchased from Abgent. A monoclonal phospho-(Ser/Thr) Akt substrate antibody (110B7), a histone H3 (D1H2) antibody, and a phospho-Chk1 (Ser345) antibody were purchased from Cell signaling. FLAG (F7425) antibody was purchased from Sigma. A monoclonal 6×His antibody was purchased from Clontech. Phosopho-CDK1 (Tyr15) antibody was purchased from NEB.
Fast protein liquid chromatography.
Both the purified proteins and the isolated proteins from cells were run on a fast protein liquid chromatography (FPLC) system (AktaPurifier UPC10; GE Healthcare) to analyze protein oligomeric states. For in vitro assays, 1.8 μg of Akt kinase was used for incubation with 20 μg of purified TopBP1-BRCT6/7/8 protein or its K1317M mutant at 30°C for 30 min. The proteins were loaded onto a high-performance column prepacked with Superdex 200 10/300 GL medium and resolved at a flow rate of 0.2 ml/min to separate the different oligomeric protein species. The fractionation volume for each collection tube was set at 400 μl. One-tenth of each fraction volume was subjected to 10% SDS-PAGE and analyzed by Western blotting. For in vivo assays, FLAG-TopBP1-transfected H1299 cells were treated with dimethyl sulfoxide (DMSO) or a specific Akt inhibitor, MK-2206 (1 μM; ChemieTek), for 24 h. The cells were incubated with a hypotonic buffer to isolate nuclei. The nuclei were then lysed with TNN buffer, and 400 μl of cell lysate was loaded and run through the column with the same settings described above. TopBP1 protein in each collected fraction was immunoprecipated with anti-FLAG beads (M2). The beads were washed, subjected to 10% SDS-PAGE, and analyzed by Western blotting.
Luciferase assay.
The expression constructs (2 μg for E2F1 and 10 μg for TopBP1 and its K1317M and S1273A mutants; the empty vector pcDNA3 was used as a control and added to E2F1-only and TopBP1-only samples so that every transfection reaction had the same amount of total DNA in the transfection mixtures), 1 μg of p14ARF promoter-luciferase plasmid, and 1 μg of β-galactosidase plasmid were cotransfected into HEK293 cells. Cells were harvested 2 days later with phosphate-buffered saline (PBS). A one-tenth sample was lysed with SDS lysis buffer for Western blotting. The other cell extracts were lysed with reporter lysis buffer (Promega), and luciferase activity and β-galactosidase activity were measured according to the manufacturer's instructions. The luciferase activity was normalized against the β-galactosidase activity. All transient expressions in this assay were carried out in triplicate.
Recombinant adenovirus construction and purification.
Adenovirus (Ad)-HAE2F1 and Ad-TopBP1 were described previously (14). The cDNAs of TopBP1-K1317M and TopBP1-S1273A were constructed in adenovirus with the AdEasy system as described previously (26). The viruses were purified by CsCl banding. Virus titers were measured with an Adeno-X rapid titer kit purchased from Clontech.
Bromodeoxyuridine incorporation assay and apoptosis assay.
REF52 cells were starved in 0.25% FBS for 2 days, followed by adenovirus infection or addition of serum according to the experimental design. Ad-cytomegalovirus (CMV) empty vector was used as a control and added to Ad-E2F1-only samples and Ad-TopBP1-only samples so that each plate received the same amount of virus. Eighteen hours later, cells were labeled with 5-bromo-2-deoxyuridine (BrdU) (10 μM) for 22 h, and then the cells were fixed with 4% formaldehyde. The incorporated BrdU was detected with fluorescein isothiocyanate-conjugated anti-BrdU antibody following the manufacturer's instructions (BD Biosciences). Images were captured on a Zeiss fluorescence microscope (Axio Observer inverted microscope). To assay apoptosis, the starved REF52 cells were harvested after 4 days of adenovirus infection and then stained with annexin V-phycoerythrin (PE) (Pharmingen) according to the manufacturer's instructions. At least 5,000 cells were gated for each sample, and the annexin profile of the cells was analyzed by flow cytometry. Annexin-positive cells were scored as apoptotic.
Chromatin binding assay.
We followed a published protocol to perform the chromatin binding assay (27). Briefly, the transfected cells were first resuspended for 5 min on ice in 150 μl of fractionation buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA) containing 0.2% NP-40 and supplemented with protease inhibitors described previously. Following centrifugation at 1,000 × g for 5 min, the supernatant was collected (fraction I), and the pellets were washed with the same buffer. The wash solution was collected as before (fraction II), and the nuclear pellets were further extracted for 40 min on ice with 150 μl of fractionation buffer containing 0.5% NP-40. The extracts were clarified by centrifugation at 16,000 × g for 15 min (fraction III). The pellets were finally lysed in 150 μl of 10% SDS-PAGE sample buffer and boiled for 5 min (fraction IV). Equal aliquots of each fraction, derived from equivalent cell numbers, were separated on 10% SDS-PAGE gels, and Western blotting was carried out.
Cross-linking.
Cells were lysed in borate buffer (50 mM Na-borate, 100 mM potassium acetate, 2 mM MgCl2, 1 mM EGTA, 1% Triton X-100, and protease inhibitors [pH 8.57]) on ice for 10 min. The samples were cleared of insoluble material by 10 min of high-speed centrifugation and then incubated with 20 mM freshly prepared dimethyl pimelimidate-2HCl (DMP) (Thermo Scientific) for 30 min. An equal volume of 50 mM NH4Cl2 in PBS was added for 10 min to quench the reaction. The cell lysates were subjected to SDS-PAGE and Western blot analysis.
Real-time RT-PCR.
RNA was extracted using TRIzol reagent (Invitrogen). Quantitative reverse transcription (RT)-PCR was performed in triplicate on an MX3005P thermal cycler using the SYBR green dye method to track the progress of the reactions, with ROX reference dye added as a reference. GAPDH (17) was run in parallel with the test genes. The following primer pairs were used: Apaf-1, forward, 5′-AATGGACACCTTCTTGGACG-3′, and reverse, 5′-GCACTTCATCCTCATGAGCC-3′; cyclin E, forward, 5′-CTCCAGGAAGAGGAAGGCAA-3′, and reverse, 5′-TCGATTTTGGCCATTTCTTCA-3′; caspase 3, forward, 5′-TCGGTCTGGTACAGATGTCG-3′, and reverse, 5′-CATACAAGAAGTCGGCCTCC-3′; thymidine kinase (TK), forward, 5′-ATGAGCTGCATTAACCTGCCCACT-3′, and reverse, 5′-ATGTGTGCAGAAGCTGCTGC-3′.
RESULTS
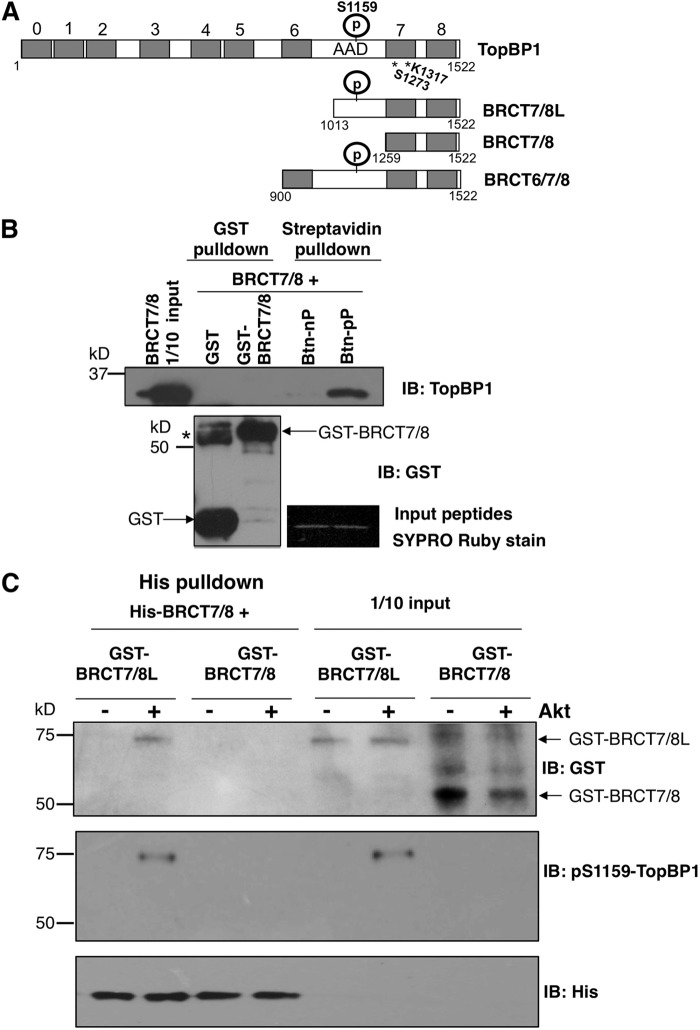
TopBP1-BRCT7/8 binds Akt-phosphorylated Ser-1159 residues and causes oligomerization.
