Abstract
RNA helicases are involved in almost every aspect of RNA metabolism, yet very little is known about the regulation of this class of enzymes. In Saccharomyces cerevisiae, the stability and translational fidelity of nonsense-containing mRNAs are controlled by the group I RNA helicase Upf1 and the proteins it interacts with, Upf2 and Upf3. Combining the yeast two-hybrid system with genetic analysis, we show here that the cysteine- and histidine-rich (CH) domain and the RNA helicase domain of yeast Upf1 can engage in two new types of molecular interactions: an intramolecular interaction between these two domains and self-association of each of these domains. Multiple observations indicate that these molecular interactions are crucial for Upf1 regulation. First, coexpression of the CH domain and the RNA helicase domain in trans can reconstitute Upf1 function in both promoting nonsense-mediated mRNA decay (NMD) and preventing nonsense suppression. Second, mutations that disrupt Upf1 intramolecular interaction cause loss of Upf1 function. These mutations weaken Upf2 interaction and, surprisingly, promote Upf1 self-association. Third, the genetic defects resulting from deficiency in Upf1 intramolecular interaction or RNA binding are suppressed by expression of Upf2. Collectively, these data reveal a set of sequential molecular interactions and their roles in regulating Upf1 function during activation of NMD and suggest that cis intramolecular interaction and trans self-association may be general mechanisms for regulation of RNA helicase functions.
INTRODUCTION
Eukaryotic cells have evolved multiple quality control mechanisms to ensure the fidelity of gene expression (1–3). One of these mechanisms, nonsense-mediated mRNA decay (NMD), which operates during mRNA translation, targets transcripts containing a premature termination codon (PTC) (4). This mRNA decay pathway ensures rapid degradation of PTC-containing transcripts and thus prevents the cell from accumulating truncated and potentially deleterious polypeptides (5, 6). NMD also targets a subset of functionally relevant wild-type mRNAs (7–9), suggesting that this decay pathway has a substantial role in posttranscriptional gene regulation and likely controls important cellular functions.
From yeast to humans, NMD requires a set of conserved regulatory factors, the Upf proteins: Upf1, Upf2, and Upf3 (4, 7). These factors interact with each other and appear to constitute the core NMD machinery in eukaryotic cells (10–13). Deletion or silencing of each of the genes encoding these factors selectively stabilizes PTC-containing transcripts and other NMD substrates (9, 11, 13–15). In multicellular organisms, NMD also requires additional regulatory factors, including Smg1 and Smg5 to Smg7 (4, 7). These factors control Upf1 phosphorylation and dephosphorylation, a cycle that, in turn, controls several important Upf1 functions during NMD, including translation repression (16), remodeling of terminating messenger ribonucleoprotein particles (mRNPs) (17), and recruitment of the decay enzymes (18, 19).
In addition to their roles in promoting NMD, yeast Upf1, Upf2, and Upf3 also control the fidelity of translation termination, as deletion of these factors causes nonsense suppression (i.e., translational readthrough of stop codons) of several yeast alleles (20–24). The nonsense suppression phenotype of upf mutants was originally thought to reflect a direct role of the Upf factors in translation termination. However, this interpretation was challenged by the results obtained from a recent genetic screen which sought to identify mutations that reverse the readthrough phenotype in upf1Δ cells (25). This study indicated that nonsense suppression in yeast upf mutants was caused at least in part by increased intracellular levels of Mg2+ occurring as an indirect consequence of stabilizing the ALR1 mRNA, an NMD substrate that codes for the yeast principal Mg2+ transporter (25).
Upf1 is the central regulator of the NMD pathway (4). This protein is a superfamily I RNA helicase and contains a cysteine- and histidine-rich (CH) region at its N terminus and a helicase region toward its C terminus (26–28). Structural analysis reveals that these Upf1 regions form two major modular domains: the CH domain and the RNA helicase domain (29, 30). The CH domain contains two zinc knuckle modules that are similar to the ring- and U-box domains of ubiquitin ligases (31). The RNA helicase domain consists of four subdomains, two core helicase domains, RecA1 and RecA2, formed mainly by conserved helicase sequences, and two regulatory domains, 1B and 1C, formed by additional sequences inserted into the RecA1 subdomain (29, 30, 32). In vitro, yeast and human Upf1 bind both ATP and RNA and exhibit RNA-dependent ATPase and 5′-to-3′ RNA helicase activities (33, 34). Upf1's ATPase and helicase activities are essential for NMD, as mutations that eliminate ATP binding or hydrolysis abolish Upf1 function in NMD (35). Current evidence suggests that Upf1's ATPase and helicase activities are required for the final steps of NMD and are most likely involved in disassembling a terminating mRNP to recycle components of the translation and NMD machineries (36, 37).
Upf1 function in NMD is most likely controlled through its interacting factors. Consistent with their role in NMD, yeast and human Upf1 show direct interaction with the core NMD factor Upf2 (12, 38). This interaction is mediated through the CH domain of Upf1 and the C terminus of Upf2 and is essential for activation of NMD (11, 30, 31, 39). Yeast and human Upf1 also exhibit physical interaction with the eukaryotic translation termination release factors eRF1 and eRF3 (40, 41) and the Dcp1/Dcp2 decapping enzyme (38, 42, 43). The precise roles of these molecular interactions in Upf1 regulation during NMD have just begun to be elucidated.
Recent structural analysis reveals that the Upf1 CH domain can also engage in an intramolecular interaction with its RNA helicase domain (29). This intramolecular interaction appears to promote more extensive Upf1 binding to RNA and to inhibit Upf1's ATPase and helicase activities (10, 29). Upf2 binding to the Upf1 CH domain weakens Upf1 binding to RNA, stimulates Upf1's ATPase and helicase activities, and triggers a dramatic conformational change of the CH domain relative to the RNA helicase domain (10, 29, 30). Based on these observations, a mechanistic model for Upf1 activation during NMD was proposed (29). In this model, binding of Upf2 to the CH domain of Upf1 is thought to disrupt Upf1 intramolecular interaction and weaken Upf1 binding to RNA, thereby triggering Upf1's ATPase and helicase activities and switching Upf1 from an RNA-clamping mode to an unwinding mode.
While this mechanistic model elegantly explains some biochemical observations described above, its relevance to Upf1 in vivo regulation was not tested. Further, it is important to note that the biochemical and structural studies on which the model is based have used truncated fragments of Upf1 and Upf2 (10, 29–32, 44). These truncated Upf1 and Upf2 fragments largely lack amino acid residues that are essential for NMD in vivo (20, 39). In addition, this model also appears to contradict other biochemical observations. For example, using the same truncated Upf1 fragment but a smaller Upf2 fragment, binding of Upf2 to the CH domain was shown to have little or no effect on Upf1's ATPase and helicase activities (10, 30).
In this study, we have further investigated the potential intra- and intermolecular interactions of yeast Upf1 in vivo. Combining the two-hybrid system with genetic analyses, we show that the Upf1 CH and RNA helicase domains can engage in two new types of molecular interactions, an intramolecular interaction between these domains and self-association by each of these domains. These molecular interactions exhibit a mutually exclusive relationship and control several important Upf1 functions during premature translation termination and NMD. Contrary to the prevailing model, our genetic data suggest that intramolecular interactions between the Upf1 CH and RNA helicase domains promote Upf2's binding to Upf1 and that Upf2 activates Upf1 function in NMD most likely by stabilizing, not destabilizing, Upf1 binding to its target RNA.
MATERIALS AND METHODS
Yeast strains.
Saccharomyces cerevisiae GGY1::171 (his3 leu2 URA3::GAL1-lacZ gal4Δ gal80Δ) was used for two-hybrid assays. Strains HFY114 (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 UPF3), HFY871 (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 upf1::HIS3 NMD2 UPF3), and HFY466 (MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 upf1::URA3 nmd2::HIS3 UPF3) were used to assay the functions of different upf1 alleles in NMD.
Plasmids.
The yeast vectors used in this study included the following: (i) pMA424, (ii) pACTII* (11), (iii) YEplac112, and (iv) pYX142, a low-copy-number yeast expression vector that contains the LEU2 gene and TPI1 promoter-driven expression cassette. The previously constructed plasmids included pMA424-UPF1 (38), pACTII*-UPF1(1-289), pACTII*-UPF1(290-971), pRS314-UPF1, and pACTII-NMD2 (11).
