Abstract
The current pathogen-typing methods have suboptimal sensitivities and specificities. DNA sequencing offers an opportunity to type pathogens with greater degrees of discrimination using single nucleotide polymorphisms (SNPs) than with pulsed-field gel electrophoresis (PFGE) and other methodologies. In a recent cluster of Escherichia coli O157:H7 infections attributed to salad bar exposures and romaine lettuce, a subset of cases denied exposure to either source, although PFGE and multiple-locus variable-number tandem-repeat analysis (MLVA) suggested that all isolates had the same recent progenitor. Interrogation of a preselected set of 3,442,673 nucleotides in backbone open reading frames (ORFs) identified only 1 or 2 single nucleotide differences in 3 of 12 isolates from the cases who denied exposure. The backbone DNAs of 9 of 9 and 3 of 3 cases who reported or were unsure about exposure, respectively, were isogenic. Backbone ORF SNP set sequencing offers pathogen differentiation capabilities that exceed those of PFGE and MLVA.
INTRODUCTION
Pathogen typing (1) can identify food-borne infections with common origins, enable the removal of contaminated foods still in commerce, and facilitate tracing contaminations back to their sources. The typing is usually performed in public health laboratories where the endonuclease-cleaved bacterial DNA is separated using pulsed-field gel electrophoresis (PFGE) (2) to produce isolate-specific patterns. The retrospective determination that Escherichia coli O157:H7 isolates from different individuals in a 1993 outbreak had identical PFGE patterns (3) prompted adoption of this technique in North America and elsewhere (2–5). While PFGE is widely used as a tool in outbreak management, its limitations are emerging. Bands of similar size comigrate, which can obscure differentiating fragments (6). A few point mutations (single nucleotide polymorphisms [SNPs]) reflect considerable phylogenetic distances between isolates but do not change the PFGE patterns. For these two reasons, isolates might appear identical when they are not. Conversely, PFGE patterns can change during subculturing, presumably because of the mobilization of elements of the genome such as bacteriophages (7, 8). Such alterations can also occur in the course of human infections (9, 10). Therefore, the progeny of the same recent bacterial progenitor can appear to be different. Furthermore, the comparisons are best performed using DNA separated on the same gels, necessitating cumbersome transfers of materials (pathogens or their DNA) to central laboratories.
When the PFGE patterns and epidemiologic data are at variance, it can be difficult to take well-informed action. To help resolve such scenarios, multiple-locus variable-number tandem-repeat analysis (MLVA), which exploits the patterns of amplicons generated by primers that flank variable-length regions of the genome, is sometimes employed (11, 12). However, as with PFGE, isolates of different origins can have identical MLVA patterns, because this technique will not resolve single nucleotide differences. Furthermore, MLVA is not widely available.
The ideal typing method would use easily conveyed data generated at decentralized facilities, not be prone to interpretation artifacts, and be unaffected by bacterial mutations that occur ex vivo or in the host (10). Additionally, this ideal method would produce sufficiently powerful data such that if two different isolates produce identical readouts, then there would be considerable confidence that they have the same recent progenitor. Whole-genome bacterial sequencing can convey unambiguous data, but pathogenicity islands, representing approximately one-fourth of the E. coli O157:H7 chromosome (13), are unstable. Hence, pathogens with the same recent progenitor acquired from the same vehicle might appear allogeneic after passing through and replicating in humans, which is one of the problems that diminishes the utility of PFGE, as noted above.
By focusing on the set of largely invariant backbone open reading frames (ORFs) in pathogen chromosomes, it is theoretically possible that isolate-specific SNPs (14) can be used to precisely type the pathogens. Indeed, the value of SNPs for identifying outbreak-related E. coli O157:H7 was suggested by Underwood et al. (15), who used whole-genome sequencing to generate a set of four SNPs, which were then used to SNP type additional isolates with identical MLVA patterns. However, the backbone chromosome ORFs common to all E. coli O157:H7 isolates contain 3,442,673 different nucleotides for analysis, and SNP typing would not be sufficiently sensitive to identify the mutations that provide a greater level of phylogenetic resolution. Specifically, one can state that two strains are highly likely to have descended from a recent common ancestor only if such a large set is interrogated, in view of the low rate with which backbone SNPs occur (approximately once every 10 years [14]) in this lineage.
