Abstract
Screening of 1,750 pneumococcal isolates for common serotypes by PCR was followed by Quellung reaction analysis of PCR-negative isolates with a comparison to the conventional (Quellung reaction only) approach. PCR agreed with Quellung reaction results for 99% of isolates. The sequential PCR/Quellung reaction algorithm is accurate and more cost-effective than the conventional approach.
TEXT
Streptococcus pneumoniae is a common cause of meningitis and pneumonia. The morbidity and mortality associated with invasive pneumococcal disease may be prevented by vaccination (1). Accurate pathogen serotyping provides important epidemiologic information to guide vaccine composition. The relative increase in nonvaccine serotypes since the introduction of the heptavalent pneumococcal conjugate vaccine underscores the need for ongoing surveillance (2–5). The Quellung reaction is the gold standard for distinguishing among the 93 known pneumococcal serotypes. Serotyping with the Quellung reaction is labor-intensive, requires expensive reagents, and can be difficult to interpret.
An alternative to the Quellung reaction is serotype determination using a PCR approach targeting polysaccharide capsule synthesis genes at the cps locus (6–8). The Centers for Disease Control and Prevention (CDC) has developed a multiplex PCR protocol with sequential reactions to determine serotypes for large collections of pneumococcal isolates causing invasive disease (6). The CDC approach targets short sequences of the cps locus that discriminate between serotypes, and primers have been adjusted over time to optimize specificity (6, 9).
The purpose of this study was to evaluate the utility of PCR-based serotyping as part of a longitudinal pneumococcal surveillance program in the United States with centralized laboratory testing (10–12). A sequential PCR/Quellung reaction serotyping algorithm was employed with initial detection of common serotypes by one multiplex PCR followed by Quellung reaction analysis to assign remaining serotypes. The serotypes chosen for the multiplex reaction reflected the predominant serotypes among invasive and noninvasive isolates in the United States tested during the prior surveillance period. The accuracy and workflow of the algorithm were compared to those of serotype determination using the Quellung reaction only.
(This work was presented in part at the 112th General Meeting of the American Society for Microbiology, San Francisco, CA, 17 June 2012 [13].)
Serotyping by the Quellung reaction using antisera from the Statens Serum Institut (Copenhagen, Denmark) was performed on a collection of 1,750 pneumococcal isolates obtained from different patients in 43 U.S. medical centers during October 2010 through March 2011 (11, 12). The isolate specimen sources represented invasive and noninvasive infections (lower respiratory tract [48%], blood [24%], sinus [8%], middle ear fluid [6%], cerebrospinal fluid [1%], and other or unknown [12%]). The serotypes were compared to those determined using the following sequential algorithm.
First, all isolates were screened by multiplex PCR using 5 primer sets, each targeting one of the common serotypes (3, 6A/B/C/D, 7F, 19A, and 22F/22A) and electrophoresis detection methods adapted from the CDC Streptococcus laboratory (http://www.cdc.gov/ncidod/biotech/strep/pcr.htm). This approach requires that the product sizes included in the multiplex reaction are different enough to be easily distinguished. A 1.0 McFarland suspension in 250 μl of Tris-EDTA (TE) buffer was prepared using pneumococcal growth from Trypticase soy agar with 5% sheep blood after 24 h of incubation in 5% CO2 at 37°C. After being heated at 100°C for 5 min, the suspension was immediately frozen for 5 min at −20°C and then stored at this temperature until ready to use.
PCR was performed on batches of 96 isolates by using previously published primer sequences (6). A 2.5-μl aliquot of extracted DNA was added to 22.5 μl of master mix (2.5 μl of 25 mM MgCl2 [Applied Biosystems], 2 μl of 10 mM deoxynucleoside triphosphate mix [Invitrogen], 2.5 μl of 10× Taq buffer [New England BioLabs], 0.5 μl [2.0 U] of Taq DNA polymerase [New England BioLabs], and 1.5 μl of each primer in the following concentrations: 8.3 μM 6A/B/C/D-f, 8.3 μM 6A/B/C/D-r, 25 μM 3-f, 25 μM 3-r, 33 μM 7F-f, 33 μM 7F-r, 25 μM 22F/22A-f, 25 μM 22F/22A-r, 16.6 μM 19A-f, and 16.6 μM 19A-r). Running conditions were 94°C for 4 min, 30 cycles of 94°C for 45 s, 54°C for 45 s, and 65°C for 2 min 30 s. After PCR, the products were detected by electrophoresis on 2% agarose gels run at 100 V for 120 min. After staining of gels with ethidium bromide, serotypes were determined by comparison with positive controls and a standard ladder included on each gel (Fig. 1).
