Abstract
Human rhinoviruses (HRVs) and human enteroviruses (HEVs) are significant respiratory pathogens. While HRV infections are restricted to the respiratory tract, HEV infections may spread to secondary target organs. The method of choice for sensitive specific detection of these viruses is reverse transcription (RT)-PCR with primers targeting the conserved 5′ noncoding region of the viral RNA. On the other hand, sequence similarities between HRVs and HEVs complicate their differential detection. In this study, we describe the use of locked nucleic acid (LNA) analogues in short double-dye probes which contained only two selectively HRV- or HEV-specific bases. The double-stranded DNA dye BOXTO (4-[6-(benzoxazole-2-yl-(3-methyl-)-2,3-dihydro-(benzo-1,3-thiazole)-2-methylidene)]-1-methyl-quinolinium chloride) was used with the LNA probes in a tricolor real-time PCR assay to allow specific detection of HRVs (probes labeled with 6-carboxyfluorescein [FAM] [green]) and HEVs (Cy5 [red]) with additional melting curve analysis (BOXTO [yellow]). The functionality of the probes was validated in PCR and RT-PCR assays using plasmids containing viral cDNA, quantified viral RNA transcripts, cultivated rhino- and enterovirus prototypes, and clinical specimens. Of 100 HRV and 63 HEV prototypes, the probes correctly identified all HEVs except one that produced only a BOXTO signal. Among 118 clinical specimens with sequencing results, concordant results were obtained for 116 specimens. Two specimens were reactive with both probes, but sequencing yielded only a single virus. Real-time PCR with LNA probes allowed sensitive group-specific identification of HRVs and HEVs and would enable relative copy number determination. The assay is suitable for rapid and accurate differential detection of HRVs and HEVs in a diagnostic laboratory setting.
INTRODUCTION
Human rhinoviruses (HRVs) and human enteroviruses (HEVs) are the most common cause of infections in people worldwide. They belong to the genus Enterovirus within the family Picornaviridae. They are small viruses (30 nm in diameter) with an infectious single-stranded RNA genome of 7,000 to 7,500 nucleotides enclosed in an icosahedral capsid. HRVs include 153 currently known types divided into three species, i.e., A (n = 77 types), B (n = 25), and C (n = 51), while HEVs consist of 104 types belonging to four species, i.e., A (n = 18), B (n = 58), C (n = 23), and D (n = 5) (www.picornaviridae.com) (1–3). HRVs are the usual cause of common cold but are also frequently found in otitis media, sinusitis, bronchitis, pneumonia, and asthma exacerbations (4, 5). There is a strong association between HRV bronchiolitis in early infancy and recurrent wheezing or the development of asthma (6, 7), which may be preventable by treatment with corticosteroids during the first episode of wheezing (8). Several HRV types circulate continuously, with typical fall and spring peaks of incidence. In contrast to HRVs, replication of HEVs is not restricted to the respiratory tract but also can take place in the small intestine and spread to various target organs. Most HEV infections are asymptomatic or manifest common cold-like symptoms. However, HEV infections can be more severe, causing poliomyelitis, meningitis, encephalitis, myocarditis, exanthema, acute hemorrhagic conjunctivitis, and severe generalized infections in newborns (9). In addition to polioviruses, some other enterovirus types are often associated with certain clinical manifestations. For example, coxsackievirus A16 (CVA16), enterovirus 71 (EV71), and CVA6 have strong associations with outbreaks of hand, foot, and mouth disease and CVA24 and EV70 with hemorrhagic conjunctivitis (9–11). Especially in the Asia-Pacific region, severe EV71 outbreaks have involved cases of fatal encephalitis in infants and children (9). Differential diagnosis of HRV and HEV infections is epidemiologically important. Specific identification of these viruses already has implications for the supportive management of patients and will become more significant when specific antiviral drugs become available.
Nucleic acid amplification techniques have replaced the isolation of viruses in cell cultures as the method of choice for the detection of picornaviruses, partly due to the outstanding sensitivity, specificity, and rapidity of such techniques. The recently identified species C HRVs cannot be cultivated in standard cell lines but can be amplified by reverse transcription (RT)-PCR (12). Both HRVs and HEVs have conserved 5′ noncoding regions (NCRs) and a few nearly identical sequence motifs, allowing the design of universal primers for their amplification in RT-PCR assays. For differentiation between HRVs and HEVs, however, these assays require a nested format, post-PCR hybridization, agarose gel electrophoresis, or sequencing (13–19), making them prone to contamination or cumbersome for diagnostic use. Recently, RT-PCR assays using real-time amplification have been developed for specific detection of HRVs and/or HEVs (20–22).
In this study, we first evaluated the feasibility of a diagnostic RT-PCR assay with real-time amplification using SYBR green I double-stranded DNA (dsDNA) dye and determination of the melting temperature (Tm), but the differentiation between HRV and HEV amplicons on the basis of Tm values was only suggestive. We then used sequences from recent strains and those available from GenBank to design group-specific locked nucleic acid (LNA) double-dye probes for real-time PCR. The novel LNA probes were used together with the dsDNA dye BOXTO (4-[6-(benzoxazole-2-yl-(3-methyl-)-2,3-dihydro-(benzo-1,3-thiazole)-2-methylidene)]-1-methyl-quinolinium chloride), allowing specific selective detection of HRVs and HEVs together with information from the melting curve analysis.
MATERIALS AND METHODS
Virus strains.