Here, we dissect a time-space cluster of E. coli O157:H7 infections using PFGE, MLVA, and backbone ORF SNPs. The gel-based methods did not distinguish the isolates of cases who denied exposure to the incriminated source from those who claimed exposure, whereas SNP profiling reconciled a subset of the discrepancies.
MATERIALS AND METHODS
Characteristics of the outbreak.
On 24 October 2011, the St. Louis County Department of Health identified a cluster of E. coli O157:H7-infected patients, many of whom had recently consumed salad bar items at different supermarket chain A stores. In the following days, cases from other counties were identified, and the Missouri Department of Health and Senior Services and the Centers for Disease Control and Prevention joined local officials in investigating this growing cluster. A matched case-control study was conducted on 2 November 2011 to identify the likely source of infection. The interview questionnaire included questions about exposures historically associated with E. coli O157:H7 and consisted of items on the Standardized National Hypothesis-Generating Questionnaire developed with the Centers for Disease Control and Prevention to which ≥50% of the cases claimed exposure in the 7 days before illness onset. The 22 cases for the case-control study were defined as Missouri residents from the metropolitan St. Louis area whose illness was identified as of 2 November, whose diarrhea began after 7 October, and whose stool cultures yielded E. coli O157:H7 isolates with PFGE and MLVA outbreak patterns. These patterns were XbaI pattern EXH01.0047 and BlnI pattern EXHA26.0015 on PFGE (pattern A/C) and MLVA pattern A1 (8-7-10-4-4-7-10-7), if MLVA was performed prior to the case-control study. The 82 controls were individuals who were neighborhood matched to the cases using a reverse directory of landline telephones, were frequency matched by age category (<18, 18 to 50, and >50 years), and reported no diarrhea in the month preceding the interview. Controls were questioned on 2 November about their exposures during the third week of October 2011 (to correspond to the case exposure period).
This cluster and its investigation were recently detailed (16). This publication included as cases three individuals who had been excluded from the case-control study. Isolates from two of these three excluded cases had the outbreak isolate PFGE and MLVA patterns, but these cases resided in Boone County, Missouri, and were excluded because, for logistic reasons, controls were selected only in the St. Louis metropolitan area. The isolate from the third excluded case, a St. Louis resident infected in October 2011, had PFGE XbaI pattern EXH01.0047and BlnI pattern EXHA26.1381 (pattern A/D).
Cases and isolates analyzed.
We genome sequenced the isolates from 22 of the 37 Missouri cases (designated cases M1 to M22) and each of the 2 Kansas cases (designated cases K1 and K2) (16). We also sequenced five isolates that were believed to be completely unrelated to the outbreak, even though four had the outbreak PFGE pattern A/C. Two of these isolates were from Washington State cases (Wa1 and Wa2) who had no linkage to each other and were infected in October 2011 with E. coli O157:H7 that had non-A1 MLVA patterns. The other two isolates were from Webster County cases, one of whom was a historic case (July 2011) (WC1), while the second isolate, which was from a case whose illness occurred during the outbreak (WC2), had a non-A1 MLVA pattern. The fifth unrelated isolate was from a St. Louis patient (case M23) who denied exposure to the salad bar or to romaine lettuce. This isolate had a nonoutbreak PFGE pattern that completely differed from that of the outbreak pattern.
Gel-based pathogen typing.
All study isolates were subjected to XbaI and Bln1 PFGE at the Missouri State Public Health, Kansas Health and Environmental, or Washington State Public Health laboratories, and stx and stx-bacteriophage insertion site genotyping (17) was done at Washington University. Most study isolates underwent MLVA at the Minnesota Department of Health Public Health Laboratory, using a validated eight-locus interrogation on a Beckman Coulter CEQ 8000 electrophoresis platform (18).
Sequencing and backbone ORF SNP set assignments.
DNA from the study strains was sequenced, and SNPs were identified, validated, and confirmed, as described in the supplemental material.
Statistics.
