Abstract
Hematopoietic stem cell transplant patients are highly susceptible to viral infections. Follow-up after transplantation includes weekly screening using single, virus-specific real-time PCR tests, mainly for viruses in the families Herpesviridae and Adenoviridae that contribute to a high morbidity, especially in pediatric populations. The Abbott PLEX-ID platform combines broad-range PCR with electrospray ionization mass spectrometry to enable the simultaneous detection of multiple pathogens in a single assay. The Viral IC Spectrum assay detects human adenoviruses, viruses from the family Herpesviridae (herpes simplex virus 1 [HSV-1], HSV-2, cytomegalovirus [CMV], Epstein-Barr virus [EBV], varicella-zoster virus [VZV], and human herpesvirus 8 [HHV-8]), human enterovirus, polyomaviruses (BK and JC), and parvovirus B19. We evaluated the performance of the Viral IC Spectrum assay with samples from 16 adult and 36 pediatric stem cell transplant patients. The sensitivity of the Viral IC Spectrum assay compared to real-time PCR quantification using the adenovirus Rgene kit for the detection of adenovirus was 96.7% from plasma samples (n = 92) and 78% from stool samples (n = 100). No adenovirus was detected in samples from noninfected patients (n = 30). PLEX-ID species identification was perfectly concordant with species-specific real-time PCR assays. In plasma and stool samples, the level of amplified products measured by PLEX-ID and the quantity in copies/ml (r = 0.82 and 0.78, respectively) were correlated up to 6 log10 copies/ml. In 67.4% of adenovirus-positive plasma samples, at least one other viral infection was detected; these included BK virus (n = 41), CMV (n = 30), EBV (n = 26), JC virus (n = 9), and HSV-1 (n = 6). The results of this study suggest that the Viral IC Spectrum assay performed on the PLEX-ID platform is reliable for adenovirus infection diagnosis in immunocompromised patients.
INTRODUCTION
Viral infections are often critical complications after human stem cell transplantation (1). Adenoviruses (AdV) and herpesviruses (mainly cytomegalovirus [CMV], Epstein-Barr-Virus [EBV], herpes simplex virus [HSV], and human herpesvirus 6 [HHV-6]) are the most frequent and severe causes of opportunistic infections in these immunocompromised patients (2–4). Adenoviruses are ubiquitous viruses of the family Adenoviridae, consisting of seven species (A to G), which include more than 60 types (5, 6). In hematopoietic stem cell transplant (HSCT) patients, adenovirus infection contributes to high morbidity and mortality and has been shown to cause pneumonia, hemorrhagic cystitis, colitis, pancreatitis, meningoencephalitis, and disseminated disease (7–9). The incidence of adenoviral infection ranges from 8 to 47% in HSCT patients and has been increasing in recent years (9). Progression to disseminated AdV disease occurs in an estimated 10 to 20% of patients with AdV infection, with a high mortality rate of 20 to 80% (4, 9, 10). The detection of adenovirus in blood is highly predictive of disseminated AdV disease (11).
In pediatric patients, adenoviral infections are even more problematic than in adults, due to the permanent circulation of viral particles in the patient population and the frequent asymptomatic persistence of adenoviruses in the gastrointestinal tract for weeks or months with or without any detectable replication (9, 12). Following pediatric HSCT, immunosuppression may lead to uncontrolled reactivation of digestive adenoviruses that often precede blood dissemination (13, 14). Thus, in pediatric patients, the ability to rapidly detect AdV in stool is crucial for monitoring and preemptive treatment. Real-time PCR assays have therefore become widely accepted for detection and quantitation of adenovirus in plasma and stool for monitoring infections following HSCT (9).
Stool from pediatric transplant patients is a source of environmental contamination (15) and serves as a reservoir for indirect transmission. Nosocomial outbreaks of adenovirus have been reported in pediatric hematologic wards, reinforcing the need for hygiene and control measures (16, 17). The ability to type adenoviruses is useful during outbreaks for epidemiological and clinical monitoring. In particular, typing of circulating viruses allows determination of whether viruses responsible for successive or contemporary infections are related to each other or are independent cases. Species and type identification is usually achieved using microneutralization assays with serotype-specific antisera or by sequencing (1). These assays require several days and must be carried out by highly trained personnel.
The PLEX-ID platform, which combines broad-range PCR with electrospray ionization mass spectrometry (PCR/ESI-MS), recently became available. The Viral IC Spectrum assay has been developed for use on the PLEX-ID for the detection and identification of viruses that cause opportunistic infections in immunocompromised patients, including herpesviruses, adenoviruses, parvovirus B19, the polyomaviruses BK and JC, and enteroviruses. In the present study, we evaluated the performance of the PLEX-ID Viral IC Spectrum assay in stool and plasma samples from HSCT adult and pediatric patients.
MATERIALS AND METHODS
Samples.
Samples were collected for routine laboratory testing between 3 September 2010 and 5 March 2012 from 52 HSCT recipients with adenovirus infection, including 36 children and 16 adults, who were hospitalized at the pediatric Robert Debré and Saint-Louis hospitals in Paris. One hundred stool samples and 92 plasma samples were tested to assess the sensitivity of PLEX-ID for the detection of adenovirus. To assess the specificity, 30 adenovirus-negative samples (22 plasma and eight stool samples) from 19 HSCT patients with no active adenovirus infection were tested.
