Abstract
Enterococci are a major cause of bloodstream infections in hospitalized patients and have limited antimicrobial treatment options due to their many resistance mechanisms. Molecular technologies have significantly shortened the time to enterococcal isolate identification compared with conventional methods. We evaluated the impact of rapid organism identification and resistance detection with the Verigene Gram-positive blood culture microarray assay on clinical and economic outcomes for patients with enterococcal bacteremia. A single-center preintervention/postintervention quasiexperimental study compared inpatients with enterococcal bacteremia from 1 February 2012 to 9 September 2012 (preintervention period) and 10 September 2012 to 28 February 2013 (postintervention period). An infectious disease and/or critical care pharmacist was contacted with the microarray assay results, and effective antibiotics were recommended. The clinical and economic outcomes for 74 patients were assessed. The mean time to appropriate antimicrobial therapy was 23.4 h longer in the preintervention group than in the postintervention group (P = 0.0054). A nonsignificant decrease in the mean time to appropriate antimicrobial therapy was seen for patients infected with vancomycin-susceptible Enterococcus isolates (P = 0.1145). For patients with vancomycin-resistant Enterococcus bacteremia, the mean time to appropriate antimicrobial therapy was 31.1 h longer in the preintervention group than in the postintervention group (P < 0.0001). In the postintervention group, the hospital length of stay was significantly 21.7 days shorter (P = 0.0484) and mean hospital costs were $60,729 lower (P = 0.02) than in the preintervention group. The rates of attributed deaths in the two groups were not statistically different. Microarray technology, supported by pharmacy and microbiology departments, can decrease the time to appropriate antimicrobial therapy, the hospital length of stay, and health care costs.
INTRODUCTION
Enterococci are facultative anaerobic Gram-positive cocci that exist as a major component of normal colonic flora and represent the third most common type of health care-associated pathogen in the United States (1, 2). Although enterococci possess intrinsic resistance to several antibiotics, they have acquired additional resistance determinants, such as that for vancomycin resistance (3, 4). Vancomycin-resistant Enterococcus (VRE) is now considered one of the most common health care-associated multidrug-resistant organisms. In 2006, the Infectious Diseases Society of America highlighted VRE as one of six particularly dangerous microbes, due to its level of resistance and limited treatment options (4).
The time from blood culture positivity to the identification of Enterococcus isolates and the availability of susceptibility testing results ranges from 48 to 72 h for most microbiology laboratories using traditional culture and susceptibility testing methods (5). Because bloodstream infections (BSIs) caused by VRE have been associated with increased mortality rates, the time to appropriate therapy is crucial (6). Empirical antimicrobial therapy for enterococcal bacteremia may include agents such as vancomycin, daptomycin, or linezolid, with subsequent streamlining or de-escalation as appropriate. In addition, costs associated with such antimicrobial therapy can vary significantly depending on the agents used. Timely initiation of appropriate antimicrobial therapy permits more-effective targeting of the causative pathogens, decreases antimicrobial exposure, and can result in cost savings (5, 7, 8).
Rapid organism identification via molecular diagnostic assays can help decrease the time to appropriate antimicrobial therapy (5, 7, 8). One such assay that utilizes molecular technologies is the Verigene Gram-positive blood culture (BC-GP) test (Nanosphere, Inc., Northbrook, IL). The BC-GP test is a qualitative in vitro diagnostic test that utilizes microarray technology to detect specific bacterial DNA from positive patient blood cultures. Within 2.5 hours, the BC-GP test can rapidly recognize the presence of the following organisms: Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus lugdunensis, Streptococcus anginosus group, Streptococcus agalactiae, Streptococcus pneumoniae, Streptococcus pyogenes, Enterococcus faecalis, Enterococcus faecium, and Listeria spp. Additionally, the BC-GP test identifies three resistance markers, i.e., mecA, vanA, and vanB; the latter two confer resistance to vancomycin in Enterococcus spp. (9).
The purpose of this study was to evaluate the impact of rapid organism identification and resistance detection with the BC-GP test on clinical and economic outcomes for patients with enterococcal bacteremia. The primary objective was to determine the extent to which the BC-GP test could improve the time to appropriate antimicrobial therapy for inpatients with enterococcal bacteremia, in comparison with conventional microbiological methods, while minimizing inappropriate antibiotic use. Secondary objectives included assessing the effects of the BC-GP test on hospital length of stay (LOS), infection-related length of stay (I-LOS), rates of infection-related readmission within 90 days, costs associated with hospital admission, and mortality rates.