GAL4(DB) fusion plasmids carrying different full-length or truncated UPF1 alleles were all constructed in the same way. In each case, a DNA fragment was amplified using a pair of primers containing an EcoRI site in the forward primer and a SalI site in the reverse primer. The DNA fragment was digested with EcoRI and SalI and ligated into pMA424 digested previously with EcoRI and SalI. Plasmids (pMA424) containing the full-length C62Y, C84S, K436E, DE572AA, or RR793AA mutant UPF1 alleles and truncated UPF1 fragments were constructed for the experiments. The truncated UPF1 fragments were 1-289 (encoding amino acids 1 to 289 of Upf1), 1-289(C62Y) (encoding amino acids 1 to 289 of Upf1 but with the C62Y change), 1-289(C84S), 1-420, 1-555, 1-666, 290-971, 290-971(K436E), 290-971(DE572AA), 290-971(RR793AA), 421-971, 556-971, 667-971, 1-207, 1-181, 1-153, 62-289, 62-207, 62-181, 80-289, 780-971, 780-914, and 780-868.
GAL4(AD) fusion plasmids carrying different full-length or truncated UPF1 alleles fused to the activation domain of GAL4 [GAL4(AD)] were all constructed in the same way. In each case, an EcoRI-SalI DNA fragment was isolated from the corresponding GAL4(DB) (DNA-binding domain of GAL4) fusion plasmid and ligated into pACTII* digested previously with EcoRI-XhoI. Plasmids (pACTII*) containing the full-length wild-type UPF1 fragment, the C62Y, C84S, K436E, DE572AA, or RR793AA UPF1 alleles, and the truncated 1-289(C62Y),1-289(C84S), 1-420, 1-555, 1-666, 290-971(K436E), 290-971(DE572AA), 290-971(RR793AA), 421-971, 556-971, 667-971, 780-971, 290-789, 1-181, 1-153, 62-181, 780-971, 780-914, and 780-868 UPF1 fragments were constructed for the experiments.
Plasmids carrying different full-length or truncated UPF1 alleles for functional analyses were constructed in pYX142 or YEplac112. Plasmids carrying the N-UPF1-[1-289] fragment or the full-length wild-type, C62Y, C84S, K436E, DE572AA, RR793AA, C62Y/K436E, C62Y/DE572AA, C62Y/RR793AA, C84S/K436E, C84S/DE572AA, and C84S/RR793AA alleles of UPF1 were constructed in the same way. In each case, an EcoRI-SalI DNA fragment was isolated from the corresponding GAL4(AD) fusion plasmid and ligated into pYX142 digested previously by EcoRI and SalI. The plasmid carrying the sequence for the C-terminally FLAG-tagged Upf1-[1-289] fragment was constructed in two steps. A 570-bp DNA fragment was amplified using a pair of oligonucleotides, N-UPF1-FLAG-r (CATAGATCTCTCGACTTACTTGTCATCGTCGTCCTTGTAATCGTTAGATTCGAAAGTAG) and UPF1-TH5′-3 (CCGGAATTCGATACCGTTTTGGAATGTTATAAC), and ligated into the TOPO TA cloning vector (Invitrogen). A 183-bp BglII DNA fragment was then isolated from the resulting plasmid and ligated into pYX142-UPF1-[1-289] digested by BglII and treated with calf intestinal alkaline phosphatase. The plasmid carrying sequence for the C-Upf1-[290-971] fragment was constructed through a three-way ligation. A 0.6-kb XbaI-EcoRI fragment containing the ADH1 promoter and a 2.0-kb EcoRI-SalI UPF1 fragment from pACTII*-UPF1(290-971) were ligated into YEplac112 digested previously by XbaI and SalI. The plasmid carrying the C-HA-UPF1-[290-971] fragment was constructed in the same way but used a 0.7-kb XbaI-EcoRI fragment containing the ADH1 promoter, initiator ATG, and DNA coding sequences of the triple hemagglutinin (HA) epitope.
Yeast two-hybrid system.
The two-hybrid tester strain GGY1::171 was used to assay Upf1 intramolecular interactions, self-association, and interaction with Upf2. In each case, a GAL4(DB) fusion and a GAL4(AD) fusion were cotransformed into the tester strain. Transformants were incubated for 3 to 5 days at 30°C on selective medium. Qualitative assays (see Fig. 1A and B) and quantitative assays (see Fig. 3 and 4) for β-galactosidase activity were performed as described previously (39).
Fig 1.
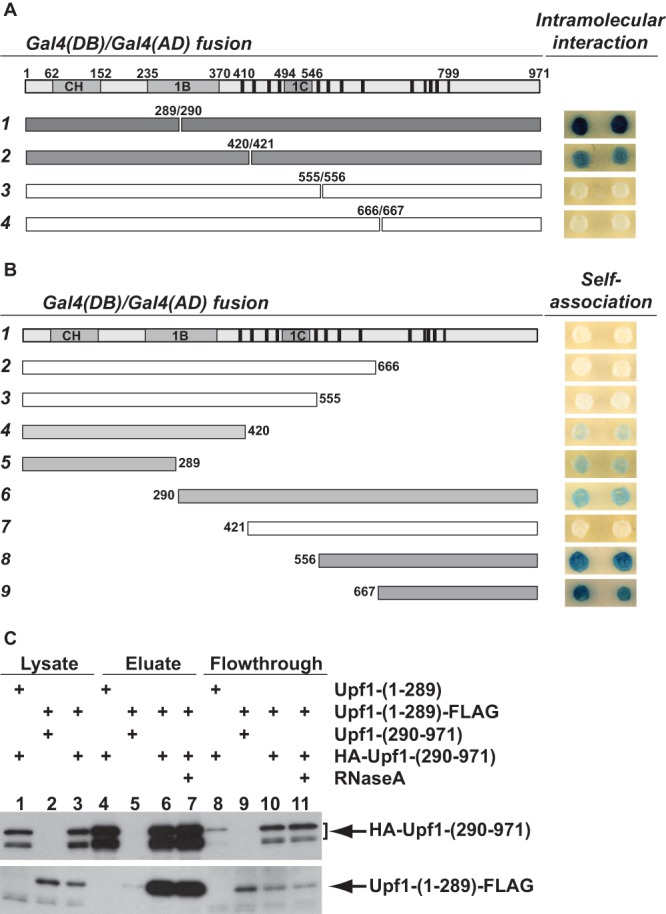
Upf1 CH domain and RNA helicase domain can engage in both intramolecular interaction and self-association. (A) Heterotypic two-hybrid interactions between the Upf1 CH domain and RNA helicase domain. DNA fragments encoding N-terminal Upf1 segments of increasing size were fused to GAL4(DB), and the complementary set of DNA fragments encoding the corresponding C-terminal Upf1 segments were fused to GAL4(AD). The interactions between each pair of Upf1 N- and C-terminal segments were assayed in the tester strain GGY1::171. Individual transformants were selected, and qualitative β-galactosidase activity was determined on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-containing plates. A dark blue colony color indicates a strong interaction, and a white colony color indicates no interaction. (B) Homotypic two-hybrid interactions of the Upf1 CH domain and RNA helicase domain. Each UPF1 DNA fragment was separately fused to GAL4(DB) and GAL4(AD). Self-association of each Upf1 segment was assayed in the two-hybrid system as described above for panel A. In both panels A and B, a schematic representation of Upf1 structural features and DNA fragments used in the two-hybrid analysis is shown on the left side of the figure. At the top of panels A and B, the CH, 1B, and 1C domains of Upf1 are indicated by gray boxes, and the 13 motifs conserved in the Upf1 RNA helicase group are indicated by black bars. In the numbered rows, gray shading within the rectangles depicting the respective fragments indicates positive interaction, and white rectangles indicate no interaction. (C) Upf1 CH and RNA helicase domains coprecipitate from cell lysates. Cell lysates were prepared from yeast strains expressing different combinations of tagged or untagged Upf1-[1-289] and Upf1-[290-971] fragments. Agarose beads conjugated with anti-HA antibodies were used to pull down the HA-Upf1-[290-971] fragment from cell lysates, and coprecipitation of Upf1-[1-289]-FLAG was determined by Western blotting. To assess the efficiency of precipitation and coprecipitation of these Upf1 fragments, samples from the input cell lysates (1:20 dilution), the flowthrough fractions (1:20), and the eluate fractions (undiluted) were analyzed on the same gel. (Top) The blot was probed with anti-HA antibody (12CA5). Two bands for HA-Upf1-[290-971] were detected, the lower of which may result from C-terminal degradation. (Bottom) The blot was probed with anti-FLAG antibody (M2).
Fig 3.
Mapping of sequence elements involved in Upf1 intramolecular interaction and self-association. A set of deletions was generated from the UPF1 CH domain (amino acids 1 to 289) or RNA helicase domain (amino acids 290 to 971). The effects of these deletions on Upf1 intramolecular interaction and self-association were analyzed in the two-hybrid system. In each case, a GAL4(DB) fusion and a GAL4(AD) fusion were cotransformed into the tester strain GGY1::171. Individual transformants were selected, and β-galactosidase activity was determined in a liquid assay. Values (means ± standard deviations) represent the respective β-galactosidase activities (in Miller units) and were derived from at least three independent cultures of individual colonies. Values less than 0.1 indicate no interaction. The schematic Upf1 structure and the color coding for DNA fragments are the same as in Fig. 1. (A) Effects of deletions from the CH domain on interaction with the RNA helicase domain. (B) Effects of deletions from the RNA helicase domain on interaction with the CH domain. (C) Effects of deletions from either the CH domain or the RNA helicase domain on self-association by each of these domains.
