Abstract
Enteric illness affects millions of individuals annually in the United States and results in >50,000 hospitalizations. The rapid and accurate identification of bacterial pathogens associated with gastroenteritis can aid acute patient management decisions, including the use of antibiotic therapy and infection control. This study compared the ProGastro SSCS multiplex real-time PCR assay (Gen-Probe Prodesse, San Diego, CA) to culture for the identification of Campylobacter spp. (Campylobacter jejuni and Campylobacter coli), Salmonella spp., and Shigella spp. and to broth enrichment followed by an FDA-cleared enzyme immunoassay (EIA) for the identification of Shiga toxin-producing Escherichia coli (STEC) isolates in stool specimens. Stool samples submitted in preservatives for routine culture and EIA were prospectively enrolled and tested at four clinical centers. Discrepancies between the ProGastro SSCS assay and culture or EIA were resolved using bidirectional sequencing. The overall prevalence of the pathogens as detected by culture was 5.6% (1.8% Campylobacter, 1.8% Salmonella, 1.3% Shigella, and 0.8% STEC). When results based on the ProGastro SSCS assay and bidirectional sequencing were applied, the overall prevalence increased to 8.3% (2.3% Campylobacter, 2.6% Salmonella, 1.8% Shigella, and 1.6% STEC). Following resolution of the discrepant results, the sensitivity of the ProGastro SSCS assay was 100% for all pathogens, and the specificities ranged from 99.4% to 100%. The sensitivity of culture compared to sequence-confirmed ProGastro SSCS results ranged from 52.9% to 76.9%, with the specificities ranging from 99.9% to 100%. Overall, these results suggest that the ProGastro SSCS assay is highly sensitive and specific in a clinical setting.
INTRODUCTION
Food-borne illness in the United States accounts for an estimated 9.4 million cases of gastroenteritis, ≥50,000 hospitalizations, and 1,351 deaths each year (1). Salmonella spp. and Campylobacter spp. (Campylobacter coli and Campylobacter jejuni) are recognized as the leading causes of bacterial gastroenteritis, followed by Shigella spp. and Shiga toxin-encoding Escherichia coli (STEC) (1, 2). Combined, these pathogens account for up to 82% of laboratory-confirmed food-borne bacterial enteritis cases (1). The vast majority of gastroenteritis cases are self-resolving, with symptoms persisting for <1 day (3). However, in at-risk populations, including infants, elderly, and immunocompromised patients, gastroenteritis can progress to severe or disseminated disease and become a significant cause of mortality (4–6). Of particular concern are the STEC organisms, which are defined as E. coli organisms of various serotypes harboring at least one of two Shiga toxin genes, stx1 and stx2. The expression of Shiga toxins by E. coli is associated with the development of hemolytic-uremic syndrome (HUS) in 2 to 10% of STEC infections (7, 8). Although HUS can develop in patients of any age, this potentially life-threatening condition is most frequently observed in children (9–11). Because of the relatively low prevalence of STEC and the significant additional cost of screening all stool specimens for Shiga toxins or genes, many laboratories routinely screen stool specimens from pediatric populations and forgo testing on adults (12, 13).
Routine clinical laboratory detection of Campylobacter (C. jejuni and C. coli), Salmonella, Shigella, and STEC includes the use of selective and differential culture media coupled with biochemical and serologic tests for identification, as well as the use of enzyme immunoassays (EIAs). While culture isolation and identification of pathogens are the current gold standard, these practices are laborious and time-consuming and may not be sufficiently sensitive. Specifically, EIAs for the detection of stx1 and stx2 may detect Shiga toxin in as little as 29% of STEC-positive specimens (14). Multiplex real-time PCR assays have been shown to be useful for the simultaneous detection of multiple respiratory tract pathogens, and a number of such assays have been cleared by the FDA for use in clinical laboratory settings. Similar assays for the detection of the agents of gastroenteritis have generally been limited to those that are for research use only (RUO) or for laboratory-developed tests (LDTs). Commercially available multiplexed molecular assays for the identification of enteric pathogens remain limited; however, assays, including the Seeplex Diarrhea-V ACE (Seegene, Seoul, South Korea), BD MAX enteric bacterial panel (BD, Sparks, MD), and xTAG GPP (Luminex, Austin, TX), have received mandatory conformity marking for products sold in the European Union (CE marking). Of these, only the xTAG GPP has also attained FDA clearance, and there are few published studies evaluating the performance of these multiplex tests (15–17).
The aim of this study was to prospectively compare the performance of the ProGastro SSCS assay (Gen-Probe Prodesse, San Diego, CA) to those of routine diagnostic methods for the identification of Campylobacter (C. jejuni and C. coli), Salmonella spp., Shigella spp., and stx1 and/or stx2 genes in stool specimens. An analysis of 1,139 prospectively tested patient specimens collected across four clinical centers serving different patient populations also provided epidemiological information regarding the nature of the pathogens detected.
(The results of this study were presented, in part, at the 113th General Meeting of the American Society for Microbiology in Denver, CO, 18 to 21 May 2013.)
MATERIALS AND METHODS
Collection of specimens.