Picornavirus prototype strains, obtained from the American Type Culture Collection and from the Finnish National Institute for Health and Welfare (THL) (Helsinki, Finland), were cultured in LLC, HeLa, or CaCo cells and stored as culture supernatants at −70°C. Poliovirus vaccine strains were obtained from the THL. The prototype strain collection contained 100 HRV types (types 1 to 100, including 75 HRV species A types and 25 species B types) and 63 HEVs, including 12 HEV species A types (CVA2 to CVA8, CVA10, CVA12, CVA14, CVA16, and enterovirus 71 [EV71]), 35 HEV species B types (CVB1 to CVB6, CVA9, and echovirus 1 [E1] to E9, E11 to E21, E24 to E27, E29, E30, E32, and E33), 15 HEV species C types (polioviruses 1 to 3, CVA1, CVA11, CVA13, CVA15, CVA17 to CVA19, CVA20a, CVA20b, CVA21, CVA22, and CVA24), and 1 HEV species D type (EV68).
Recombinant plasmids with cDNA clones of HRV-A1b, HRV-B14, HRV-A85, CVA9, CVB4, CVA16, and E11 have been described elsewhere (23–27). RNA transcripts representing HEV species A to D and HRV species A to C were obtained from Peter Simmonds, University of Edinburgh (Edinburgh, United Kingdom) (28). Clinical isolates of human parechoviruses (HPEVs) 1 to 6 were provided by Katja Wolthers, Academic Medical Center (Amsterdam, The Netherlands) (29).
Clinical specimens.
LNA probes were validated with a total of 118 HRV- and HEV-positive clinical specimens, including 61 respiratory specimens that were subjected to post-PCR hybridization and Tm determination in the SYBR green assay. The clinical specimens were originally sent to the laboratory for diagnostic purposes, and they cover a wide range of specimen types (see Table 2). When the specimens were collected during a clinical research project, the ethics committee of the Turku University Hospital approved the study protocols. At least the partial 5′ NCR sequence, confirming the differentiation between HRV and HEV, was available for all specimens. Most of the HEVs were also typed by VP1 gene sequencing, or the typing results for the corresponding isolates were obtained from the THL Enterovirus Laboratory.
Table 2.
Clinical specimens (n = 118) tested by real-time PCR assay with LNA probes
| Specimen type (n, if >1) | Reference resultsa |
Test result(s) | |
|---|---|---|---|
| Species | Virus type (n, if >1) | ||
| Biopsy | HEV-A | EV71b | HEV |
| Bronchoalveolar lavage | HRV-A | Untyped | HRV |
| Cerebrospinal fluid (5) | HEV-A | EV71 | HEV |
| HEV-B | E30 (3), E30b | HEV | |
| Conjunctival fluid | HEV-C | CVA24b | HEV |
| Feces (43) | HEV-A | CVA2,b CVA6 (3), CVA16 (3), CVA16,b EV71b | HEV |
| HEV-B | CVA9,b CVB2, CVB3, CVB4b (2), CVB5 (2), E2,b E5, E6, E7, E9 (3), E9,b E13, E18,b E19, E20,b E25 | HEV | |
| HEV-C | CVA13,b CVA24 (2), CVA24b (6), EV96b | HEV | |
| HEV-Cb | PV2 and PV3d | HEV | |
| Dualb | CVA9 and CVA24 | HEV | |
| Dual | E18 and untyped HRV-B | HEV and HRV | |
| HRV-C | Untyped | HRV | |
| Nasal swab (54) | HEV-A | CVA2, CVA16 | HEV |
| HEV-D | Untyped | HEV and HRVc | |
| Dual | CVA6 and untyped HRV-C | HRV and HEV | |
| HRV-A | Untyped (24) | HRV | |
| HRV-B | Untyped (3) | HRV | |
| HRV-C | Untyped (22) | HRV | |
| HRV-C | Untyped | HRV and HEVc | |
| Nasal wash | HRV-C | Untyped | HRV |
| Nasopharynx aspirate (3) | HRV-A | Untyped | HRV |
| HRV-C | Untyped (2) | HRV | |
| Throat swab (4) | HRV-A | Untyped | HRV |
| HEV-A | CVA6, CVA16 | HEV | |
| HRV-B | Untyped | HRV | |
| Vesicular fluid (5) | HEV-A | CVA6 (5) | HEV |
Type refers to the VP1 genotyping result; for HRV, species determination was by 5′ NCR sequencing.
Cultured specimen.
Result not confirmed by sequencing.
Sabin poliovirus vaccine strains.
RNA extraction and RT.
Nucleic acids were extracted using an automated NucliSense easyMAG nucleic acid extractor (bioMérieux, Boxtel, The Netherlands) and stored at −70°C. RT reactions with 10 μl of extracted RNA and 1.2 μM reverse primer ENRI4− (Table 1) were performed in a total volume of 40 μl, as described previously (30), and cDNAs were stored at −70°C. HRV-A1b and HEV-E11 RNAs were used as positive controls for RT. A control without any RNA was included in each experiment.
Table 1.
Primers and probes
| Oligonucleotide | Sequencea | Reference no. |
|---|---|---|
| Reverse primer ENRI4− | 5′-GAA ACA CGG ACA CCC AAA GTA-3′ | 3, 32, 34 |
| Forward primer ENRI3+ | 5′-CGG CCC TGA ATG CGG CTA A-3′ | 3, 32, 34 |
| HRV LNA probe RIp1 | 5′-FAM-TYG GTY CCA TCC C-DQ1-3′ | |
| HRV LNA probe RIp2 | 5′-FAM-TCG GTY CCG TCC C-DQ1-3′ | |
| HEV LNA probe ENp1 | 5′-Cy5-TCG GTT CCG CTG C-DQ3-3′ | |
| HEV LNA probe ENp2 | 5′-Cy5-TCG GTT CCG CCA C-DQ3-3′ |
LNA bases are underlined; Y indicates T or C. FAM, 6-carboxyfluorescein; Cy5, indodicarbocyanine; DQ, dark quencher.