We used Fisher's exact test (2-tailed P values) to determine the statistical significance of differences in the ratios of SNPs that are synonymous (mutations that do not change the corresponding amino acid structures) to SNPs that are nonsynonymous (mutations that do change the corresponding amino acids) in different strain sets.
Nucleotide sequence accession numbers.
This whole-genome shotgun project has been deposited at DDBJ, EMBL, and GenBank under accession no. AWPM00000000, AWPN00000000, AWPO00000000, AWPP00000000, AWPQ00000000, AWPR00000000, AWPS00000000, AWPT00000000, AWPU00000000, AWPV00000000, AWPW00000000, AWPX00000000, AWPY00000000, AWPZ00000000, AWQA00000000, AWQB00000000, AWQC00000000, AWQD00000000, AWQE00000000, AWQF00000000, AWQG00000000, AWQH00000000, AWQI00000000, AWQJ00000000, AWQK00000000, AWQL00000000, AWQM00000000, AWQN00000000, and AWQO00000000 (isolates M1 to M23, K1, K2, WC1, WC2, Wa1, and Wa2, respectively). The versions described in this paper are versions AWPM01000000, AWPN01000000, AWPO01000000, AWPP01000000, AWPQ01000000, AWPR01000000, AWPS01000000, AWPT01000000, AWPU01000000, AWPV01000000, AWPW01000000, AWPX01000000, AWPY01000000, AWPZ01000000, AWQA01000000, AWQB01000000, AWQC01000000, AWQD01000000, AWQE01000000, AWQF01000000, AWQG01000000, AWQH01000000, AWQI01000000, AWQJ01000000, AWQK01000000, AWQL01000000, AWQM01000000, AWQN01000000, and AWQO01000000, respectively.
RESULTS
Epidemiologic summary and correlation with gel-based pathogen typing.
The publication that detailed the outbreak (16) reported 58 cases from Missouri (n = 37), Arizona (n = 1), Arkansas (n = 2), Georgia (n = 1), Illinois (n = 9), Indiana (n = 2), Kansas (n = 2), Kentucky (n = 1), Minnesota (n = 2), and Nebraska (n = 1) as being infected with E. coli O157:H7 that displayed the outbreak PFGE and MLVA patterns. The definitions of the outbreak strain pattern in that publication included PFGE patterns A/C and A/D. PCR genotyping demonstrated that each of the 39 Missouri and Kansas case isolates belonged to E. coli O157:H7 phylogenetic cluster 1. Cluster 1 has two main branches (14), the 87-14 lineage, named for an E. coli O157:H7 strain recovered in Washington State in 1987, and the TW14359 lineage, named for an E. coli O157:H7 strain associated with a nationwide outbreak attributed to spinach in 2006 (19).
Of the 37 Missouri cases, 23 reported (“exposure claimed”) and 11 denied (“exposure denied”) consuming items from the salad bar. Five of the 11 exposure-denied cases also denied consuming any romaine lettuce. Three Missouri residents neither claimed nor denied exposure to the salad bar or to romaine lettuce (“exposure unsure”). The two Kansas residents were classified as exposure denied.
SNP patterns and epidemiologic associations.
