Abstract
Staphylococcus aureus is a common cause of bacteremia, with a substantial impact on morbidity and mortality. Because of increasing rates of methicillin-resistant Staphylococcus aureus, vancomycin has become the standard empirical therapy. However, beta-lactam antibiotics remain the best treatment choice for methicillin-susceptible strains. Placing patients quickly on the optimal therapy is one goal of antimicrobial stewardship. This retrospective, observational, single-center study compared 33 control patients utilizing only traditional full-susceptibility methodology to 22 case patients utilizing rapid methodology with CHROMagar medium to detect and differentiate methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains hours before full susceptibilities were reported. The time to targeted therapy was statistically significantly different between control patients (mean, 56.5 ± 13.6 h) and case patients (44.3 ± 17.9 h) (P = 0.006). Intensive care unit status, time of day results emerged, and patient age did not make a difference in time to targeted therapy, either singly or in combination. Neither length of stay (P = 0.61) nor survival (P = 1.0) was statistically significantly different. Rapid testing yielded a significant result, with a difference of 12.2 h to targeted therapy. However, there is still room for improvement, as the difference in time to susceptibility test result between the full traditional methodology and CHROMagar was even larger (26.5 h). This study supports the hypothesis that rapid testing plays a role in antimicrobial stewardship by getting patients on targeted therapy faster.
INTRODUCTION
Staphylococcus aureus is frequently associated with nosocomial and community-acquired bacteremia worldwide (1, 2). Furthermore, S. aureus bacteremia is associated with a high incidence of morbidity and mortality (3), with methicillin-resistant S. aureus (MRSA) bacteremia having a substantially higher rate of mortality than methicillin-susceptible S. aureus (MSSA) bacteremia (4). Because of increasing rates of MRSA and the severity of infection it produces, vancomycin has become standard empirical therapy for Gram-positive bacteremia.
Despite using the same empirical therapy until methicillin resistance is excluded, targeted treatments for MRSA and MSSA bacteremia differ. The treatment of choice for MSSA bacteremia is an intravenous (IV) penicillinase-resistant penicillin (e.g., oxacillin or nafcillin) or a first-generation cephalosporin (e.g., cefazolin), which have demonstrated superiority over vancomycin (5). The use of vancomycin also has the burden and expense of monitoring serum drug concentrations, in addition to the concerns of MIC creep with overuse (6). Placing patients on targeted therapy as quickly as possible is a goal of antimicrobial stewardship that has traditionally been limited by the time to identification of the organism and determination of antimicrobial susceptibilities. Therefore, rapid detection of MRSA and MSSA may prove to be an effective tool for antimicrobial stewardship.
The traditional methodology used for S. aureus identification and determination of full susceptibilities at our institution, UNC Health Care, was the VITEK2 system (bioMérieux, Durham, NC) or disk diffusion, taking up to 48 h to obtain results. In September 2011, we implemented a culture-based rapid susceptibility screen using BBL CHROMagar MRSA II (Becton, Dickinson, Sparks, MD). We used CHROMagar MRSA II for the detection and differentiation of MSSA and MRSA directly from positive blood culture bottles (BacT/Alert, bioMérieux, Durham, NC). The manufacturer recommends that CHROMagar MRSA II results be interpreted after 24 h of incubation, with 92% positive agreement and 99.9% negative agreement for the determination of MRSA. However, it was later independently demonstrated that results could be obtained after only 12 to 16 h of incubation with the same high degree of sensitivity and specificity as after 24 h of incubation (7). Therefore, in January 2012, our institution began interpreting results after 12 to 16 h of incubation.
Because of the added expense associated with rapid testing, it is important to assess the impact rapid testing has on the overall care to the patient. The objective of this study was to determine whether a difference in clinical outcomes exists when comparing traditional and rapid testing for MRSA and MSSA in routine blood cultures. Clinical endpoints, including time to targeted therapy, length of stay, and survival, were examined.
MATERIALS AND METHODS
Study design.
This was a retrospective, observational, single-center, institutional review board-approved study completed from October 2010 to July 2012 in an 803-bed academic medical center. Patients were potentially included if they were retrospectively identified as having a routine blood culture positive for MSSA. The patients were designated as either a potential control or a potential case patient. Control patients were those for whom only traditional blood culture methodology was utilized for identification and susceptibility testing. Case patients were those for whom CHROMagar medium was utilized in addition to the traditional culture work-up.