Primers.
Universal HRV/HEV forward primer ENRI3+ and reverse primer ENRI4− (Table 1) have been described and extensively used previously (19, 28, 31, 32). They amplify a ∼120-bp-long product at the 3′ end of the 5′ NCR of HRVs and HEVs. All of the standard DNA oligonucleotides were obtained from Oligomer (Espoo, Finland).
SYBR green-Tm and post-PCR hybridization assays.
PCR with SYBR green detection and melting curve analysis was performed as described previously (19). PCR products were captured on streptavidin-coated microtiter plate wells and measured with time-resolved fluorometry, using lanthanide chelate-labeled probes (32).
Sequence analyses.
Partial 5′ NCR sequences were amplified by PCR using the ENRI4− primer with a forward primer generating a ∼397-bp-long product (19, 30). HRV species were named according to the 5′ NCR sequence (33). For genotyping of HEVs, VP1 amplicons were generated as described previously (34). The amplicons were purified with a QIAquick PCR purification kit (Qiagen) and sequenced at the DNA Sequencing Service of the Turku Centre for Biotechnology (Turku, Finland). The species or HEV genotype of the virus corresponding to the studied sequence was assigned by using the NCBI Basic Local Alignment Tool (BLAST) (www.ncbi.nlm.nih.gov).
Real-time PCR with LNA probes and BOXTO.
Conditions for the real-time PCR for simultaneous amplification, HRV/HEV probe typing, and Tm determination were optimized in preliminary experiments. The final reaction mixture consisted of QuantiTect Probe master mix (Qiagen), 600 nM primers ENRI3+ and ENRI4−, 100 nM each of the probes RIp1, RIp2, ENp1, and ENp2 (Table 1), and 0.3 μM BOXTO (TATAA Biocenter, Gothenburg, Sweden) in a total volume of 25 μl. Amplifications were carried out in a Rotor-Gene 6000 instrument (Qiagen) with the following cycling parameters: 15 min at 95°C and then 50 cycles of 15 s at 95°C, 30 s at 65°C to 56°C with 1°C/cycle touchdown during the first 10 cycles, and 40 s at 72°C; melting curves were then generated by raising the temperature from 72°C to 95°C by 1°C every 5 s. Amplifications were monitored using green (6-carboxyfluorescein [FAM]), yellow (BOXTO), and red (Cy5) channels, with melting curves on the yellow channel of the instrument. Threshold levels were manually set to about one-tenth of the maximum signal. An exponential curve crossing the threshold within 42 cycles was considered a positive test result. If the amplification was measurable with BOXTO alone, then a Tm that was not lower than 1.5°C below the Tm of HRV-A1b (in this data set, ≥84°C) was considered to indicate product specificity. LNA oligonucleotides were obtained from Eurogentec (Seraing, Belgium).
RESULTS
Preliminary results with the SYBR green-Tm assay.
Target cDNAs from selected HRV and HEV strains were efficiently amplified with real-time PCR with SYBR green, and the melting temperature (Tm) was determined for each of them. The analysis included 17 different HRV A/B types and 25 different HEV types. Of the HRVs, 13/17 (76%) had an HRV-like Tm, while 22/25 (88%) HEVs had an HEV-like Tm. All except two of them were correctly grouped in the post-PCR hybridization assay.
The SYBR green-Tm assay was further evaluated with 61 clinical specimens. Partial 5′ NCR sequences were produced, compared with GenBank sequences using BLAST searches, and identified as HRVs or HEVs. The SYBR green-Tm assay correctly identified 51/61 (84%) clinical specimens. In contrast, the post-PCR hybridization assay gave correct unambiguous results for only 17 of 61 (28%) clinical specimens. Especially HRV species C strains were hybridization assay negative or gave false-positive reactions with the HEV probe. Thus, the results of the SYBR green-Tm assay for clinical specimens could not be accurately confirmed by the previous hybridization test.
LNA probe design.
HRV and HEV sequences of the 5′ NCR amplicons obtained from prototypes and clinical specimens were aligned with corresponding sequences available from GenBank (Fig. 1). According to the alignments, bases at two positions adjacent to the reverse primer were exclusively HRV or HEV specific; while most other bases were highly conserved, there was still too much variability for standard real-time PCR probe design. In order to reduce the number of variable bases within the probes, the probes were shortened to 13 bases in length by replacement of 4 or 5 DNA nucleotides with LNA nucleotides, which increases the Tm of the oligonucleotides (Fig. 1 and Table 1). To decrease the likelihood of incorrect binding, LNA bases included the two that differentiated HRVs from HEVs. Degenerate bases at specific positions in the sequence were designed for certain variable positions. Tm predictions for LNA probes, aiming at 70°C, were carried out with the Tm calculator provided by Exiqon (http://lna-tm.com), with conditions set to a salt concentration of 115 mM and a concentration of target plus probe of 0.5 μM. Reverse-strand sequences were selected for the probes because they had less G than C and their 5′ ends contained no G. The HRV LNA probes (RIp) were labeled at the 5′ end with FAM and HEV LNA probes (ENp) with Cy5, and both types of probes were labeled at the 3′ end with appropriate dark quencher (DQ).
Fig 1.