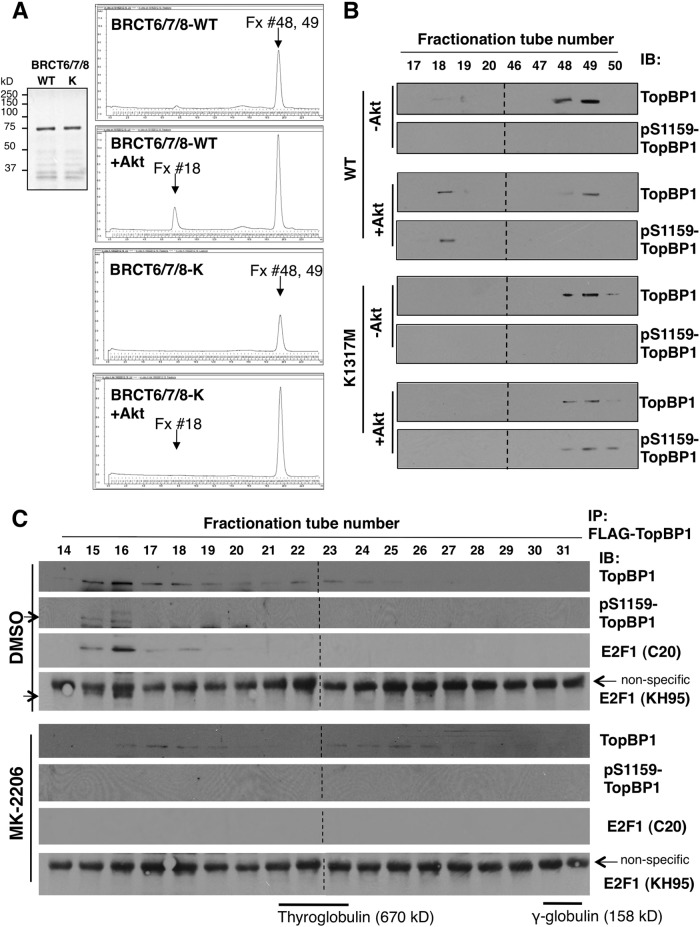
In our previous study, we mapped the oligomerization domain of TopBP1 to the carboxyl terminus (amino acids [aa] 1013 to 1522, referred to here as BRCT7/8L [Fig. 1A]), which is comprised of the 7th and 8th BRCT domains (aa 1259 to 1522) and a fragment between the 6th and 7th BRCT domains containing the Ser-1159 Akt phosphorylation site (16). To further narrow down the oligomerization domain, we tested the interaction between the TopBP1-BRCT 7th and 8th domains (aa 1259 to 1522, named BRCT7/8) and a biotinylated peptide flanking Ser-1159 (REERARLAS1159NLQWPS) or a biotinylated phosphopeptide (REERARLApS1159NLQWPS, referred to here as pSer-1159 peptide) by streptavidin-Sepharose pulldown assay. As shown previously for BRCT7/8L (16), BRCT7/8 could specifically bind the pSer-1159 peptide (Fig. 1B). In the same experiment, purified BRCT7/8 protein failed to oligomerize with GST-BRCT7/8. BRCT7/8 also could not interact with unphosphorylated BRCT7/8L (Fig. 1C). However, when BRCT7/8L was phosphorylated by Akt at Ser-1159, it could be pulled down by His-tagged BRCT7/8 in a His pulldown assay (Fig. 1C). Together with the data published previously (16), these data strongly support the notion that the interaction between the 7th and 8th BRCT domains and the pSer-1159 residue mediates Akt-dependent oligomerization of TopBP1.
Fig 1.
TopBP1-BRCT7/8 binds to Ser-1159-phosphorylated TopBP1-BRCT7/8L. (A) Schematic diagram of the TopBP1 constructs used in this study. S1159, Akt phosphorylation (P) site; AAD, ATR-activating domain. The numbers at the top indicate the designation of each BRCT domain, 0 to 8 (7). The numbers below are amino acid positions. S1273 and K1317 are the predicted amino acids in BRCT7/8 for phosphopeptide binding. * indicates the positions of both residues. (B) Purified TopBP1-BRCT7/8 was incubated with purified GST, GST-TopBP1-BRCT7/8, biotin-pS1159 peptide (biotin-REERARLApSer1159NLQWPS derived from the TopBP1 sequence, labeled Btn-pP), or non-biotin-phosphorylated peptide (Btn-nP) for 3 h. Glutathione-Sepharose GST pulldown (for GST and GST-BRCT7/8) and streptavidin-Sepharose pulldown (for Btn-nP and Btn-pP) were performed, followed by immunoblotting (IB) with anti-TopBP1 carboxyl terminus antibody, which recognizes TopBP1-BRCT7/8. The membrane was also probed with GST antibody. The asterisk represents a dimeric form of GST. GST is known to have dimerizing ability and occasionally is not completely resolved to monomeric GST on SDS-PAGE. To ensure the loading of Btn-nP and Btn-pP peptides, the input peptides were separated in SDS-PAGE and visualized by Sypro Ruby staining. (C) GST-TopBP1-BRCT7/8L and GST-TopBP1-BRCT7/8 proteins were first treated with purified Akt kinase. The proteins were then incubated with His-TopBP1 protein. The His-TopBP1 complex was then pulled down with nickel-Sepharose, followed by SDS-PAGE. The membrane was probed with the indicated antibodies.
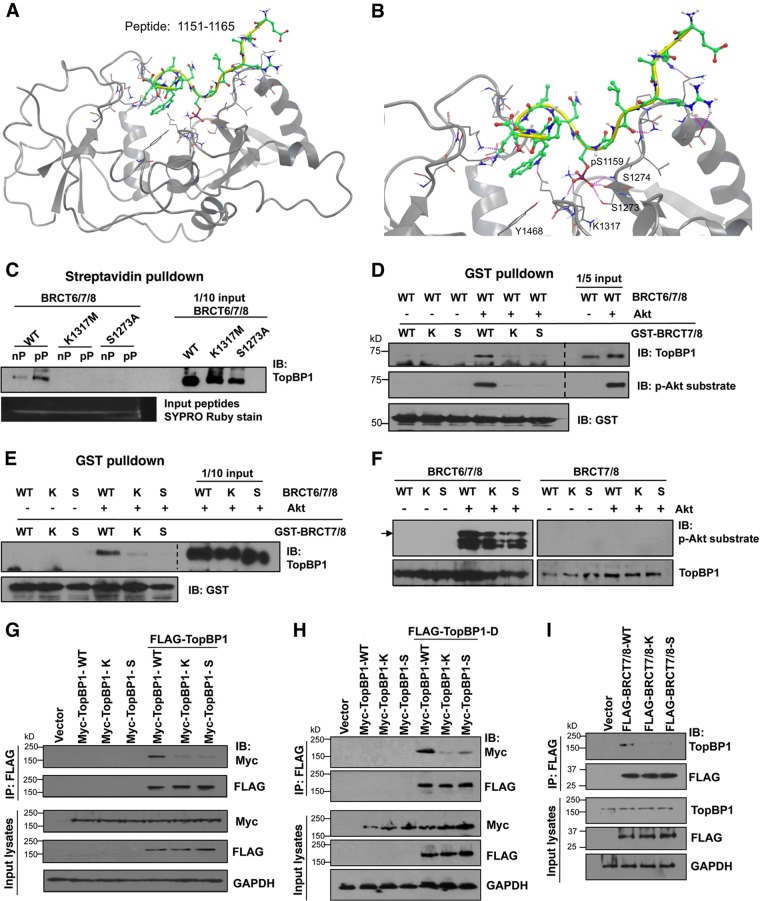
Mutations within BRCT7/8 that impede binding to pSer-1159 also hamper oligomerization.