We excluded patients less than 2 years old, since antibiotic selection and dosing in this population are different. Patients were also excluded if they had a beta-lactam allergy that inhibited the ability to change therapy to a beta-lactam antibiotic after MSSA was reported. The growth of any other organism in a routine blood culture during treatment for the MSSA bacteremia was also an exclusion criterion. Lastly, patients were excluded if their first Gram stain from a positive blood culture was done at an outside hospital or if it was unclear when targeted therapy was initiated.
The primary endpoint of the study was the time to targeted therapy, reported in hours. We defined time to targeted therapy as the time from report of Gram-positive cocci on Gram stain of the first positive blood culture to the time of pharmacy verification of a beta-lactam antibiotic. Pharmacy verification was necessary for dispensation of the antibiotic from the pharmacy to the patient and was a point in time we could easily capture, unlike medication administration time, which was not documented electronically. Secondary endpoints that were examined in this study included length of stay, survival, and time to susceptibility test result.
Culture methods.
Blood cultures were collected from patients using standard procedures and incubated on the BacT/Alert 3D system (bioMérieux, Durham, NC). Cultures flagged as positive by the instrument were Gram stained, and the results reported to the responsible physician. Following routine laboratory protocol, positive bottles were subcultured to chocolate, colistin-naladixic acid, and MacConkey agars (Becton Dickson, Sparks, MD). Organisms were identified using standard techniques, and full susceptibility testing was performed with Kirby Bauer disk diffusion or VITEK2 (bioMérieux, Durham, NC) as specified by the Clinical and Laboratory Standards Institute. During the study period, CHROMagar MRSA II medium (Becton Dickson, Sparks, MD) was added to the subculture setup for the first bottle demonstrating Gram-positive cocci in clusters on Gram stain for each new episode of bacteremia. In-house studies were performed to verify that the determination of MRSA and MSSA could be accurately made as early as 12 h after inoculation from a positive blood culture.
Statistics.
The study was powered to detect a minimum difference of 8 h in time to targeted therapy, a difference we considered meaningful since this would save at least one dose of vancomycin. We computed the necessary sample size assuming that the number of controls would be approximately twice the number of cases. Historical data suggested that time to targeted therapy was approximately normally distributed, with a mean of 53.8 h and a standard deviation (SD) of 9.8. Thus, 80% power would be achieved with 38 control patients and 19 case patients.
Power calculations and final analyses were conducted with R 3.0.0 (2013) for Windows (R Foundation for Statistical Computing, Vienna, Austria). The primary analysis was assessed with the t test after the normality assumption had been checked with the Shapiro-Wilk test. Secondary analyses were assessed with t tests with the Satterthwaite-Welch adjustment for nonconstant variance, Mann-Whitney U tests for unpaired nonnormal data, and the Wilcoxon signed-rank test for paired nonnormal data. The chi-squared test and Fisher's exact test were used for some analyses where appropriate. We determined that the significant primary result would hold after adjusting for possible confounding factors with a multiple regression. We considered P values of <0.05 to be significant.
RESULTS
A review of 68 controls and 40 cases (CHROMagar) yielded 33 and 22 patients, respectively, meeting the inclusion criteria. A breakdown of the exclusion criteria met in each group is presented in Table 1. Patient characteristics at the time of infection are presented in Table 2. The only characteristic analyzed that reached a statistically significant difference between groups was patient age (P < 0.001), a characteristic that is not expected to have a major effect on the primary endpoint but may have an effect on secondary endpoints. While there was not a statistically significant difference in characteristics, including gender, dialysis, and total parenteral nutrition, there was a noticeable difference in the percentage of patients in each group. Theoretically, gender should not make a difference in any endpoint, but a need for dialysis and total parenteral nutrition would require the patient to have a catheter in place and potentially affect treatment response and secondary endpoints. Also of note, 100% of control and case patients were initially started on empirical therapy with vancomycin. A change to a beta-lactam antibiotic, which was a criterion for inclusion, occurred in 100% of the control group and 95.5% of the case group. One patient in the case group was switched to trimethoprim-sulfamethoxazole instead of a beta-lactam antibiotic but was included since this was still considered targeted therapy.
Table 1.