Alignment of HRV and HEV 5′ NCR cDNA sequences adjacent to the reverse primer. Different variants are shown for sequences of the LNA probe site corresponding to nucleotides 529 to 541 of the HRV-A1b genome (GenBank accession no. D00239). Nucleotides corresponding to the 3′ end of the reverse primer (ENRI4−) are underlined. Positions with either HRV- or HEV-specific bases at the probe site are shown in bold.
Real-time RT-PCR with LNA probes and BOXTO.
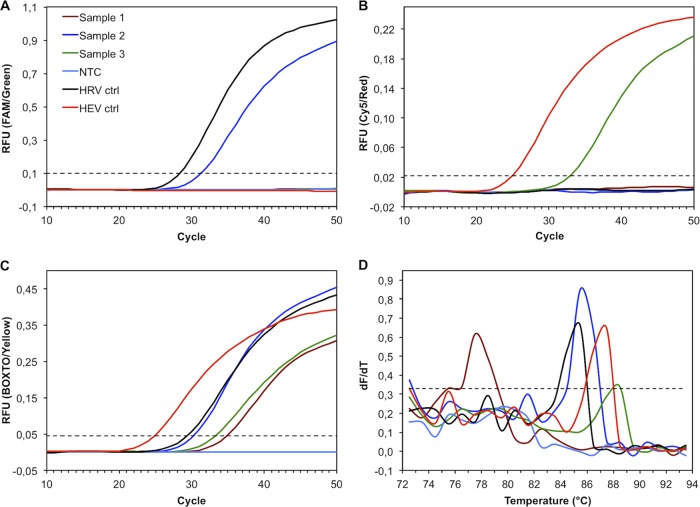
In the assay, HRV-specific amplicons reacting with RIp were monitored on the green channel (FAM) (Fig. 2A), HEV-specific amplicons reacting with ENp on the red channel (Cy5) (Fig. 2B), and any amplification (Fig. 2C) and the melting curve (Fig. 2D) on the yellow channel (BOXTO). The specificity of the differentiation was assessed by testing 100 HRV and 63 HEV prototypes. Probe fluorescence correlated well with that of BOXTO, indicating that it was mainly dependent on the amount of cDNA and not on the virus type (Fig. 3A and B). All of the HRV prototypes and all except one of the HEV prototypes were detected and correctly identified with the LNA probes. From the prototype sequence, CVA1 was predicted to have a potential mismatch (G to A) (Fig. 1) and was not detected with ENp, but it should be noted that the threshold cycle (CT) observed with BOXTO indicated a small amount of virus in the sample (Fig. 3B). On the other hand, 8 HRV prototypes with a potential mismatch combination of variable bases (variant 7) (Fig. 1) were detected with RIp as well as with BOXTO. HPEV prototypes 1 to 6 were negative in the assay.
Fig 2.
Real-time PCR for detection and discrimination of human rhinovirus (HRV) and human enterovirus (HEV) using double-dye LNA probes and the dsDNA dye BOXTO. (A) Amplification plots monitored on the green channel, measuring the relative fluorescence unit (RFU) signals generated by the FAM-labeled HRV probes. (B) Amplification plots monitored on the red channel, measuring the RFU signals generated by the Cy5-labeled HEV probes. (C) Amplification plots monitored on the yellow channel, measuring the RFU signals generated by the dsDNA dye BOXTO. (D) Melting curves plotted as the first derivative of fluorescence versus temperature (dF/dT). All panels have the same data set as indicated in panel A, including clinical specimens producing a nonspecific amplicon (Tm of <84°C; sample 1), an HRV-specific amplicon (sample 2), or an HEV-specific amplicon (sample 3), a no-template control (NTC), an HRV control (ctrl), and an HEV control. Dotted lines, manually set analytical threshold.
Fig 3.
Distribution of real-time PCR threshold cycle (CT) values for cDNA samples from cell culture supernatants of human rhinovirus (HRV) and human enterovirus (HEV) prototype strains and clinical specimens. Scatter plots of CT values observed with virus-specific probes over dsDNA dye BOXTO are shown. (A) HRV prototype strains (types 1 to 100, including 75 HRV-A and 25 HRV-B strains) detected with FAM-RIp. All strains were negative with the HEV-specific probe. (B) HEV prototype strains (12 HEV-A, 35 HEV-B, 15 HEV-C, and 4 HEV-D strains) detected with Cy5-ENp. CVA1 (black circle; value on the x axis for visualization only) was detected with BOXTO alone. All strains were negative with the HRV-specific probe. (C) Clinical specimens with HRVs (n = 58). (D) Clinical specimens with HEVs (n = 58). A conjunctival fluid specimen with CVA24 (black circle) was detected by ENp only. Clinical specimens that were tested after culture are shown as gray circles. In each plot, the dotted line represents a linear regression line with the indicated equation and coefficient of determination, R2.
The differentiation specificity was further validated with 118 different HRV- or HEV-positive clinical specimens, of which 95 were tested directly and 23 after culture (Table 2). The specimens included 58 specimens containing HEV strains, 58 specimens containing HRV strains, and 2 specimens containing both HEV and HRV strains. For two nasal swabs, mixed findings could not be confirmed; one produced only HEV-like 5′ NCR sequence and the other only HRV-like 5′ NCR sequence. The correlations between probe and dsDNA dye signals in single-virus-containing clinical specimens are shown in Fig. 3C and D. A CVA24-positive conjunctival fluid specimen produced a signal with ENp (red channel) and not with BOXTO (yellow channel), suggesting a low concentration of the virus in the specimen (Fig. 3D). Compared with the results verified by sequencing, the specificity of the LNA probes in differentiating between HRV and HEV in clinical specimens was 98% (116/118 specimens). The analytical specificity of the LNA probes was further demonstrated with clinical specimens containing other viruses, including adenovirus, influenza viruses A and B, respiratory syncytial virus, metapneumovirus, parainfluenza viruses 1 to 4, coronaviruses 229E, NL63, and OC43, bocavirus, herpes simplex viruses 1 and 2, cytomegalovirus, and Epstein-Barr virus. RT-PCR results with the LNA probes were negative (no CT or CT of >42) for all of these specimens. For negative specimens, nonspecific late amplifications showing no or low Tm (<84°C) were sometimes observed with the dsDNA dye BOXTO (Fig. 2C and D).