A total of 1,244 specimens submitted for routine stool culture were tested at four U.S. clinical laboratories in accordance with site-specific institutional review board (IRB)-approved protocols. The inclusion criteria for samples included that stools had been diluted into a preservative transport medium, Cary-Blair (BD, Sparks, MD) or Para-Pak C&S (Meridian Bioscience, Cincinnati, OH) within 2 h of collection, that there was adequate residual volume for testing and archiving (≥0.5 ml), and that the samples had completed testing or were frozen at −70°C within 5 days of collection. Of the 1,244 specimens, 1,139 specimens were collected on a prospective basis over a 6-month time period (July through November 2011). This included 250/1,139 (22%) specimens that were extracted and tested within 5 days of collection and 889/1,139 (78%) specimens that were frozen prior to extraction for testing at a later time. An additional 105 previously characterized (via culture and EIA) frozen specimens collected between 2007 and 2011 were tested retrospectively to enrich them for positive specimens.
Culture and EIA methods.
Each stool protocol-compliant specimen was tested for Campylobacter (C. jejuni and C. coli), Salmonella, Shigella, and STEC using the clinical center standard of care culture method, which included the use of various selective and differential media, including xylose-lysine-deoxycholate agar, Hektoen enteric agar, Campylobacter blood agar, MacConkey agar, MacConkey agar with sorbitol, Trypticase soy agar with 5% sheep blood, Campy-thio broth (Remel and BD), and CHROMagar O157 (CHROMagar, Paris, France) for presumptive identification of the enteric pathogens. Campylobacter plates were incubated at 42°C for up to 72 h under microaerophilic conditions, and all other media were incubated at 35°C and were held up to 48 h before being regarded as negative. The presumptive colonies were fully identified using standard biochemical tests, including oxidase, hippurate hydrolysis, API 20 (bioMérieux, Durham, NC), RapID NF (Remel, Lenexa, KS), or a fully automated identification system, Phoenix (BD, Sparks, MD) or Vitek2 (bioMérieux, Marcy l'Etoile, France). The detection of stx1 and stx2 was accomplished using a toxin-specific enzyme immunoassay (Premier EHEC; Meridian Bioscience) following 18 to 24 h of enrichment in MacConkey broth (Remel, Lenexa, KS), according to the manufacturer's instructions. Samples that were positive for STEC by broth/EIA and/or the ProGastro SSCS assay underwent PCR followed by bidirectional sequencing to confirm the presence of the stx1 and/or stx2 genes. Two PCR/sequencing assays were used that targeted different regions of the stx1 and stx2 genes from those targeted by the ProGastro SSCS assay. A ProGastro SSCS assay result was considered to be true positive for STEC if that sample tested positive for STEC by the broth/EIA method and true negative if the sample tested negative for STEC by the broth/EIA method. A result was considered to be true positive for stx1 or stx2 if the sample tested positive for STEC by the broth/EIA method and by PCR/sequencing
Nucleic acid extraction and ProGastro SSCS PCR.
Stool specimens submitted in transport medium were processed according to the ProGastro SSCS package insert. Briefly, specimens in transport medium were further diluted 1:10 into Cary-Blair medium to a final concentration of approximately 1:40. A 100-μl aliquot of the dilution was combined with 10 μl of an internal control (included with kit). Nucleic acid extraction was conducted using the NucliSENS easyMAG system (bioMérieux) using the Specific A 1.0.2 protocol with an input and elution volume of 110 μl. An aliquot of extracted nucleic acid (5 μl) was combined with 20 μl of SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) or STEC mastermix and was analyzed using the SmartCycler II quantitative PCR (qPCR) system and Dx software version 1.7b or 3.0 (Cepheid, Sunnyvale, CA). The ProGastro SSCS multiplex PCR assay is composed of 7 different primer sets divided between two premixed mastermixes; the SSC mastermix detects and differentiates Salmonella, Shigella, and Campylobacter species (however, C. jejuni and C. coli are not differentiated), and the STEC mastermix detects and differentiates the Shiga toxin 1 (stx1) and Shiga toxin 2 (stx2) genes as an indicator of Shiga toxin-producing E. coli. The presence of each target was detected and interpreted using the SmartCycler Dx software, with threshold cycle (CT) values ranging from 13 to 45 for Salmonella spp., C. coli, C. jejuni, stx1, and stx2 and 13 to 37 for Shigella spp., indicating a positive result. External positive (provided with kit) and negative (uninoculated Cary-Blair or Para-Pak C&S media) controls were analyzed in parallel with each group of clinical specimens. Batches of up to 22 specimens (plus external controls) were completed in a total turnaround time (TAT) of 3 to 4 h, including the time for automated extraction.
Bidirectional sequencing for discrepant resolution and STEC verification.