To further establish the mechanism for TopBP1 oligomerization, we sought to identify point mutations within the 7th and 8th BRCT domains that would destroy its interaction with the pSer-1159 residue. We then investigated the effects of these point mutations on the oligomerization of TopBP1 and the repression of E2F1 transcriptional activity by TopBP1. To facilitate the identification of the critical residues within BRCT7/8 for phosphopeptide binding before the structure of TopBP1-BRCT7/8 was available (28), we constructed a homology model of TopBP1-BRCT7/8 (amino acids 1267 to 1488) with the pSer-1159 peptide based on the structure of BRCA1 BRCT repeats with MODELER, using the Insight II graphic environment (data not shown). This model predicts that pSer-1159 is positioned in a groove comprised of Lys-1317, Ser-1273, Ser-1274, and Asn-1315. A simulation of the pSer-1159 peptide with the known structure of TopBP1-BRCT7/8 also shows hydrogen bonding between pSer-1159 and Ser-1273, Ser-1274, and Lys-1317 (Fig. 2A and B). Ser-1273 and Lys-1317 are conserved among BRCT repeat-containing proteins, such as BRCA1, BARD1, MDC1, and 53BP1. We therefore generated S1273A and K1317M mutations and tested their interactions with the pSer-1159 peptide. Indeed, both S1273A and K1317M mutant TopBP1-BRCT6/7/8 failed to bind the pSer-1159 peptide (Fig. 2C). We then tested whether these mutations affected Akt-dependent oligomerization of TopBP1. We performed an established in vitro TopBP1 self-association assay (16) in which GST-BRCT7/8 was incubated with Akt-phosphorylated BRCT6/7/8 and the complex was then pulled down with glutathione-Sepharose beads. As shown in Fig. 2D and E, wild-type GST-BRCT7/8 could associate with BRCT6/7/8 only after BRCT6/7/8 was phosphorylated by Akt. Importantly, mutation of Ser-1273 or Lys-1317 in GST-BRCT7/8 attenuated its association with Akt-phosphorylated BRCT6/7/8. These mutations did not affect phosphorylation of TopBP1 by Akt (Fig. 2F and 3A). We also transfected FLAG- and Myc-tagged TopBP1 (WT, S1273A, or K1317M) into HEK293 cells and tested TopBP1 self-association within the cells (16). Indeed, S1273A or K1317M greatly attenuated TopBP1 self-association in vivo (Fig. 2G). We also tested an S1159 phosphorylation mimicking mutant S1159D-TopBP1 (16). This mutant (TopBP1-D) associated with WT TopBP1, but the interaction was much weaker with the S1273A or K1317M mutant (Fig. 2H). Previously, we showed FLAG-TopBP1-BRCT6/7/8 interacted with endogenous TopBP1 in an Akt-dependent manner (16). We performed a similar assay to test whether S1273A or K1317M mutation disrupts this interaction with endogenous TopBP1 in cells. Indeed, WT TopBP1-BRCT7/8 bound endogenous TopBP1 in HEK293 cells, and S1273A and K1317M mutations abrogated this interaction (Fig. 2I). Thus, mutations of the residues important for BRCT7/8 binding to pSer-1159 block Akt-dependent TopBP1 oligomerization. These data further support the idea that the binding between BRCT7/8 and the pSer-1159 residue is the molecular basis for TopBP1 oligomerization.
Fig 2.
K1317M and S1273A mutations within TopBP1-BRCT7/8 block its binding to the pS1159 peptide and inhibit TopBP1 oligomerization. (A) Molecular dynamic simulation between the pS1159 peptide (aa 1151 to 1165) and TopBP1-BRCT7/8 structure. (B) Enlarged image of the simulation in panel A to show the interactions between the pS1159 residue and S1273, S1274, and K1317. (C) WT, K1317M, or S1273A TopBP1-BRCT6/7/8 protein was incubated with Btn-pS1159 peptide (pP) or non-Btn-phosphorylated peptide (nP), followed by streptavidin-Sepharose pulldown and immunoblotting with anti-TopBP1 carboxyl-terminus antibody to detect BRCT6/7/8 bound to Btn-pS1159 peptide or Btn-S1159 peptide. To ensure equal loading of peptides, the input peptides were run in SDS-PAGE and visualized by Sypro Ruby staining. (D) Purified TopBP1-BRCT6/7/8-WT protein was first phosphorylated with recombinant Akt kinase and then incubated with purified GST-TopBP1-BRCT7/8-WT or its mutant K1317M (labeled K) or S1273A (labeled S). GST pulldown with glutathione-Sepharose was performed, resolved in SDS-PAGE, and immunoblotted with the indicated antibodies, including a phospho-Akt substrate-specific antibody, which has been shown to recognize pS1159-TopBP1 (16). The space between the images from input and GST pulldown was excised for brevity (dashed lines). (E) Purified TopBP1-BRCT6/7/8 proteins (WT, K1317M, or S1273A) were first phosphorylated by Akt kinase and then incubated with GST-TopBP1-BRCT7/8-WT or its K or S mutant, respectively. GST pulldown and Western blot analysis were performed as for panel B. The space between the images from input and GST pulldown was excised for brevity (dashed line). (F) Purified TopBP1-BRCT6/7/8 (WT, K1317M, or S1273A) and purified TopBP1-BRCT7/8 (WT, K1317M, or S1273A) were incubated with Akt kinase at 30°C for 30 min and then subjected to 10% SDS-PAGE to check the protein phosphorylation status by probing with a phospho-Akt substrate-specific antibody. The arrow indicates the full-length BRCT6/7/8. The smaller bands represent degradative products. (G) HEK293 cells were cotransfected with WT, K1317M, or S1273A Myc-TopBP1 alone and in combination with WT FLAG-TopBP1. After 48 h, the lysates were immunoprecipitated (IP) with anti-FLAG beads, followed by immunoblotting. (H) The same procedure was performed as for panel G, except for transfection of the FLAG-TopBP1-S1159D mutant. (I) HEK293 cells were transfected with WT, K1317M, or S1273A FLAG-TopBP1-BRCT7/8. After 48 h, the lysates were immunoprecipitated with anti-FLAG beads, followed by immunoblotting.
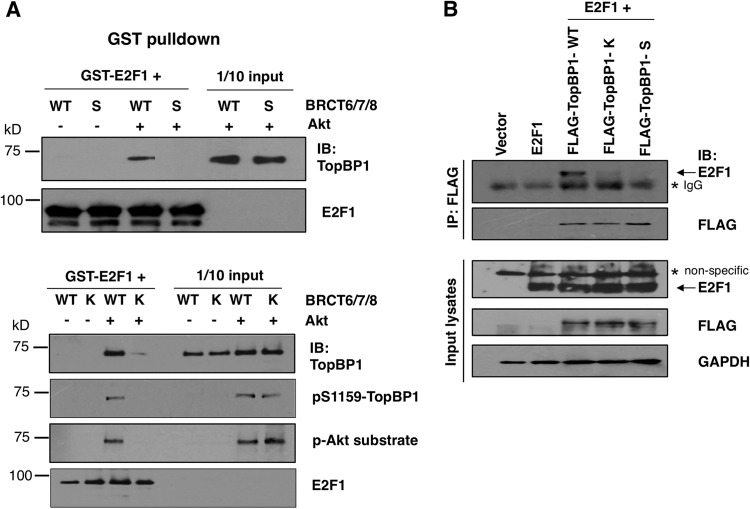
Fig 3.
K1317M and S1273A mutant TopBP1 failed to interact with E2F1 upon Akt activation in vitro and in vivo. (A) (Top) Both purified TopBP1-BRCT6/7/8-WT and TopBP1-BRCT6/7/8-S1273A proteins were first phosphorylated with Akt kinase and then incubated with purified GST-E2F1 for 3 h. GST pulldown with glutathione-Sepharose was performed, followed by immunoblotting with TopBP1 and E2F1 antibodies. (Bottom) The same procedure was carried out, but using TopBP1-BRCT6/7/8-K1317M protein to replace S1273A for GST pulldown. The status of protein phosphorylation was detected with either phospho-TopBP1 or phospho-Akt substrate antibody. (B) HEK293 cells were transfected with 2 μg of the plasmid expressing E2F1, along with 10 μg of FLAG-TopBP1 (WT, K1317M, or S1273A); 48 h later, cells were harvested for immunoprecipitation with anti-FLAG beads. Western blotting was performed with the indicated antibodies.
pSer-1159 binding-defective TopBP1 mutants are faulty in E2F1 binding.
Our prior study established that Akt-dependent oligomerization is crucial for TopBP1 to interact with and repress E2F1 (16). Thus, we expected that the S1273A and K1317M mutants are also defective in E2F1 binding. Indeed, in the in vitro assay, WT TopBP1 bound to E2F1 in an Akt-dependent manner, but S1273A and K1317M mutant TopBP1 could not (Fig. 3A). Using a pS1159-specific antibody and a p-Akt substrate-specific antibody, we showed K1317M mutant TopBP1 could still be phosphorylated by purified Akt kinase at Ser-1159 (Fig. 3A, bottom right). Thus, the impact of these mutations on E2F1 interaction is due to their defect in Akt-dependent oligomerization. To further determine whether S1273A and K1317M mutations impair the interaction between TopBP1 and E2F1 in vivo, we transfected WT or mutant FLAG-TopBP1 with E2F1 in HEK293 cells and performed coimmunoprecipitation assays. As shown in Fig. 3B, the S1273A and K1317M mutations significantly decreased the interaction between TopBP1 and E2F1 inside the cells.