All stool and plasma samples were processed by traditional methods used routinely. When requested by the treating physician, whole-blood samples were tested for CMV and EBV, and plasma samples were tested for HSV-1, HSV-2, and varicella-zoster virus (VZV). Nucleic acids were purified from 200 μl of sample and eluted in 100 μl using the MagNA Pure LC system (Roche Diagnostics, Mannheim, Germany) with the MagNA Pure DNA isolation kit (Roche Diagnostics). Before extraction, stool specimens were prepared by dilution of 0.5 g or 500 μl of stool in 8 ml of phosphate-buffered saline (PBS). The resulting suspension was subject to three −20°C freeze-thaw cycles followed by a centrifugation step. The supernatant was passed through a 0.45-μm filter (Minisart Plus syringe filters; Sartorius Stedim Biotech GmBH, Goettingen, Germany). Nucleic acids were extracted from the supernatant.
Ethics statement.
The study protocol was approved by the review board of Hôpital Saint Louis, and the study was carried out in accordance with the Declaration of Helsinki. This study was a noninterventional study with no addition to usual procedures. Biological material and clinical data were obtained only for standard viral diagnostics following physicians' prescriptions (no specific sampling and no modification of the sampling protocol). Data analyses were carried out using an anonymized database. According to the French Health Public Law (CSP Art L 1121-1.1), such protocols are exempt from the need for informed consent.
Adenovirus detection and typing.
Adenoviruses were detected and quantified with the Adenovirus Rgene kit (bioMérieux/Argene, Varhilles, France) according to the manufacturer's instructions (threshold for quantification, 200 copies/ml). For positive samples, identification of AdV species A to F was performed using six individual real-time PCR assays as previously described (18). These assays were carried out on an ABI 7500 thermocycler (Life Technologies, Carlsbad, CA). Adenovirus type identification was performed by sequencing hypervariable region 7 (HVR7) of the hexon gene (18).
Herpesvirus detection.
HSV-1, HSV-2, and VZV were detected on plasma samples by using real-time PCR assays on an ABI 7500 thermocycler as previously described (19). Two hundred microliters of plasma was extracted and eluted in 110 μl using the Qiasymphony system (Qiagen, Courtaboeuf, France), and 5 μl of nucleic acid eluate was used for the amplification. CMV and EBV were detected and quantified using the CMV kit and the EBV kit (Qiagen Hamburg GmbH, Germany), respectively, on the m2000 RealTime platform (Abbott Molecular, Des Plaines, IL). Extraction and amplification are automated on the platform. DNA was extracted from 300 μl of EDTA-whole blood and eluted in a final volume of 150 μl. According to the manufacturer's instructions, PCR was carried out in a 96-well plate with a reaction volume of 50 μl containing 20 μl of DNA extract and 30 μl of master mixture. CMV and EBV quantification thresholds are, respectively, 200 and 1,000 copies/ml of whole blood.
PLEX-ID analysis.
Samples used for PLEX-ID testing were stored at −80°C. Nucleic acids were extracted from 300 μl of plasma and stool supernatant and recovered in 200 μl by using a magnetic-bead-based method with the PLEX-ID SP instrument (extractor) and PLEX-ID FH instrument (fluid handler), using the PLEX-ID viral total nucleic acid preparation kit (all from Abbott Molecular). For each sample, 80 μl of nucleic acids was distributed by the PLEX-ID FH into 8 reaction wells of 96-well assay plates. Amplification was performed with the PLEX-ID Viral IC Spectrum amplification reagent kit (Abbott Molecular, Des Plaines, IL), which was designed to detect the presence of viral nucleic acids from the major taxonomic groups associated with opportunistic infections of immunocompromised patients. The assay employs 14 primer pairs in 8 wells of the assay plate. Of these, the second well contains primer pair 943, which is specific for the variable region of the adenovirus hexon gene, and the fourth well contains primer pair 5155, which is specific for the conserved region of the adenovirus penton gene (Table 1). Other wells contain primers that specifically amplify polyomaviruses, human herpesviruses, parvoviruses, enteroviruses, and the extraction control, pumpkin DNA. After mass spectrometry, the PLEX-ID software converted the mass information into base compositions and determined the virus(es) present in each sample by comparing the base composition signature to a database. For this study, PLEX-ID results were generated in the detailed research mode, which allows raw data analysis and differentiation of human adenovirus species and genotypes. In contrast, results generated in the diagnostic mode identify human adenovirus only at the family level, with no differentiation of species or genotypes. Though the assay has been validated only for plasma samples, we also evaluated the performance in stool specimens, as the monitoring of adenovirus infection in HSCT patients often includes quantification in stool.
Table 1.
Primer pairs of the Viral IC Spectrum assay targeting adenovirus gene sequences
| Primer pair | Gene targeted | Direction | Sequence |
|---|---|---|---|
| PP943 | hexon | Forward | 5′ TTGCAAGATGGCCACCCCATCGAT 3′ |
| Reverse | 5′ TGTGGCGCGGGCGAACTGCA 3′ | ||
| PP5155 | penton | Forward | 5′TCGTTCCTGCCCTCACAGATCACG 3′ |
| Reverse | 5′TAGGTCCGGCGACTGGCGTCAGT 3′ |
Statistical analysis.