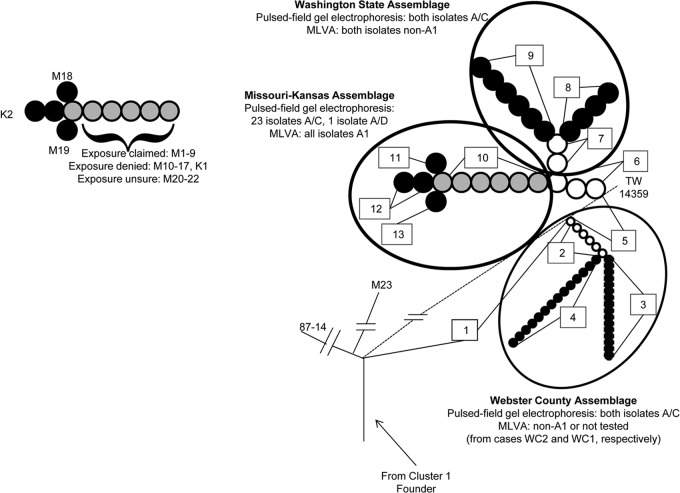
Figure 1 portrays the phylogeny of the study isolates. The case M23 isolate, an offshoot of the 87-14 lineage, was not further analyzed. The remaining study isolates share 93 (backbone ORF SNP set 1) of the 106 radial SNPs with strain TW14359 (14) before forming three assemblages, defined by SNPs that are not found in strain TW14359. The Webster County assemblage, consisting of two isolates with the proximal backbone ORF SNP set 2 and the backbone terminal ORF SNP sets 3 or 4, branches first from the TW14359 lineage. The remaining 26 isolates share six proximal SNPs (backbone ORF SNP set 5) with strain TW14359 before they diverge from the TW14359 lineage. After this divergence, these 26 isolates share three proximal SNPs (backbone ORF SNP set 6) and then bifurcate into the Washington State assemblage (isolates from the two cases from that state) and the Missouri-Kansas assemblage (the remaining isolates, all from Missouri and Kansas cases). The specific radial SNPs that underlie the phylogeny in Fig. 1 are detailed in Table S1 in the supplemental material. Notably, none of these SNPs were found in any other sequenced cluster 1 E. coli O157:H7 strain according to the GenBank database when our analysis was performed. However, a SNP (2041566CA) in isolate Wa1 is also present in strain EDL933, a cluster 3 E. coli O157:H7 strain (14), and probably represents an independent mutation. Because the comparisons in this investigation related the case isolates to strain TW14359, this newly identified polyphyletic SNP does not affect our findings.
Fig 1.
Phylogenetic topology of study isolates. Numbers within rectangles denote single nucleotide polymorphism (SNP) sets. Alphanumeric symbols denote cases in whom isolates (except for reference strain TW14359) originated. White, black, and gray circles signify proximal, terminal, and dual proximal-terminal SNP sets, respectively. Cases from whom E. coli O157:H7 isolates were recovered are described in the text and in Table 1. (Right) The topology relates to specific SNP sets in study strains, reference strains TW14359 and 87-14, and the isolate from the negative-control case M23. Pulsed-field gel electrophoresis and multiple-locus variable-number tandem-repeat analysis (MLVA) patterns are provided for isolates within each assemblage. The dashed line portrays the radial SNPs in strain TW14359 that are not in strain 87-14. (Left) SNP sets correspond to strains from specific cases within the Missouri-Kansas assemblage. The exposure status is provided for each case with an isolate in which SNP set 10 is a terminal SNP set. For cases K2, M18, and M19, each of whom denied exposure, SNP set 10 is a proximal SNP set.
Table 1.
Outbreak versus nonoutbreak classifications of study isolates based on PFGE and MLVA or SNP-based typing methodologies
| Exposure | No. of isolates with: |
|||
|---|---|---|---|---|
| PFGE/MLVAa patterns |
SNPb set patterns |
|||
| Outbreakc | Nonoutbreak | Outbreakd | Nonoutbreak | |
| Claimed | 8e (9f) | 1g | 9f | 0 |
| Denied | 12h | 0 | 9i | 3j |
| Unsure | 3k | 0 | 3k | 0 |
| Unrelated | 0 | 5l | 0 | 5l |
PFGE, pulsed-field gel electrophoresis; MLVA, multiple-locus variable-number tandem-repeat analysis.
SNP, single nucleotide polymorphism.
Outbreak patterns defined as PFGE pattern A/C and MLVA pattern A1 (if performed prior to 2 November 2011) for the case-control study and as PFGE pattern A/C or A/D and MLVA pattern A1 for the publication that detailed the outbreak (16).
The outbreak pattern consists of backbone open reading frame SNP sets 1, 5, 6, and 10 (i.e., SNP set 10 was the terminal SNP set).
Isolates from cases M1 to M8 (all from Missouri).
Isolates from cases M1 to M9 (all from Missouri). The isolate from case M9 had PFGE pattern A/D and was designated as a case in the publication that detailed the outbreak (16) but not in the case-control study. The value in the leftmost column is in parentheses to note that the single subject whose isolate had a variant PFGE pattern was included in one analysis (the paper that detailed the outbreak [16]) but not the other (the case-control study).