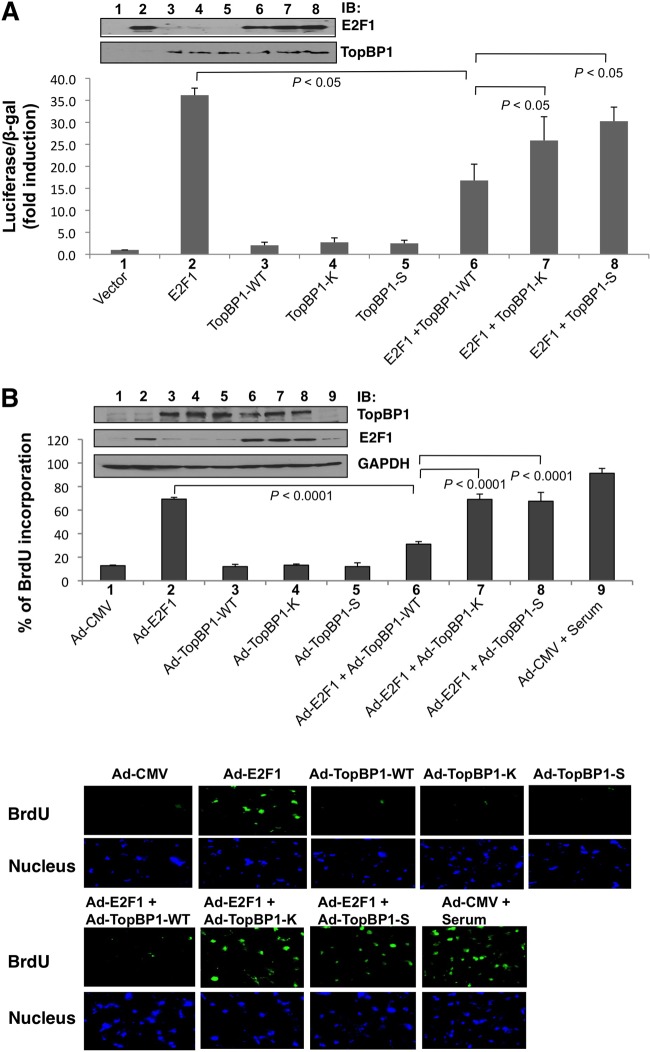
S1273A and K1317M mutants are defective in repressing E2F1 activities.
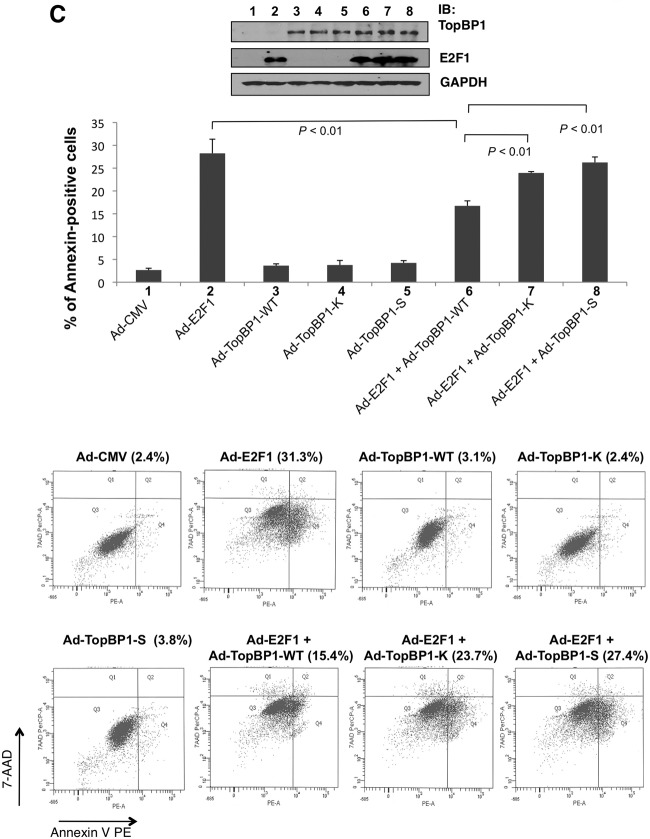
To gain additional biological evidence for how the mutations at the pSer-1159 binding sites impact the regulation of E2F1 function by TopBP1, we performed an E2F1 activity reporter assay (14). Both S1273A and K1317M mutations impaired TopBP1's ability to repress E2F1 transcriptional activity (Fig. 4A). This is consistent with their defect in binding to E2F1.
Fig 4.
K1317M and S1273A mutant TopBP1 failed to repress E2F1 activities. (A) E2F1 luciferase reporter assay. E2F1 transcriptional activity was measured with a p14ARF promoter-luciferase activity assay. E2F1-expressing plasmid (2 μg) was cotransfected with 10 μg of the plasmid expressing TopBP1 (WT, K1317M, or S1273A) into HEK293 cells. An empty vector, pcDNA3, was used as a control and added to E2F1-only and TopBP1-only samples so that each transfection reaction had the same amount of total DNA in transfection mixtures. The luciferase activity of transfected E2F1 was determined as the fold induction relative to that of the empty-vector control. The assay was performed in three independent transfection replicates (biological replicates). The data represent means and standard deviations. The P values are based on a two-tailed t test. One-tenth of each sample was lysed with SDS lysis buffer for Western blot analysis (top). (B) BrdU incorporation. REF52 cells were brought to quiescence by serum starvation for 48 h and then infected with Ad-CMV (empty vector), Ad-E2F1, Ad-TopBP1-WT, Ad-TopBP1-K1317M, Ad-TopBP1-S1273A, or Ad-E2F1 and Ad-TopBP1 (WT, K1317M, or S1273A) as indicated at a multiplicity of infection (MOI) of 400. Ad-CMV was used as a control and added to Ad-E2F1-only samples and Ad-TopBP1-only samples so that each plate received the same amount of virus. Some Ad-CMV-infected cells were stimulated with 20% fetal bovine serum; 18 h later, a BrdU incorporation assay was performed, and the incorporated BrdU was detected with fluorescein isothiocyanate-conjugated anti-BrdU antibody. The nuclei were stained with Hoechst 33258. At least 300 nuclei were counted for each sample under a microscope for BrdU incorporation. The data shown represent means and standard deviations from six replicates. The P values are based on a two-tailed t test. (Top) Equal expression of Ad-TopBP1-WT and its K1317M and S1273A mutants, as well as equal expression of Ad-E2F1, in each sample was verified by immunoblotting. (Bottom) Representative images at ×200 magnification for each indicated group. (C) Apoptosis assay. REF52 cells were serum starved for 48 h and then infected with Ad-E2F1, Ad-TopBP1-WT, or the K1317M or S1273A mutant as indicated at an MOI of 400. After infection, the cells were grown in 0.25% serum for 4 days, and apoptosis was scored by annexin V-PE–7-AAD staining. The data shown are means and standard deviations from three independent samples. The P values are based on a two-tailed t test. (Top) An aliquot of cell lysates was analyzed by Western blotting with the indicated antibodies. (Bottom) Representative profiles of apoptosis. The percentages of annexin-positive cells are shown above each profile.
Infection by recombinant adenovirus expressing E2F1 in serum-starved REF52 cells has been established to quantify E2F1-induced DNA synthesis and apoptosis (29). Coinfection of Ad-TopBP1 but not S1159A mutant Ad-TopBP1 can repress E2F1 function in these assays (14, 16). To examine whether S1273A and K1317M mutants could regulate E2F1 function in S phase and apoptosis induction, we constructed S1273A and K1317M mutant TopBP1 in recombinant adenoviral vectors and coinfected serum-starved REF52 cells with Ad-E2F1. Indeed, unlike WT TopBP1, both mutants failed to suppress E2F1 function in the induction of BrdU incorporation (Fig. 4B) and apoptosis (Fig. 4C). Thus, oligomerization is important for TopBP1 to regulate E2F1 function.
Akt phosphorylation stimulates oligomerization of wild-type but not K1317M BRCT6/7/8.
To gain further biochemical evidence for the structural change (oligomerization) induced by Akt phosphorylation in TopBP1, we purified wild-type and K1317M BRCT6/7/8 proteins (Fig. 5A, left). These purified products were subsequently phosphorylated with purified Akt kinase. The BRCT6/7/8 proteins were then run on a Superdex 200 10/300 GL column (Fig. 5A). BRCT6/7/8 eluted at fractions 48 and 49. After Akt phosphorylation, a new peak emerges at a much earlier elution (fraction 18) in BRCT6/7/8-WT but not in the K1317M mutant. The earlier eluted fraction on size exclusion chromatography corresponds to a higher-molecular-weight species. We collected these fractions and performed Western blot analysis using a TopBP1 antibody and a pS1159-TopBP1-specific antibody (Fig. 5B). The TopBP1 antibody recognized fractions 48 to 49, as well as fraction 18, but the pS1159-specific antibody recognized only fraction 18, indicating that all the pS1159-phosphorylated BRCT6/7/8 had formed a larger complex and thus eluted earlier. The K1317M mutant BRCT6/7/8 could also be phosphorylated at Ser-1159, as recognized by pS1159-specific antibody, but it failed to form high-molecular-weight species. Thus, we conclude that Akt phosphorylation induces oligomerization of BRCT6/7/8. The estimated size of fraction 18 is greater than 670 kDa and is likely bigger than a 10-mer of BRCT6/7/8. However, the detailed molecular character of this high-molecular-weight species will require additional biophysical determination.