Analyses were performed using the statistical R package (2.15.0) (R Development Core Team, Vienna, Austria; http://www.R-project.org). All tests were two-sided at the 0.05 significance level. Means of PLEX-ID levels were compared using Student's t test, and rates of PLEX-ID positivity were compared using chi-squared tests. Correlations between viral loads in copies/ml and PLEX-ID levels were assessed using Pearson's correlation coefficient.
RESULTS
Population description.
A total of 192 adenovirus-positive (>200 copies/ml) specimens (92 plasma and 100 stool) from 52 HSCT recipients with adenovirus infection (36 children and 16 adults) were tested using the PLEX-ID Viral IC Spectrum assay. One to 17 samples were collected per patient (median, 2 samples/patient; mean, 3.7 samples/patient). The viral loads determined by real-time PCR ranged from 2.56 to 9.76 log10 copies/ml for plasma samples and from 2.57 to 9.8 log10 copies/ml for stool samples (Table 2). The adenovirus species represented included types A (n = 20), B (n = 14), C (n = 119), D (n = 6), F (n = 6), A+C (n = 9), B+C (n = 9), and undetermined (n = 9). A list of specific types represented in plasma and stool samples is shown in Table 3.
Table 2.
Frequencies of adenovirus loads in plasma and stool specimens using a quantitative real-time PCR assaya
| Viral load (log10 copies/ml) | % of specimens |
|
|---|---|---|
| Plasma | Stool | |
| <3 | 6.5 | 7.0 |
| ≥3 and <4 | 28.0 | 23.0 |
| ≥4 and <5 | 29.0 | 17.0 |
| ≥5 | 36.5 | 53.0 |
Data were obtained with an adenovirus Rgene kit (Argene, Varhilles, France).
Table 3.
PLEX_ID results in adenovirus-infected plasma and stool samples by viral species and typea
| Species | Type | No. of specimens |
||||
|---|---|---|---|---|---|---|
| Plasma |
Stool |
Totalc | ||||
| Total | PLEX-ID +b | Total | PLEX-ID +b | |||
| A | 12 | 4 | 4 | 4 | ||
| 31 | 4 | 4 | 12 | 12 | 16 | |
| B | 3 | 1 | 1 | 1 | ||
| 7 | 4 | 4 | 4 | |||
| 11 | 1 | 1 | 1 | |||
| Undetd | 6 | 6 | 2 | 1 | 8 | |
| C | 1 | 11 | 11 | 16 | 9 | 27 |
| 2 | 40 | 38 | 27 | 21 | 67 | |
| 5 | 8 | 8 | 8 | 6 | 16 | |
| 6 | 3 | 3 | 1 | 1 | 4 | |
| Undet | 1 | 1 | 4 | 2 | 5 | |
| D | 56 | 1 | 1 | 1 | ||
| Undet | 5 | 5 | 5 | |||
| F | 41 | 4 | 4 | 2 | 1 | 6 |
| A+C | A31+C1 | 6 | 5 | 6 | ||
| A31+C2 | 3 | 3 | 3 | |||
| B+C | B3+C1 | 7 | 7e | 2 | 2 | 9 |
| Undet | 2 | 1f | 7 | 9 | ||
| Total | 92 | 89 | 100 | 78 | 192 | |
The species were determined with 6 single real-time PCR assays, one each targeting species A, B, C, D, E, and F (18, 19). The types were determined using sequencing of hypervariable region 7 (HVR7) of the hexon gene (18, 21).
Plasma or stool samples were positive for human adenovirus with the Viral IC Spectrum assay on the PLEX-ID system.
Total of plasma and stool samples analyzed in the study.
Undet, undetermined. Either the amplification for species identification was negative, or it was positive but HVR7 sequencing was negative or was positive but unable to discriminate between several types.
Only adenovirus species C was identified by PLEX-ID.
Adenovirus species C was identified by PLEX-ID.
Analyses of plasma samples.
Of the 92 positive plasma samples tested, with a median viral load of 4.47 log10 copies/ml (interquartile range [IQR], 3.62 to 5.69), 96.7% (89/92) were positive by PLEX-ID (Table 3). The median viral load for PLEX-ID-negative samples was 3.1 log10 copies/ml, with a range of 2.6 to 3.2 (Fig. 1). The positivity rates were 62.2% for the adenovirus primer pair 943 and 100% for the adenovirus primer pair 5155. The mean PLEX-ID quantification based on primer pair 5155 in samples that were amplified by primer pair 943 was significantly higher than adenovirus in samples that were not amplified by primer pair 943 (648.8 [n = 56] versus 214.3 [n = 33]) (P = <1e−4). Primer pair 943 amplified adenovirus C less effectively (55.1%; 38/69) than other species (90.0%; 18/20) (P = 0.004).
Fig 1.