Breakdown of exclusion criteria
| Exclusion criterion | No. in control group (%) | No. in case group (% tested by CHROMagar) |
|---|---|---|
| Age <2 yrs old | 5 (7.4) | 3 (7.5) |
| Beta-lactam allergy | 4 (5.9) | 3 (7.5) |
| Another organism in blood | 5 (7.4) | 0 |
| Outside-hospital Gram stain | 6 (8.8) | 5 (12.5) |
| Targeted-therapy time unclear | 15 (22.1) | 7 (17.5) |
Table 2.
Patient characteristics at time of infection
| Characteristic | No. in control group (%) | No. in case group (% tested by CHROMagar) | P value |
|---|---|---|---|
| Avg age (yr) | 56 | 31 | <0.001 |
| Male | 19 (57.6) | 18 (81.8) | 0.08 |
| Dialysis | 5 (15.2) | 2 (9.1) | 0.69 |
| Total parenteral nutrition | 1 (3.0) | 2 (9.1) | 0.56 |
| Catheter in place | 9 (27.3) | 10 (45.5) | 0.25 |
| Intensive care unit | 5 (15.2) | 4 (18.2) | 1 |
| Empirical vancomycin | 33 (100) | 22 (100) | NAa |
| Change to a beta-lactam antibiotic | 33 (100) | 21 (95.5) | NA |
NA, not applicable.
Another patient characteristic examined was the presence of a central venous catheter at the time of infection. For patients who had a central venous catheter at the time of infection, we aimed to determine whether this catheter had no intervention, was replaced over a guide wire, was removed with a new catheter inserted, or was removed with no new catheter inserted. In the control group, 3 (33.3%) patients had no intervention to the catheter and 6 (66.7%) patients had the catheter removed and a new catheter inserted. In the case group, 3 (30%) patients had no intervention to the catheter, 2 (20%) patients had the catheter replaced over a guide wire, 4 (40%) patients had the catheter removed and a new catheter inserted, and 1 (10%) patient had the catheter removed with no new catheter inserted. While this characteristic may not affect the primary endpoint, it is important to mention since having a central venous catheter that is not removed or replaced when it is a likely source of bacteremia can affect the success of bacteremia treatment and patient outcome. About 30% of both groups had no intervention to a catheter that was present at the time of infection.
The primary endpoint, time to targeted therapy in hours, was significantly different between controls, with a mean ± SD of 56.5 ± 13.6 h, and cases, with a mean ± SD of 44.3 ± 17.9 h (P = 0.006). Intensive care unit status (P = 0.21), time of day results emerged (P = 0.54), and patient age (P = 0.17) did not make a difference in time to targeted therapy in isolation or in combination. Time to targeted therapy remained significant (P = 0.01) even after adjusting for these potential covariates. While the number of patients in the intensive care unit and the number of patients whose test results emerged after first shift were very small, we adjusted for these two covariates because they had the potential to affect the primary endpoint by altering the time it took for the physician to see that the test had produced results.
The secondary endpoints evaluated included length of stay and survival. The mean length of stay in days was not significantly different, with a mean ± SD of 13.4 ± 11.5 days in the control group and 10.0 ± 5.0 days in the case group (P = 0.61). Survival was also not significantly different between groups, with 97% survival in the control group and 100% in the case group (P = 1.0).
An additional secondary endpoint that was assessed was the time to susceptibility test result. The time to full susceptibility results in the control group (46.1 ± 10.9 h) compared to that in the case group (48.0 ± 12.1 h) was not significantly different (P = 0.62). When we compared the time to full susceptibility determination in the control group (46.1 ± 10.9 h) to the time to CHROMagar result in the case group (19.6 ± 5.9 h), we found a significant difference (P < 0.001). When we compared the case group time to full susceptibility results (48.0 ± 12.1 h) to the case group time to CHROMagar results (19.6 ± 5.9 h), we similarly found a significant difference (P < 0.001). Figure 1 presents a graph comparing time to targeted therapy with time to susceptibility test result.
Fig 1.