Fig 5.
Size exclusion chromatography analysis of TopBP1. (A) (Left) Coomassie blue staining of purified TopBP1-BRCT6/7/8-WT and BRCT6/7/8-K1317M proteins for FPLC. (Right) A representative FPLC chromatogram for each indicated group (the x axis is elution volume; the y axis is UV absorbance at 280 nm). Purified TopBP1-BRCT6/7/8-WT or -K1317M (20 μg) was mock phosphorylated or phosphorylated with 1 μl of recombinant Akt kinase (1.8 μg). It was then loaded in 100 μl of PBS into an AktaPurifier UPC10 system and run through a Superdex 200 column. The protein fraction (Fx) volume collected for each tube was 400 μl. (B) Forty-microliter aliquots from the selected collection tubes corresponding to the protein peak on the chromatogram were subjected to 10% SDS-PAGE and analyzed by Western blotting with TopBP1 and pS1159-TopBP1 antibodies. Fractions 17 to 20 were run on the same gels (in the order 46 to 50 and 17 to 20, from left to right) and blotted together. To match the order of the fractions on the FPLC profile in panel A for easy readability, the images of fractions 17 to 20 were rearranged to the left of fractions 46 to 50, with dashed lines indicating the junctions. (C) FPLC fractionation of cellular lysates. H1299 cells were transfected with 10 μg of FLAG-TopBP1. After 48 h, the cells were treated with either DMSO or MK-2206 (1 μM) for 24 h. Nuclear extracts were prepared and run through a Superdex 200 column. FLAG-TopBP1 in each fraction was immunoprecipitated with anti-FLAG (M2) beads, subjected to 10% SDS-PAGE, and analyzed by Western blotting with TopBP1, pS1159-TopBP1, and E2F1 antibodies. The arrows indicate pS1159-TopBP1 or E2F1 (recognized by KH95 anti-E2F1 antibody; the identity of E2F1 was confirmed by another E2F1 [C20] antibody, as indicated). The immunoblotting assays between DMSO and MK-2206 samples were performed together and exposed on the same films (the exposure times were identical). Fractions 14 to 22 and 23 to 31 were blotted together and exposed on the same films, with the space excised (dashed lines) for clarity of presentation. The elution peaks of the gel filtration standard (Bio-Rad), including the molecular mass markers thyroglobulin and γ-globulin, were determined on a parallel run under the same conditions, as indicated at the bottom.
E2F1/TopBP1 interacts within a high-molecular-weight complex in an Akt-dependent manner inside the cells.
We wanted to investigate if the in vitro results could be recapitulated in vivo. We transfected FLAG-TopBP1 to a p53-null non-small-cell lung carcinoma cell line, H1299, and treated the cells with an allosteric Akt inhibitor, MK-2206. We then prepared nuclear extracts and ran them on a Superdex 200 10/300 GL column. The fractions were then immunoprecipitated with anti-FLAG beads (Fig. 5C). In DMSO control cells, FLAG-TopBP1 was mainly found in fractions 15 to 18 (peak at 16), and there were some weaker signals in 22 and 23. In the MK-2206-treated cells, FLAG-TopBP1 was equally distributed in two peaks, the first at fractions 17 and 18 and the second at fractions 23 to 26. Thus, Akt inhibitors decreased the sizes of TopBP1-containing complexes and also redistributed it from a big complex to a smaller one. The bigger-complex species contains pS1159-TopBP1, as revealed by anti-pS1159-TopBP1 immunoblotting. We also examined the presence of E2F1 in TopBP1 immunoprecipitates from each fraction. Importantly, E2F1 interacted with TopBP1 in fractions 15 to 18, with a peak at 16. The interaction could not be seen in fractions 22 to 24 (although the signals of TopBP1 were equivalent between fractions 18 and 22). The interaction also could not be detected in MK-2206-treated cells. Thus, we conclude that Akt can regulate the TopBP1 complex status and consequently the interaction of E2F1 with TopBP1, which is exclusively in the high-molecular-weight complex.
Oligomerization of TopBP1 inhibits its chromatin binding under replication stress.
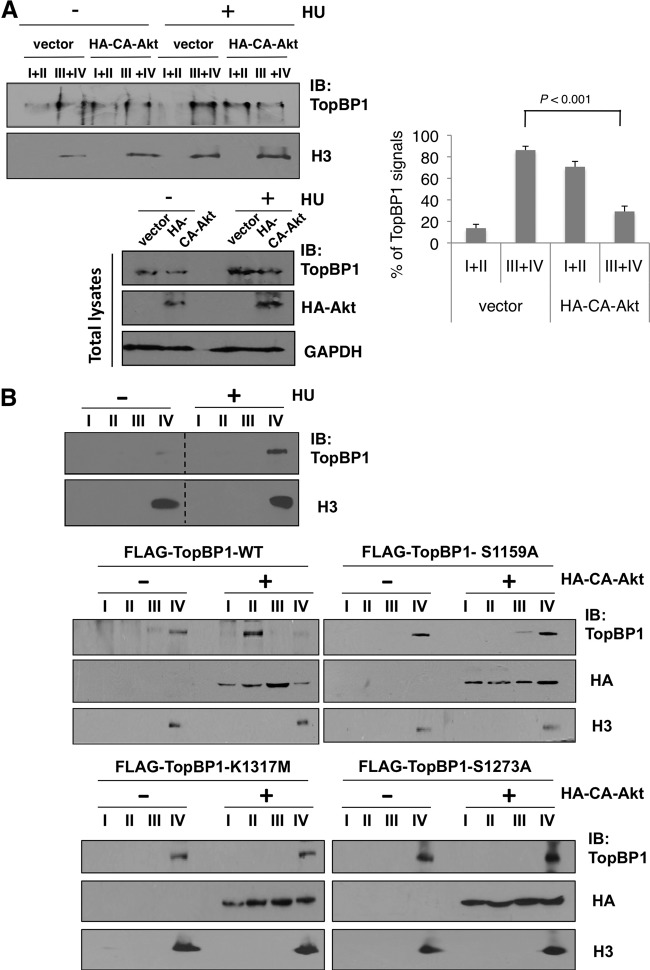
When cells suffer from replicative stress, TopBP1 is recruited to chromatin for activation of ATR/Chk1. With the regulation of TopBP1 quaternary structure by Akt, we wanted to investigate whether Akt regulates TopBP1 function in checkpoint activation. Since chromatin binding is essential for TopBP1 to perform its checkpoint activation function, we first tested whether Akt could affect recruitment of TopBP1 to chromatin upon replicative stress. Hydroxyurea (HU) treatment in H1299 cells induced accumulation of TopBP1 on chromatin (fractions III and IV [27]) (Fig. 6A). Importantly, overexpression of constitutively active Akt (CA-Akt) released TopBP1 from the chromatin-bound fractions to soluble fractions (I and II) (Fig. 6A; the relative intensities of TopBP1 signals from three independent experiments are shown in the graph on the right). This result indicated that overactivation of Akt impaired chromatin recruitment of TopBP1 upon replicative stress. To test whether phosphorylation of Ser-1159 mediated the release of TopBP1 from chromatin upon Akt activation, we examined the chromatin binding of S1159A mutant TopBP1. As shown in Fig. 6B, after HU treatment, FLAG-TopBP1-WT bound tightly with chromatin (fraction IV, which is resistant to 0.5% NP-40 extraction), like endogenous TopBP1 (Fig. 6B, top). With cotransfection of HA-CA-Akt, FLAG-TopBP1-WT was released to the soluble fraction II. However, FLAG-TopBP1-S1159A remained at fraction IV even with cotransfection of HA-CA-Akt. In addition to the S1159A mutant, K1317M and S1273A mutant TopBP1 also remained at chromatin fractions even with cotransfection of HA-CA-Akt (Fig. 6B). Thus, we conclude that Akt induces TopBP1 oligomerization and inhibits its chromatin binding, which is necessary for TopBP1 checkpoint activation function.
Fig 6.
Hyperactivation of Akt inhibits chromatin recruitment of TopBP1 after HU treatment, and this effect is mediated through Ser-1159 phosphorylation-induced TopBP1 oligomerization. (A) Chromatin binding assay of endogenous TopBP1. H1299 cells were transfected with either an empty vector or 10 μg of HA-CA-Akt. After 48 h, the cells were treated with vehicle control or hydroxyurea (2 μM) for 12 h and then harvested and fractionated following the protocol described in Materials and Methods. (Top left) The combined aliquots from fractions I and II and aliquots from fractions III and IV for each group were separated on a 10% SDS-PAGE gel and immunoblotted with the indicated antibodies. (Right) The assay was repeated independently three times. TopBP1 signals in each fraction from the HU-treated samples were quantified using image J software and are shown as means and standard deviations. (Bottom left) Aliquots of total cell lysates were also analyzed by Western blotting with TopBP1, HA, and GAPDH antibodies. (B) (Top) H1299 cells were treated with vehicle control or hydroxyurea (2 μM) for 12 h and then harvested and fractionated as for panel A. The immunoblotting of samples with and without HU treatment was performed together. The space between the images from these two sets of samples was excised for brevity (dashed lines). (Middle and bottom) H1299 cells were transfected with 10 μg of HA-CA-Akt or an empty vector, along with 10 μg of FLAG-TopBP1 (WT or S1159A, K1317M, or S1273A mutant). After 48 h, the cells were treated with hydroxyurea (2 μM) for 12 h and then harvested and fractionated as described for panel A. An aliquot from each fraction was subjected to 10% SDS-PAGE and probed with the indicated antibodies.