Specimens with discordant culture and ProGastro SSCS results were resolved using PCR, followed by bidirectional sequencing. Additionally, since the Premier EHEC EIA does not differentiate between Stx1 and Stx2, all samples determined to be STEC positive via one or both testing methods (EIA or ProGastro assay) were verified by bidirectional sequencing for the detection and differentiation of stx1 and stx2. PCR amplification was conducted using multiple target-specific single-plex reactions on an ABI 2720 thermocycler using the following cycling conditions: 1 cycle at 95°C for 10 min (stage 1), 45 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min (stage 2), and 1 cycle at 72°C for 7 min (stage 3). The primers used in these assays targeted either a different region of the same gene used in the ProGastro SSCS assay or an alternative species-specific gene (Table 1). Following PCR, all samples and controls were analyzed by capillary electrophoresis using the Qiagen QIAxcel and QIAxcel BioCalculator software version 3.2 (Qiagen, Hilden, Germany). Samples demonstrating sufficient PCR product were prepared for sequencing by using ExoSap-IT (Affymetrix) and sent to Eurofins MWG Operon (Huntsville, AL) for bidirectional sequencing using the same primers used to generate the PCR product. The resulting sequence was analyzed using Geneious version 5.6.3, and the regions of the sequence with the most contiguous bases with a quality value of ≥20 were selected. Specifically, for all targets except Campylobacter, the sequencing product was 329 to 376 nucleotides, which provided just under 300 consecutive nucleotides for sequence analysis. The Campylobacter sequencing target was larger (820 nucleotides) and provided 600 to 700 nucleotides for sequence analysis. Selected segments of the sequencing data were run through the NCBI basic nucleotide BLAST program (http://www.ncbi.nlm.nih.gov/blast/), and the top three matches having E values of ≤1e−3 for the particular targets were reported as positive.
Table 1.
ProGastro SSCS targets and sequencing primers used for discrepant analysis
| Genus or gene target | Primer direction and sequence (5′ to 3′) | Sequencing target amplicon size (bp) | Sequencing assay gene target | ProGastro SSCS gene target |
|---|---|---|---|---|
| Salmonella | Forward, CCTGTCAGCCAAATATTACG | 344 | ttRSBCA | orgC |
| Reverse, ACGACGGGTTAAATTAGCCA | ||||
| Shigella | Forward, TGAAGTTTCTCTGCGAGCAT | 342 | ipaH | ipaH |
| Reverse, CAATACCTCCGGATTCCG | ||||
| Campylobacter | Forward, GGATGACACTTTTCGGAGC | 820 | 16S rRNA | glyA (C. jejuni) |
| Reverse, CATTGTAGCACGTGTGTC | cadF (C. coli) | |||
| stx1 | Forward, TGACAGTAGCTATACCACGT | 329 | stx1 | stx1 |
| Reverse, GAACAGAGTCTTGTCCATGA | ||||
| stx2 | Forward, GGACCTCACTCTGAACTG | 376 | stx2 | stx2 |
| Reverse, CCGCCATTGCATTAACAGAA |
Calculations and statistics.
Results from the ProGastro SSCS assay (the gold standard) were compared to those of the culture and EIA methods. Sensitivities and specificities were calculated using standard methods. Ninety-five-percent confidence intervals were calculated according to the efficient score method (corrected for continuity) as described by Newcombe (18).
RESULTS
Epidemiology.
Based on culture and EIA methodologies, the total prevalence of the four pathogens detected by the ProGastro SSCS assay in prospectively collected specimens was 5.64% (1.76% Campylobacter, 1.76% Salmonella, 1.32% Shigella, and 0.80% STEC); however, when combined molecular techniques (ProGastro SSCS assay with bidirectional sequencing confirmation) were applied, the total prevalence increased to 8.33% (2.28% Campylobacter, 2.63% Salmonella, 1.84% Shigella, and 1.58% STEC) (Fig. 1). Consistent with previously published epidemiologic studies, the highest prevalence of positive results (18.1% [27/149]) was observed in patients 2 to 5 years of age, with Shigella spp. as the primary pathogen detected in this age group (40.7% [11/27]). STEC was most prevalent in 12- to 18-year-old patients (5/18) and it was the second most common pathogen in patients ≤2 years of age (4/18). Campylobacter was the most prevalent pathogen detected (33/102); however, this was similar to the overall prevalence of Salmonella (30/102). No differences in prevalence were identified between the genders or between any of the age groups.
Fig 1.
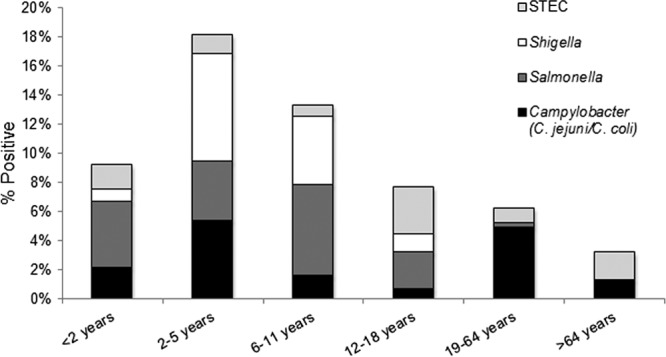
Positivity rate for ProGastro SSCS targets in different age groups. The total prevalence for all targets detected by the ProGastro SSCS assay in each age group is noted at the top of each bar graph. The shaded portions of each bar correspond to the proportion of each of the 4 pathogens in each group. The overall prevalence of positive stools in prospectively tested specimens was 5.64% using culture and EIA methods and 8.33% using combined molecular methods (ProGastro SSCS results confirmed using bidirectional sequencing).