Fig 4.
Effects of amino acid substitutions in Upf1 on intramolecular interaction, self-association, and interaction with Upf2. Specific amino acid substitutions were introduced into full-length Upf1 or different fragments of Upf1. The effects of these mutations on intramolecular interaction, self-association, and Upf2 interaction were analyzed in the two-hybrid system as described in the legend to Fig. 3. The schematic Upf1 structure is the same as in Fig. 1. Specific amino acid substitutions are marked by a small circle in each case. (A) Effects of Upf1 amino acid substitutions on the intramolecular interaction between the CH domain and the RNA helicase domain. (B) Effects of Upf1 amino acid substitutions on self-association of full-length Upf1 and interaction with Upf2. (C) Effects of Upf1 amino acid substitutions on self-association of the isolated Upf1 CH or RNA helicase domains.
Protein analysis.
Yeast whole-cell extracts were prepared as previously described (45). Immunoprecipitation of epitope-tagged Upf1 fragments was performed using the ProFound HA tag IP/Co-IP (IP/Co-IP stands for immunoprecipitation or coimmunoprecipitation) kit (catalog no. 23610; Pierce). HA-tagged protein was eluted from the agarose beads with 2× nonreducing sample buffer in the presence of 1% β-mercaptoethanol. The cell lysate and flowthrough samples were diluted with 2× nonreducing sample buffer at the ratio of 1:20 prior to loading, and 12-μl samples were loaded per lane on a 10% SDS-polyacrylamide gel. After electrophoresis, gels were blotted onto Immobilon-P transfer membrane (Millipore) using a semidry transfer apparatus (Bio-Rad). Blots were probed with monoclonal antibodies against either the FLAG epitope (M2; Sigma) or the HA epitope (12CA5; Roche). Polyclonal antibodies against Upf1 from rabbit were used to assess the levels of Upf1 protein expression in different strains. Proteins were detected using enhanced chemiluminescence (ECL) Western blotting detection reagents (GE Healthcare).
Functional analysis of UPF1.
Plasmids harboring individual UPF1 alleles were introduced into different yeast strains. The functions of each UPF1 allele in promoting NMD and in preventing nonsense suppression were determined by analyzing the steady-state levels of nonsense-containing transcripts (CYH2 pre-mRNA and can1-100 mRNA) and can1-100 suppression, respectively. Total RNA isolation and Northern blotting analysis were performed as previously described (38). Random primed DNA probes made from the 0.6-kb EcoRI-HindIII CYH2 fragment, the 0.6-kb NdeI-EcoRI CAN1 fragment, or the 0.5-kb EcoRI-EcoRI SCR1 fragment were used to detect the CYH2 pre-mRNA, can1-100 mRNA, and SCR1 RNA, respectively. The can1-100 suppression assay was carried out as previously described (23).
RESULTS
Two-hybrid assays reveal Upf1 intramolecular and self-association interactions.
Yeast Upf1 binds RNA and exhibits nucleic acid-dependent ATPase and helicase activities in vitro (33). To understand the regulation of these Upf1 activities, we utilized the yeast two-hybrid system (46) and evaluated potential intramolecular interactions between the protein's CH and RNA helicase domains. Four different DNA fragments encoding N-terminal Upf1 segments of increasing size were fused to the GAL4 DNA-binding domain (DB) and a complementary set of DNA fragments encoding the corresponding C-terminal Upf1 segments were fused to the GAL4 activation domain (AD). The interactions between each pair of Upf1 N- and C-terminal segments were then tested in the two-hybrid system. As shown in Fig. 1A, strong interaction was observed between the Upf1 N- and C-terminal segments demarcated at amino acids 289 and 290 (construct 1), fairly strong interaction was observed between the Upf1 segments demarcated at amino acids 420 and 421 (construct 2), but no interaction was observed between the Upf1 segments demarcated at amino acids 555 and 556 or 666 and 667 (constructs 3 and 4). Since full-length wild-type Upf1 showed no detectable self-association (Fig. 1B, construct 1), the observed interactions between the Upf1 N- and C-terminal segments most likely reflect intramolecular interactions between the CH and RNA helicase domains of the same Upf1 molecule, not intermolecular interactions between two different full-length Upf1 molecules.
To assess whether Upf1 may contain latent dimerization motifs, we also used the two-hybrid system to test for self-association of each of the Upf1 segments depicted in Fig. 1A. Fairly strong interactions were detected for the Upf1 N-terminal segments 1-289 and 1-420, and the Upf1 C-terminal segments 290-971, 556-971, and 667-971 (Fig. 1B). These results indicate that although full-length Upf1 does not self-associate, fragments of the protein containing the CH or RNA helicase domain can self-associate, i.e., engage in intermolecular interactions.
Upf1, Upf2, and Upf3 are capable of forming a complex in vivo (11). To rule out the possibility that the observed Upf1 intra- and intermolecular interactions are bridged by the other Upf factors, we also assessed each of these interactions in yeast tester strains containing deletions of the UPF1, UPF2, and UPF3 genes. All the interactions observed in the wild-type tester strain still occurred in these deletion strains (data not shown). These data indicate that the observed intramolecular interactions between the Upf1 CH and RNA helicase domains, and the self-association of each of these domains, are likely to be direct and not bridged by Upf1, Upf2, or Upf3. The possibility that the observed Upf1 intra- and intermolecular interactions are bridged by other Upf1-interacting proteins, such as the translation termination factors Sup35 and Sup45 or the decapping enzyme subunits Dcp1 and Dcp2, could not be ruled out because each of these factors is encoded by an essential gene that cannot be deleted from two-hybrid tester strains.
To validate the observed two-hybrid interaction between the Upf1 CH and RNA helicase domains by an independent method, we tested whether Upf1-[1-289] and Upf1-[290-971] fragments could coimmunoprecipitate. Plasmids encoding the C-terminally FLAG-tagged Upf1-[1-289] and the N-terminally HA-tagged Upf1-[290-971] fragments were introduced into a upf1Δ strain. Cell lysates were prepared from the resulting strain, and agarose beads conjugated with anti-HA antibodies were utilized to pull down the HA-Upf1-[290-971] fragment. Cell lysates from the upf1Δ strain expressing untagged Upf1-[290-971] and Upf1-[1-289]-FLAG serve as specificity controls. As demonstrated by Western blotting, anti-HA beads efficiently precipitated Upf1-[1-289]-FLAG from the cell lysate containing HA-Upf1-[290-971] (Fig. 1C, lane 6) but not from the cell lysate containing untagged Upf1-[290-971] (Fig. 1C, lane 5). Upf1-[1-289]-FLAG also coimmunoprecipitated with HA-Upf1-[290-971] from the cell lysate treated with RNase A (Fig. 1C, lane 7). These data validate our two-hybrid results and further demonstrate that the observed interaction between the Upf1 CH and RNA helicase domains is not bridged by RNA.
Coexpression of Upf1 N- and C-terminal fragments reconstitutes the function of the native protein in NMD and translation termination.
To investigate the functional significance of the Upf1 intramolecular interactions detected in the experiments of Fig. 1A, we tested whether coexpression of the N-Upf1-[1-289] and C-Upf1-[290-971] fragments could reconstitute Upf1 function in NMD. DNA fragments encoding N-Upf1-[1-289] or C-Upf1-[290-971] (Fig. 2A) were cloned into yeast expression vectors, and the resulting plasmids were introduced into upf1Δ cells either individually or in combination. The empty vectors and a plasmid carrying the wild-type UPF1 gene were included as controls. Northern blotting analyses showed that the mRNAs encoding N-Upf1-[1-289] and C-Upf1-[290-971] fragments were expressed (data not shown) and that the steady-state level of the CYH2 pre-mRNA, an endogenous substrate of the NMD pathway (6, 9), was very high in upf1Δ cells (Fig. 2B, lane 1) and barely detectable in cells expressing full-length UPF1 (Fig. 2B, lane 5). Expression of either N-Upf1-[1-289] or C-Upf1-[290-971] alone did not affect the level of the CYH2 pre-mRNA in upf1Δ cells (Fig. 2B, lanes 2 and 3). In contrast, coexpression of both N-Upf1-[1-289] and C-Upf1-[290-971] markedly reduced the abundance of this nonsense-containing transcript to a level only slightly higher than that in wild-type UPF1 cells (Fig. 2C, compare lanes 4 and 5). These results show that coexpression of the CH and RNA helicase domains reconstitutes Upf1's NMD function in trans.
Fig 2.