Akt inhibits HU-induced checkpoint activation via phosphorylation of TopBP1 at Ser-1159.
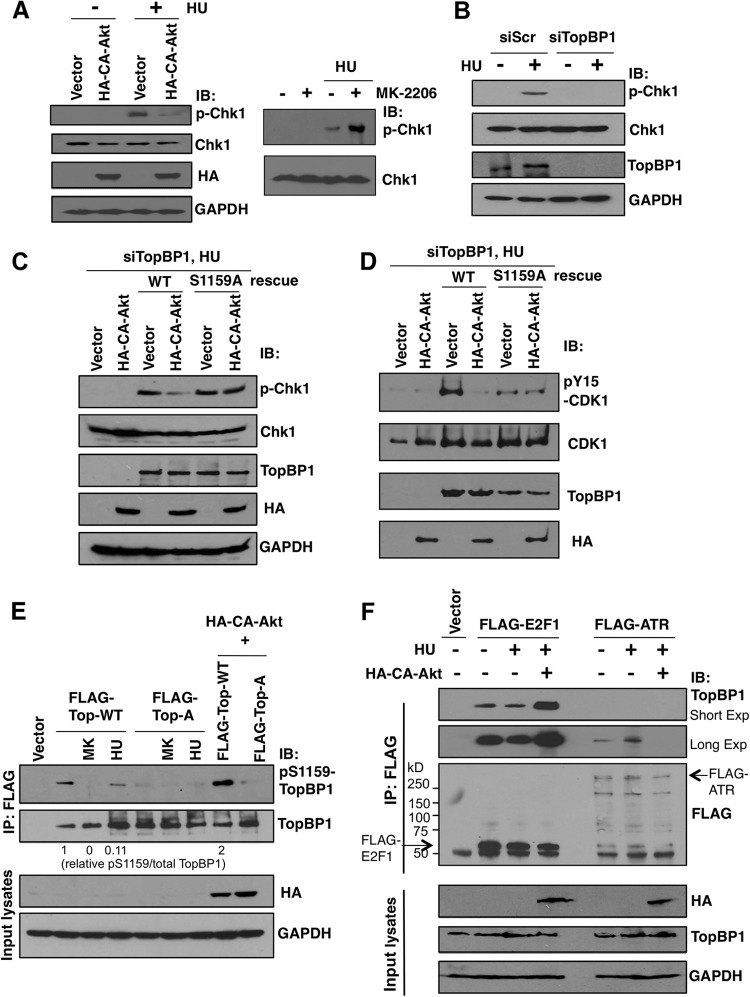
We then tested whether hyperactivation of Akt could inhibit Chk1 phosphorylation upon HU treatment. Indeed, expression of CA-Akt in H1299 cells blocked HU-induced Chk1 activation (Fig. 7A, left). Conversely, treatment with MK-2206 augmented Chk1 activation (Fig. 7A, right). To further investigate whether checkpoint inhibition by Akt was mediated through phosphorylation of TopBP1 at Ser-1159, we used established TopBP1-depleted H1299 cells (Fig. 7B) (20) reconstituted with either WT or S1159A mutant TopBP1. As expected from the role of TopBP1 in ATR/Chk1 activation, depletion of TopBP1 blocked activation of Chk1 upon HU treatment (Fig. 7B). Reconstitution of TopBP1 with the WT or S1159A mutant restored Chk1 activation (Fig. 7C), indicating that Ser-1159 phosphorylation is not required for ATR/Chk1 activation. Significantly, while CA-Akt inhibited Chk1 phosphorylation in WT TopBP1-reconstituted cells, it failed to inhibit Chk1 phosphorylation in the cells that were reconstituted with S1159A-TopBP1 (Fig. 7C). We also measured phosphorylation of Y15-CDK1 as an independent assay for the checkpoint function. When Chk1 is activated, it phosphorylates and inhibits the function of Cdc25C (30, 31), a phosphatase for pY15-CDK1, thus leading to accumulation of pY15-CDK1. As shown in Fig. 7D, rescue of the TopBP1 level with WT or S1159A TopBP1 restored pY15-CDK during HU treatment. A lower level of pY15-CDK in an S1159A-rescued sample than in a WT-rescued sample is consistent with lower expression of S1159A-TopBP1 than WT in this experiment. Again, while CA-Akt inhibited pY15-CDK1 in WT TopBP1-reconstituted cells, it failed to inhibit CDK1 phosphorylation in S1159A-TopBP1-reconstituted cells. Thus, Akt inhibits HU-induced checkpoint activation by phosphorylating TopBP1 at Ser-1159.
Fig 7.
Akt inhibits HU-induced Chk1 activation through Ser-1159 phosphorylation. The interactions of TopBP1 with E2F1 versus with ATR are inversely regulated by replicative stress and Akt activation. (A) (Left) Ten micrograms of either empty vector or HA-CA-Akt was transfected into H1299 cells; 48 h later, the cells were treated with hydroxyurea (2 μM) for 12 h or left untreated and then subjected to 10% SDS-PAGE and immunoblotted with the indicated antibodies. (Right) H1299 cells were treated with hydroxyurea (2 μM) with or without MK-2206 (1 μM), and the lysates were analyzed. (B) H1299 stable cell lines expressing either a control scrambled small interfering RNA (siRNA) (siScr) or siTopBP1 were left untreated or treated with hydroxyurea (2 μM) for 12 h. The cells were harvested, and the cellular lysates were analyzed by Western blotting as indicated. (C) H1299 stable cells expressing TopBP1 siRNA were transfected or cotransfected with 2 μg of HA-CA-Akt and FLAG-TopBP1-WT or FLAG-TopBP1-S1159A. After 2 days, the cells were treated with hydroxyurea (2 μM) for 12 h. The same procedure as described for panel B was then performed. (D) The same experiment as in panel C was repeated, and the membrane was probed with different antibodies (pY15-CDK1, CDK1, TopBP1, and HA) for Western blot analysis. (E) An empty vector, FLAG-TopBP1 WT (labeled FLAG-Top-WT), or S1159A mutant (labeled FLAG-Top-A), with and without HA-CA-Akt, was transfected into H1299 cells. Two days later, the cells were left untreated or treated with hydroxyurea (2 μM) for 12 h or with MK-2206 (labeled MK) (1 μM) for 24 h. The cells were harvested with TNN buffer and immunoprecipitated with anti-FLAG (M2) beads, followed by Western blotting with the indicated antibodies. The signals of pS1159-TopBP1 and total TopBP1 were measured by Image J densitometry, and the relative values of pS1159/total TopBP1 are shown below the gel. (F) H1299 cells were transfected with FLAG-E2F1 or FLAG-ATR alone or in combination with HA-CA-Akt; 48 h later, the cells were left untreated or treated with hydroxyurea (2 μM) for 12 h. Immunoprecipitation with anti-FLAG (M2) beads was performed, followed by Western blotting as indicated.
Thus far, our data indicate that TopBP1 exists in cells as at least two distinct states: state I, S1159 phosphorylated when Akt is active, oligomerized, and binding to E2F1; state II, S1159 unphosphorylated when Akt is inactive, monomeric, and binding to chromatin to activate ATR/Chk1. These two states are controlled by Akt. We suspected that upon replicative stress, such as HU treatment, phosphorylation of Ser-1159 in TopBP1 might be inhibited and thus allow TopBP1 to be recruited to stalled replication forks. Indeed, in normally growing H1299 cells, a pS1159-TopBP1 phosphospecific antibody easily recognized pS1159-TopBP1. This signal disappeared upon treatment of MK-2206 or in the S1159A mutant and was enhanced upon cotransfection with CA-Akt (Fig. 7E). Importantly, although HU treatment induced TopBP1 expression, it significantly inhibited Ser-1159 phosphorylation. Thus, HU treatment modulates the TopBP1 phosphorylation status (probably by inhibiting Akt) and shifts it from state I to state II.
To further demonstrate the regulation of TopBP1 binding partners—E2F1 versus ATR—by Akt or HU treatment, we transfected FLAG-E2F1 or FLAG-ATR into H1299 cells. We then performed anti-FLAG immunoprecipitation to examine the association of TopBP1 with FLAG-E2F1 versus FLAG-ATR and to investigate how HU and CA-Akt influence the association. As shown in Fig. 7F, TopBP1 could be identified in both FLAG-E2F1 and FLAG-ATR immunoprecipitates. Importantly, HU treatment induced its association with ATR but slightly inhibited the association with E2F1 whereas CA-Akt enhanced its association with E2F1 but eliminated the association with ATR.