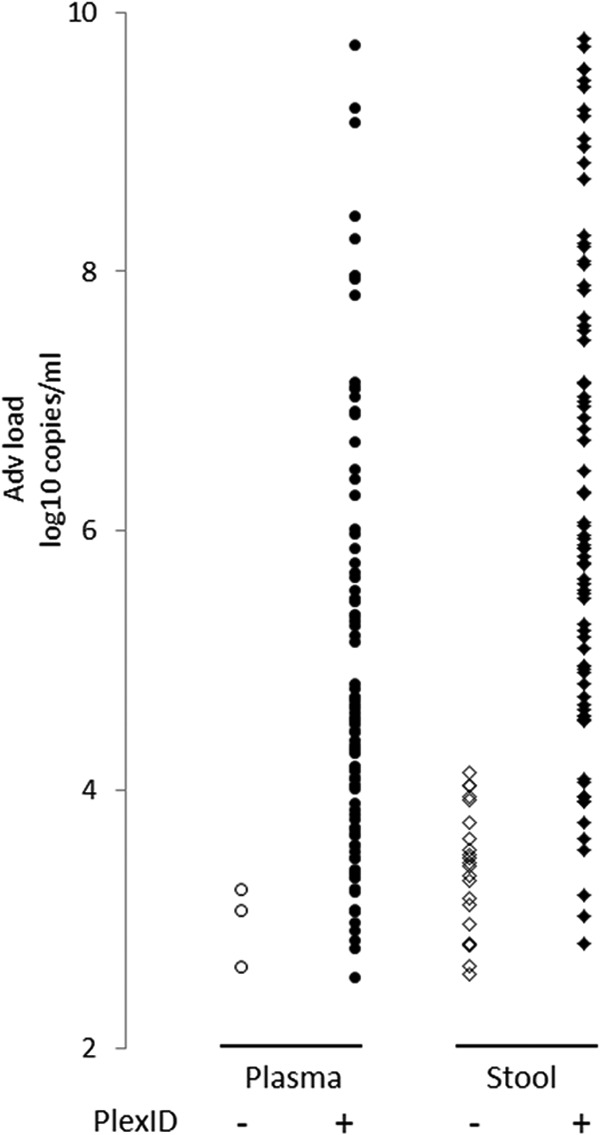
Adenovirus load for PLEX-ID-negative and -positive samples. Adenovirus loads, in log10 copies/ml, determined by quantitative real-time PCR (bioMérieux/Argene, Varhilles, France) are plotted against negative (open symbols) and positive (closed symbols) PLEX-ID results in plasma (circles) and stool samples (diamonds).
PLEX-ID analysis showed 100% concordance with a single test at the species level (A, B, C, D, and F) for positive samples with single infections. The base compositions obtained with the primer pairs according to type identification in plasma and in stool are listed in Table 4. Unequivocally correct type identifications were made for types 41 (n = 4) and 31 (n = 4). For AdV species B, two patterns of base composition were found the two primer pairs that matched more than one type (either 11, 34, and 35 or 3, 7, and 16) (Table 4). For plasma samples positive for AdV species C by PLEX-ID (n = 69), a single base composition (A21 G26 C34 T16) was observed with the primer pair 5155, targeting the penton gene, for all samples amplified; thus, type could not be distinguished with this primer. In contrast, of the 38 samples amplified with primer pair 943, targeting the hexon gene, several base compositions were obtained. Of 19 samples that were identified as type 2 using standard sequence analysis, 8 were correctly typed by the PLEX-ID assay. Type 5 samples (n = 4) were all correctly identified by PLEX-ID. In other samples, the base compositions matched several types. PLEX-ID was able to identify AdV C species in a plasma sample with a low viral load for which the species could not be determined by routine techniques. Three type C6 samples were identified as C5 by PLEX-ID, as the base compositions were identical, showing that discrimination between types C5 and C6 is not accurate with this PLEX-ID assay. One of several samples from a patient infected with type C1 was identified as C2 by PLEX-ID. One of several samples from a patient infected with type C1 was identified as C2 by PLEX-ID.
Table 4.
Base composition of amplified products obtained with the adenovirus primer pairs 943 and 5155
| Typea | PLEX-ID result | Base composition obtained with: |
|
|---|---|---|---|
| 943 | 5155 | ||
| A12 | A12 | A20 G32 C37 T23 | A19 G25 C34 T19 |
| A31 | A31 | A20 G32 C38 T22 | A19 G25 C34 T19 |
| B | B11, -34, -35 | A22 G32 C37 T21 | A17 G27 C34 T19 |
| B | B3, -7, -16 | A23 G32 C36 T21 | A17 G27 C35 T18 |
| F41 | F41 | A21 G33 C37 T21 | A19 G25 C36 T17 |
| C-undetb | A21 G26 C34 T16 | ||
| C1 | C | A21 G26 C34 T16 | |
| C1 | C | A20 G33 C39 T20 | A21 G26 C34 T16 |
| C1 | C2 | A20 G33 C38 T21 | A21 G26 C34 T16 |
| C2 | C | A21 G26 C34 T16 | |
| C2 | C | A20 G33 C39 T20 | A21 G26 C34 T16 |
| C2 | C2 | A20 G33 C38 T21 | A21 G26 C34 T16 |
| C5 | C | A21 G26 C34 T16 | |
| C5 | C5 | A21 G32 C39 T20 | A21 G26 C34 T16 |
| C5c | C5 | A20 G33 C39 T20 | A21 G26 C34 T16 |
| C6c | C5 | A20 G33 C39 T20 | A21 G26 C34 T16 |
| D-undet | D19, -36 | A20 G36 C38 T18 | A17 G27 C36 T17 |
| D56 | D | A20 G36 C38 T18 | A18 G27 C36 T16 |
| Undet | C | A21 G26 C34 T16 | |
The types were determined using sequencing of hypervariable region 7 (HVR7) of the hexon gene (18, 21).