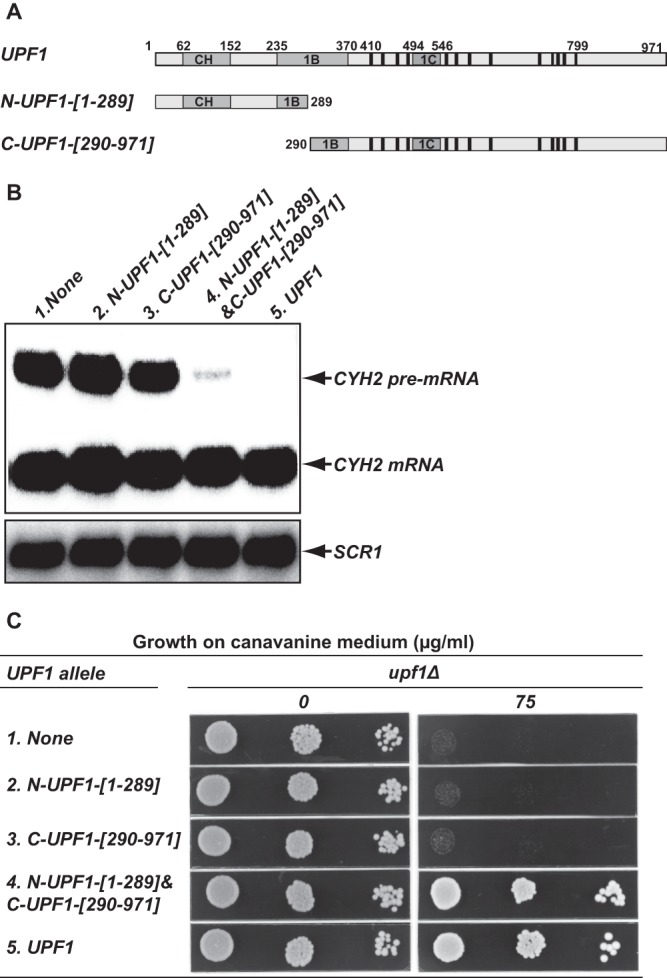
Coexpression of the N-terminal CH domain and the C-terminal RNA helicase domain reconstitutes Upf1 function. The yeast upf1Δ strain (HFY871) was transformed with plasmids harboring the indicated UPF1 alleles, and the resulting strains were analyzed for NMD activity and the ability to prevent nonsense suppression. (A) Schematic representation of the N- and C-terminal Upf1 fragments used in this experiment. (B) Coexpression of the CH domain and the RNA helicase domain reconstitutes Upf1 function in promoting NMD. Total RNA was isolated from each strain, and the steady-state level of the CYH2 pre-mRNA in each strain was analyzed by Northern blotting, using a random-primed probe specific for CYH2. (C) Coexpression of the CH domain and the RNA helicase domain reconstitutes Upf1 function in preventing nonsense suppression. Cells were grown in selective liquid medium to mid-log phase. Aliquots (10 μl) of serial dilutions from each yeast strain were spotted on plates containing synthetic complete medium (SC) without arginine and with no (0) or 75 μg/ml canavanine and grown at 30°C for 2 days. No growth on canavanine-containing plates indicates suppression of the can1-100 allele, and growth on these plates indicates a lack of nonsense suppression.
In addition to its role in NMD, Upf1 also controls the fidelity of translation termination, a function manifested by the promotion of nonsense suppression in upf1Δ cells (20, 22, 23). The nonsense suppression phenotype in upf1Δ cells was originally thought to reflect a direct role of Upf1 in controlling the efficiency of translation termination (20, 22, 23), but it was recently shown to at least be partially attributable to an indirect consequence of NMD regulation of ALR1 mRNA expression, and thus intracellular Mg2+ levels (25). To assess whether coexpression of the N-Upf1-[1-289] and C-Upf1-[290-971] fragments could reconstitute Upf1 function in preventing nonsense suppression, we analyzed the effects of expression of N-Upf1-[1-289], C-Upf1-[290-971], or both on suppression of the can1-100 nonsense allele. CAN1 encodes a yeast arginine permease that is also capable of transporting the toxic arginine analog canavanine into cells (23). Suppression of the can1-100 nonsense allele in upf1Δ cells leads to the production of a functional arginine permease that, in turn, renders cells sensitive to canavanine in the growth medium (23). As shown in Fig. 2C, wild-type UPF1 cells are resistant to 75 μg/ml canavanine (row 5), but upf1Δ cells are sensitive to this concentration of the drug (row 1). Sensitivity of the upf1Δ cells to canavanine was not affected by the expression of either N-Upf1-[1-289] or C-Upf1-[290-971] (rows 2 and 3), but coexpression of both these Upf1 fragments in upf1Δ cells promoted canavanine resistance (row 4). All cells grew equally well on medium containing no canavanine (Fig. 2C, left panel). These results demonstrate that coexpression of the CH and RNA helicase domains also reconstitutes Upf1 activity in preventing nonsense suppression, a function that must include reconstitution of NMD for the ALR1 mRNA (25).
Mapping sequence elements involved in Upf1 intramolecular interaction and self-association.
To delineate sequence elements involved in Upf1 intramolecular interaction and self-association, we generated deletions from the N-Upf1-[1-289] and C-Upf1-[290-971] fragments and analyzed the effects of these deletions on the types of two-hybrid interactions originally assessed in the experiments shown in Fig. 1A and B. For intramolecular interactions, we evaluated the ability of a panel of truncated Upf1-[1-289] or Upf1-[290-971] fragments to interact with nontruncated versions of the “complementary” fragment (Fig. 3A and B). Our analysis revealed that intramolecular interaction between the Upf1 CH and helicase domains is mediated through at least two regions, including one region from the CH domain, amino acids 62 to 289 (Fig. 3A), and a large region of the helicase domain that could not sustain substantive deletions from either its N or C terminus (Fig. 3B). The region spanning amino acids 62 to 289 includes the three zinc knuckle modules of the CH domain and the N-terminal part of regulatory domain 1B (29). Consistent with these two-hybrid results, recent structural analyses revealed that amino acid residues from the regions from amino acids 62 to 289 and amino acids 290 to 421 of yeast Upf1 make multiple contacts in the crystals (29). However, the Upf1 fragment used for structural analysis only covered the region from amino acids 54 to 851 (29) and lacks most residues from the 789-971 region that are critical for Upf1 intramolecular interaction in our two-hybrid experiments.
Our analysis revealed that efficient self-association of the isolated CH domain is largely dependent on Upf1 amino acids 153 to 289, located immediately downstream of the three zinc knuckle modules (Fig. 3C) (29). Efficient self-association of the isolated RNA helicase domain requires Upf1 amino acids 868 to 971 (Fig. 3C). This region is located downstream of the RNA helicase motif VI in the RecA2 domain of Upf1. Notably, the published biochemical and structural experiments carried out thus far all utilized Upf1 fragments lacking this important region critical for Upf1 self-association (see introduction).
Upf1 intramolecular interaction and self-association are mutually exclusive events.
The trans complementation observed between Upf1 fragments encompassing the CH and RNA helicase domains (Fig. 2B and C) indicates that both domains are essential for Upf1 function and that, while structurally separable, they interact physically. To delineate the role of this intramolecular interaction in regulating Upf1 activities, we sought to identify specific mutations that disrupt it. We analyzed five previously characterized upf1 loss-of-function alleles in our two-hybrid assay to determine whether these alleles may code for mutant proteins that are deficient in intramolecular interaction. The C62Y and C84S alleles contain mutations in the CH domain (20, 47), whereas the K436E, DE572AA, and RR793AA alleles, respectively, contain mutations in the ATP-binding, ATP hydrolysis, and RNA-binding motifs of the RNA helicase domain (35). The mutant proteins encoded by the latter three alleles are completely defective in the corresponding biochemical activities (35). Each of these five mutations had no effect on Upf1 expression in vivo (20, 35). As shown in Fig. 4A, intramolecular interaction between the CH domain and the RNA helicase domain was decreased about 3-fold by the C62Y and C84S mutations and more than 20-fold by the K436E and DE572AA mutations. The RR793AA mutation affected this interaction only modestly.
Since the Upf1 CH and RNA helicase domains are capable of both intra- and intermolecular interactions, we considered the possibility that these two types of interactions are mutually exclusive and that mutations that weaken one promote the other. As shown in Fig. 4B, the C62Y, C84S, K436E, and DE572AA mutant proteins, which were partially or completely defective in intramolecular interaction (Fig. 4A), all demonstrated detectable self-association, i.e., intermolecular interaction (Fig. 4B). In contrast, the RR793AA mutant protein, which had only a modest defect in intramolecular interaction, did not self-associate (Fig. 4B). These data support our interpretation of the intramolecular interaction assays and provide further evidence that the C62Y, C84S, K436E, and DE572AA mutations impair intramolecular interactions within Upf1. Since the C62Y, C84S, K436E, and DE572AA mutations abolish self-association of the isolated CH or RNA helicase domain (Fig. 4C), the self-association that we observed for full-length C62Y, C84S, K436E, and DE572AA mutant proteins is most likely mediated by a single functional dimerization motif still present on these mutant proteins. The failure of the wild-type protein to self-associate (Fig. 4B) indicates that intramolecular interactions are likely to predominate in full-length Upf1.
