Abstract
Chytridiomycosis is a lethal fungal disease contributing to declines and extinctions of amphibian species worldwide. The currently used molecular screening tests for chytridiomycosis fail to detect the recently described species Batrachochytrium salamandrivorans. In this study, we present a duplex real-time PCR that allows the simultaneous detection of B. salamandrivorans and Batrachochytrium dendrobatidis. With B. dendrobatidis- and B. salamandrivorans-specific primers and probes, detection of the two pathogens in amphibian samples is possible, with a detection limit of 0.1 genomic equivalent of zoospores of both pathogens per PCR. The developed real-time PCR shows high degrees of specificity and sensitivity, high linear correlations (r2 > 0.995), and high amplification efficiencies (>94%) for B. dendrobatidis and B. salamandrivorans. In conclusion, the described duplex real-time PCR can be used to detect DNA of B. dendrobatidis and B. salamandrivorans with highly reproducible and reliable results.
INTRODUCTION
Chytridiomycosis causes worldwide declines and extinctions of amphibian populations and is one of the most important infectious diseases in amphibians (1–3). Batrachochytrium dendrobatidis was the sole Chytridiomycetes taxon known to infect vertebrate hosts and to be able to cause this devastating disease (4) until a second chytrid species was isolated from a mortality event that drove the Dutch fire salamander (Salamandra salamandra) population nearly to extinction (5, 6). This novel species, Batrachochytrium salamandrivorans, cannot be detected with the B. dendrobatidis-specific PCR described by Annis et al. (7) or the B. dendrobatidis-specific real-time PCR described by Boyle et al. (8). Because both B. dendrobatidis and B. salamandrivorans are able to cause amphibian chytridiomycosis, the development of a test that would allow fast reliable detection and quantification of these two pathogens is necessary. This test could aid in rapid diagnosis of chytridiomycosis in diseased amphibians but also could be used to map the worldwide distribution of the novel pathogen. Therefore, the aim of this study was to develop a duplex real-time PCR that allows detection of B. dendrobatidis and B. salamandrivorans in amphibian samples with high sensitivity and specificity.
MATERIALS AND METHODS
Chytrid strains and culture conditions.
B. dendrobatidis and B. salamandrivorans were grown in TGhL broth (16 g tryptone, 4 g gelatin hydrolysate, 2 g lactose per liter of distilled water) in 25-cm3 cell culture flasks and incubated at 20°C (B. dendrobatidis) or 15°C (B. salamandrivorans). Homolaphlyctis polyrhiza, Gaertneriomyces semiglobifer, Geranomyces variabilis, Rhizophlyctis rosea, Rhizoclosmatium globosum, Polychytrium aggregatum, Monoblepharis polymorpha, and Podochytrium dentatum (Table 1) were grown in PmTG broth (0.5 g peptonized milk, 0.5 g tryptone, 2.5 g glucose per liter of distilled water) in 25-cm3 cell culture flasks, with incubation at 23°C. To obtain zoospores of B. dendrobatidis and B. salamandrivorans, 2 ml of a 5-day-old culture was transferred to TGhL broth with 1% agar plates and incubated for 5 to 7 days at 20°C (for B. dendrobatidis) or 15°C (for B. salamandrivorans). Zoospores were subsequently collected by flooding the agar plates with 2 ml of filtered (0.2-μm filter) pond water and collecting the fluid. The number of zoospores present in the suspension was determined using a hemocytometer.
Table 1.
Overview of the Chytridiomycota isolates used to verify the specificity of the real-time duplex PCR for B. dendrobatidis and B. salamandrivorans
| Species | Class | Order | Isolate | Amplificationa |
|---|---|---|---|---|
| Batrachochytrium dendrobatidis | Chytridiomycetes | Rhizophydiales | JEL423 | Yes (B. dendrobatidis) |
| Batrachochytrium salamandrivorans | Chytridiomycetes | Rhizophydiales | AMFP13/1 | Yes (B. salamandrivorans) |
| Homolaphlyctis polyrhiza | Chytridiomycetes | Rhizophydiales | JEL142 | No |
| Rhizophlyctis rosea | Chytridiomycetes | Rhizophlyctidales | JEL532 | No |
| Gaertneriomyces semiglobifer | Chytridiomycetes | Spizellomycetales | JEL384 | No |
| Rhizoclosmatium globosum | Chytridiomycetes | Chytridiales | JEL791 | No |
| Podochytrium dentatum | Chytridiomycetes | Chytridiales | JEL30 | No |
| Polychytrium aggregatum | Chytridiomycetes | Polychytriales | JEL109 | No |
| Geranomyces variabilis | Chytridiomycetes | Spizellomycetales | JEL518 | No |
| Monoblepharis polymorpha | Monoblepharidomycetes | Monoblepharidales | JEL486 | No |
The component of the duplex real-time PCR that showed amplification in positive samples is indicated in parentheses.