Performance of the ProGastro SSCS assay using prospectively collected specimens.
Of the 1,153 specimens tested, 14 failed to generate a valid result upon initial or repeat test, resulting in a final call rate of 98.8%. The sensitivity and specificity values of the ProGastro SSCS assay for the detection of each target at each of the four clinical sites are presented in Table 2. Compared to culture and EIA, the overall sensitivity of the ProGastro SSCS assay was 98.5% (64/65) for all targets. This included 100% sensitivity for C. coli and C. jejuni (20/20), Shigella spp. (15/15), and stx1 and/or stx2 (9/9). The sensitivity for the identification of Salmonella spp. was a combined 95.2% (20/21) but was 100% at 3 of the 4 clinical centers. The combined specificity of the ProGastro SSCS assay for each target ranged from 98.9% to 99.4%.
Table 2.
Comparison of ProGastro SSCS assay to culture/EIA for prospectively tested specimens
| Species or gene target and clinical site | Resultsa |
Performance (% [95% CI])b |
||||
|---|---|---|---|---|---|---|
| TP | FP | TN | FN | Sensitivity | Specificity | |
| C. coli/C. jejuni | ||||||
| A | 9 | 8 | 327 | 0 | 100 (63–100) | 97.6 (95–99) |
| B | 5 | 1 | 382 | 0 | 100 (46–100) | 99.7 (98–100) |
| C | 6 | 2 | 277 | 0 | 100 (52–100) | 99.3 (97–100) |
| D | 0 | 2 | 120 | 0 | NA | 98.3 (94–100) |
| Total | 20 | 13c | 1,106 | 0 | 100 (80–100) | 98.9 (98–99) |
| Salmonella spp. | ||||||
| A | 5 | 0 | 339 | 0 | 100 (46–100) | 100 (99–100) |
| B | 7 | 4 | 377 | 0 | 100 (56–100) | 98.9 (97–100) |
| C | 1 | 0 | 284 | 0 | 100 (55–100) | 100 (98–100) |
| D | 7 | 6 | 108 | 1 | 87.5 (47–99) | 94.7 (88–98) |
| Total | 20 | 10d | 1,108 | 1e | 95.2 (74–100) | 99.1 (98–100) |
| Shigella spp. | ||||||
| A | 2 | 1 | 341 | 0 | 100 (5–100) | 99.4 (97–100) |
| B | 13 | 4 | 371 | 0 | 100 (72–100) | 98.9 (97–100) |
| C | 0 | 0 | 285 | 0 | NA | 100 (98–100) |
| D | 0 | 1 | 121 | 0 | NA | 99.1 (95–100) |
| Total | 15 | 6f | 1,118 | 0 | 100 (75–100) | 99.4 (99–100) |
| stx1 and/or stx2 | ||||||
| A | 0 | 2 | 341 | 0 | NA | 92.9 (90–95) |
| B | 4 | 2 | 383 | 0 | 100 (40–100) | 99.5 (98–100) |
| C | 4 | 3 | 278 | 0 | 100 (40–100) | 98.9 (97–100) |
| D | 1 | 2 | 119 | 0 | 100 (5–100) | 98.3 (94–100) |
| Total | 9 | 9g | 1,121 | 0 | 100 (63–100) | 99.2 (98–100) |
TP, true positive; FP, false positive, TN, true negative; FN, false negative;
CI, confidence interval; NA, not applicable.
Six of 13 were positive for C. jejuni or C. coli following bidirectional sequencing analysis.
Ten of 10 were positive for Salmonella spp. following bidirectional sequencing analysis.
One of 1 was negative for Salmonella spp. following bidirectional sequencing analysis.
Six of 6 were positive for Shigella spp. following bidirectional sequencing analysis.
Eight of 9 were positive for stx1, stx2, or both following bidirectional sequencing analysis.
Discrepant results between the ProGastro SSCS assay and culture and EIA methods were resolved using bidirectional sequencing (Table 3). Care was taken to select targets for discrepant resolution that were different from those utilized by the ProGastro SSCS assay. Following resolution, sensitivity and specificity for the detection of Salmonella spp. and Shigella spp. were 100%. Eight of nine false-positive stx1 and/or stx2 results were resolved as true positive (final specificity, 99.9%) and six of 13 false-positive C. coli and C. jejuni results were resolved as true positive (final specificity, 99.4%) following sequence analysis.
Table 3.
Performance of ProGastro SSCS assay for prospectively tested specimens following discrepant resolution
| Species or gene target | Resultsa |
Performance (% [95% CI])b |
||||
|---|---|---|---|---|---|---|
| TP | FP | TN | FN | Sensitivity | Specificity | |
| C. coli/C. jejuni | 26 | 7 | 1,106 | 0 | 100 (84–100) | 99.4 (99–100) |
| Salmonella spp. | 30 | 0 | 1,109 | 0 | 100 (86–100) | 100 (99–100) |
| Shigella spp. | 21 | 0 | 1,118 | 0 | 100 (81–100) | 100 (99–100) |
| stx1 and/or stx2 | 17 | 1c | 1,121 | 0 | 100 (78–100) | 99.9 (99–100) |
TP, true positive; FP, false positive, TN, true negative; FN, false negative.