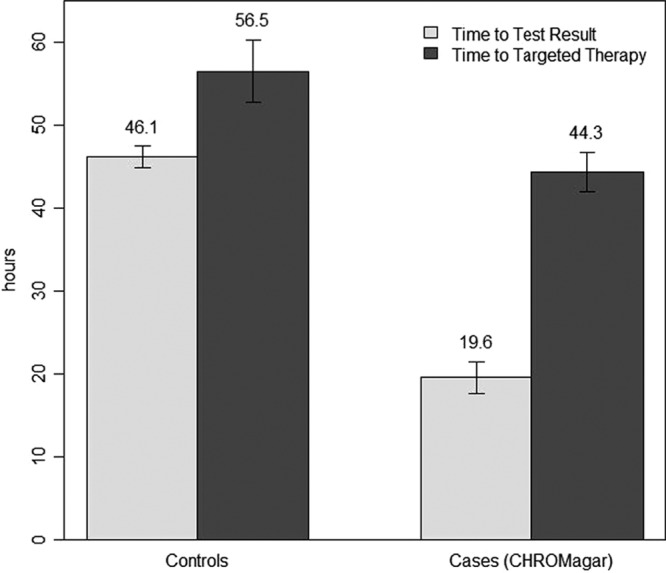
Time to susceptibility test result compared to time to targeted therapy reported in hours (error bars show ± 1 standard error).
DISCUSSION
Results from the CHROMagar assay were reported in our electronic medical record as a comment that reads “Oxacillin-susceptible S. aureus predicted by rapid resistance testing. Approximately 2% of S. aureus may be resistant to oxacillin due to mechanisms not detected by this method.” This comment was based on historical data on MRSA isolates from our laboratory; however, all of the isolates reported as MSSA by CHROMagar in this study were confirmed to be MSSA upon full susceptibility results. Full susceptibility results were reported in our electronic medical record as a list of susceptibility test results for oxacillin, gentamicin, vancomycin, erythromycin, clindamycin, tetracycline, and trimethoprim-sulfamethoxazole. The medical team was not alerted that the CHROMagar or full susceptibility results had been posted in the medical record. Therefore, the change from empirical to targeted therapy was reliant on the medical team noticing that the results were available.
We found that there was a significant decrease in time to targeted therapy in the CHROMagar group, while there was not a significant difference in the time it took for full susceptibilities to be reported. This suggests that the decrease in time to targeted therapy is attributed to the utilization of the CHROMagar medium. Despite the fact that we found a significant difference in time to targeted therapy in the CHROMagar group, the results presented in Fig. 1 demonstrate that there is still room for improvement, as the difference in time to targeted therapy was 12.2 h but the difference in time to susceptibility test result was even larger, at 26.5 h. Theoretically, the difference in time to targeted therapy could have been up to 26.5 h, which would provide another 14.3 h of targeted therapy, a difference that could be clinically relevant. The change in treatment occurred within 24 h of full susceptibility results in 84.8% of the control group. The change in treatment occurred within 24 h of the CHROMagar result in only 31.8% of the case group. However, 54.5% in the CHROMagar group were changed within 24 h of the full susceptibility results instead. The failure to change antibiotics within 24 h of CHROMagar result suggests that the medical staff may not have trusted the rapid results enough to change therapy or that they did not rapidly notice the results in the medical record. Both of these theories provide an opportunity for intervention that could further improve the time to targeted therapy.
There are many tests available for the rapid determination of MRSA from MSSA in positive blood cultures, including molecular methodologies. With the many rapid tests that are available, choosing which test to use at an institution will depend on a variety of factors, including cost, reported specificity and sensitivity, time to test result, U.S. Food and Drug Administration-approved indications, and the data available to support their use with regard to improvement of patient outcomes (8). Their role in antimicrobial stewardship ultimately relates to how they promote the goals of stewardship, namely, improving patient care and health care outcomes (8). For this study, we utilized CHROMagar medium, which, although not as rapid as molecular tests, can cost 5 to 20 times less than a molecular assay. Importantly, the utilization of a culture-based approach requires no extra equipment or specialized expertise, making it accessible to any institution.
We acknowledge that our study has several limitations. We performed a retrospective, observational, single-center study. We had to rely on the accuracy of documentation in medical records to derive some of our data. We also chose to base time to targeted therapy on pharmacy verification time instead of drug administration time, because our medication administration records were not available in an electronic format and were more difficult to access. Moreover, we were unable to fully evaluate cost differences. Unlike oxacillin treatment, which costs $78 per day for an adult patient with MSSA bacteremia at our institution (receiving 2 g IV every 4 h), the cost of vancomycin treatment is variable. Since vancomycin dosage is based on age, weight, renal function, and patient-specific serum trough concentrations, it was not possible to calculate an expected daily cost of vancomycin. Further analysis would be needed to determine whether total drug costs would be reduced with the use of rapid testing.