Mutations weakening Upf1 intramolecular interaction also negatively affect interaction with Upf2.
The activity of the NMD pathway is dependent on an interaction between Upf1 and Upf2 (39). To better understand the functional role of the Upf1-Upf2 interaction and its possible relationship to Upf1 intramolecular interaction, we sought to identify upf1 alleles defective in Upf2 interaction. Two-hybrid analyses showed that Upf1-Upf2 interactions were unaffected by the RR793AA mutation, partially impaired by the C84S, K436E, and DE572AA mutations, and substantially impaired by the C62Y mutation (Fig. 4B). Since the Upf2-interacting domain of Upf1 was previously mapped to the CH domain (11), it was surprising that mutations within both the CH domain (C62Y and C84S) and the RNA helicase domain (K436E and DE572AA) resulted in partially defective interaction with Upf2. One possible explanation for this result is that efficient Upf1-Upf2 interaction requires a prior Upf1 intramolecular interaction and that these mutations all cause a general defect in Upf1 intramolecular interaction. Consistent with this idea, the C62Y, C84S, K436E, and DE572AA mutations all impaired Upf1 intramolecular interaction (Fig. 4A).
The C62Y and C84S mutations map to a region in the CH domain that is required for binding to the RNA helicase domain (Fig. 3A). Further, structural analysis of yeast Upf1 revealed that the C62 and C84 residues bind to a common Zn2+ ion and form part of a two zinc knuckle module that makes direct contacts with the stalk region on the surface of the RecA1 domain (29). Thus, these two residues probably make a direct contribution to Upf1 intramolecular interaction. In contrast, the K436E and DE572AA mutations map outside the two regions in the RNA helicase domain that are required for binding to the CH domain (see above). In RNA helicases, the conserved K436 and DE572 residues are located near the catalytic center and are mainly involved in ATP binding and hydrolysis (48). Hence, it is unlikely that these two residues make a direct contribution to Upf1 intramolecular interaction and their apparent impaired intramolecular interactions most likely result from indirect effects of these mutations on the conformation of the RNA helicase domain.
ATP-mediated conformational changes in Upf1 are likely to regulate many aspects of its molecular interaction and function. As demonstrated in our two-hybrid experiments, the K436E mutation previously shown to impair ATP binding drastically impaired intramolecular interaction between the CH domain and the RNA helicase domain (Fig. 4A) and self-association of the isolated helicase domain (Fig. 4C) and substantially inhibited the interaction with Upf2 (Fig. 4B). Furthermore, although the C62Y and C84S mutations impaired Upf1 intramolecular interaction to similar extents (Fig. 4A), they affected Upf2 interaction differentially, with the C62Y mutation having a much more sizeable reduction in Upf2 interaction (Fig. 4B). These results indicate that the C62Y mutation specifically impairs not only intramolecular interaction and self-association but also Upf1-Upf2 interaction.
Distinct functional roles of Upf1 and Upf2 in promoting NMD and controlling translation termination.
To further elucidate the regulatory roles of Upf1 intra- and intermolecular interactions, we examined genetic interactions between UPF2 and the different upf1 alleles described above. In these experiments, single-copy plasmids carrying upf1 alleles expressed from the strong TPI1 promoter were individually introduced into upf1Δ UPF2 or upf1Δ upf2Δ cells. The function of each upf1 allele in promoting NMD and in preventing nonsense suppression in these cells was analyzed by measuring the steady-state levels of the nonsense-containing CYH2 pre-mRNA and can1-100 mRNA and by assessing can1-100 nonsense suppression, respectively.
Functional analysis of the wild-type, K436E, and DE572AA alleles revealed differential abilities of Upf1 to control the efficiency of translation termination and promote NMD. As shown in Fig. 5, expression of the wild-type UPF1 gene in upf1Δ UPF2 cells promoted NMD (i.e., led to reductions of the CYH2 pre-mRNA and can1-100 mRNA) and prevented nonsense suppression (i.e., allowed full growth on canavanine-containing medium), as would be expected. In upf1Δ upf2Δ cells, however, expression of wild-type UPF1 did not promote NMD but still prevented nonsense suppression. The latter results are consistent with earlier observations (23, 49) and suggest that Upf1's ability to promote NMD absolutely requires Upf2, whereas its role in preventing nonsense suppression can compensate for the absence of Upf2. In addition, although the K436E and DE572AA alleles were completely defective in NMD in upf1Δ UPF2 cells, these alleles prevented nonsense suppression as effectively as wild-type UPF1 in upf1Δ upf2Δ cells (Fig. 5). This result shows that Upf1's roles in promoting NMD and in preventing nonsense suppression also have different requirements for its ATP binding and hydrolysis activities. Its function in NMD requires both ATP binding and hydrolysis, whereas its role in preventing nonsense suppression can be independent of these activities. Although these observations suggest that Upf1 function in preventing nonsense suppression, i.e., controlling the efficiency of translation termination, can be completely separated from and be independent of its NMD-promoting activity, at least one alternative explanation must be considered. Our recent experiments demonstrated that NMD control of the ALR1 mRNA, a transcript encoding yeast's principal Mg2+ transporter, substantially influenced the degree of nonsense suppression (25). Since the ability of cells to grow well on canavanine-containing medium could be attributable to reductions in the abundance of the ALR1 mRNA (25), it was important to assess the levels of this mRNA in each of the strains under consideration. Figure 5A and Table 1 show that the abundance of the ALR1 mRNA is reduced by the UPF1 gene, but not by the K436E and DE572AA alleles, in upf1Δ UPF2 cells and that the abundance of the mRNA is essentially unchanged by the UPF1 gene or the K436E and DE572AA alleles in upf1Δ upf2Δ cells. These results indicate that the observed nonsense suppression phenotypes are unlikely to be solely attributable to the inactivation of NMD and imply that Upf1 has a direct role in controlling translation termination. The data also suggest that these two functions of Upf1 must be ordered and that its function in controlling translation termination precedes the function in promoting NMD.
Fig 5.
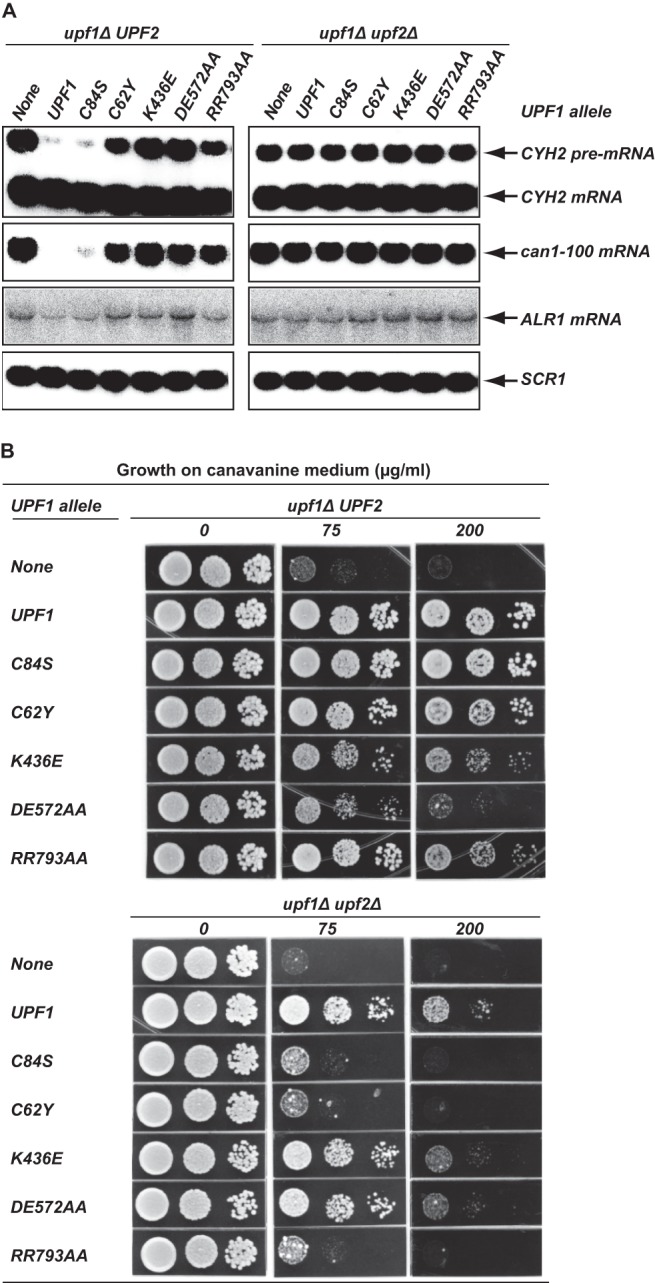
Genetic interaction between UPF2 and upf1 alleles impaired in different Upf1 activities. Plasmids (pYX142) carrying the indicated UPF1 alleles were individually transformed into the upf1Δ UPF2 strain (HFY871) or the upf1Δ upf2Δ strain (HFY466). The resulting strains were analyzed for NMD activity and the ability to prevent nonsense suppression. The amount of canavanine in the medium (0, 75, and 200 μg/ml) is shown. (A) Function of different upf1 alleles in promoting NMD in the presence or absence of UPF2. Total RNA was isolated from strains harboring different upf1 alleles. The steady-state levels of the CYH2 pre-mRNA and the can1-100 and ALR1 mRNAs were analyzed by Northern blotting, using random-primed probes specific for CYH2, CAN1, ALR1, or SCR1 transcripts with the latter serving as a loading control. (B) Function of different upf1 alleles in preventing can1-100 suppression in the presence or absence of UPF2. Strains harboring different upf1 alleles were assayed for their ability to prevent can1-100 suppression as described in the legend to Fig. 2C.