CI, confidence interval.
Identified as stx1 by the ProGastro SSCS assay and was negative by EIA and bidirectional sequencing analysis.
Performance of the ProGastro SSCS assay using retrospectively analyzed specimens.
In addition to the prospective clinical study, two clinical sites performed retrospective analyses of 105 stool samples previously characterized using routine culture and EIA methods. This set of specimens was artificially enriched with a higher percentage of stools that were positive for each target, including 30.5% C. coli and C. jejuni, 2.86% Salmonella spp., 3.81% Shigella spp., and 18.1% stx1 and/or stx2. The sensitivity and specificity of the ProGastro SSCS assay for the identification of Salmonella spp., Shigella spp., and stx1 and/or stx2 were 100% in these specimens. The sensitivity and specificity for C. coli and C. jejuni were 96.4% and 93.5%, respectively; however, all discrepant results were resolved in support of the ProGastro SSCS results following bidirectional sequencing analysis (Table 4).
Table 4.
Comparison of ProGastro SSCS assay to routine culture/EIA for retrospectively tested specimens
| Species or gene target | Resultsa |
Performance (% [95% CI])b |
||||
|---|---|---|---|---|---|---|
| TP | FP | TN | FN | Sensitivity | Specificity | |
| C. coli/C. jejuni | 27 | 5c | 72 | 1d | 96.4 (80–100) | 93.5 (85–98) |
| Salmonella spp. | 3 | 0 | 102 | 0 | 100 (31–100) | 100 (95–100) |
| Shigella spp. | 4 | 0 | 101 | 0 | 100 (40–100) | 100 (95–100) |
| stx1 and/or stx2 | 19 | 0 | 86 | 0 | 100 (79–100) | 100 (95–100) |
TP, true positive; FP, false positive, TN, true negative; FN, false negative.
CI, confidence interval.
Five of 5 were positive for C. jejuni or C. coli following bidirectional sequencing analysis.
One of 1 was negative for C. jejuni or C. coli following bidirectional sequencing analysis.
Detection and differentiation of stx1 and stx2 in clinical specimens.
A total of 37 specimens (18 prospective, 19 retrospective) tested positive for stx1 and/or stx2 using the ProGastro SSCS assay. This included 17 (45.9%) positive for stx1, 9 (24.3%) positive for stx2, and 11 (29.7%) positive for both stx1 and stx2. Bidirectional sequence analysis confirmed 36/37 results. The single discrepancy was a specimen that had a positive result for stx1 by the ProGastro SSCS assay but a negative result for stx1 and/or stx2 by EIA and bidirectional sequencing (Table 5).
Table 5.
Differentiation of stx1 and stx2 in clinical specimens compared to bidirectional sequencing
| Gene target(s) | Resultsa |
Performance (% [95% CI])b |
||||
|---|---|---|---|---|---|---|
| TP | FP | TN | FN | Sensitivity | Specificity | |
| stx1 | 16 | 1c | 1,227 | 0 | 100 (76–100) | 99.9 (99–100) |
| stx2 | 9 | 0 | 1,235 | 0 | 100 (63–100) | 100 (99–100) |
| stx1 + stx2 | 11 | 0 | 1,233 | 0 | 100 (68–100) | 100 (99–100) |
TP, true positive; FP, false positive, TN, true negative; FN, false negative.
CI, confidence interval.
Identified as stx1 by the ProGastro SSCS assay and was negative by EIA and bidirectional sequencing analysis.
Culture/EIA sensitivity and specificity for detection of enteric pathogens.
A comparison of sequence-resolved ProGastro SSCS results to routine culture methods demonstrated culture sensitivities of 66.7% to 76.9% for detection of Salmonella spp., Shigella spp., and C. coli and C. jejuni in prospectively tested specimens (Table 6). Additionally, EIA-based detection of stx1 and/or stx2 following broth culture enrichment of specimens was only 50% as sensitive as the ProGastro SSCS assay in the prospectively tested specimens. In the current study, this resulted in the identification of an additional 6 (23%) cultures containing C. coli and C. jejuni, 10 (33%) cultures containing Salmonella spp., 6 (29%) cultures containing Shigella spp., and 8 (47%) cultures containing stx1 and/or stx2.
Table 6.
Culture/EIA sensitivity and specificity compared to sequence confirmed PCR results
| Species or gene target | Resultsa |
Performance (% [95% CI])b |
||||
|---|---|---|---|---|---|---|
| TP | FP | TN | FN | Sensitivity | Specificity | |
| C. coli/C. jejuni | 20 | 0 | 1,113 | 6 | 76.9 (56–90) | 100 (99–100) |
| Salmonella spp. | 20 | 1 | 1,108 | 10 | 66.6 (47–82) | 99.9 (99–100) |
| Shigella spp. | 15 | 0 | 1,118 | 6 | 71.4 (48–88) | 100 (99–100) |
| stx1 and/or stx2 | 9 | 0 | 1,122 | 8 | 52.9 (29–76) | 100 (99–100) |
TP, true positive; FP, false positive, TN, true negative; FN, false negative.