Undet, undetermined. Either the amplification for species identification was negative, or it was positive but HVR7 sequencing was negative or was positive but unable to discriminate between several types.
In stool only.
The relative quantities determined using the PLEX-ID assay correlated well with viral load determined by real-time PCR viral load within the range of 2.3 to 6 log10 copies/ml (r2 = 0.67). Above 6 log10 copies/ml, the level given by the PLEX-ID had reached a plateau (Fig. 2). In addition, the comparison of kinetics of PLEX-ID levels and copies/ml in patients with persistent plasma adenovirus infection (patients with at least 4 follow-up samples) showed similar profiles (Fig. 3). Together, these data suggest that relative quantitation obtained using the PLEX-ID assay gives an indication of the viral load in the sample.
Fig 2.
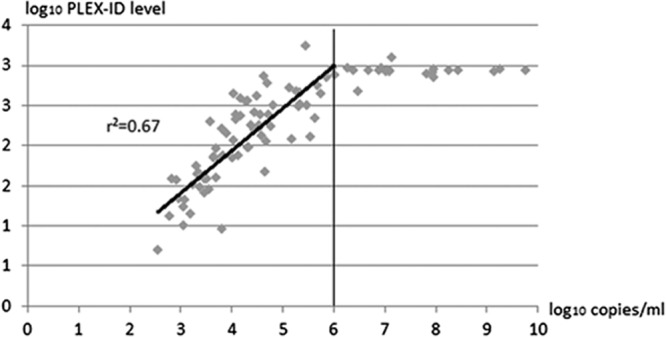
Correlation between adenovirus load in plasma determined by quantitative real-time PCR (log10 copies/ml) and adenovirus PLEX-ID levels (log10 levels). The linear regression was determined for adenovirus loads below 6 log10 copies/ml.
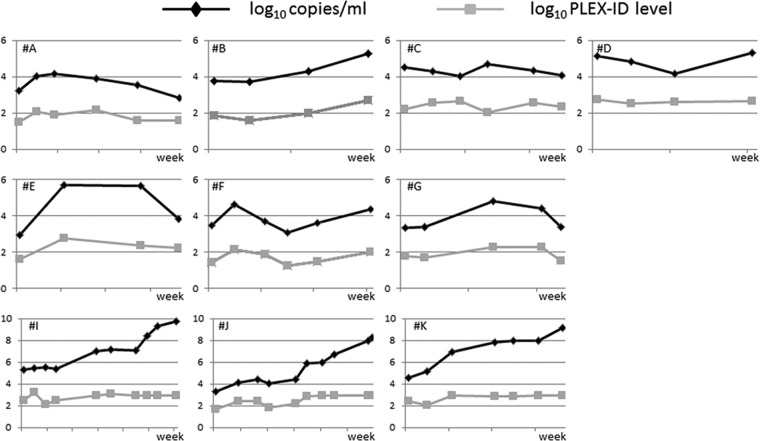
Fig 3.
Kinetic profiles of adenovirus plasma loads (in log10 copies/ml) and adenovirus PLEX-ID levels (log10) in patients (A to J) with at least 4 follow-up samples.
Analyses of stool samples.
Among the 100 stool samples analyzed with a median viral load of 5.25 log10 copies/ml (IQR, 3.92 to 7.06), 78% were positive by PLEX-ID. PLEX-ID detection in stool samples was significantly less effective than for plasma samples (P = 0.0001). The lower sensitivity in stool was independent of PCR inhibition, as the internal control failed to be amplified in none of the samples. PLEX-ID-negative samples had a mean log viral load of 3.42 (range, 2.57 to 4.13) (Fig. 1). Among the 78 positive samples, the positivity rate with primer pair 943 was 74.4% and that with primer pair 5155 was 100%. As observed for plasma samples, the rate of amplification with primer pair 943 was significantly lower for AdV C species (60.0%) than for other species (93.9%) (P = 0.0006). There was 100% agreement at the species level for adenovirus species A, B, C, D, and F in samples with a single infection. PLEX-ID gave unequivocally correct type identification for all samples positive for type 12 (n = 4), type 31 (n = 12), and type 41 (n = 1). The base compositions obtained in stool were identical to those found in plasma. In addition, 3 other base compositions were identified for AdV species C types 5 and 6 and for AdV species D. A good correlation between viral load and PLEX-ID level (log level) within the range of 2.3 to 6 log10 copies/ml (r2 = 0.61) was found for stool specimens (data not shown).
Specificity.
To assess specificity, we analyzed 30 negative samples (22 plasma and 8 stool) from HSCT patients with no active adenovirus infection. All negative samples tested were negative by PLEX-ID as well, confirming the specificity of the adenoviral detection.
Detection of coinfections with multiple adenoviruses.
The ability of the PLEX-ID Viral IC Spectrum assay to detect coinfections with two adenoviruses was evaluated by testing plasma and stool specimens collected from three patients confirmed to be coinfected with two adenovirus types by the real-time PCR assays used for species identification. In 10 of the 11 clinical samples, only the more abundant virus was detected by PLEX-ID (Table 5).
Table 5.