Fig 1.
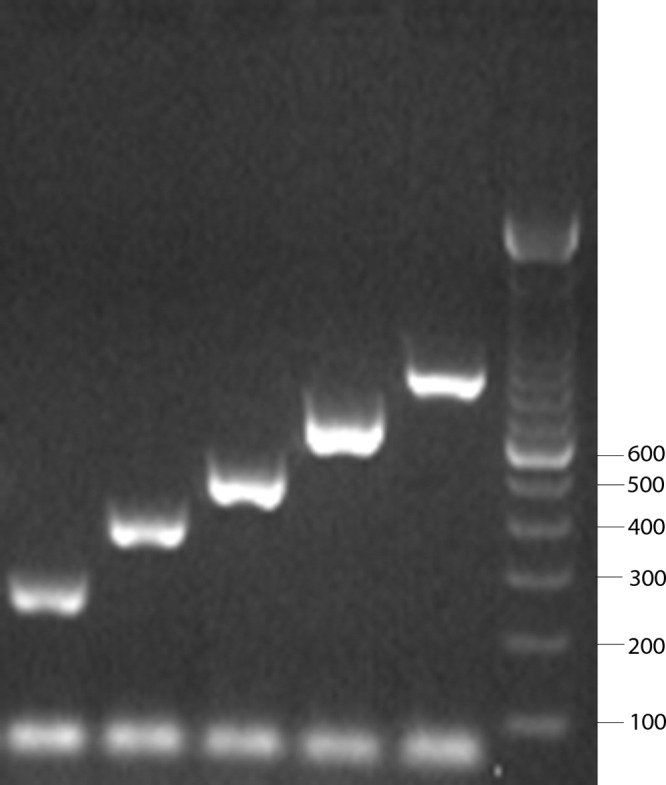
The leftmost five lanes show band sizes expected for (from left to right) serotype 6A/B/C/D (250 bp), 3 (371 bp), 19A (478 bp), 22F/22A (643 bp), and 7F (826 bp) isolates (6). The rightmost lane is a standard ladder, and the values shown to the right are in bp. The band at the bottom of each lane represents unused primers.
The PCR-negative isolates were then analyzed by the Quellung method for assignment to one of the 93 serotypes. The Quellung reaction was also used to further assign isolates identified by PCR as serogroup 6 to serotype 6A, 6B, 6C, or 6D. The algorithm results were compared to those of the conventional approach (serotyping of all isolates using the Quellung reaction). Isolates with discordant serotyping results were analyzed a second time by PCR and Quellung reaction. Reagent and labor costs were determined for both methods.
The PCR result agreed with the Quellung reaction result for 1,733 isolates (99%). The serotype (or serogroup) for 850 of the 1,750 isolates (48.6%) was included in the multiplex PCR (19A, n = 350, 20%; 6, n = 167, 9.5%; 3, n = 163, 9.3%; 7F, n = 86, 4.9%; 22F, n = 84, 4.8%). Although primers for 22F also target 22A, no isolates with the latter serotype were detected by the Quellung reaction. To address changes in serotype distribution observed in 2010-2011 (12), primers for serotype 22F/22A (643-bp product) will be replaced with those targeting the more prevalent serotype 35B (n = 123; 677-bp product) when the next group of surveillance isolates are analyzed.
The 17 isolates with discordant PCR and Quellung reaction results (1%) are described in Table 1. For four isolates (isolates 8, 9, 10, and 17), repeat PCR provided the same result but the repeat Quellung reaction result was different and concordant with PCR. This suggests an error in the initial Quellung reaction observation. Repeat PCR on seven isolates (isolates 6, 7, 11 to 14, and 16) changed to a result that was concordant with the initial and repeat Quellung reaction observations. Review of the initial gels did not reveal any errors in PCR interpretation. Technical error in labeling specimens is the likely explanation for the seven initial discordant PCR results and the one isolate (isolate 15) for which both PCR and Quellung reaction results changed. Encouraging technologists to double-check labels and recorded results or a second observer of Quellung reactions may help to prevent these errors.