CI, confidence interval.
DISCUSSION
This study examines the clinical performance (sensitivity and specificity) of a novel multiplex PCR assay compared to those of the gold standard culture and EIA techniques for the rapid detection of four bacterial pathogens commonly associated with gastroenteritis: Salmonella, Shigella, Campylobacter (C. jejuni and C. coli), and STEC.
The identification of bacterial pathogens associated with acute gastroenteritis is accomplished primarily through the culture of stool specimens on various selective and differential screening media, followed by confirmatory identification using manual or automated biochemical tests (API20, RapID NF, BD Phoenix, Vitek2, etc.). This process requires initial inoculation of the stool specimen onto numerous pieces of media, followed by 18 to 24 h of incubation, and inspection of plates for characteristic colony morphology and often requires further subculture and incubation before a final identification can be made. These steps are labor-intensive and can require a minimum of 48 to 72 h before results can be reported to the physician. This delay in turnaround time (TAT) often results in the use of empirical antibiotic therapy, which may be unnecessary or even detrimental to patient care (19–23). Specifically, the use of fluoroquinolones has been associated with prolonged shedding of Salmonella spp. in stool and an increased rate of resistance in Campylobacter spp., and the use of β-lactams, sulfonamides, and quinolones has been associated with an increased risk of HUS when used to treat STEC-associated enteritis (19–23). Therefore, the use of antibiotics for enteritis attributed to Salmonella spp. or Campylobacter spp. should be reserved for patients at risk for severe or invasive disease, and antibiotic therapy is not appropriate for the treatment of STEC (4, 5). Conversely, antibiotic therapy is recommended for all patients infected with Shigella spp. because of the ease of transmission of these organisms and impact of the drugs on infection control (5).
Currently available methods for the rapid identification of enteric bacterial pathogens include enzyme immunoassays (EIAs) and nucleic acid amplification tests (NAATs). Among the commonly used EIAs are those that detect Campylobacter spp. and Stx1/2. A study comparing three commercially available Campylobacter species EIAs found sensitivity values of 98.5% to 99.3% and specificity values of 98.0 to 98.2% compared to culture methods (24). These assays were used to directly interrogate stool specimens submitted in Cary-Blair medium, and the results were available within 2 h. The detection of STEC can be accomplished using culture or EIA. Culture-based methods utilize MacConkey agar containing sorbitol as a screen and are limited to the detection of E. coli O157 serotypes, which are typically unable to ferment sorbitol. The use of EIAs for the direct detection of Stx1/2 results in increased sensitivity compared to that of culture because of the ability to detect Shiga toxins in both O157 and non-O157 serotypes (14, 25). In spite of this advantage, recent data suggest that Stx1/2 EIAs may be only 29% as sensitive as molecular methods (14).
There are currently 3 commercially available molecular tests (in addition to the ProGastro SSCS assay) aimed at the identification of enteric pathogens in stool specimens. The Seeplex Diarrhea-V ACE (Seegene, Seoul, South Korea) and the BD MAX enteric bacterial panel (BD, Sparks, MD) have received CE marking for use in Europe, and the xTAG GPP (Luminex, Austin, TX) has received both CE marking and FDA clearance. The Seeplex test is composed of 3 independent panels that if run together are capable of identifying 9 bacterial and 3 viral pathogens associated with gastroenteritis. The xTAG GPP test identifies 9 bacterial, 3 viral, and 3 parasitic pathogens. Both assays can be completed in approximately 5 h. Compared to routine culture, EIA, and singleplex PCR methods, the Seeplex test was 100% sensitive for 6/9 targets but was only 84.2%, 50.0%, and 87.5% sensitive for Salmonella spp., Clostridium difficile, and rotavirus, respectively (15). The sensitivity of the xTAG GPP was >95% for all targets except Norovirus strain GII (92.5%), Salmonella spp. (82.7%), enterotoxigenic E. coli (90.6%), and Cryptosporidium (91.7%) (16). A similar study reported significantly more positive results using xTAG GPP for rotavirus, norovirus, Campylobacter spp., Salmonella spp., and C. difficile than with routine methods (17). A potential weakness of highly multiplexed panels is in the interpretation of results that indicate infection with multiple pathogens or in positive results that do not correlate with patient presentation. A positive result for C. difficile in an otherwise healthy individual from the community with no recent exposure to antibiotics or a health care environment may be the result of asymptomatic carriage rather than C. difficile disease. Likewise, the use of a multiplexed panel to test a symptomatic individual who has been hospitalized for >3 days and has a low risk of community-acquired enteritis may not be cost-effective.