Isolate from case M9.
Isolates from cases M10 to M19 (all from Missouri) and cases K1 and K2 (both from Kansas).
Isolates from cases M10 to M17 and K1.
Isolates from cases M18, M19, and K2, which possess SNP sets 1, 5, 6, 10, and 11 (from case M18), 1, 5, 6, 10, and 12 (from case K2), and 1, 5, 6, 10, 13 (from case M19).
Isolates from cases M20 to M22 (all from Missouri).
Isolates from cases unrelated to the outbreak. Isolates from cases WC1 and WC2 have SNP sets 1, 2, and 3 or 1, 2, and 4, respectively, and isolates from cases Wa1 and Wa2 have SNP sets 1, 5, 6, 7, and 8 or 1, 5, 6, 7, and 9, respectively. Isolates from cases WC1, Wa1, and Wa2 have nonoutbreak MLVA patterns. All isolates in these groups, except the isolate from case M23, have the PFGE pattern A/C.
The 24 isolates in the Missouri-Kansas assemblage have four different terminal backbone ORF SNP set patterns. Most isolates possess the backbone ORF SNP sets 1, 5, 6, and 10, including the isolates from each of the 9 exposure-claimed cases, isolates from 9 of the 12 exposure-denied cases, and isolates from each of the 3 exposure-unsure cases. The remaining isolates (from exposure-denied cases M18, K2, and M19) have, in addition to the proximal backbone ORF SNP sets 1, 5, 6, and 10, terminal backbone ORF SNP sets 11, 12, or 13, consisting of only one, two, and one unique SNPs, respectively (Fig. 1; see also Table S1 in the supplemental material).
Of the 61 SNPs newly identified in the study strains, 43 are nonsynonymous. In comparison, among the 504 subgroup C SNPs identified in the five E. coli O157:H7 strains that were extensively analyzed by Leopold et al. (14), 285 were nonsynonymous (P = 0.04).
DISCUSSION
Circumchromosomal SNP analysis of 82.6 million nucleotides in 24 isolates lent clarity to a cluster of E. coli O157:H7 infections for which the PFGE and MLVA patterns of isolates were identical, but the cases denied uniform exposure histories. The backbone ORF SNP set analysis also enabled the confident assignment of case M9 to the outbreak (exposed) group, despite a variant PFGE pattern in that case's isolate. Overall, the backbone ORF SNP set analysis confirmed that 4 of the 24 putative outbreak cases were misclassified by PFGE, including cases whose isolates were subjected to MLVA for corroboration. Additionally, the backbone ORF sequences were identical among isolates from cases who claimed exposure or were unsure about their exposure. This technique, therefore, offers a precise and durable way to differentiate isolates. Recent reports demonstrated the value of whole-pathogen sequencing in controlling nosocomial antibiotic-resistant Klebsiella pneumoniae and Staphylococcus aureus and nosocomial neonatal E. coli outbreaks (20–23). We now demonstrate that whole bacterial genome sequencing can dissect E. coli O157:H7 infection clusters and reduce the proportion of variant epidemiologic histories by refining the assignment of isolates as being outbreak or nonoutbreak. This added precision can be critical when investigations are conducted on a product that might still be in commerce, because E. coli O157:H7 cases come to light 7 to 10 days after exposure (22).