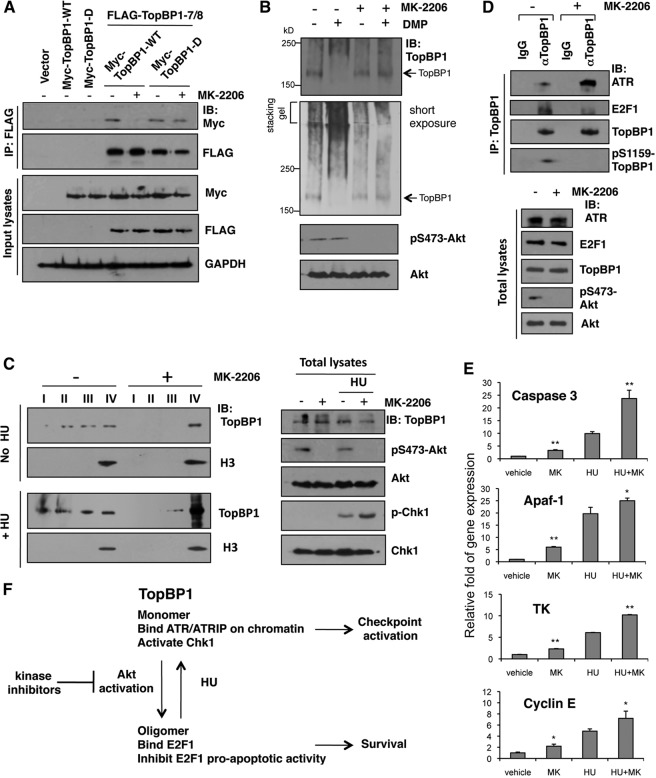
Validation of the proposed TopBP1 regulation in a PTEN-mutated cell line upon treatment with an Akt inhibitor, MK-2206.
To translate the information from our model to be relevant to cancer and to investigate how kinase inhibitors used in the clinic might impact a key TopBP1 pathway, we used a PTEN-null MDA-MB468 breast cancer cell line and treated the cells with MK-2206 to validate our proposed TopBP1 regulation. First, we confirmed oligomerization between Myc-TopBP1 and FLAG-TopBP1 in this cell line (Fig. 8A). Moreover, oligomerization with WT TopBP1 was inhibited by MK-2206, but an S1159D phosphomimetic mutant still bound to other TopBP1 molecules, even upon MK-2206 treatment. We then analyzed the status of endogenous TopBP1 oligomerization by chemical cross-linking. MDA-MB468 cells were treated with MK-2206 or vehicle, and then the lysates were incubated with buffer alone or buffer containing DMP, followed by SDS-PAGE (Fig. 8B). In the presence of DMP, TopBP1 migrated as a high-molecular-weight complex in MDA-MB468 cells. Significantly, upon MK-2206 treatment, it was reduced to the monomeric form corresponding to the non-cross-linked TopBP1. Thus, TopBP1 in MDA-MB468 cells exists in a high-molecular-weight form in an Akt-dependent manner.
Fig 8.
An Akt inhibitor in PTEN-mutated cancer cells blocked TopBP1 S1159 phosphorylation and inhibited TopBP1 oligomerization and E2F1 binding but enhanced TopBP1 chromatin binding, ATR interaction, and Chk1 activation. (A) MDA-MB468 cells were transfected with WT or S1159D Myc-TopBP1 alone and in combination with FLAG-TopBP1-BRCT7/8, and 24 h later, some cells were treated with an Akt inhibitor (MK-2206; 1 μM) for 24 h. Then, the cells were lysed and immunoprecipitated with anti-FLAG beads, followed by immunoblotting as indicated. (B) TopBP1 oligomerization in vivo. MDA-MB468 cells were left untreated or treated with 1 μM MK-2206 for 24 h and then lysed and incubated with 20 mM DMP for 30 min following the protocol for cross-linking described in Materials and Methods. The cell lysates were analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. (C) Chromatin binding assay. MDA-MB468 cells were left untreated or treated with 1 μM MK-2206 for 12 h, and all of the cells were also serum starved for 12 h. FBS (20%) was then added to the culture medium. Two hours later, the cells were left untreated or treated with 2 μM HU for 2 h and then harvested and fractionated. (Left) Aliquots from fractions I, II, III, and IV were separated on a 10% SDS-PAGE gel and immunoblotted with the indicated antibodies. (Right) Aliquots of total cell lysates were analyzed by Western blotting. (D) MDA-MB468 cells were left untreated or treated with 1 μM MK-2206 for 5 h and then treated with 2 μM HU for 5 h. The cell lysates were immunoprecipates with TopBP1 antibody, followed by immunoblotting with the indicated antibodies. The same amount of normal mouse IgG was used as a control for immunoprecipitation. (E) MDA-MB468 cells received 1 μM MK-2206 for 5 h and then were harvested or further treated with HU as for panel D before harvest. RNA was extracted, and quantitative real-time RT-PCR was performed using primers specific for caspase 3, Apaf-1, TK, and cyclin E. The results were normalized to GAPDH levels and are expressed relative to the expression of the control cells. The experiment was performed in triplicate. *, P < 0.05, and **, P < 0.005 compared with the no-MK-2206 group (two-tailed t test). (F) Model depicting the transition between two distinct states of TopBP1 complexes under Akt activation or replicative stress, such as hydroxyurea treatment. Kinase inhibitors used in cancer therapy may impact the TopBP1 pathway to achieve their effects.
We also examined TopBP1 chromatin binding. Consistent with the data presented in Fig. 6, a significant portion of TopBP1 was released from chromatin in Akt-activated MDA-MB468 cells. Upon MK-2206 treatment, TopBP1 accumulated mainly in the chromatin fraction (Fig. 8C). This shift is consistent with the enhancement of Chk1 phosphorylation during HU treatment by MK-2206 (Fig. 8C, right).
We performed TopBP1 immunoprecipitation to examine its complex formation with ATR versus E2F1 upon MK-2206 treatment. MK-2206 treatment did block phosphorylation of TopBP1 Ser-1159 (Fig. 8D, top). Correspondingly, the interaction between TopBP1 and ATR was enhanced by MK-2206 (Fig. 8D, top). At the same time, its interaction with E2F1 was inhibited. In line with the data indicating that TopBP1 interacts with E2F1 and represses E2F1 transcriptional activity, the expression of several E2F1 target genes, such as the caspase 3, Apaf-1, thymidine kinase, and cyclin E genes, was enhanced by MK-2206 (Fig. 8E). Thus, the kinase inhibitors may exert their clinical effects by rectifying TopBP1 function (Fig. 8F).
DISCUSSION
Using point mutations within BRCT7/8 of TopBP1 that specifically block its binding to the pS1159 residue, we provide compelling evidence to support the idea that the binding between BRCT7/8 and pS1159 forms the molecular basis for Akt-induced oligomerization of TopBP1. These BRCT7/8 point mutants are defective in E2F1 binding and repression, similar to the S1159A mutant TopBP1. The oligomerization of TopBP1 after Akt phosphorylation was directly shown by size exclusion chromatography. Binding of E2F1 was seen only in the high-molecular-weight form of TopBP1. Intriguingly, upon Akt phosphorylation, the high-molecular-weight species of TopBP1-BRCT6/7/8 was fractionated to a single peak in size exclusion chromatography. These data demonstrate that TopBP1 oligomerizes to a distinct structure (e.g., 10-mer or larger) when Ser-1159 is phosphorylated instead of forming many different numbers of oligomers, such as dimers, trimers, etc. The multimeric nature also suggests that BRCT7/8 of the first TopBP1 molecule binds to the pS1159 residue of a second TopBP1, the BRCT7/8 of which binds to the pS1159 residue of a third TopBP1, and so on in a cooperative manner, instead of two reciprocal BRCT7/8-pS1159 interactions between two TopBP1 molecules. It would be very interesting in the future to determine the structural details of this high-molecular-weight species.
Phosphorylation of Ser-1159 has been shown to be required for TopBP1 to interact with and regulate other transcription factors, such as Miz1, human papillomavirus 16 (HPV16) E2 (16), and SPBP (19). Thus, oligomerization is likely important for a more general transcriptional function of TopBP1 and is not restricted to E2F1 regulation. On the other hand, the Akt-dependent structural regulation not only induces TopBP1 binding to E2F1, it also inhibits TopBP1 functioning in checkpoint activation. This is clearly demonstrated by the fact that rescuing TopBP1 expression in TopBP1-depleted cells with an S1159A mutant TopBP1 restored HU-induced Chk1 activation in the presence of constitutively active Akt (Fig. 7C). Thus, it appears that the Akt-dependent structural regulation functions as a molecular switch to flip back and forth between two alternative functions of TopBP1: (i) oligomers binding E2F1 to repress apoptosis when Akt is activated and (ii) monomers ready to be recruited to damaged chromatin to activate ATR/Chk1 when Akt activity is low (Fig. 8F).