Table 1.
Relative ALR1 mRNA levels in yeast upf1Δ UPF2 or upf1Δ upf2Δ cells harboring different UPF1 allelesa
| UPF1 allele | Relative ALR1 mRNA level |
|
|---|---|---|
| upf1Δ UPF2 cells | upf1Δ upf2Δ cells | |
| None | 1.00 | 1.00 |
| UPF1 | 0.47 | 1.00 |
| C84S | 0.60 | 1.00 |
| C62Y | 0.94 | 1.12 |
| K436E | 1.02 | 1.10 |
| DE572AA | 1.16 | 1.10 |
| RR793AA | 0.83 | 1.10 |
Data were taken from the Northern blots shown in Fig. 5A. ALR1 mRNA signals in each of these strains were first adjusted based on the SCR1 signals. The adjusted ALR1 mRNA signals in upf1Δ UPF2 or upf1Δ upf2Δ cells harboring different UPF1 alleles were then compared to those of the corresponding cells harboring the empty vector.
Similar analyses of the C84S, C62Y, and RR793AA alleles revealed a positive role for Upf2 in regulating Upf1 function in both translation termination and NMD. As demonstrated in can1-100 suppression assays, the upf1 C84S, C62Y, and RR793AA alleles were defective in preventing nonsense suppression in upf1Δ upf2Δ cells but appeared to be almost fully functional in upf1Δ UPF2 cells (Fig. 5B). This result suggests that the presence of Upf2 can suppress the defects caused by these upf1 mutations. In addition, although the C62Y and C84S alleles impaired Upf1 intramolecular interactions to similar extents (Fig. 4A), they differed in their respective abilities to interact with Upf2, with the C62Y allele manifesting severely impaired Upf2 interaction (Fig. 4B). These two upf1 alleles also exhibited different activities in upf1Δ UPF2 cells. The C84S allele was fully functional in promoting NMD, whereas the C62Y allele was only partially functional for this activity (Fig. 5A). Since the C62Y mutation in UPF1 that specifically impairs Upf2 interaction results in the loss of Upf1 function, Upf2 must have a positive role in regulating Upf1 activities.
Upf1 intramolecular interaction is controlled through its ATPase activity.
To further elucidate the roles of intramolecular interactions in the control of Upf1 function, we investigated whether such interactions are subject to additional regulation. We introduced each of the upf1 C62Y, C84S, K436E, DE572AA, and RR793AA alleles analyzed in Fig. 5 into wild-type yeast cells and examined whether expression of any allele could exert a dominant-negative effect on NMD. Western blotting indicated that cells with each of these upf1 alleles yielded polypeptide levels comparable to those of cells with wild-type UPF1 (Fig. 6A). We found that overexpression of the C62Y and C84S alleles had no effect on the accumulation of the CYH2 pre-mRNA and can1-100 mRNA but that comparable experiments with the K436E, DE572AA, and RR793AA alleles caused 4- to 6-fold increases in the levels of both transcripts (Fig. 6B). These results indicate that upf1 alleles harboring mutations in the helicase domain but not in the CH domain can inhibit NMD in a dominant-negative manner. To determine whether the NMD inhibitory activity of the K436E, DE572AA, or RR793AA allele may result from Upf1 intramolecular interaction, we analyzed the consequence of weakening Upf1 intramolecular interaction on the NMD inhibitory activity of these alleles. We combined the C62Y or C84S mutation with the K436E, DE572AA, or RR793AA mutation and examined the inhibitory activity of the resulting double mutant alleles. These experiments showed that the C62Y or C84S mutation, when integrated within the same molecule of Upf1, eliminate the dominant-negative NMD inhibitory activity of the K436E, DE572AA, and RR793 alleles (Fig. 5B). These results indicate that the NMD inhibitory activity of K436E, DE572AA, or RR793AA allele may well be caused by Upf1 intramolecular interaction between the CH domain and the RNA helicase domain in these mutant proteins. Since the K436E, DE572AA, and RR793AA mutant proteins all lack the RNA-dependent ATPase activity, these results also suggest that the Upf1 ATPase activity is likely to be required for disrupting intramolecular interactions necessary for a switch to a new functional state of Upf1.
Fig 6.
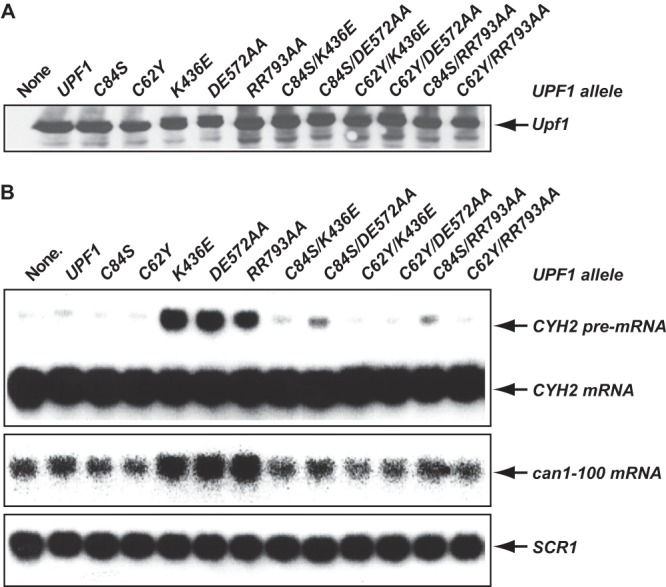
Mutations in the CH domain that weaken Upf1 intramolecular interactions eliminate the dominant-negative effects on NMD resulting from mutations in the RNA helicase domain. Specific amino acid substitutions were introduced into the CH domain or the RNA helicase domain or both domains of Upf1. The resulting upf1 alleles were cloned into pYX142 and individually transformed into the wild-type strain HFY114. Whole-cell extracts and total RNA were prepared from each of the resulting strains. (A) The levels of Upf1 protein in these strains were analyzed by Western blotting, using a polyclonal antibody against Upf1. (B) The steady-state levels of the CYH2 pre-mRNA and the can1-100 mRNA in these strains were analyzed by Northern blotting, using random-primed probes specific for CYH2, CAN1, or SCR1 transcripts as described in the legend to Fig. 5A.
DISCUSSION
New Upf1 molecular interactions.
Our two-hybrid experiments reveal that the Upf1 CH and helicase domains are capable of engaging in two distinct types of molecular interactions: intramolecular interaction between these two domains and self-association by each of these domains. These molecular interactions are crucial for Upf1 regulation, and the functional significance of these interactions was underscored by several observations. First, coexpression of the CH domain and the RNA helicase domain reconstitutes Upf1 function in promoting NMD and in preventing nonsense suppression (Fig. 2B and C). Second, mutations that disrupt Upf1 intramolecular interaction result in loss of Upf1 function (Fig. 5B). Third, these new Upf1 molecular interactions are mediated by overlapping sequence elements within the CH and RNA helicase domains and exhibit a mutually exclusive relationship (Fig. 4A and B). Finally, these molecular interactions are also largely dependent on ATP-mediated effects on the Upf1 RNA helicase domain (Fig. 4A and C).
Roles of Upf1 intramolecular interaction and interaction with Upf2.