Quantitation standards and DNA extracts.
Suspensions containing standardized numbers of B. dendrobatidis and B. salamandrivorans zoospores were prepared as described by Boyle et al. (8). Tenfold serial dilution series ranging from 1,000 to 0.01 genomic equivalents (GEs) of zoospores per real-time PCR mixture were prepared for B. dendrobatidis and B. salamandrivorans. DNA of the other described Chytridiomycota species (Table 1) was prepared from growing cultures with DNA extraction in 100 μl of Prepman Ultra reagent (Applied Biosystems, Foster City, CA), following the DNA extraction method described by Hyatt et al. (9).
Primer and probe design.
The previously described forward primer STerF (5′-TGCTCCATCTCCCCCTCTTCA-3′) and reverse primer STerR (5′-TGAACGCACATTGCACTCTAC-3′) were used to detect the 5.8S rRNA gene of B. salamandrivorans (6) (GenBank accession number KC762295). The B. salamandrivorans-specific Cy5-labeled probe STerC (5′-ACAAGAAAATACTATTGATTCTCAAACAGGCA-3′), based on the 5.8S rRNA gene of B. salamandrivorans, was developed using Kodon (Applied Maths, Kortrijk, Belgium) (Fig. 1). The primer set ITS1-3 Chytr (5′-CCTTGATATAATACAGTGTGCCATATGTC-3′) and 5.8S Chytr (5′-TCGGTTCTCTAGGCAACAGTTT-3′) and the TaqMan probe Chytr MGB2 (5′-CGAGTCGAAC-3′) described by Boyle et al. (8) were used to detect the ITS1 rRNA gene of B. dendrobatidis. All primers and probes were checked with BLASTN analysis, to ensure that amplification of genes from other organisms or species was unlikely. For B. salamandrivorans, the specificity of the primer set was tested in a SYBR green real-time PCR with DNA extracts of pure B. salamandrivorans culture and negative controls, with melting curve analysis and gel electrophoresis of the real-time PCR products (see below).
Fig 1.

Specific primers and probe for Batrachochytrium salamandrivorans. (A) rRNA gene sequences of the ITS1, 5.8S, and ITS2 regions used for the design of the B. salamandrivorans primers and probe. (B) B. salamandrivorans primer and probe sequences. The sequence used is from GenBank accession number KC762295.
B. salamandrivorans SYBR green real-time PCR.
The B. salamandrivorans SYBR green assay was performed on a CFX96 real-time system (Bio-Rad Laboratories, Hercules, CA). A reaction mixture composed of 12.5 μl SYBR green PCR mix (1× SensiMix SYBR No-ROX; Bioline Reagents Ltd., London, United Kingdom), B. salamandrivorans forward primer STerF at a concentration of 0.3 μM, B. salamandrivorans reverse primer STerR at a concentration of 0.3 μM, 5 μl template, and a volume of RNase- and DNase-free water to a total of 25 μl was used in each reaction. Amplification conditions consisted of 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 62°C for 15 s. A temperature gradient from 60°C to 95°C, with plate reads at every temperature increment of 0.5°C, was used to generate melting curve data.
Duplex real-time TaqMan PCR assay optimization.