Table 1.
Discrepancy analysis of isolates with discordant PCR and Quellung reaction results
| Isolate | Serogroup/serotype |
|||
|---|---|---|---|---|
| Initial result |
Repeat result |
|||
| Quellung | PCRa | Quellung | PCRa | |
| 1 | 3 | No reaction | 3 | No reaction |
| 2 | 3 | No reaction | 3 | No reaction |
| 3 | 22F | 19A | Nontypeable | 19A |
| 4 | 16F | 19A | 16F | 19A |
| 5 | 19F | 19A | 19F | 19A |
| 6 | 19A | 3 | 19A | 19A |
| 7 | 19A | 3 | 19A | 19A |
| 8 | Nontypeable | 19A | 19A | 19A |
| 9 | 22F | 19A | 19A | 19A |
| 10 | Nontypeable | 6 | 6C | 6 |
| 11 | 15C | 19A | 15C | No reaction |
| 12 | 15B | 19A | 15B | No reaction |
| 13 | 15B | 3 | 15B | No reaction |
| 14 | 31 | 7F | 31 | No reaction |
| 15 | 7F | 22 | 10A | No reaction |
| 16 | 22F | No reaction | 22F | 22 |
| 17 | Nontypeable | 22 | 22F | 22 |
Multiplex PCR detects serogroups 6 and 22 and serotypes 3, 7F, and 19A.
It is difficult to explain the reason for the five persistent discordant results. The two serotype 3 isolates with no PCR may represent strains with variations in the cps locus preventing amplification with the primers used. The other three isolates, with reproducible 19A PCR results (nontypeable, 16F, and 19F), may represent unusual strains that have cps loci resembling that of 19A. The 19A primers used in the current study target the mnaA gene, and a false-positive result for a 19F isolate (strain 2584-08) with a rare 19A-like cps locus has been reported by Pimenta et al. (9). New 19A primers targeting the wzy gene also gave false-positive results for the same 2584-08 (19F) isolate and led to redesign of primers to improve specificity for the serotype 19A wzy gene (9). Additional quality control measures, such as confirming serotype 19A PCR results with the Quellung reaction, could eliminate the primer specificity issues but would decrease the labor savings associated with the PCR serotyping approach.
A strength of this study is the large number of isolates tested from diverse geographic regions of the United States. This study confirms the reliability of the PCR approach to serotyping with contemporary clinical isolates of pneumococci from noninvasive as well as invasive sources. Clinicians may request serotyping of a pneumococcal isolate in order to assess for vaccine failure or to understand the epidemiology of disease. This study is an example of an approach that clinical laboratories may want to consider adopting with a single multiplex PCR to detect the most common serotypes and referral of negative isolates to a reference laboratory.
Testing of 1,750 isolates by using the Quellung reaction approach required 466 h of labor and $12,000 in reagents. The PCR/Quellung sequential approach (Quellung reaction testing needed for only the 900 PCR-negative and 167 serogroup 6 isolates) required 174 h of labor and $6,100 in reagents. The sequential PCR/Quellung algorithm required 292 fewer hours of technologist time, with a material cost savings of $5,900.
In conclusion, this study showed that detecting the most common pneumococcal serotypes by a multiplex PCR and then characterizing the remaining isolates with the Quellung method is a highly accurate method. The sequential PCR/Quellung algorithm offers substantial cost savings in comparison to a conventional antiserum approach when serotyping large collections of contemporary isolates.
ACKNOWLEDGMENTS
This work was supported by Forest Laboratories, Inc. (New York, NY). Forest Laboratories, Inc., was involved in the decision to present these results. Forest Laboratories, Inc., had no involvement in the design, collection, analysis, or interpretation of data. Scientific Therapeutics Information, Inc. (Madison, NJ), provided editorial assistance for the manuscript, which was funded by Forest Research Institute, Inc.