Consistent with previous data (20, 23, 35), our genetic experiments demonstrate that Upf1 has roles in both translation termination and NMD. These two Upf1 functions appear to be distinct and separable. Upf1's function in translation termination can largely bypass Upf2 and be independent of ATP binding and hydrolysis. In contrast, its function in NMD absolutely requires Upf2 and is dependent on its ATPase activity (Fig. 5A). Given the intimate connection between translation termination and NMD (4, 50), the distinct roles revealed here for Upf1 in translation termination and NMD are unlikely to reflect two physically separated activities. Rather, they most likely represent two temporally distinct Upf1 functions. On the basis of our genetic data, we propose that Upf1's role in translation termination occurs in the early phase of Upf1 function and likely represents initial Upf1 binding to its target RNA after nonsense codon recognition. In contrast, the role of Upf1 in NMD occurs in the late phase of its function and likely represents Upf1's RNP remodeling activity involving activation of its RNA-dependent ATPase.
As revealed by our genetic analyses, Upf1 intramolecular interaction and its interaction with Upf2 both play essential roles in regulating Upf1 function (Fig. 2 and 5). Upf1 intramolecular interaction likely performs two important functions, the first of which is to promote Upf1 binding to its target RNA. This conclusion follows from a genetic inference that certain upf1 mutants impaired in intramolecular interaction are defective in RNA binding, because these mutants (upf1 C62Y and C84S) share a comparable genetic defect with the RNA-binding RR793AA mutant and they are all substantially defective in preventing nonsense suppression in the absence of Upf2 (Fig. 5B). Upf1 intramolecular interaction also appears to promote efficient Upf1 binding to Upf2. This conclusion follows from our observation that all mutations that disrupt Upf1 intramolecular interaction weaken Upf1 interaction with Upf2 (Fig. 4B). Upf1 interaction with Upf2 also promotes Upf1 binding to its target RNA. This conclusion is strongly supported by our observation that the genetic defect resulting from the deficiency of Upf1 RNA binding is suppressed by expression of Upf2. The upf1 RR793AA RNA-binding mutant is completely defective in preventing nonsense suppression in the absence of Upf2 but appears to be fully functional in its presence (Fig. 5B). Genetic suppression of the Upf1 RNA binding defect by Upf2 suggests that Upf2 can recruit or stabilize Upf1 binding to its target RNA. In addition, since the genetic defect resulting from impaired Upf1 intramolecular interaction is also suppressed by Upf2 (Fig. 5B), this indicates that Upf1 intramolecular interaction and its interaction with Upf2 function redundantly in promoting Upf1 binding to its target RNA.
Multiple intramolecular interactions between the Upf1 CH and helicase domains have also been observed or inferred from recent structural and biochemical analyses showing the following. (i) The CH domain promotes more extensive Upf1 binding to RNA (29). (ii) Elimination of the CH domain or binding of Upf2 to the CH domain decreases Upf1 binding to RNA and increases Upf1 ATPase and helicase activities (10, 29). (iii) The CH domain displays dramatically different conformations relative to the RNA helicase domain when Upf1 is in a complex with RNA or Upf2 (29, 30). These observations have led to a mechanistic model (29) proposing that intramolecular interaction between the CH and RNA helicase domains promotes extensive Upf1 binding to RNA and leads to the inhibition of the Upf1 ATPase and helicase activities. Upf2 binding to the CH domain is thought to weaken Upf1 intramolecular interaction and Upf1 binding to RNA and to lead to activation of the Upf1 ATPase and helicase activities.
While our genetic data generally agree with the proposed role for the CH domain in promoting Upf1 binding to RNA, they disagree with many of the proposed Upf1 regulatory mechanisms suggested by previous biochemical and structural experiments. First, previous biochemical experiments suggested an inhibitory role of the CH domain in regulating Upf1 ATPase and helicase activities (10, 29). In contrast, our Upf1 trans-complementation experiment and mutational analyses all revealed a positive role of the CH domain in regulating Upf1 function (Fig. 2 and 5). Second, previous structural analyses suggested that Upf1 intramolecular interaction inhibits or competes with Upf2 binding (29). In contrast, our two-hybrid data indicated that Upf1 intramolecular interaction promotes Upf2 binding (Fig. 4). Finally, previous biochemical and structural analyses suggested that Upf2 binding to the CH domain weakens Upf1 binding to RNA (10, 29). In contrast, our genetic suppression experiment indicated that Upf2 binding most likely promotes Upf1 binding to its target RNA (Fig. 5B). The discrepancies between our genetic data and previous biochemical data are surprising but may well be attributable to the use of truncated Upf1 fragments in previous biochemical and structural studies (10, 29–32, 44). These Upf1 fragments all lack the C-terminal residues critical for Upf1 intramolecular interaction (Fig. 3A) and essential for Upf1 function in NMD (20). It is possible that, in these truncated Upf1 fragments, the CH domain forms nonproductive molecular interactions with the RNA helicase domain that inhibit the biochemical activities of the helicase domain.
Potential role of Upf1 self-association.
Our two-hybrid analyses revealed that Upf1 contains specific sequence elements that promote self-association. One element maps to the N-terminal CH domain, and another element maps to the C-terminal RNA helicase domain. Our observations that these self-association elements largely overlap those involved in Upf1 intramolecular interaction and that Upf1 self-association is inhibited by its intramolecular interaction argue that Upf1 self-association likely plays an important role in regulating Upf1 function. In addition to forming a complex with Upf2 and Upf3 (10, 11), Upf1 also appears to form a separate complex with the Dcp1/Dcp2 decapping enzyme in yeast and mammalian cells (38, 42). These results raise the possibility that Upf1 self-association functions at the late stage of NMD and serves to recruit the Dcp1/Dcp2 decapping enzyme to transcripts targeted for NMD. It is possible, for example, that Upf1 is partitioned into two different pools, one of which is associated with translating ribosomes and another with the Dcp1/Dcp2 decapping enzyme. ATP hydrolysis by Upf1 may trigger the dissociation of Upf2 from the target RNA and lead to a conformational switch in Upf1 that maintains the protein's ability to bind its target RNA, but with the CH and helicase domains no longer interacting, the dimerization motifs in these domains may become accessible for self-association. This form of Upf1 could then recruit the Dcp1/Dcp2 decapping enzyme to the target RNA via the Upf1 subunit in the enzyme complex.
Our finding that Upf1 can self-associate is surprising in light of available evidence that Upf1 exists as a monomer both in solution and in crystals (29, 30, 32, 33, 51). One possible explanation for this difference is that Upf1 self-association represents a transient molecular interaction that has thus far escaped detection by conventional biochemical experiments but not by the two-hybrid methods used here. Much like Upf1, several helicases that are members of the non-ring-forming SF1 and SF2 families, including the hepatitis C virus (HCV) NS3 RNA helicase and the Rep or UvrD DNA helicases from Escherichia coli, also exist as monomers in solution or crystals. However, for each of these helicases, transient interactions between different monomers are required to form an active complex for in vitro DNA or RNA unwinding (52–55). Further, a new dimerization motif has recently been identified in the DEAD box helicase Hera from Thermus thermophilus (56). On the basis of these results and our observations, we speculate that transient self-association may be a general mechanism for regulating helicase function.
A model for Upf1 regulation.
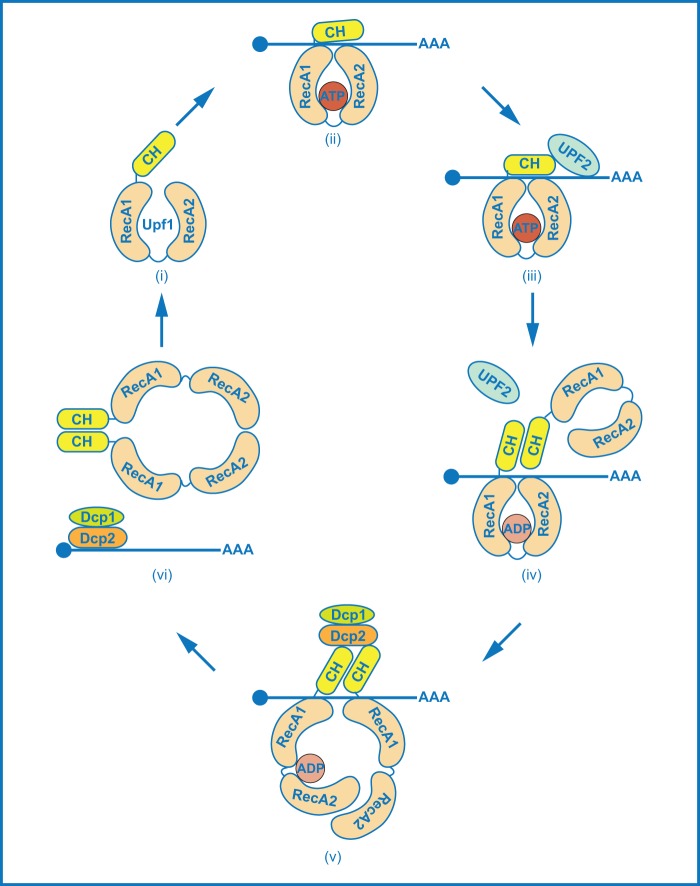
The data presented here suggest that Upf1 cycles between monomeric and dimeric forms as it executes its multiple functions, leading to the following revised model for Upf1 regulation (Fig. 7). Upf1 initially associates with translating ribosomes (57). At a premature termination codon, conformational changes in the ribosome and interactions with release factors (4) result in ATP binding to Upf1 (step i). This triggers intramolecular interaction between the CH and RNA helicase domains and promotes Upf1 binding to its target mRNA (step ii). Upf2 joins the Upf1-RNA complex and further stabilizes Upf1 binding to its target mRNA (step iii). Stable association of Upf1 to its target activates Upf1's ATPase activity to remodel the terminating RNP. ATP hydrolysis by Upf1 triggers the dissociation of Upf2 from the target mRNA and dissociation of the mRNP from ribosomes and leads to a conformational switch in Upf1 (step iv). In the new conformation, Upf1 maintains the ability to bind its target RNA, but the CH and helicase domains no longer interact and the dimerization motifs in these Upf1 domains are accessible for self-association. This form of Upf1 likely recruits the Dcp1/Dcp2 decapping enzyme to the target mRNA (step v). Upf1 dimerization triggers the release of dimeric Upf1 from its target mRNA and promotes the delivery of the target mRNA to the decapping enzyme (step vi). The resulting dimeric form of Upf1 is unstable and dissociates into its monomeric form.