Herein, we report 100% postresolution sensitivity for the identification of C. coli and C. jejuni, Salmonella spp., Shigella spp., and STEC (stx1 and/or stx2) in 1,139 prospectively tested and 105 retrospectively tested stool specimens using the ProGastro SSCS assay. When the prevalences of these four pathogens were analyzed by culture methods alone, 5.64% (1.76% Campylobacter, 1.76% Salmonella, 1.32% Shigella, and 0.80% STEC) of 1,139 prospective patient specimens were positive. The prevalence increased to 8.33% (2.28% Campylobacter, 2.63% Salmonella, 1.84% Shigella, and 1.58% STEC) when molecular techniques were applied. Of particular interest was the identification of nearly double the number of specimens that were positive for stx1 and/or stx2 using the ProGastro SSCS assay compared to the number detected using culture-enriched EIA. HUS secondary to STEC gastroenteritis is most commonly associated with E. coli serotype O157; however, stx1 and/or stx2 have been identified in >150 non-O157 serotypes (26) and may cause up to 20 to 50% of STEC-related illness and 33% of STEC-related deaths (2, 6, 26). For this reason, the CDC recently recommended culture-based screening for E. coli O157 as well as direct antigen or nucleic acid testing for stx1 and/or stx2 in all specimens submitted for stool culture (8). Despite this recommendation, many laboratories do not routinely screen stool samples because of the relatively low prevalence of E. coli O157 and high cost of performing EIA or NAAT on all submitted stool specimens (12, 13). Multiplex nucleic acid amplification-based panels facilitate the routine testing of stool specimens for stx1 and/or stx2, along with other enteric pathogens, thereby enabling compliance with the CDC recommendation and the Joint Commission's updated standard (http://www.jointcommission.org/standards_information/standards.aspx) and aiding patient care by identifying significantly more stx1- and/or stx2-positive stools than culture or EIA methods.
The specificity of the ProGastro SSCS assay following discrepant resolution was 100% for Salmonella spp. and Shigella spp., 99.9% for stx1 and/or stx2, and 99.4% for C. coli and C. jejuni. Seven of 13 specimens that were culture negative and ProGastro SSCS positive (i.e., false positive) for C. coli and C. jejuni were not resolved as positive by bidirectional sequencing. These false-positive results might be due to (i) low nucleic acid concentrations in the specimens that were not detected during bidirectional sequence analysis, (ii) cross-reactivity of the ProGastro SSCS primer-probe set with closely related Campylobacter spp., or (iii) laboratory contamination with template or amplicon. Eight of nine specimens that were EIA negative and ProGastro SSCS positive for stx1 or stx2 were confirmed to be positive by bidirectional sequence analysis. Despite the apparent increase in sensitivity for the detection of stx1 and/or stx2, it is important to note that the detection of nucleic acid does not necessarily correlate with the presence of toxin, and the results of this study were not correlated with any specific clinical syndrome. Specimens that are culture negative but test positive using a nucleic acid amplification assay may be the result of superior test sensitivity; however, these results might also be due the presence of nonviable organisms, free DNA, or a clinically insignificant quantity of a potential pathogen. Therefore, it is important to correlate any nonquantitative nucleic acid test result with clinical symptoms and patient history.
One benefit of the ProGastro SSCS assay is the use of real-time PCR for the amplification and detection of targets. This reduces the risk of amplicon contamination that is inherent in molecular assays that require the manipulation of postamplification products prior to detection. Another advantage is the lack of a requirement for culture confirmation of results, as indicated in the FDA-cleared product insert. A drawback to the assay is its requirement for offline extraction of nucleic acids prior to analysis and the necessity of setting up duplicate real-time PCRs to accommodate the detection of all targets. These steps add labor and can complicate the assay setup. Another drawback is the lack of additional high-prevalence targets, such as norovirus, which accounts for up to 58% of food-borne enteritis cases (1); however, the ability to rapidly rule out the 4 most common causes of bacterial gastroenteritis may allow for a more focused workup of submitted specimens for viral or less common bacterial or protozoan pathogens.
The strengths of this study include the large number of prospectively collected stools tested, along with enrichment for infrequent targets (Shigella spp., STEC) using a retrospectively tested cohort of specimens. Comparable sensitivity and specificity characteristics across 4 clinical centers highlight the reproducibility of the results across different regions and between different laboratories and operators. A potential weakness of the study includes the use of laboratory-specific reference culture and EIA methods rather than a standardized protocol; however, this also substantiates the increased sensitivity of the ProGastro SSCS assay compared to the reference method used at each study site, regardless of the culture method or medium.
ACKNOWLEDGMENTS
Gen-Probe Prodesse provided materials and support for this study. L.C. received travel support to present the results, in part, at the 113th Meeting of the American Society for Microbiology in Denver, CO.
We thank Milena Pitashny Hoffman, Baylor College of Medicine and Texas Children's Hospital, for performing STEC EIAs for the specimens tested at that site.