The B. dendrobatidis and B. salamandrivorans real-time PCR assays were first optimized as simplex assays. Assays were performed on a CFX96 real-time system (Bio-Rad Laboratories, Hercules, CA). Amplification conditions for the simplex and duplex assays consisted of 10 min at 95°C followed by 40 cycles of melting (95°C for 15 s) and annealing/extension (62°C for 1 min). Primer concentrations were optimized in a checkerboard system, with a standard probe concentration of 250 nM. Subsequently, the probe concentrations were optimized with the previously determined optimal primer concentrations. After optimization of the simplex assays, the two PCRs were combined to form the duplex real-time PCR. The precision of the developed duplex real-time PCR assay was evaluated by determining intra- and interassay variability, expressed as the mean coefficient of variation. For the interassay variability, three replicates of the quantitation standard were run in three separate assays; for the intra-assay variability, three replicates were run in one assay. The specificity of the duplex real-time PCR was evaluated by assaying DNA extracts of a wide range of Chytridiomycota species (Table 1). Real-time PCR efficiency, slope, and r2 values were calculated with Bio-Rad CFX Manager v1.6 (Bio-Rad Laboratories, Hercules, CA), with the baseline-subtracted curve-fit setting. Slope and r2 were calculated with the standard curves determined with the quantitation standards described earlier. Efficiency was calculated as 10−1/slope − 1. After optimization of the duplex real-time PCR, a protocol that includes adding bovine serum albumin (BSA) to the PCR mixture was validated as an alternative to diluting samples, in order to alleviate PCR inhibition that could arise due to the nature of amphibian samples (9, 10). BSA (Sigma-Aldrich Inc., Bornem, Belgium) was added to the PCR mixture at a concentration of 400 ng μl−1, as this is the optimal concentration to relieve PCR inhibition (11). Four replicates of the described quantitation standards of B. dendrobatidis and B. salamandrivorans with added BSA and four replicates without added BSA were run. Mean quantification cycle (Cq) values generated for the two conditions were compared to evaluate any significant effect of the addition of BSA on basic PCR results.
Amphibian samples.
To validate the use of the real-time PCR to detect B. dendrobatidis and B. salamandrivorans in skin samples from amphibians, we applied the optimized protocol to samples from (i) 10 fire salamanders (S. salamandra) experimentally inoculated with B. salamandrivorans, (ii) 41 fire salamanders (S. salamandra) from the declining population in the Netherlands, (iii) 51 fire salamanders (S. salamandra) from a stable Belgian population, and (iv) 27 yellow-bellied toads (Bombina variegata) from a healthy Dutch population with known B. dendrobatidis infection (see Table 4). The B. salamandrivorans infection experiment with fire salamanders was carried out with approval of the ethics committee of the Faculty of Veterinary Medicine, Ghent University (approval no. EC2013/10). Skin swabs were collected by gently rubbing a sterile cotton-tipped swab 10 times across the ventral abdomen, inner thigh, and hind limb digits (9, 12). From dead amphibians, pieces of skin taken from the ventral abdomen (approximate size, 0.25 cm2) were collected for analysis. DNA was extracted from skin swabs in 100 μl Prepman Ultra (Applied Biosystems, Foster City, CA) (9). DNA was extracted from skin tissue using proteinase K digestion, following the protocol of Bandi et al. (13). After DNA extraction, 1:10 dilutions were prepared, to minimize possible PCR inhibition (9), and were stored at −20°C until further use. All tested samples were run in both simplex and duplex real-time PCR assays, for comparison of the variability in Cq values between the simplex and duplex runs. Samples that did not generate a signal were assigned a Cq value of 40, corresponding to the maximum number of cycles run in this real-time PCR setup. For positive samples, the number of GEs per swab or total skin tissue was calculated with the quantitation standards. A sample was considered positive when the number of GEs per swab/skin tissue exceeded 20, which, because of the dilution of the sample in the process of DNA extraction, corresponded to a detection limit of 0.1 GE per real-time PCR.
Table 4.
Overview of the amphibian samples used to validate the real-time duplex PCRs for B. dendrobatidis and B. salamandrivorans
| Amphibian species | Origin (coordinates and/or reference) and yr | Health status | Skin sample type (n) | No. PCR positive (species) | Mean GEs/swab for positive samples (range)a |
|---|---|---|---|---|---|
| Fire salamander (Salamandra salamandra) | Bunderbos, Netherlands (N50°55′, E5°45′), 2010 | Declining | Swab (33) | 13 (B. salamandrivorans) | 219.8 (60–1,750) |
| Tissue (8) | 4 (B. salamandrivorans) | 2,398.3 (242–10,180) | |||
| Merelbeke, Belgium (N50°57′, E3°43′), 2012 | Healthy | Swab (51) | 0 | NA | |
| B. salamandrivorans infection experiment (6), 2013 | Diseased | Swab (5) | 5 (B. salamandrivorans) | 6,920 (1,572–10,740) | |
| Tissue (5) | 5 (B. salamandrivorans) | 10,915 (3,420–19,380) | |||
| B. salamandrivorans infection experiment (6), 2013 | Healthy | Swab (5) | 0 | NA | |
| Tissue (5) | 0 | NA | |||
| Yellow-bellied toad (Bombina variegata) | ‘t Rooth, Netherlands (N50°50′, E5°47′) (A. Spitzen-van der Sluijs, A. Martel, C. A. Hallmann, W. Bosman, T. W. J. Garner, P. van Rooij, R. Jooris, F. Haesebrouck, and F. Pasmans, submitted for publication), 2013 | Healthy | Swab (27) | 14 (B. dendrobatidis) | 175.4 (20–1,488) |
Mean values of genomic equivalents (GEs) of zoospores per swab of B. dendrobatidis and B. salamandrivorans for the positive samples were calculated with the included quantitation standards for B. dendrobatidis and B. salamandrivorans. NA, not applicable.