Sensitivity and repeatability.
The sensitivity and efficiency of the amplification steps with LNA probes and/or dsDNA dyes were determined using a dilution series of plasmid cDNA clones. Plasmid cDNAs were detected at a concentration of 5 copies/reaction, and the amplifications were linear to at least 5 × 107 copies/reaction with all methods of detection, with mean ± standard deviation (SD) efficiencies of 0.99 ± 0.03 (Table 3). The SD of the CT values for the different cDNAs, calculated at the standard curve intercept, was lower with the LNA probes (SD, 0.63) than with either SYBR green (SD, 1.42) or BOXTO (SD, 1.47). This might be due to sequence variations of the amplicons and differences in the amounts of dsDNA dye bound to them.
Table 3.
Standard curve parameters and amplification efficiencies for different plasmid virus cDNAs in PCRa
| Plasmid virus | Intercept parameters with: |
Slope parameters with: |
Efficiency parameters with: |
||||||
|---|---|---|---|---|---|---|---|---|---|
| FAM-RIp | Cy5-ENp | BOXTO | FAM-RIp | Cy5-ENp | BOXTO | FAM-RIp | Cy5-ENp | BOXTO | |
| HRV-A1b | 40.17 | 39.15 | −3.43 | −3.31 | 0.96 | 1.00 | |||
| HRV-B14 | 38.92 | 41.25 | −3.48 | −3.35 | 0.94 | 0.99 | |||
| HRV-A85 | 40.04 | 41.93 | −3.37 | −3.30 | 0.98 | 1.01 | |||
| CVA16 | 39.97 | 39.82 | −3.36 | −3.32 | 0.98 | 1.00 | |||
| CVB4 | 40.71 | 39.20 | −3.33 | −3.24 | 1.00 | 1.03 | |||
| EV11 | 39.46 | 42.60 | −3.33 | −3.24 | 1.00 | 1.04 | |||
Linear regression curves (y = mx + b, where y = CT, m = slope, x = log10[copies/PCR], and b = intercept when x = 0) and efficiencies (E = 10−1/m − 1) were determined with 10-fold dilutions containing 5 × 107 to 5 plasmid copies/reaction.
The sensitivity and repeatability of the RT-PCR assay with LNA probes were further assessed by using dilution series of in vitro transcribed HEVs representing species A to D and HRVs representing species A to C (28). Transcripts were tested in three separate assays at 10 to 10,000 copies of RNA/μl, corresponding to 12.5 to 12,500 cDNA copies/reaction in the PCR. Of the eight different transcripts, four were constantly detected at levels of ≤10 RNA copies/μl and four at ≤100 RNA copies/μl (Table 4). To assess the intra-assay repeatability, five replicates of each transcript at 100 RNA copies/μl were determined within the same assay. The SDs of the CT values varied from 0.2 to 1.8 cycles (Table 4).
Table 4.
Limits of detection and intra-assay repeatability of HRV and HEV transcripts
| Virus type | RNA LOD (copies/μl)a |
CT (mean ± SD) forb: |
|
|---|---|---|---|
| FAM-RIp | Cy5-ENp | ||
| CVA16 | ≤10 | NR | 36.8 ± 0.6 |
| E30 | 10–100 | NR | 37.6 ± 0.3 |
| EV70 | 10–100 | NR | 39.4 ± 0.9 |
| CVA21 | ≤10 | NR | 40.3 ± 0.7 |
| HRV-A16 | ≤10 | 36.0 ± 0.3 | NR |
| HRV-B14 | 10–100 | 38.2 ± 0.5 | NR |
| HRV-C40 | 10–100 | 41.0 ± 1.8 | NR |
| HRV-C pat19c | ≤10 | 37.0 ± 0.2 | NR |
LOD, limit of detection reproduced in three separate assays; 10 RNA copies/μl corresponds to 12.5 cDNA copies/reaction in the PCR.
Five replicates of 100 RNA copies/μl. NR, no reaction.
Provisionally assigned type (28).
DISCUSSION
In this study, a real-time RT-PCR assay with double-dye LNA probes unambiguously detected and differentiated HRVs and HEVs for nearly all cultivated prototypes and in different clinical samples. In addition, the assay was compatible with the use of the dsDNA dye BOXTO, which would facilitate detection of the few strains not covered by the probes.
The 5′ NCR of picornaviruses, due to its high rate of conservation, is an optimal target for highly sensitive diagnostic assays (4, 13–19). We have compared several combinations of primers corresponding to different conserved stretches in the 5′ NCR of HRVs and HEVs, and we obtained the most sensitive RT-PCR assay with the primers for the sites utilized here (30). Conventional assays with these universal primers detected both HRVs and HEVs, but their discrimination was not possible with amplicon length analyses. To overcome this problem, post-PCR hybridization assays were developed (32). Since then, we have found an increasing number of picornaviruses that could not be correctly identified with those probes. Sequence analysis usually revealed them to be HRVs. In this study, HRV-C strains were found to give false-positive reactions with the HEV hybridization probe. When using real-time PCR with melting curve analysis, it was possible to directly differentiate HRVs from HEVs with about 84% certainty. Unfortunately, the differentiation by Tm could be only arbitrarily defined, without a sharp line between HRVs and HEVs, and some strains would be misidentified.