Footnotes
Published ahead of print 18 September 2013
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. MMWR Morb. Mortal. Wkly. Rep. 59:418–422 [PubMed] [Google Scholar]
- 3.Herikstad H, Yang S, Van Gilder TJ, Vugia D, Hadler J, Blake P, Deneen V, Shiferaw B, Angulo FJ. 2002. A population-based estimate of the burden of diarrhoeal illness in the United States: FoodNet, 1996-7. Epidemiol. Infect. 129:9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon MA. 2008. Salmonella infections in immunocompromised adults. J. Infect. 56:413–422 [DOI] [PubMed] [Google Scholar]
- 5.Guerrant RL, Van Gilder T, Steiner TS, Thielman NM, Slutsker L, Tauxe RV, Hennessy T, Griffin PM, DuPont H, Sack RB, Tarr P, Neill M, Nachamkin I, Reller LB, Osterholm MT, Bennish ML, Pickering LK, Infectious Diseases Society of America 2001. Practice guidelines for the management of infectious diarrhea. Clin. Infect. Dis. 32:331–351 [DOI] [PubMed] [Google Scholar]
- 6.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J. Infect. Dis. 192:1422–1429 [DOI] [PubMed] [Google Scholar]
- 8.Gould LH, Bopp C, Strockbine N, Atkinson R, Baselski V, Body B, Carey R, Crandall C, Hurd S, Kaplan R, Neill M, Shea S, Somsel P, Tobin-D'Angelo M, Griffin PM, Gerner-Smidt P, Centers for Disease Control and Prevention (CDC) 2009. Recommendations for diagnosis of Shiga toxin-producing Escherichia coli infections by clinical laboratories. MMWR Recommend. Rep. 58(RR-12):1–14 [PubMed] [Google Scholar]
- 9.Gyles CL. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85(13 Suppl):E45–E62 [DOI] [PubMed] [Google Scholar]
- 10.Karmali MA. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karmali MA, Petric M, Lim C, Fleming PC, Arbus GS, Lior H. 2004. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. 1985. J. Infect. Dis. 189:556–563 [DOI] [PubMed] [Google Scholar]
- 12.Kiska DL, Riddell SW. 2011. Counterpoint: should all stools be screened for Shiga toxin-producing Escherichia coli? J. Clin. Microbiol. 49:2394–2397 [PubMed] [Google Scholar]
- 13.Marcon MJ. 2011. Point: should all stools be screened for Shiga toxin-producing Escherichia coli? J. Clin. Microbiol. 49:2390–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vallières E, Saint-Jean M, Rallu F. 2013. Comparison of three different methods for detection of Shiga toxin-producing Escherichia coli in a tertiary pediatric care center. J. Clin. Microbiol. 51:481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coupland LJ, McElarney I, Meader E, Cowley K, Alcock L, Naunton J, Gray J. 2012. Simultaneous detection of viral and bacterial enteric pathogens using the Seeplex diarrhea ACE detection system. Epidemiol. Infect. 141:2111–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claas E, Burnham CA, Mazulli T, Templeton K, Topin F. 2013. Performance of the xTAG gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J. Microbiol. Biotechnol. 23:1041–1045 [DOI] [PubMed] [Google Scholar]
- 17.Mengelle C, Mansuy JM, Prere MF, Grouteau E, Claudet I, Kamar N, Huynh A, Plat G, Benard M, Marty N, Valentin A, Berry A, Izopet J. 2013. Simultaneous detection of gastrointestinal pathogens with a multiplex Luminex-based molecular assay in stool samples from diarrhoeic patients. Clin. Microbiol. Infect. 10.1111/1469-0691.12255 [DOI] [PubMed] [Google Scholar]
- 18.Newcombe RG. 1998. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat. Med. 17:857–872 [DOI] [PubMed] [Google Scholar]
- 19.Wiström J, Jertborn M, Ekwall E, Norlin K, Söderquist B, Strömberg A, Lundholm R, Hogevik H, Lagergren L, Englund G, Norrby SR. 1992. Empiric treatment of acute diarrheal disease with norfloxacin. A randomized, placebo-controlled study. Swedish Study Group. Ann. Intern. Med. 117:202–208 [DOI] [PubMed] [Google Scholar]
- 20.Carlstedt G, Dahl P, Niklasson PM, Gullberg K, Banck G, Kahlmeter G. 1990. Norfloxacin treatment of salmonellosis does not shorten the carrier stage. Scand. J. Infect. Dis. 22:553–556 [DOI] [PubMed] [Google Scholar]
- 21.Goodman LJ, Trenholme GM, Kaplan RL, Segreti J, Hines D, Petrak R, Nelson JA, Mayer KW, Landau W, Parkhurst GW, Levin S. 1990. Empiric antimicrobial therapy of domestically acquired acute diarrhea in urban adults. Arch. Intern. Med. 150:541–546 [PubMed] [Google Scholar]
- 22.Wong CS, Jelacic S, Habeeb RL, Watkins SL, Tarr PI. 2000. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, McDaniel AD, Wolf LE, Keusch GT, Waldor MK, Acheson DW. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664–670 [DOI] [PubMed] [Google Scholar]
- 24.Granato PA, Chen L, Holiday I, Rawling RA, Novak-Weekley SM, Quinlan T, Musser KA. 2010. Comparison of premier CAMPY enzyme immunoassay (EIA), ProSpecT Campylobacter EIA, and ImmunoCard STAT! CAMPY tests with culture for laboratory diagnosis of Campylobacter enteric infections. J. Clin. Microbiol. 48:4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehl KS, Havens P, Behnke CE, Acheson DW. 1997. Evaluation of the premier EHEC assay for detection of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 35:2051–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson KE, Thorpe CM, Sears CL. 2006. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 43:1587–1595 [DOI] [PubMed] [Google Scholar]