The backbone ORF SNP set analysis can serve as an adjunct to gel-based typing of pathogens. However, there are barriers to immediate widespread adoption of this technology, such as cost, time to result, access to sequencing, and widespread availability of the standardized analytical software, although these barriers are falling. Furthermore, our postsequencing analysis was aided by knowledge of the phylogeny of E. coli O157:H7, which is not as extensive as that for other pathogens. Specifically, we were able to build on whole-genome sequence data from multiple strains (14, 24–28), the recognition that SNPs accrue slowly in the backbone ORFs of E. coli O157:H7 (14, 29), and the prior demonstration of the value of limiting the phylogenetic analysis to stringently defined SNPs and ORFs in the backbone (14, 29). We were also aware of the pitfalls in the database, such that when the polyphyletic SNP at 5137547 was identified, we immediately diverted it from analysis. Such knowledge is growing for other pathogens. Indeed, Zhou et al. recently proposed that whole-genome sequencing provides data that are superior to those from PFGE for the purpose of typing Salmonella enterica serovar Agona strains (30). Nonetheless, despite our familiarity with the population structure of E. coli O157:H7, a considerable amount of manual analysis was needed to generate our phylogeny. Ultimately, any migration from gel-based typing, currently performed largely at state and regional laboratories, to sequencing-based technology will require careful thought as to how to optimize the hardware and analytical capacity and to integrate them with existing capabilities to use public health laboratory resources most strategically. Factors that will warrant consideration include the processes and infrastructure for isolating, identifying, and taking possession of pathogens as expeditiously as possible after cases present for care, ways to transport specimens (live organism or DNA) to an appropriate facility for sequencing, and phylogenetic characterization and data dissemination after sequencing.
It is important to note that at least at the present time, de novo sequencing, rather than categorizing an isolate based on the presence or absence of known SNPs, is needed to differentiate E. coli O157:H7. If we had sought only the SNPs that had been discovered when our work commenced, we would not have differentiated within or between the members of the three assemblages. Also, if we performed SNP typing based on the sequencing of a limited number of pilot strains (15), we would not have identified the differentiating mutations in SNP sets 11, 12, and 13. Hence, until databases become sufficiently “rarefied” that they contain all the SNPs in all the wild-type isolates in existence, sequence-based analysis will remain critical for maximizing confidence (short of certainty) that two isolates are identical.
Interestingly, almost all backbone SNPs in the study isolates (except those in the case M23 isolate) were also present in isolate TW14359. This means that the offshoots from the strain TW14359 lineage, i.e., SNP sets 2 and 6, occurred relatively recently in phylogenetic terms, reinforcing the concept that extant E. coli O157:H7 strains have a small effective population size (14, 29). Finally, the proportion of the nonsynonymous to synonymous mutations among the SNPs that were discovered in the study strains was even greater than that in the strains sequenced by Leopold et al. (14), again demonstrating the tolerance of this clade for diversifying selection.
In summary, the backbone ORF SNP set analysis provided greater pathogen-differentiating power than did a combination of PFGE and MLVA. Sequence-based typing of pathogens is phylogenetically sound, epidemiologically useful, as suggested by Underwood et al. (15), who used a somewhat different strategy, and increasingly feasible. We emphasize that pathogen typing requires bacterial isolates, which reinforces the critical role of clinical microbiologists in the initial recovery of pathogens from infected cases. Outbreak investigations integrate clinical and microbiologic case definitions, hypothesis generation and testing, and molecular studies to identify sources. Whole-genome sequencing and analysis focused on the backbone chromosomes of pathogens can improve the efficiencies of these iterative processes by increasing the precision of statistical estimates and confidence in epidemiological conclusions. Until such precise differentiation technology is widely available, caution should be exercised before one assumes that isolates with indistinguishable gel-based types have common sources, even if the illnesses that they cause are clustered in time and space.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Children's Discovery Institute of Washington University and St. Louis Children's Hospital (NIH grant U54 AI057160), the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE) (NIH grant U54 AI057160), the Washington University Biobank of the DDRCC (NIH grant P30 DK052574), The Genome Institute at Washington University (NIH grants U54 HG003079 and U54 HG004968), and the Melvin E. Carnahan Professorship (P. I. Tarr). P. I. Tarr has received an honorarium for a lecture at the headquarters of Cepheid on E. coli O157:H7 infections.
We thank Ariana Jasarevic for her assistance with manuscript preparation, Alexander Weymann for his comments on the text, Stephanie Schildknecht for performing the PFGE, Michael Kiwala for NCBI data deposition, Charlie Hunt for supplying the Kansas isolates, the clinical microbiologists who isolated and then forwarded the E. coli O157:H7 strain that we analyzed, and the epidemiologists for their efforts in gathering data used to identify the source of the cluster.
Footnotes
Published ahead of print 18 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01930-13.