In addition to the above-described limitations, there were two secondary endpoints that we hoped to evaluate in this study, but we were unable to do so. Time to culture clearance was a patient outcome where we expected we might see a difference if patients were placed on targeted therapy faster. However, samples for repeat cultures were drawn at different times from every patient based on physician preferences. The other endpoint was time to defervescence, but this was also too difficult to accurately examine in a retrospective study. We found that most patients had their first fever at home, which was not documented in our medical record. We also found that some patients were started on antibiotics before they ever had a documented fever in the hospital. This variability made these endpoints too unreliable to analyze and report.
This is one of a limited number of studies that has looked at the impact rapid testing may have on clinical outcomes and on antimicrobial stewardship. It is also one of the first studies that specifically evaluated these outcomes with CHROMagar detection of MRSA and MSSA. In this study, patients were placed on targeted therapy 12.2 h faster with CHROMagar utilization. However, as noted, there is further room for improvement given the larger difference of 26.5 h between times to traditional and to rapid test results. To try to further improve the time to targeted therapy, we will implement an antimicrobial stewardship intervention in which CHROMagar results will be paged to an on-call pharmacist who will then collaborate with the medical team to switch to targeted therapy. This approach has previously been successful at our institution (9). Regardless of our future endeavors, this study supports the idea that rapid culture-based detection of MRSA and MSSA plays a role in antimicrobial stewardship by decreasing the time that it takes to place patients on targeted antimicrobial therapy.
ACKNOWLEDGMENTS
This research was partly supported by a grant from the National Institutes of Health (UL1TR000083) for biostatistical support through the North Carolina Translational and Clinical Sciences Institute.
The authors declare no conflicts of interest.
Footnotes
Published ahead of print 2 October 2013
REFERENCES
- 1.Biedenbach DJ, Moet GJ, Jones RN. 2004. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveillance Program (1997-2002). Diagn. Microbiol. Infect. Dis. 50:59–69 [DOI] [PubMed] [Google Scholar]
- 2.Styers D, Sheehan DJ, Hogan P, Sahm DF. 2006. Laboratory-based surveillance of current antimicrobial resistance patterns and trends among Staphylococcus aureus: 2005 status in the United States. Ann. Clin. Microbiol. Antimicrob. 5:2–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Hal SJ, Jensen SO, Vaska VL, Espedido BA, Paterson DL, Gosbell IB. 2012. Predictors of mortality in Staphylococcus aureus bacteremia. Clin. Microbiol. Rev. 25:362–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blot SI, Vandewoude KH, Hoste EA, Colardyn FA. 2002. Outcome and attributable mortality in critically ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Arch. Intern. Med. 162:2229–2235 [DOI] [PubMed] [Google Scholar]
- 5.Schweizer ML, Furuno JP, Harris AD, Johnson JK, Shardell MD, McGregor JC, Thom KA, Cosgrove SE, Sakoulas G, Perencevich EN. 2011. Comparative effectiveness of nafcillin or cefazolin versus vancomycin in methicillin-susceptible Staphylococcus aureus bacteremia. BMC Infect. Dis. 11:279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould IM. 2013. Treatment of bacteraemia: methicillin-resistant Staphylococcus aureus (MRSA) to vancomycin-resistant S. aureus (VRSA). Int. J. Antimicrob. Agents 425:S17–S21 [DOI] [PubMed] [Google Scholar]
- 7.Chihara S, Hayden MK, Minogue-Corbett E, Singh K. 2009. Shortened time to identify Staphylococcus species from blood cultures and methicillin resistance testing using CHROMagar. Int. J. Microbiol. 2009:636502. 10.1155/2009/636502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geiger K, Brown J. 2013. Rapid testing for methicillin-resistant Staphylococcus aureus: implications for antimicrobial stewardship. Am. J. Health Syst. Pharm. 70:335–342 [DOI] [PubMed] [Google Scholar]
- 9.Heil EL, Daniels LM, Long DM, Rodino KG, Weber DJ, Miller MB. 2012. Impact of a rapid peptide nucleic acid fluorescence in situ hybridization assay on treatment of Candida infections. Am. J. Health Syst. Pharm. 69:1910–1914 [DOI] [PubMed] [Google Scholar]