G.V.D. has received research funding from Abbott Laboratories, Schering-Plough, Bayer Pharmaceutical, Merck, Shionogi, Cubist, and Astra-Zeneca. He has been on the speakers' bureaus of Abbott Laboratories, Aventis, Astra-Zeneca, Forest Laboratories, Pfizer, Astellas, and Schering-Plough. D.J.D has received research funding from Forest Laboratories, Merck, Pfizer, Schering-Plough, Astellas, and bioMérieux. S.S.R. has received research funding from bioMérieux, Forest Laboratories, Pocared, and Nanosphere. All other authors have no conflict.
Footnotes
Published ahead of print 11 September 2013
REFERENCES
- 1.Rodgers GL, Klugman KP. 2011. The future of pneumococcal disease prevention. Vaccine 29S:C43–C48 [DOI] [PubMed] [Google Scholar]
- 2.Hsu HE, Shutt KA, Moore MR, Beall BW, Bennett NM, Craig AS, Farley MM, Jorgensen JH, Lexau CA, Petit A, Reingold A, Schaffner W, Thomas A, Whitney CG, Harrison LH. 2009. Effect of pneumococcal conjugate vaccine on pneumococcal meningitis. N. Engl. J. Med. 360:244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyaw MH, Lynfiekld R, Schaffner W, Craig AS, Hadler J, Reingold A, Thomas AR, Harrison LH, Bennett NM, Farley MM, Facklam RR, Jorgensen JH, Besser J, Zell ER, Schuchat A, Whitney CG. 2006. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N. Engl. J. Med. 354:1455–1463 [DOI] [PubMed] [Google Scholar]
- 4.Moore MR, Gertz RE, Woodbury RL, Barkocy-Gallagher GA, Schaffner W, Lexau C, Gershman K, Reingold A, Farley M, Harrison LH, Hadler JL, Bennett NM, Thomas AR, McGee L, Pilishvili T, Brueggemann AB, Whitney CG, Jorgensen JH, Beall B. 2008. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197:1016–1027 [DOI] [PubMed] [Google Scholar]
- 5.Richter SS, Heilmann KP, Dohrn CL, Riahi F, Beekmann SE, Doern GV. 2009. Changing epidemiology of antimicrobial-resistant Streptococcus pneumoniae in the United States, 2004–05. Clin. Infect. Dis. 48:e23–e33 [DOI] [PubMed] [Google Scholar]
- 6.Pai R, Gertz RE, Beall B. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 44:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brito DA, Ramirez M, de Lencastre H. 2003. Serotyping Streptococcus pneumoniae by multiplex PCR. J. Clin. Microbiol. 41:2378–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence ER, Griffiths DB, Martin SA, George DB, Hall LMC. 2003. Evaluation of semiautomated multiplex PCR assay for determination of Streptococcus pneumoniae serotypes and serogroups. J. Clin. Microbiol. 41:601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pimenta FC, Gertz RE, Roundtree A, Yu J, Nahm MH, McDonald RR, Carvalho MDG, Beall BW. 2009. Rarely occurring 19A-like cps locus from a serotype 19F pneumococcal isolate indicates continued need of serology-based quality control for PCR-based serotype determinations. J. Clin. Microbiol. 47:2353–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter SS, Heilmann KP, Coffman SL, Huynh HK, Brueggemann AB, Pfaller MA, Doern GV. 2002. The molecular epidemiology of penicillin-resistant Streptococcus pneumoniae in the United States, 1994–2000. Clin. Infect. Dis. 34:330–339 [DOI] [PubMed] [Google Scholar]
- 11.Doern GV, Diekema DJ, Heilmann KP, Dohrn CL, Riahi F, Richter SS. 2012. In vitro activity of ceftaroline against clinical isolates of Streptococcus pneumoniae recovered in 43 U.S. medical centers during 2010–2011. Antimicrob. Agents Chemother. 56:3406–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richter SS, Heilmann KP, Dohrn CL, Riahi F, Diekema DJ, Doern GV. 2013. Pneumococcal serotype before and after introduction of conjugate vaccines, United States, 1999–2011. Emerg. Infect. Dis. 19:1074–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter SS, Dohrn CL, Heilmann KP, Riahi F, Diekema DJ, Doern GV. 2012. Evaluation of sequential PCR/Quellung reaction pneumococcal serotyping algorithm, abstr C-104 Abstr. 112th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC [Google Scholar]