REFERENCES
- 1.Tauxe RV. 2006. Molecular subtyping and the transformation of public health. Foodborne Pathog. Dis. 3:4–8 [DOI] [PubMed] [Google Scholar]
- 2.Karama M, Gyles CL. 2010. Methods for genotyping verotoxin-producing Escherichia coli. Zoonoses Public Health 57:447–462 [DOI] [PubMed] [Google Scholar]
- 3.Barrett TJ, Lior H, Green JH, Khakhria R, Wells JG, Bell BP, Greene KD, Lewis J, Griffin PM. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swaminathan B, Gerner-Smidt P, Ng LK, Lukinmaa S, Kam KM, Rolando S, Gutierrez EP, Binsztein N. 2006. Building PulseNet International: an interconnected system of laboratory networks to facilitate timely public health recognition and response to foodborne disease outbreaks and emerging foodborne diseases. Foodborne Pathog. Dis. 3:36–50 [DOI] [PubMed] [Google Scholar]
- 5.Terajima J, Izumiya H, Tamura K, Watanabe H. 2002. PulseNet Japan—network system for the utilization of epidemiological information and the results of pulsed-field gel electrophoresis. Nihon Rinsho 60:1070–1076 (In Japanese) [PubMed] [Google Scholar]
- 6.Davis MA, Hancock DD, Besser TE, Call DR. 2003. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J. Clin. Microbiol. 41:1843–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murase T, Yamai S, Watanabe H. 1999. Changes in pulsed-field gel electrophoresis patterns in clinical isolates of enterohemorrhagic Escherichia coli O157:H7 associated with loss of Shiga toxin genes. Curr. Microbiol. 38:48–50 [DOI] [PubMed] [Google Scholar]
- 8.Iguchi A, Osawa R, Kawano J, Shimizu A, Terajima J, Watanabe H. 2002. Effects of repeated subculturing and prolonged storage at room temperature of enterohemorrhagic Escherichia coli O157:H7 on pulsed-field gel electrophoresis profiles. J. Clin. Microbiol. 40:3079–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedrich AW, Zhang W, Bielaszewska M, Mellmann A, Kock R, Fruth A, Tschape H, Karch H. 2007. Prevalence, virulence profiles, and clinical significance of Shiga toxin-negative variants of enterohemorrhagic Escherichia coli O157 infection in humans. Clin. Infect. Dis. 45:39–45 [DOI] [PubMed] [Google Scholar]
- 10.Mellmann A, Bielaszewska M, Karch H. 2009. Intrahost genome alterations in enterohemorrhagic Escherichia coli. Gastroenterology 136:1925–1938 [DOI] [PubMed] [Google Scholar]
- 11.Lindstedt BA, Tham W, Danielsson-Tham ML, Vardund T, Helmersson S, Kapperud G. 2008. Multiple-locus variable-number tandem-repeats analysis of Listeria monocytogenes using multicolour capillary electrophoresis and comparison with pulsed-field gel electrophoresis typing. J. Microbiol. Methods 72:141–148 [DOI] [PubMed] [Google Scholar]
- 12.Izumiya H, Pei Y, Terajima J, Ohnishi M, Hayashi T, Iyoda S, Watanabe H. 2010. New system for multilocus variable-number tandem-repeat analysis of the enterohemorrhagic Escherichia coli strains belonging to three major serogroups: O157, O26, and O111. Microbiol. Immunol. 54:569–577 [DOI] [PubMed] [Google Scholar]
- 13.Ohnishi M, Tanaka C, Kuhara S, Ishii K, Hattori M, Kurokawa K, Yasunaga T, Makino K, Shinagawa H, Murata T, Nakayama K, Terawaki Y, Hayashi T. 1999. Chromosome of the enterohemorrhagic Escherichia coli O157:H7; comparative analysis with K-12 MG1655 revealed the acquisition of a large amount of foreign DNAs. DNA Res. 6:361–368 [DOI] [PubMed] [Google Scholar]