Fig 7.
A model for Upf1 regulation during the activation of NMD. During premature translation termination, monomeric Upf1 initially associated with translating ribosomes (not shown in the figure) binds ATP (step i). ATP binding to Upf1 tightens intramolecular interactions between the two RecA domains and also triggers intramolecular interactions between the CH and helicase domains. These molecular interactions promote Upf1 binding to its target mRNA (step ii). Upf2 joins the mRNA complex, further stabilizes Upf1 binding to the target mRNA, and activates Upf1's ATPase activity (step iii). As a consequence of ATP hydrolysis by Upf1, Upf2 dissociates from the mRNP complex and Upf1 switches to a new conformation (step iv). In this new conformation, Upf1 still binds to its target mRNA, but the CH and helicase domains no longer interact, and the dimerization motifs in these domains are accessible. Upf1 dimerizes on the target mRNA and recruits the Dcp1/Dcp2 decapping enzyme (step v). Upf1 dimers deliver the target mRNA to the decapping enzyme and then dissociate from the transcript (step vi). The dimeric form of Upf1 is unstable, and the monomeric conformation of Upf1 is favored. Upf2, Dcp1, Dcp2, ATP, and ADP are not drawn to scale.
The new Upf1 cis and trans molecular interactions reported here are all mediated by amino acid sequences outside its two core helicase domains. Given the fact that, in addition to the conserved core helicase domains, most helicases contain variable N- and/or C-terminal extensions (48), the cis and trans regulatory mechanisms revealed here for Upf1 regulation are likely shared by other helicases.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grant R37GM27757 to A.J.
We thank Stuart Peltz for plasmids carrying the C84S, K436E, DE572AA, and RR793AA alleles of UPF1.
Footnotes
Published ahead of print 7 October 2013
REFERENCES
- 1.Isken O, Maquat LE. 2007. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21:1833–1856 [DOI] [PubMed] [Google Scholar]
- 2.Parker R. 2012. RNA degradation in Saccharomyces cerevisiae. Genetics 191:671–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoemaker CJ, Green R. 2012. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 19:594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kervestin S, Jacobson A. 2012. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13:700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pulak R, Anderson P. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7:1885–1897 [DOI] [PubMed] [Google Scholar]
- 6.He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc. Natl. Acad. Sci. U. S. A. 90:7034–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. 2010. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 67:677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehwinkel J, Raes J, Izaurralde E. 2006. Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem. Sci. 31:639–646 [DOI] [PubMed] [Google Scholar]
- 9.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12:1439–1452 [DOI] [PubMed] [Google Scholar]
- 10.Chamieh H, Ballut L, Bonneau F, Le Hir H. 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 15:85–93 [DOI] [PubMed] [Google Scholar]
- 11.He F, Brown AH, Jacobson A. 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17:1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. 2001. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol. 21:209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lykke-Andersen J, Shu MD, Steitz JA. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103:1121–1131 [DOI] [PubMed] [Google Scholar]
- 14.Page MF, Carr B, Anders KR, Grimson A, Anderson P. 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19:5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. 2003. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 22:3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. 2008. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133:314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. 2012. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 40:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai T, Cho H, Liu Z, Bowler MW, Piao S, Parker R, Kim YK, Song H. 2012. Structural basis of the PNRC2-mediated link between mRNA surveillance and decapping. Structure 20:2025–2037 [DOI] [PubMed] [Google Scholar]
- 19.Cho H, Kim KM, Kim YK. 2009. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol. Cell 33:75–86 [DOI] [PubMed] [Google Scholar]
- 20.Weng Y, Czaplinski K, Peltz SW. 1996. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol. 16:5491–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM. 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10:691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Czaplinski K, Rao Y, Peltz SW. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20:880–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maderazo AB, He F, Mangus DA, Jacobson A. 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol. 20:4591–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bidou L, Stahl G, Hatin I, Namy O, Rousset JP, Farabaugh PJ. 2000. Nonsense-mediated decay mutants do not affect programmed −1 frameshifting. RNA 6:952–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson MJ, Jacobson A. 2010. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 24:1491–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leeds P, Wood JM, Lee BS, Culbertson MR. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2165–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altamura N, Groudinsky O, Dujardin G, Slonimski PP. 1992. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J. Mol. Biol. 224:575–587 [DOI] [PubMed] [Google Scholar]
- 28.Koonin E. 1992. A new group of putative RNA helicases. Trends Biochem. Sci. 17:495–497 [DOI] [PubMed] [Google Scholar]
- 29.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. 2011. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 41:693–703 [DOI] [PubMed] [Google Scholar]
- 30.Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S. 2009. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 28:2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadlec J, Guilligay D, Ravelli RB, Cusack S. 2006. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA 12:1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. 2007. Structural and functional insights into the human Upf1 helicase core. EMBO J. 26:253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czaplinski K, Weng Y, Hagan KW, Peltz SW. 1995. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1:610–623 [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6:1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weng Y, Czaplinski K, Peltz SW. 1996. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol. 16:5477–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh S, Ganesan R, Amrani N, Jacobson A. 2010. Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA 16:1832–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franks TM, Singh G, Lykke-Andersen J. 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143:938–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He F, Jacobson A. 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 9:437–454 [DOI] [PubMed] [Google Scholar]
- 39.He F, Brown AH, Jacobson A. 1996. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA 2:153–170 [PMC free article] [PubMed] [Google Scholar]
- 40.Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12:1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lykke-Andersen J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22:8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lejeune F, Li X, Maquat LE. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12:675–687 [DOI] [PubMed] [Google Scholar]
- 44.Kadlec J, Izaurralde E, Cusack S. 2004. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat. Struct. Mol. Biol. 11:330–337 [DOI] [PubMed] [Google Scholar]
- 45.Dong S, Jacobson A, He F. 2010. Degradation of YRA1 pre-mRNA in the cytoplasm requires translational repression, multiple modular intronic elements, Edc3p, and Mex67p. PLoS Biol. 8:e1000360. 10.1371/journal.pbio.1000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fields S, Song O. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245–246 [DOI] [PubMed] [Google Scholar]
- 47.Cui Y, Dinman JD, Peltz SW. 1996. Mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 15:5726–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fairman-Williams ME, Guenther UP, Jankowsky E. 2010. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 20:313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He F, Jacobson A. 2001. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol. 21:1515–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amrani N, Sachs MS, Jacobson A. 2006. Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 7:415–425 [DOI] [PubMed] [Google Scholar]
- 51.Weng Y, Czaplinski K, Peltz SW. 1998. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA 4:205–214 [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng W, Hsieh J, Brendza KM, Lohman TM. 2001. E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J. Mol. Biol. 310:327–350 [DOI] [PubMed] [Google Scholar]
- 53.Levin MK, Patel SS. 1999. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 274:31839–31846 [DOI] [PubMed] [Google Scholar]
- 54.Maluf NK, Ali JA, Lohman TM. 2003. Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J. Biol. Chem. 278:31930–31940 [DOI] [PubMed] [Google Scholar]
- 55.Serebrov V, Pyle AM. 2004. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature 430:476–480 [DOI] [PubMed] [Google Scholar]
- 56.Klostermeier D, Rudolph MG. 2009. A novel dimerization motif in the C-terminal domain of the Thermus thermophilus DEAD box helicase Hera confers substantial flexibility. Nucleic Acids Res. 37:421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Min EE, Roy B, Amrani N, He F, Jacobson A. 2013. Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit. RNA 19:1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]