Adenovirus detection and identification with the Viral IC Spectrum assay in clinical samples with AdV coinfections
| Patient | Sample | Viral loada | AdV identificationb |
CTc |
PLEX-ID identificationd | ||
|---|---|---|---|---|---|---|---|
| A | B | C | |||||
| 1 | Plasma 1 | 4.01 | B3, C1 | 42.6 | 37.7 | C | |
| Plasma 2 | 4.45 | B3, C1 | 42.8 | 36.6 | C | ||
| Plasma 3 | 5.86 | B3, C1 | 40.1 | 32.0 | C | ||
| Plasma 4 | 5.98 | B3, C1 | 40.0 | 30.2 | C | ||
| Plasma 5 | 6.68 | B3, C1 | 38.2 | 25.8 | C | ||
| Stool 1 | 7.14 | B3, C1 | 27.0 | 28.0 | B3 | ||
| Stool 2 | 9.43 | B3, C1 | 28.7 | 19.3 | B3, C | ||
| 2 | Stool 1 | 4.05 | A31, C1 | 34.3 | 34.2 | C | |
| Stool 2 | 7.65 | A31, C1 | 18.8 | 31.7 | A31 | ||
| 3 | Stool 1 | 6.28 | A31, C1 | 21.7 | 33.3 | A31 | |
| Stool 2 | 9.03 | A31, C1 | 15.6 | 33.7 | A31 | ||
The viral load was determined with quantitative real-time PCR as described in Materials and Methods. Results are expressed in log10 copies/ml.
The species were determined with 6 single real-time PCR assays, one each targeting species A, B, C, D, E, and F (18, 19). The types were determined using sequencing of hypervariable region 7 (HVR7) of the hexon gene (20).
Cycle threshold (CT) values in single real-time PCR assays to detect adenoviruses A, B, and C.
Species and type given by the PLEX-ID using the Viral IC Spectrum assay.
Detection of coinfections with other viruses.
Among the 92 adenovirus-positive plasma samples, PLEX-ID detected coinfections with other viruses in 67.4%. Other viruses detected included BK virus (n = 41), cytomegalovirus (n = 30), Epstein-Barr virus (n = 26), JC virus (n = 9), and HSV-1 (n = 6). No other viruses were detected in 32.6% (30/92) of the samples. Twenty-seven samples contained only one other virus, 23 were positive for two other viruses, 10 were positive for three other viruses, and 1 each was positive for four and five other viruses. A total of 42 samples were positive for CMV or EBV (45.6%), and 14 samples (15.2%) were positive for both. Sixty whole-blood samples taken at the same time as the 92 plasma samples were sent for routine testing for CMV and EBV, including 27 that were positive for CMV and 28 that were positive for EBV. Among the corresponding plasma samples, PLEX-ID identified 23 positive for CMV (85%) and 17 positive for EBV (61%). Specificities for CMV and EBV were 100% (23/23) and 94.4% (17/18), respectively. Of 32 samples with no routine test requested for CMV and EBV, seven and eight were positive for CMV and EBV, respectively. The samples that were positive for HSV-1 collected from four patients were checked with an HSV-1-specific real-time PCR assay. Two patients, including one having clinical evidence of ulcerative infection in the mouth, had two samples each with levels of HSV-1 determined by PLEX-ID ranging from 36 to 127. HSV-1-specific real-time PCR confirmed HSV-1 infection for these two patients. The last two samples had low levels of HSV-1 (8 and 14) determined by PLEX-ID and were found to be negative with the HSV-1-specific real-time PCR. No related diseases were attributed to the detection of polyomaviruses BK or JC.
DISCUSSION
In this group of immunocompromised patients, the PLEX-ID Viral IC Spectrum assay showed sensitivity similar to that of quantitative real-time PCR for the detection of adenovirus in plasma samples (96.8%). The sensitivity was lower in stool samples (78%), but the optimal extraction procedure for such samples has not been determined, and the assay has not been validated in this matrix. The lower AdV loads observed in AdV PLEX-ID-negative samples than in AdV PLEX-ID-positive samples indicated a sensitivity of the Viral IC Spectrum assay lower than that of the quantitative real-time PCR assay used in this study. Previous reports showed that up to one-third of HSCT pediatric patients' stool samples were positive for adenovirus within the first 100 days following HSCT (14). In most instances the virus is cleared spontaneously. Almost all patients who experience systemic adenovirus infection have detectable viral DNA in stool specimens before the onset of viremia, suggesting that the intestinal tract may be a common source of virus dissemination and that virus detection in stool could be used to identify patients with adenovirus reactivation. However, only patients with high viral loads in stool (exceeding 106 copies per gram of stool) are at risk for systemic infection (13, 14). In the analyses of HSCT patient samples reported here, the Viral IC Spectrum assay run on the PLEX-ID system detected adenovirus in stool samples far below this threshold and may be sensitive enough for screening of patients at risk for adenovirus infection. The data presented here indicate that the assay system warrants further clinical validation in a controlled prospective study.
The Viral IC Spectrum assay was designed to be qualitative. The use of an internal calibrator allows semiquantitative analysis; the number of amplicons of viral genome relative to the number of amplicons of the internal standard is determined. The levels obtained for adenovirus using the PLEX-ID assay correlated well with the copy numbers determined with quantitative real-time PCR. A plateau was observed for viral loads above 6 log10 copies/ml. This was expected, as the PLEX-ID technology is based on endpoint PCR analysis. The kinetic profiles of PLEX-ID levels and copies/ml were also in agreement, suggesting that quantitative data obtained using the PLEX-ID assay provide a preliminary estimation of the viral load. Whether PLEX-ID levels can be used as a surrogate for viral load or as threshold values to initiate follow-up with quantitative assays deserves further validation. An evaluation of reproducibility over time should be undertaken first, and then an analysis of the assay should be performed in controlled prospective studies.