MATERIALS AND METHODS
Study location.
This was a preintervention/postintervention study with nonequivalent groups and a quasiexperimental design that was performed at the University of Florida Health Jacksonville over 13 months. The University of Florida Health Jacksonville is a 695-bed academic health center located in Jacksonville, Florida, that is affiliated with the University of Florida. The study was approved by the University of Florida Health Science Center Jacksonville institutional review board.
Verigene BC-GP test.
The BC-GP test is performed with the Verigene system (Nanosphere), a sample-to-result molecular diagnostics workstation. Each BC-GP test cartridge contains a microarray, i.e., a solid-phase support with capture probes (oligonucleotides) representing nucleic acid sequences that complement those of bacterial DNA. When a perfect match is identified, gold nanoparticles and silver deposits are used to amplify the signal for interpretation by the workstation (9).
Study design.
All patients with documented enterococcal bacteremia between 1 February 2012 and 9 September 2012 (pre-BC-GP period) and between 10 September 2012 and 28 February 2013 (post-BC-GP period) were included in this study. Exclusion criteria included polymicrobial bacteremia, death before the culture results, incarceration, and involvement with other investigational protocols.
Prior to implementation of the BC-GP test, Gram staining was performed for positive blood cultures, and results were called to the patient's nurse stationed in the patient care area. Gram staining, organism identification, and susceptibility results for blood cultures positive for Enterococcus spp. were reported in the electronic health record (EHR) as they became available. Following implementation, the BC-GP test was performed 24 h per day, 7 days per week, by the clinical microbiology laboratory. The BC-GP test was performed on all initially positive blood cultures that demonstrated Gram-positive cocci in the Gram stain smear. Following implementation of the BC-GP test, the patient's nurse was contacted in a similar fashion regarding positive blood culture results. To facilitate reporting of BC-GP test results, microbiology personnel paged the antimicrobial stewardship team on weekdays (Monday through Friday) between the hours of 7:30 a.m. and 5:00 p.m. with BC-GP tests that resulted in the identification of Enterococcus spp. and detection of the vanA or vanB gene. The antimicrobial stewardship team consisted of pharmacists with specialty training in either infectious diseases or critical care. Pharmacists contacted physicians with pharmacotherapeutic recommendations, as appropriate, based on BC-GP test results and were able to enter verbal medication orders as dictated by the physicians. BC-GP test results were confirmed by conventional microbiological methods.
Data collection.
A retrospective report that identified patients with a previous blood culture that was positive for Enterococcus spp. was generated by the clinical microbiology laboratory. The EHR was used to identify the time, in hours, from blood culture collection to administration of appropriate antimicrobial therapy. For vancomycin-susceptible Enterococcus (VSE), appropriate antimicrobial therapy was defined as an antibiotic regimen to which the enterococcal isolate was susceptible; it could have included ampicillin (with or without gentamicin or ceftriaxone), ampicillin-sulbactam, piperacillin-tazobactam, or vancomycin. For VRE, appropriate antimicrobial therapy was defined as an antibiotic regimen to which the enterococcal isolate was susceptible; it could have included daptomycin or linezolid. Data collected from the EHR included age, gender, comorbidities, total parenteral nutrition (TPN) status, organism identification and susceptibility results, antibiotic treatment duration, antibiotic therapeutic monitoring data, and hospital charges. Hospital LOS, I-LOS, and crude mortality rates were also assessed. A post hoc analysis of attributed deaths among deceased patients was performed by an infectious disease physician. The primary investigator retrospectively gathered information only after subjects were discharged or expired, whichever occurred first. Research associates validated 10% of the data.
Statistical analysis.
Qualitative variables were compared using chi-square analysis or Fisher's exact test (as appropriate). Quantitative variables were compared using analysis of variance. A 2-tailed P value of 0.05 was used to determine statistical significance.
RESULTS
During the study period, 107 patients were identified and screened for study inclusion, and 74 patients met the criteria. Forty-six and 28 patients were included in the pre-BC-GP and post-BC-GP groups, respectively. Rates of VRE bacteremia were 37% in the pre-BC-GP group and 50% in the post-BC-GP group (P = 0.33). There were no statistically significant differences between study groups with regard to baseline demographics or risk factors for enterococcal BSIs (Table 1).
Table 1.