To solve these problems, we designed new double-dye LNA probes that were shown to be highly specific for either HRVs or HEVs. The advantage of the LNA nucleotides, compared to DNA nucleotides, is that their higher affinity toward complementary nucleotides increases the Tm of an oligonucleotide, thus making the use of shorter probes feasible (35, 36). This comes with the obvious disadvantage of increased probability of probe reactions with identical but irrelevant sequences, underlining the importance of specific primers. To obtain further assurance of the assay specificity, we used LNA probes in combination with the dsDNA dye BOXTO (Fig. 2). Addition of the dsDNA-binding dye to the PCR mixture reveals the formation of primer dimers or other nonspecific products with deviating Tm values (37). Furthermore, specific amplicons with mismatches preventing probe binding or hydrolysis are likely to be detected. This was demonstrated in the case of the HEV type CVA1, which was detected only with BOXTO (Fig. 3). However, the observed Tm values differ from virus to virus and, as with any dsDNA dye, the Tm determination is a relative measure that depends on the assay conditions. A combination of double-dye probes with a dsDNA dye in real-time PCR would be a valuable tool also for diagnostic tests for many highly variable infectious disease agents other than HRV and HEV.
Real-time RT-PCR assays have been developed for HRVs and/or HEVs (20, 22, 38, 39). Tapparel et al. (18) also sequenced recent clinical strains to design two double-dye DNA probes that could specifically detect HRVs. However, since the HRV primers had a limited ability to detect HEVs, a secondary assay was used to identify HEVs. Kares et al. (22) described a SYBR green assay for the detection of HRVs and HEVs, without distinguishing between them, and a separate real-time PCR with LightCycler probes for the detection of HEVs. To our knowledge, PCR assays with LNA probes have not been used previously for the detection of HRVs and HEVs.
The sensitivity and repeatability of our RT-PCR assay were determined using RNA transcripts representing all HEV and HRV species. The limits of detection varied from ≤10 copies of RNA/μl (≤12.5 cDNA copies/reaction in the PCR) to 10 to 100 copies of RNA/μl among the transcripts (Table 4). The assay showed consistent repeatability at low concentrations of RNA transcripts, confirming the reliability of the assay and its suitability for diagnostic use. The amplification efficiency of the PCR was determined with plasmids with viral cDNAs, and technical plasmid replicates demonstrated excellent assay stability and linearity (Table 3). However, both transcripts and plasmids still represented only a few examples of all HRV or HEV strains, and variant viruses may affect the sensitivity of the assay and its use for quantification.
In our study, we validated the new LNA probes with 163 HRV or HEV prototype strains and 118 clinical specimens, to cover different sample and virus types. All enterovirus prototype strains except one (CVA1) were detected and correctly identified with LNA probes, and cross-reactions with other viruses were not found. Most of the clinical specimens were from the respiratory tract and included several representatives of newly discovered species C HRVs, which were clearly identified as HRVs. Thus, the 5′ NCR location of the primers and probes used in this study is optimal for the differential detection of HRVs and HEVs in the clinical material.
HRV is the most common cause of diverse acute respiratory infections in the human population. In addition, HRV has been frequently detected in stool specimens from children (40, 41). HEV is a frequent etiological agent in infections of the central nervous system; in such cases, cerebrospinal fluid samples are the best source for specific diagnosis. However, HEV can be detected much more often in the stool or respiratory secretions, and the finding can give valuable information for clinicians (42). Therefore, it is often important and beneficial to combine the specific detection of HRV and HEV in samples from the respiratory and gastrointestinal tracts. The specificity of the LNA probes for circulating strains of HEVs was demonstrated in a variety of clinical specimens assayed both directly and after cell culture isolation (Table 2).
The main purpose of our investigation was to improve the real-time, group-specific detection of HRVs and HEVs in clinical specimens. Real-time PCR also allows quantitative analysis with, for example, comparison of CT values or determination of copy numbers relative to a standard curve of known concentrations. Assessment of the viral load may be helpful in the clinical determination of whether HRV is causing the illness, in view of the fact that HRV findings are common also for asymptomatic subjects (43). In this study, a slightly higher mean CT was obtained with the LNA probes and BOXTO, compared to SYBR green (data not shown). There are two factors that might have influenced this, i.e., (i) the use of probe mixtures increases the fluorescence background and decreases the relative fluorescence intensity of the reactive probe and (ii) the concentration of BOXTO was relatively low, to avoid interference with probe reactions. Since the reaction efficiency was not affected, the shifts in CT values were insignificant and were compensated for by the use of 5 additional cycles in the PCR with LNA probes.
Detection of HRV and HEV, as well as differentiation between them, is challenging and time-consuming without straightforward methods. The real-time RT-PCR presented here takes advantage of double-dye LNA probes and BOXTO dye for rapid, sensitive, and specific detection and differentiation of HRV and HEV in a single assay format suitable for diagnostic laboratories.
ACKNOWLEDGMENTS
M.W., R.Ö., and T.H. are inventors in an application for a patent (M. Waris, R. Österback, T. Hyypiä, 28 May 2010, European Patent Office, patent application WO2010/136652) owned by the University of Turku and covering the application of the probes used in this study. During preparation of the manuscript, R.Ö. was supported by the Finnish Cultural Foundation and the Maud Kuistila Memorial Foundation.