It is conceivable that oligomerization of TopBP1 might block its direct binding to the stalled replication forks (10, 11) or interfere with the interactions between its BRCT1/2 and the 9-1-1 clamp (12) and between BRCT7/8 and autophosphorylated ATR (13), thus impairing its chromatin recruitment. There is also mutual regulation between these two states. For example, HU treatment can inhibit Ser-1159 phosphorylation while it increases total levels of TopBP1 (Fig. 7E), thus ensuring accumulation of TopBP1 monomers competent for ATR activation. It is conceivable that these two forms of TopBP1 are kept in balance during normal cellular proliferation. Since Akt is frequently activated in many tumor types, the elucidated mechanism of checkpoint perturbation by Akt has important implications for the genomic instability seen in the Akt-activated tumors. In fact, we show that an allosteric Akt inhibitor, MK-2206, currently being tested in cancer treatment, can reverse the effect of Akt hyperactivation on TopBP1 in a PTEN-mutated cancer cell line. Thus, the pathway of regulation elucidated here is clinically significant. As both Akt activation and TopBP1 overexpression are common in many types of cancer, selective blockade of TopBP1 oligomerization may serve as a potential therapeutic strategy in cancer treatment. The fact that we can demonstrate TopBP1 oligomerization in vitro with purified proteins is compelling evidence that TopBP1 oligomerization is mediated by phosphorylation of Ser-1159 and its binding to BRCT7/8 and that no other, unknown factors are required. Thus, the binding of BRCT7/8 and pSer-1159 may be a rational therapeutic target to block the inhibitory activity of TopBP1 toward E2F1 in cancer and to reactivate E2F1-mediated apoptosis during chemotherapy.
The distinct structural states of TopBP1 uncovered in this study pose additional intriguing questions deserving future investigation. TopBP1-BRCT1/2 binds Cdk2-phosphorylated treslin/TICRR (TopBP1-interacitng, checkpoint and replication regulator) to facilitate loading of Cdc45 onto replication origins (32, 33). It would be interesting to investigate how oligomerization affects TopBP1 function in DNA replication. TopBP1 interacts with the 9-1-1 clamps and treslin/TICRR through the same BRCT1/2; thus, TopBP1 binding to checkpoint proteins and binding to treslin/TICRR are likely mutually exclusive. In light of this, a mechanism must exist to turn off checkpoint response and allow TopBP1 to interact with treslin/TICRR for reinitiation of previously stalled replication forks. Akt is known to play an important role during S phase progression (34). It is tempting to suggest that Akt-mediated inhibition of TopBP1 checkpoint function might be important for switching the TopBP1 function from checkpoint activation to DNA replication. Experiments to test this hypothesis are under way. Another question is whether Akt can affect the “nontranscriptional” functions of E2F1 in response to DNA damage (35). E2F1 has been shown to directly contribute to DNA repair by promoting the recruitment of DNA repair factors, such as RPA, to sites of DNA double-strand breaks (DSB) (36). Expression of CA-Akt blocked RPA recruitment to sites of radiation-induced damage, whereas Akt inhibitors enhanced RPA foci (3). These data in general are consistent with our observations regarding the effect of Akt in inhibiting TopBP1 checkpoint function and suggest that Akt might inhibit the nontranscriptional functions of E2F1. Future investigation will be warranted to elucidate this further.
ACKNOWLEDGMENTS
We gratefully acknowledge Fannie Lin for insightful discussion.
This work was supported by the National Institutes of Health (RO1CA100857, RO1CA138641, and ARRA 3 P30CA125123-03S5) and the Department of Defense Breast Cancer Research Program (W81XWH-09-1-0338).
We have no financial interests that pose a conflict of interest regarding this article.
Footnotes
Published ahead of print 30 September 2013
REFERENCES
- 1.King FW, Skeen J, Hay N, Shtivelman E. 2004. Inhibition of Chk1 by activated PKB/Akt. Cell Cycle 3:634–637 [PubMed] [Google Scholar]
- 2.Tonic I, Yu WN, Park Y, Chen CC, Hay N. 2010. Akt activation emulates Chk1 inhibition and Bcl2 overexpression and abrogates G2 cell cycle checkpoint by inhibiting BRCA1 foci. J. Biol. Chem. 285:23790–23798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu N, Hegarat N, Black EJ, Scott MT, Hochegger H, Gillespie DA. 2010. Akt/PKB suppresses DNA damage processing and checkpoint activation in late G2. J. Cell Biol. 190:297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyn RE. 2009. Linking PTEN with genomic instability and DNA repair. Cell Cycle 8:2322–2323 [PubMed] [Google Scholar]
- 5.Shtivelman E, Sussman J, Stokoe D. 2002. A role for PI 3-kinase and PKB activity in the G2/M phase of the cell cycle. Curr. Biol. 12:919–924 [DOI] [PubMed] [Google Scholar]
- 6.Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L, Mansukhani M, Murty VV, Gaciong Z, Meek SE, Piwnica-Worms H, Hibshoosh H, Parsons R. 2005. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell 7:193–204 [DOI] [PubMed] [Google Scholar]
- 7.Rappas M, Oliver AW, Pearl LH. 2011. Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res. 39:313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia V, Furuya K, Carr AM. 2005. Identification and functional analysis of TopBP1 and its homologs. DNA Repair 4:1227–1239 [DOI] [PubMed] [Google Scholar]
- 9.Kumagai A, Lee J, Yoo HY, Dunphy WG. 2006. TopBP1 activates the ATR-ATRIP complex. Cell 124:943–955 [DOI] [PubMed] [Google Scholar]
- 10.Yan S, Michael WM. 2009. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle 8:2877–2884 [DOI] [PubMed] [Google Scholar]
- 11.Yan S, Michael WM. 2009. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J. Cell Biol. 184:793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. 2007. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 21:1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC, Glover JN, Yang XH, Zou L. 2011. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol. Cell 43:192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu K, Lin FT, Ruppert JM, Lin WC. 2003. Regulation of E2F1 by BRCT-domain containing protein TopBP1. Mol. Cell. Biol. 23:3287–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu K, Luo Y, Lin FT, Lin WC. 2004. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 18:673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu K, Paik JC, Wang B, Lin FT, Lin WC. 2006. Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J. 25:4795–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu K, Bellam N, Lin HY, Wang B, Stockard CR, Grizzle WE, Lin WC. 2009. Regulation of p53 by TopBP1: a potential mechanism for p53 inactivation in cancer. Mol. Cell. Biol. 29:2673–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M. 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell 10:509–521 [DOI] [PubMed] [Google Scholar]
- 19.Sjottem E, Rekdal C, Svineng G, Johnsen SS, Klenow H, Uglehus RD, Johansen T. 2007. The ePHD protein SPBP interacts with TopBP1 and together they co-operate to stimulate Ets1-mediated transcription. Nucleic Acids Res. 35:6648–6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K, Ling S, Lin WC. 2011. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol. Cell. Biol. 31:4464–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedram A, Razandi M, Evinger AJ, Lee E, Levin ER. 2009. Estrogen inhibits ATR signaling to cell cycle checkpoints and DNA repair. Mol. Biol. Cell 20:3374–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Case DA, Darden TA, Cheatham ITE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. 2006. AMBER 9. University of California, San Francisco, CA [Google Scholar]
- 23.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. 2003. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24:1999–2012 [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–934 [Google Scholar]
- 25.Lin WC, Lin FT, Nevins JR. 2001. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15:1833–1844 [PMC free article] [PubMed] [Google Scholar]
- 26.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 95:2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andegeko Y, Moyal L, Mittelman L, Tsarfaty I, Shiloh Y, Rotman G. 2001. Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 276:38224–38230 [DOI] [PubMed] [Google Scholar]
- 28.Leung CC, Gong Z, Chen J, Glover JN. 2011. Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control. J. Biol. Chem. 286:4292–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. U. S. A. 94:7245–7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. 1997. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277:1501–1505 [DOI] [PubMed] [Google Scholar]
- 31.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. 1997. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277:1497–1501 [DOI] [PubMed] [Google Scholar]
- 32.Kumagai A, Shevchenko A, Dunphy WG. 2010. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140:349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA. 2010. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 24:183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu N, Lao Y, Zhang Y, Gillespie DA. 2012. Akt: a double-edged sword in cell proliferation and genome stability. J. Oncol. 2012:951724. 10.1155/2012/951724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biswas AK, Johnson DG. 2012. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 72:13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Zhu F, Weaks RL, Biswas AK, Guo R, Li Y, Johnson DG. 2011. E2F1 promotes the recruitment of DNA repair factors to sites of DNA double-strand breaks. Cell Cycle 10:1287–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]