Comparison of baseline characteristics for the pre- and post-BC-GP groups
| Characteristic | Data for patients in group |
|
|---|---|---|
| Pre-BC-GP | Post-BC-GP | |
| No. | 46 | 28 |
| Mean age (yr) | 53.2 | 60.6 |
| Male (no. [%]) | 20 (43) | 17 (61) |
| History of diabetes mellitus (no. [%]) | 22 (48) | 13 (46) |
| History of cancer (no. [%]) | 6 (13) | 3 (11) |
| TPNa (no. [%]) | 4 (9) | 3 (11) |
TPN, total parenteral nutrition.
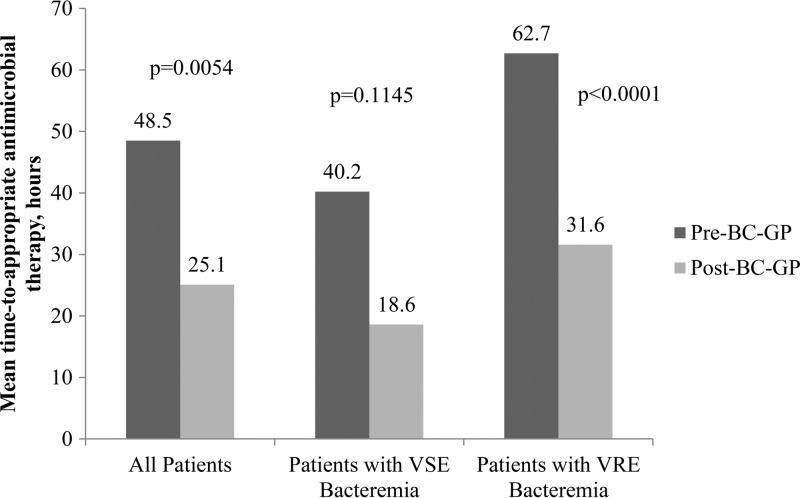
The mean time to appropriate antimicrobial therapy was 23.4 h less for patients in the post-BC-GP group than for patients in the pre-BC-GP group (48.5 versus 25.1 h; P = 0.005). There was a nonsignificant decrease in the time to appropriate antimicrobial therapy for patients with VSE bacteremia (40.2 versus 18.6 h; P = 0.115). For patients with VRE bacteremia, a 31.1-hour decrease in the mean time to appropriate antimicrobial therapy between the pre- and post-BC-GP groups (62.7 versus 31.6 h; P < 0.0001) was observed (Fig. 1).
Fig 1.
Mean times from blood culture collection to administration of the appropriate antimicrobial regimen, grouped by infecting organism. BC-GP, Gram-positive blood culture test; VSE, vancomycin-susceptible Enterococcus; VRE, vancomycin-resistant Enterococcus.
The mean hospital LOS was 21.7 days greater for the pre-BC-GP group than for the post-BC-GP group (43.2 versus 21.5 days; P = 0.048). When deceased patients were removed from the analysis, statistical significance was not maintained (43.5 versus 22.2 days; P = 0.141). There was no difference with regard to I-LOS (13.3 versus 9.8 days; P = 0.096). Similarly, rates of infection-related readmission within 90 days were not significantly different for the pre- and post-BC-GP groups (23.9% versus 17.9%; P = 0.772). The crude mortality rate was lower in the pre-BC-GP group than in the post-BC-GP group (15.2% versus 39.3%; P = 0.026). Attributed mortality rates were not deemed to be statistically significant (2.1% versus 14.2%; P = 0.065).
The mean hospital costs were $103,075 and $42,346 for the pre- and post-BC-GP groups, respectively (P = 0.02); this benefit was maintained when deceased patients were removed from the analysis ($99,333 versus $41,139; P = 0.047). In order to evaluate the costs associated with I-LOS, we assessed total hospital costs during antimicrobial therapy, which were similar between groups ($26,426 versus $18,805; P = 0.08).
There was complete agreement between the BC-GP test results and the results derived from conventional culture and susceptibility methods. The mean time from blood culture positivity to reporting of the BC-GP test result was 3.15 h. In comparison, the mean time from blood culture positivity to reporting of susceptibilities determined via conventional culture and susceptibility methods was 50.6 h.
DISCUSSION
Delays in the availability of culture and susceptibility data for patients with BSIs can lead to unnecessary antimicrobial exposure, avoidable health care costs, and increases in hospital morbidity and mortality rates. Rapid organism identification with molecular diagnostic assays has demonstrated positive clinical and economic benefits when utilized by antimicrobial stewardship programs (ASPs) (5, 7, 8).