The Viral IC Spectrum assay showed very good concordance with our real-time PCR assays for the adenovirus species identification in samples with a single infection. The Viral IC Spectrum assay includes two primer pairs, 943 and 5155, designed to amplify the hexon gene and the penton gene, respectively. The combination of two primer pairs in two distinct regions offered advantages of sensitivity and species identification. The primer pair 5155 provided a unique base composition for each species tested except for species D. More various base compositions were obtained with the primer pair 943, but these base compositions were not distinct enough to unequivocally ascertain the type, with the exceptions of types A12, A31, and F41. Because the primer pair 943 targets a more variable region than the primer pair 5155, the primer pair 943 was less sensitive, especially for C species viruses. Although decisions regarding clinical treatment do not always require typing, the ability to obtain such detailed information is critical to epidemiological studies (17) and can play a significant role in understanding the level and form of pathogenicity that different strains may possess.
Several studies have reported simultaneous infections with more than one adenovirus type in HSCT patients (8, 14). The sensitivity of PLEX-ID to detect coinfections of more than one adenovirus in clinical samples was lower than that obtained using the combination of species-specific real-time PCR assays, likely because of competition of primers for hybridization in the Viral IC Spectrum assay. High levels of each virus were required to identify coinfected samples. Thus, the Viral IC Spectrum assay should not be used to identify coinfections with more than one adenovirus type.
The samples tested were collected from deeply immunocompromised patients, who frequently present several active viral infections or reactivation. The Viral IC Spectrum assay includes different primers targeting other viruses found commonly in immunocompromised patients, including CMV, EBV, HSV-1, HSV-2, HHV-8, parvovirus B19, and polyomaviruses BK and JC. The ability of the single Viral IC Spectrum assay to simultaneously detect multiple viruses provides a significant advantage over single-species-specific PCR assays. As expected, two-thirds of patients in our study were infected with more than one viral species. Interestingly, more than one half of theses samples were positive for more than two other viruses. The virus other than adenoviruses most frequently detected was BK virus. BK virus detection in plasma is associated with nephropathy in kidney transplant patients, and it was recently shown that BK viremia preceded hemorrhagic cystitis in children undergoing allogeneic HSCT (20). Despite the lack of disease symptoms attributed to BK in our patient population, a molecular screening for BK virus in plasma could be of interest for implementation in the virological follow-up after HSCT. For this HSCT population, about half of the adenovirus-positive samples also contained CMV or EBV. Since the plasma samples were used for the Viral IC Spectrum assay and routine CMV and EBV quantifications are done with whole-blood samples in our laboratory, a direct comparison was difficult. As expected, a lower sensitivity for CMV and EBV was observed in the plasma with the Viral IC Spectrum assay than real-time PCR quantitation of corresponding whole-blood samples. Further studies testing the two strategies on plasma are required to accurately compare their respective sensitivities, as the Viral IC spectrum assay has been validated on plasma only. Four patients were also identified to have a plasma HSV-1 infection, including two with low levels detected by PLEX-ID that were not confirmed with an HSV-1-specific real-time PCR assay.
The Viral IC Spectrum assay could therefore represent an interesting screening tool to avoid multiple sampling and additional separate PCR assays and to determine which patients will require quantitative follow-up. Further investigation is required to determine accurately the sensitivity and specificity for herpesviruses and polyomaviruses. In addition, workflow analysis studies and laboratory cost evaluations should be used to assess the value of the Viral IC Spectrum assay as a screening tool in transplant patients.
In summary, our data show the potential of the PLEX-ID technology and the Viral IC Spectrum assay to detect and type adenoviruses and other viruses known to be potentially pathogenic in transplant patients. The high sensitivity of the PLEX-ID assay for adenovirus indicates the usefulness of the assay for the surveillance of high-risk patients. Our preliminary results showing that the assay sensitively detects other viruses indicate that the Viral IC Spectrum assay deserves further evaluation for use in the management of immunocompromised patients.
ACKNOWLEDGMENTS
The Viral IC Spectrum assay kits were provided by Abbott Molecular. We are grateful to Marcus Picard-Maureau from Abbott Molecular for his scientific assistance. We are indebted to Sneha Somasekar for her careful review of the manuscript and her editorial suggestions.
J. LeGoff and F. Simon have consulted for Abbott Molecular. All other authors have declared that no competing interests exist.