Footnotes
Published ahead of print 18 September 2013
REFERENCES
- 1.Hyypiä T, Hovi T, Knowles NJ, Stanway G. 1997. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 78:1–11 [DOI] [PubMed] [Google Scholar]
- 2.Simmonds P, McIntyre C, Savolainen-Kopra C, Tapparel C, Mackay IM, Hovi T. 2010. Proposals for the classification of human rhinovirus species C into genotypically assigned types. J. Gen. Virol. 91:2409–2419 [DOI] [PubMed] [Google Scholar]
- 3.Knowles NJ, Hovi T, Hyypia T, King AMQ, Lindberg AM, Pallansch MA, Palmenberg AC, Simmonds P, Skern T, Stanway G, Yamashita T, Zell R. 2012. Family picornaviridae, p 855–880 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, New York, NY [Google Scholar]
- 4.Miller EK, Lu X, Erdman DD, Poehling KA, Zhu Y, Griffin MR, Hartert TV, Anderson LJ, Weinberg GA, Hall CB, Iwane MK, Edwards KM, New Vaccine Surveillance Network 2007. Rhinovirus-associated hospitalizations in young children. J. Infect. Dis. 195:773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruuskanen O, Waris M, Ramilo O. 2013. New aspects on human rhinovirus infections. Pediatr. Infect. Dis. J. 32:553–555 [DOI] [PubMed] [Google Scholar]
- 6.Kotaniemi-Syrjänen A, Vainionpää R, Reijonen TM, Waris M, Korhonen K, Korppi M. 2003. Rhinovirus-induced wheezing in infancy: the first sign of childhood asthma? J. Allergy Clin. Immunol. 111:66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, Printz MC, Lee WM, Shult PA, Reisdorf E, Carlson-Dakes KT, Salazar LP, DaSilva DF, Tisler CJ, Gern JE, Lemanske RF. 2008. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am. J. Respir. Crit. Care Med. 178:667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehtinen P, Ruohola A, Vanto T, Vuorinen T, Ruuskanen O, Jartti T. 2007. Prednisolone reduces recurrent wheezing after a first wheezing episode associated with rhinovirus infection or eczema. J. Allergy Clin. Immunol. 119:570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palacios G, Oberste MS. 2005. Enteroviruses as agents of emerging infectious diseases. J. Neurovirol. 11:424–433 [DOI] [PubMed] [Google Scholar]
- 10.McMinn PC. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 26:91–107 [DOI] [PubMed] [Google Scholar]
- 11.Osterback R, Vuorinen T, Linna M, Susi P, Hyypiä T, Waris M. 2009. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg. Infect. Dis. 15:1485–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bochkov YA, Palmenberg AC, Lee WM, Rathe JA, Amineva SP, Sun X, Pasic TR, Jarjour NN, Liggett SB, Gern JE. 2011. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat. Med. 17:627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kämmerer U, Kunkel B, Korn K. 1994. Nested PCR for specific detection and rapid identification of human picornaviruses. J. Clin. Microbiol. 32:285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loens K, Goossens H, de Laat C, Foolen H, Oudshoorn P, Pattyn S, Sillekens P, Ieven M. 2006. Detection of rhinoviruses by tissue culture and two independent amplification techniques, nucleic acid sequence-based amplification and reverse transcription-PCR, in children with acute respiratory infections during a winter season. J. Clin. Microbiol. 44:166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wisdom A, Leitch EC, Gaunt E, Harvala H, Simmonds P. 2009. Screening respiratory samples for detection of human rhinoviruses (HRVs) and enteroviruses: comprehensive VP4-VP2 typing reveals high incidence and genetic diversity of HRV species C. J. Clin. Microbiol. 47:3958–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyypiä T, Puhakka T, Ruuskanen O, Mäkelä M, Arola A, Arstila P. 1998. Molecular diagnosis of human rhinovirus infections: comparison with virus isolation. J. Clin. Microbiol. 36:2081–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, Holloway B, Dare RK, Kuypers J, Yagi S, Williams JV, Hall CB, Erdman DD. 2008. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 46:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tapparel C, Cordey S, Van Belle S, Turin L, Lee WM, Regamey N, Meylan P, Mühlemann K, Gobbini F, Kaiser L. 2009. New molecular detection tools adapted to emerging rhinoviruses and enteroviruses. J. Clin. Microbiol. 47:1742–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peltola V, Waris M, Osterback R, Susi P, Ruuskanen O, Hyypiä T. 2008. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J. Infect. Dis. 197:382–389 [DOI] [PubMed] [Google Scholar]
- 20.Deffernez C, Wunderli W, Thomas Y, Yerly S, Perrin L, Kaiser L. 2004. Amplicon sequencing and improved detection of human rhinovirus in respiratory samples. J. Clin. Microbiol. 42:3212–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dagher H, Donninger H, Hutchinson P, Ghildyal R, Bardin P. 2004. Rhinovirus detection: comparison of real-time and conventional PCR. J. Virol. Methods 117:113–121 [DOI] [PubMed] [Google Scholar]
- 22.Kares S, Lönnrot M, Vuorinen P, Oikarinen S, Taurianen S, Hyöty H. 2004. Real-time PCR for rapid diagnosis of entero- and rhinovirus infections using LightCycler. J. Clin. Virol. 29:99–104 [DOI] [PubMed] [Google Scholar]