- 14.Leopold SR, Magrini V, Holt NJ, Shaikh N, Mardis ER, Cagno J, Ogura Y, Iguchi A, Hayashi T, Mellmann A, Karch H, Besser TE, Sawyer SA, Whittam TS, Tarr PI. 2009. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc. Natl. Acad. Sci. U. S. A. 106:8713–8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Underwood AP, Dallman T, Thomson NR, Williams M, Harker K, Perry N, Adak B, Willshaw G, Cheasty T, Green J, Dougan G, Parkhill J, Wain J. 2013. Public health value of next-generation DNA sequencing of enterohemorrhagic Escherichia coli isolates from an outbreak. J. Clin. Microbiol. 51:232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slayton RB, Turabelidze G, Bennett SD, Schwensohn CA, Yaffee AQ, Khan F, Butler C, Trees E, Ayers TL, Davis ML, Laufer AS, Gladbach S, Williams I, Gieraltowski LB. 2013. Outbreak of Shiga toxin-producing Escherichia coli (STEC) O157:H7 associated with romaine lettuce consumption, 2011. PLoS One 8:e55300. 10.1371/journal.pone.0055300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaikh N, Holt NJ, Johnson JR, Tarr PI. 2007. Fim operon variation in the emergence of enterohemorrhagic Escherichia coli: an evolutionary and functional analysis. FEMS Microbiol. Lett. 273:58–63 [DOI] [PubMed] [Google Scholar]
- 18.Hyytia-Trees E, Lafon P, Vauterin P, Ribot EM. 2010. Multilaboratory validation study of standardized multiple-locus variable-number tandem repeat analysis protocol for Shiga toxin-producing Escherichia coli O157: a novel approach to normalize fragment size data between capillary electrophoresis platforms. Foodborne Pathog. Dis. 7:129–136 [DOI] [PubMed] [Google Scholar]
- 19.Wendel AM, Johnson DH, Sharapov U, Grant J, Archer JR, Monson T, Koschmann C, Davis JP. 2009. Multistate outbreak of Escherichia coli O157:H7 infection associated with consumption of packaged spinach, August-September 2006: the Wisconsin investigation. Clin. Infect Dis. 48:1079–1086 [DOI] [PubMed] [Google Scholar]
- 20.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 4:148ra116. 10.1126/scitranslmed.3004129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 366:2267–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altmann M, Spode A, Altmann D, Wadl M, Benzler J, Eckmanns T, Krause G, An der Heiden M. 2011. Timeliness of surveillance during outbreak of Shiga toxin-producing Escherichia coli infection, Germany, 2011. Emerg. Infect. Dis. 17:1906–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherry NL, Porter JL, Seemann T, Watkins A, Stinear TP, Howden BP. 2013. Outbreak investigation using high-throughput genome sequencing within a diagnostic microbiology laboratory. J. Clin. Microbiol. 51:1396–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, Mladonicky JM, Somsel P, Rudrik JT, Dietrich SE, Zhang W, Swaminathan B, Alland D, Whittam TS. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc. Natl. Acad. Sci. U. S. A. 108:20142–20147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotewicz ML, Jackson SA, LeClerc JE, Cebula TA. 2007. Optical maps distinguish individual strains of Escherichia coli O157:H7. Microbiology 153:1720–1733 [DOI] [PubMed] [Google Scholar]
- 27.Feng P, Lampel KA, Karch H, Whittam TS. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750–1753 [DOI] [PubMed] [Google Scholar]
- 28.Eppinger M, Daugherty S, Agrawal S, Galens K, Sengamalay N, Sadzewicz L, Tallon L, Cebula TA, Mammel MK, Feng P, Soderlund R, Tarr PI, Debroy C, Dudley EG, Fraser CM, Ravel J. 2013. Whole-genome draft sequences of 26 enterohemorrhagic Escherichia coli O157:H7 strains. Genome Announc. 1:e0013412. 10.1128/genomeA.00134-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leopold SR, Shaikh N, Tarr PI. 2010. Further evidence of constrained radiation in the evolution of pathogenic Escherichia coli O157:H7. Infect. Genet. Evol. 10:1282–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z, McCann A, Litrup E, Murphy R, Cormican M, Fanning S, Brown D, Guttman DS, Brisse S, Achtman M. 2013. Neutral genomic microevolution of a recently emerged pathogen, Salmonella enterica serovar Agona. PLoS Genet. 9:e1003471. 10.1371/journal.pgen.1003471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.