Footnotes
Published ahead of print 9 October 2013
REFERENCES
- 1.Breuer S, Rauch M, Matthes-Martin S, Lion T. 2012. Molecular diagnosis and management of viral infections in hematopoietic stem cell transplant recipients. Mol. Diagn. Ther. 16:63–77 [DOI] [PubMed] [Google Scholar]
- 2.Baldwin A, Kingman H, Darville M, Foot AB, Grier D, Cornish JM, Goulden N, Oakhill A, Pamphilon DH, Steward CG, Marks DI. 2000. Outcome and clinical course of 100 patients with adenovirus infection following bone marrow transplantation. Bone Marrow Transplant. 26:1333–1338 [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti S, Mautner V, Osman H, Collingham KE, Fegan CD, Klapper PE, Moss PAH, Milligan DW. 2002. Adenovirus infections following allogeneic stem cell transplantation: incidence and outcome in relation to graft manipulation, immunosuppression, and immune recovery. Blood 100:1619–1627 [DOI] [PubMed] [Google Scholar]
- 4.Robin M, Marque-Juillet S, Scieux C, Peffault de Latour R, Ferry C, Rocha V, Molina J-M, Bergeron A, Devergie A, Gluckman E, Ribaud P, Socié G. 2007. Disseminated adenovirus infections after allogeneic hematopoietic stem cell transplantation: incidence, risk factors and outcome. Haematologica 92:1254–1257 [DOI] [PubMed] [Google Scholar]
- 5.Dehghan S, Seto J, Hudson NR, Robinson CM, Jones MS, Dyer DW, Chodosh J, Seto D. 2011. Complete genome sequence of human adenovirus prototype 17. J. Virol. 85:11540–11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seto D, Chodosh J, Brister JR, Jones MS, Members of the Adenovirus Research Community 2011. Using the whole-genome sequence to characterize and name human adenoviruses. J. Virol. 85:5701–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echavarría M. 2008. Adenoviruses in immunocompromised hosts. Clin. Microbiol. Rev. 21:704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lion T, Baumgartinger R, Watzinger F, Matthes-Martin S, Suda M, Preuner S, Futterknecht B, Lawitschka A, Peters C, Potschger U, Gadner H. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114–1120 [DOI] [PubMed] [Google Scholar]
- 9.Matthes-Martin S, Feuchtinger T, Shaw PJ, Engelhard D, Hirsch HH, Cordonnier C, Ljungman P, Fourth European Conference on Infections in Leukemia 2012. European guidelines for diagnosis and treatment of adenovirus infection in leukemia and stem cell transplantation: summary of ECIL-4 (2011). Transpl. Infect. Dis. 14:555–563 [DOI] [PubMed] [Google Scholar]
- 10.Howard DS, Phillips GL, II, Reece DE, Munn RK, Henslee-Downey J, Pittard M, Barker M, Pomeroy C. 1999. Adenovirus infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 29:1494–1501 [DOI] [PubMed] [Google Scholar]
- 11.Erard V, Huang M-L, Ferrenberg J, Nguy L, Stevens-Ayers TL, Hackman RC, Corey L, Boeckh M. 2007. Quantitative real-time polymerase chain reaction for detection of adenovirus after T cell-replete hematopoietic cell transplantation: viral load as a marker for invasive disease. Clin. Infect. Dis. 45:958–965 [DOI] [PubMed] [Google Scholar]
- 12.George D, El-Mallawany NK, Jin Z, Geyer M, Della-Latta P, Satwani P, Garvin JH, Bradley MB, Bhatia M, van de Ven C, Morris E, Schwartz J, Cairo MS. 2012. Adenovirus infection in paediatric allogeneic stem cell transplantation recipients is a major independent factor for significantly increasing the risk of treatment related mortality. Br. J. Haematol. 156:99–108 [DOI] [PubMed] [Google Scholar]
- 13.Jeulin H, Salmon A, Bordigoni P, Venard V. 2011. Diagnostic value of quantitative PCR for adenovirus detection in stool samples as compared with antigen detection and cell culture in haematopoietic stem cell transplant recipients. Clin. Microbiol. Infect. 17:1674–1680 [DOI] [PubMed] [Google Scholar]
- 14.Lion T, Kosulin K, Landlinger C, Rauch M, Preuner S, Jugovic D, Pötschger U, Lawitschka A, Peters C, Fritsch G, Matthes-Martin S. 2010. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia 24:706–714 [DOI] [PubMed] [Google Scholar]
- 15.Abad FX, Pintó RM, Bosch A. 1994. Survival of enteric viruses on environmental fomites. Appl. Environ. Microbiol. 60:3704–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faden H, Wynn RJ, Campagna L, Ryan RM. 2005. Outbreak of adenovirus type 30 in a neonatal intensive care unit. J. Pediatr. 146:523–527 [DOI] [PubMed] [Google Scholar]
- 17.Leruez-Ville M, Chardin-Ouachée M, Neven B, Picard C, Le Guinche I, Fischer A, Rouzioux C, Blanche S. 2006. Description of an adenovirus A31 outbreak in a paediatric haematology unit. Bone Marrow Transplant. 38:23–28 [DOI] [PubMed] [Google Scholar]
- 18.Sarantis H, Johnson G, Brown M, Petric M, Tellier R. 2004. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J. Clin. Microbiol. 42:3963–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watzinger F, Suda M, Preuner S, Baumgartinger R, Ebner K, Baskova L, Niesters HGM, Lawitschka A, Lion T. 2004. Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J. Clin. Microbiol. 42:5189–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskin BL, Denburg M, Furth S, Diorio D, Goebel J, Davies SM, Jodele S. 2013. BK viremia precedes hemorrhagic cystitis in children undergoing allogeneic hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 19:1175–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy T, Lebeck MG, Capuano AW, Schnurr DP, Gray GC. 2009. Molecular typing of clinical adenovirus specimens by an algorithm which permits detection of adenovirus coinfections and intermediate adenovirus strains. J. Clin. Virol. 46:80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]