- 23.Stanway G, Hughes PJ, Mountford RC, Minor PD, Almond JW. 1984. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 12:7859–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang KH, Auvinen P, Hyypiä T, Stanway G. 1989. The nucleotide sequence of coxsackievirus A9: implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269–3280 [DOI] [PubMed] [Google Scholar]
- 25.Hughes PJ, North C, Jellis CH, Minor PD, Stanway G. 1988. The nucleotide sequence of human rhinovirus 1B: molecular relationships within the rhinovirus genus. J. Gen. Virol. 69:49–58 [DOI] [PubMed] [Google Scholar]
- 26.Jenkins O, Booth JD, Minor PD, Almond JW. 1987. The complete nucleotide sequence of coxsackievirus B4 and its comparison to other members of the Picornaviridae. J. Gen. Virol. 68:1835–1848 [DOI] [PubMed] [Google Scholar]
- 27.Dahllund L, Nissinen L, Pulli T, Hyttinen VP, Stanway G, Hyypiä T. 1995. The genome of echovirus 11. Virus Res. 35:215–222 [DOI] [PubMed] [Google Scholar]
- 28.McLeish N, Witteveldt J, Clasper L, McIntyre C, McWilliam Leitch EC, Hardie A, Bennett S, Gunson R, Carman WF, Feeney SA, Coyle PV, Vipond B, Muir P, Benschop K, Wolthers K, Waris M, Osterback R, Johannessen I, Templeton K, Harvala H, Simmonds P. 2012. Development and assay of RNA transcripts of enteroviruses species A–D, rhinovirus species A–C and human parechovirus: assessment of assay sensitivity and specificity of real-time screening and typing methods. J. Clin. Microbiol. 50:2910–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benschop K, Thomas X, Serpenti C, Molenkamp R, Wolthers K. 2008. High prevalence of human parechovirus (HPeV) genotypes in the Amsterdam region and identification of specific HPeV variants by direct genotyping of stool samples. J. Clin. Microbiol. 46:3965–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santti J, Hyypiä T, Halonen P. 1997. Comparison of PCR primer pairs in the detection of human rhinoviruses in nasopharyngeal aspirates. J. Virol. Methods 66:139–147 [DOI] [PubMed] [Google Scholar]
- 31.Hyypiä T, Auvinen P, Maaronen M. 1989. Polymerase chain reaction for human picornaviruses. J. Gen. Virol. 70:3261–3268 [DOI] [PubMed] [Google Scholar]
- 32.Lönnrot M, Sjöroos M, Salminen K, Maaronen M, Hyypiä T, Hyöty H. 1999. Diagnosis of enterovirus and rhinovirus infections by RT-PCR and time-resolved fluorometry with lanthanide chelate labeled probes. J. Med. Virol. 59:378–384 [PubMed] [Google Scholar]
- 33.Piralla A, Rovida F, Campanini G, Rognoni V, Marchi A, Locatelli F, Gerna G. 2009. Severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J. Clin. Virol. 45:311–317 [DOI] [PubMed] [Google Scholar]
- 34.Nix WA, Oberste MS, Pallansch MA. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 44:2698–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen M, Nielsen CB, Nielsen KE, Jensen GA, Bondensgaard K, Singh SK, Rajwanshi VK, Koshkin AA, Dahl BM, Wengel J, Jacobsen JP. 2000. The conformations of locked nucleic acids (LNA). J. Mol. Recognit. 13:44–53 [DOI] [PubMed] [Google Scholar]
- 36.Kumar R, Singh SK, Koshkin AA, Rajwanshi VK, Meldgaard M, Wengel J. 1998. The first analogues of LNA (locked nucleic acids): phosphorothioate-LNA and 2′-thio-LNA. Bioorg. Med. Chem. Lett. 8:2219–2222 [DOI] [PubMed] [Google Scholar]
- 37.Lind K, Ståhlberg A, Zoric N, Kubista M. 2006. Combining sequence-specific probes and DNA binding dyes in real-time PCR for specific nucleic acid quantification and melting curve analysis. Biotechniques 40:315–319 [DOI] [PubMed] [Google Scholar]
- 38.Gambarino S, Costa C, Elia M, Sidoti F, Mantovani S, Gruosso V, Bergallo M, Cavallo R. 2009. Development of a RT real-time PCR for the detection and quantification of human rhinoviruses. Mol. Biotechnol. 42:350–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manayani DJ, Cherian T, Murali N, Finny GJ, Green J, Brown D, Ravi V, Abraham M, Sridharan G. 2002. Evaluation of a one-tube RT-PCR system for detection of enteroviruses. J. Clin. Virol. 24:25–30 [DOI] [PubMed] [Google Scholar]
- 40.Broberg E, Niemelä J, Lahti E, Hyypiä T, Ruuskanen O, Waris M. 2011. Human rhinovirus C-associated severe pneumonia in a neonate. J. Clin. Virol. 51:79–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honkanen H, Oikarinen S, Peltonen P, Simell O, Ilonen J, Veijola R, Knip M, Hyöty H. 2013. Human rhinoviruses including group C are common in stool samples of young Finnish children. J. Clin. Virol. 56:250–254 [DOI] [PubMed] [Google Scholar]
- 42.Kupila L, Vuorinen T, Vainionpäā R, Marttila RJ, Kotilainen P. 2005. Diagnosis of enteroviral meningitis by use of polymerase chain reaction of cerebrospinal fluid, stool, and serum specimens. Clin. Infect. Dis. 40:982–987 [DOI] [PubMed] [Google Scholar]
- 43.Jansen RR, Wieringa J, Koekkoek SM, Visser CE, Pajkrt D, Molenkamp R, de Jong MD, Schinkel J. 2011. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J. Clin. Microbiol. 49:2631–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]