Bauer and colleagues conducted a study utilizing a rapid PCR methicillin-resistant Staphylococcus aureus (MRSA)/S. aureus culture test (Cepheid, Sunnyvale, CA) on blood cultures positive for S. aureus, to detect or to rule out MRSA within 1 h (7). The researchers evaluated the clinical benefits of implementing this assay, with support from clinical pharmacists' interventions (antibiotic consultations and recommendations), compared with the use of traditional culture and susceptibility testing methods. The average time to switch from empirical vancomycin therapy to cefazolin or nafcillin was 1.7 days shorter and the average length of stay was 6.2 days shorter when the PCR test was used with pharmacist involvement than when traditional methods were used. The study also reported a decrease in mean hospital costs of approximately $21,000 with the use of the PCR test (7). Decreases in total antibiotic exposure and I-LOS were also realized with the use of rapid diagnostic technology for the management of patients with coagulase-negative staphylococci (8). Forrest and colleagues utilized peptide nucleic acid (PNA) fluorescent in situ hybridization (FISH) for identification of hospital-acquired enterococcal bacteremia, in an effort to minimize the time to effective antimicrobial therapy (5). On average, E. faecalis and E. faecium were identified significantly faster with PNA FISH than with traditional culture methods (3 and 2.4 days earlier, respectively), and the time to initiation of effective antimicrobial therapy and the 30-day mortality rate were significantly reduced (5). Although many clinical and economic benefits have been reported with implementation of rapid diagnostic technology, institutions may not fully realize the benefits of this expensive technology without active intervention from an ASP (10).
This is the first study utilizing rapid microarray technology to optimize therapy for enterococcal bacteremia. We were able to demonstrate significant decreases in the time to appropriate antimicrobial therapy for patients with enterococcal bacteremia as a whole and also for patients with VRE bacteremia. Additionally, we observed minimization of inappropriate antimicrobial use. Of the 14 patients in the post-BC-GP group with documented VSE bacteremia, 0/14 patients received inappropriate therapy based on our study definitions. In areas with high rates of endemic vancomycin resistance, ASPs can demonstrate cost avoidance by utilizing a molecular assay to minimize daptomycin or linezolid utilization for patients infected with vancomycin-susceptible isolates. Based on conventional microbiological methods, a cost avoidance of approximately $500 to $1,000 per patient may be realized if 3 days of empirical treatment with linezolid or daptomycin are avoided, based on average wholesale costs. Although the cost for the BC-GP test is greater than that for conventional methods, this rapid technology would be cost-effective when supported by appropriate interventions.
The mean hospital LOS was significantly decreased following implementation of the BC-GP test, but statistical significance was not maintained after removal of deceased patients from the analysis. There was a trend toward decreased I-LOS, but this was not found to be statistically significant. Considering that many of these patients received the majority of their antimicrobial treatment as inpatients, there was only minor variance in I-LOS between groups, due to the uniformity of recommended treatment durations for enterococcal BSIs (11, 12). The mean hospital costs were $60,729 less in the post-BC-GP group, and this benefit retained statistical significance when deceased patients were removed from the analysis. This may be attributed to early initiation of or switching to appropriate antimicrobial therapy by the ASP in response to BC-GP test results as well as ancillary recommendations on disease state management, including repeat blood cultures, echocardiograms, and infectious disease consultations. A higher crude mortality rate was identified in the post-BC-GP group. The majority of deceased patients were of advanced age, 40% had positive blood cultures collected during their admission to the intensive care unit, and 20% expired within 72 h of the positive blood culture. Because this study design included all patients who received at least one dose of appropriate antimicrobial therapy, data on the severity of illness, concomitant infections, and/or surgical procedures were not collected. Since this study did not exclude any patients based on these factors, some of these patients might have expired prior to receiving any additional benefits of earlier appropriate therapy. The study was not powered to assess mortality rates, and no difference in attributed mortality rates was found during the post hoc analysis.
There were some limitations in this study. The study was conducted at a single institution and utilized a small sample size, particularly in the post-BC-GP period. Sample size was dependent on the number of enterococcal bacteremia cases encountered during the study period, and patients with polymicrobial culture results were excluded from the study, because the BC-GP test is approved only for testing of monomicrobial blood cultures. Furthermore, the data were retrospectively extracted in a nonblinded manner from the electronic health record, and accurate documentation was assumed. Although differences in baseline demographics between groups were not identified, it is not known whether unmeasured or unreported confounders might have affected the clinical outcome results. Finally, the established pager hours might have limited the ability of the ASP to act on BC-GP test results when a switch in therapy was indicated. Expansion of pager hours and/or additional ASP coverage could have resulted in a more profound impact on the time to appropriate therapy. Despite these limitations, a significant clinical reduction in the time to appropriate therapy was achieved.
Minimizing the time to appropriate antimicrobial therapy permits more-effective targeting of the causative pathogens, decreases antimicrobial exposure, and can result in cost avoidance (5, 7, 8). Utilization of assays such as the BC-GP test, supported by antimicrobial stewardship teams, can optimize antimicrobial use, decrease unnecessary LOS and costs, and improve the time to appropriate therapy.
ACKNOWLEDGMENTS
We thank University of Florida Health Jacksonville microbiology laboratory personnel for their assistance with the BC-GP test and Ryan Butterfield and Dale Kraemer for their assistance with statistical analysis.
We declare no conflict of interest in relation to this work.
Footnotes
Published ahead of print 25 September 2013
REFERENCES
- 1.Bartlett JG, Auwaerter PG, Pham PA. 2012. Johns Hopkins ABX guide: diagnosis and treatment of infectious diseases, 3rd ed. Jones & Bartlett Learning, Burlington, MA [Google Scholar]
- 2.Hayakawa K, Marchaim D, Martin ET, Tiwari N, Yousuf A, Sunkara B, Pulluru H, Kotra H, Hasan A, Bheemreddy S, Sheth P, Lee DW, Kamatam S, Bathina P, Nanjireddy P, Chalana IK, Patel S, Kumar S, Vahia A, Ku K, Yee V, Swan J, Pogue JM, Lephart PR, Rybak MJ, Kaye KS. 2012. Comparison of clinical characteristics and outcomes associated with vancomycin-resistant Enterococcus faecalis and vancomycin-resistant E. faecium bacteremia. Antimicrob. Agents Chemother. 56:2452–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gold HS. 2001. Vancomycin-resistant enterococci: mechanisms and clinical observations. Clin. Infect. Dis. 33:210–219 [DOI] [PubMed] [Google Scholar]
- 4.Talbot GH, Bradley J, Edwards JE, Gilbert D, Scheld M, Bartlett JG. 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42:657–668 [DOI] [PubMed] [Google Scholar]
- 5.Forrest GN, Roghmann M, Toombs LS, Johnson JK, Weekes E, Lincalis DP, Venezia RA. 2008. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob. Agents Chemother. 52:3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiazGranados CA, Zimmer SM, Klein M, Jernigan JA. 2005. Comparison of mortality associated with vancomycin-resistant and vancomycin-susceptible enterococcal bloodstream infections: a meta-analysis. Clin. Infect. Dis. 41:327–333 [DOI] [PubMed] [Google Scholar]
- 7.Bauer KA, West JE, Balada-Llasat J, Pancholi P, Stevenson KB, Goff DA. 2010. An antimicrobial stewardship program's impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin. Infect. Dis. 51:1074–1080 [DOI] [PubMed] [Google Scholar]
- 8.Wong JR, Bauer KA, Mangino JE, Goff DA. 2012. Antimicrobial stewardship pharmacist interventions for coagulase-negative staphylococci positive blood cultures using rapid polymerase chain reaction. Ann. Pharmacother. 46:1484–1490 [DOI] [PubMed] [Google Scholar]
- 9.Nanosphere Inc 2012. Verigene® Gram-positive blood culture nucleic acid test (BC-GP) package insert. Nanosphere Inc., Northbrook, IL [Google Scholar]
- 10.Carver PL, Lin SW, DePestel DD, Newton DW. 2008. Impact of mecA gene testing and intervention by infectious disease clinical pharmacists on time to optimal antimicrobial therapy for Staphylococcus aureus bacteremia at a university hospital. J. Clin. Microbiol. 46:2381–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mermel LA, Allon M, Bouza E, Craven DE, Flynn P, O'Grady NP, Raad II, Rijnders BJA, Sherertz RJ, Warren DK. 2009. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 49:1–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baddour LM, Wilson WR, Bayer AS, Fowler VG, Bolger AF, Levison ME, Ferrieri P, Gerber MA, Tani LY, Gewitz MH, Tong DC, Steckelberg JM, Baltimore RS, Shulman ST, Burns JC, Falace DA, Newburger JW, Pallasch TJ, Takahashi M, Taubert KA. 2005. Infective endocarditis: diagnosis, antimicrobial therapy, and management of complications: a statement for healthcare professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, and the Councils of Clinical Cardiology, Stroke, and Cardiovascular Surgery and Anesthesia, American Heart Association: endorsed by the Infectious Diseases Society of America. Circulation 111:e394–e433 [DOI] [PubMed] [Google Scholar]