RESULTS AND DISCUSSION
Assay optimization.
The primer and probe concentrations used in the duplex real-time PCR were the lowest concentrations that yielded the highest ΔRn (defined as the Rn value [the fluorescence emission of the reporter dye, normalized to the background fluorescence] of a reaction mixture containing all the reaction components [including the template] minus the Rn value of an unreacted negative control) and lowest quantification cycle (Cq) values, respectively, in a checkerboard system. This resulted in a PCR mixture of 25 μl per reaction composed of 12.5 μl TaqMan PCR mix (1× iQ Supermix; Bio-Rad Laboratories, Hercules, CA), B. salamandrivorans forward primer STerF at a concentration of 0.3 μM, B. salamandrivorans reverse primer STerR at a concentration of 0.3 μM, B. salamandrivorans Cy5-labeled probe STerC at a concentration of 0.1 μM, B. dendrobatidis forward primer ITS1-3 Chytr at a concentration of 0.9 μM, B. dendrobatidis reverse primer 5.8S Chytr at a concentration of 0.9 μM, B. dendrobatidis FAM-labeled probe Chytr MGB2 at a concentration of 0.15 μM, and 5 μl template. The amplification conditions were identical to the conditions used in the B. dendrobatidis-specific real-time PCR (8) with the exception of an increase in the annealing/extension temperature from 60°C to 62°C. No differences between PCR results were found when a serial 10-fold dilution series of B. dendrobatidis DNA was assayed in triplicate with standard and elevated annealing/extension temperatures. All B. salamandrivorans-positive samples, as determined with the B. salamandrivorans PCR (6) or by immunohistochemistry (9), and positive controls generated a single peak in the SYBR green real-time PCR melting curve analysis, with a constant melting temperature (Tm) of 75.5°C. Gel electrophoresis of the PCR product of B. salamandrivorans-positive samples and positive controls always generated a single DNA band at the expected size of approximately 160 bp. Negative samples and negative controls did not generate a peak in the melting curve analysis or generate a visible band in gel electrophoresis, indicating that no nonspecific binding of the primers occurred.
Sensitivity and specificity.
The sensitivity of the duplex real-time TaqMan PCR assay was tested with the described quantitation standards of B. dendrobatidis and B. salamandrivorans. Triplicates of serial dilution series ranging from 0.01 GE to 1,000 GEs of zoospores of the two pathogens were assayed with the duplex real-time PCR (Fig. 2). Although some of the 0.01-GE samples did generate Cq values, these were not consistent. The remaining concentrations of the 10-fold dilution series of both pathogens were detected with the duplex real-time PCR assay in all replicates. This demonstrates that the limit of detection of the B. salamandrivorans component of this duplex real-time PCR is 0.1 GE per PCR, which is similar to that for B. dendrobatidis in the B. dendrobatidis-specific real-time PCR (8). A detection limit lower than 1 GE of B. salamandrivorans suggests the presence of a high copy number of the ITS1 region, as already demonstrated for certain B. dendrobatidis strains (14, 15). This ability of the duplex real-time PCR to detect the two pathogens at low levels makes it ideal for early pathogen detection in environmental screening and in the diagnosis of chytridiomycosis. To ensure that no interference occurred when DNA of B. dendrobatidis and B. salamandrivorans was present in a sample, quantitation standards with DNA of both pathogens were assayed. This resulted in Cq values very similar to the values obtained with single-strain standards, indicating that accurate quantification can be performed with samples containing DNA of the two pathogens. To verify the specificity of the duplex real-time PCR, a total of 10 different isolates belonging to the class Chytridiomycota, including B. dendrobatidis and the B. salamandrivorans type strain, were assayed (Table 1). The real-time PCR amplified only B. dendrobatidis and B. salamandrivorans of all included isolates, indicating a high degree of specificity.
Fig 2.
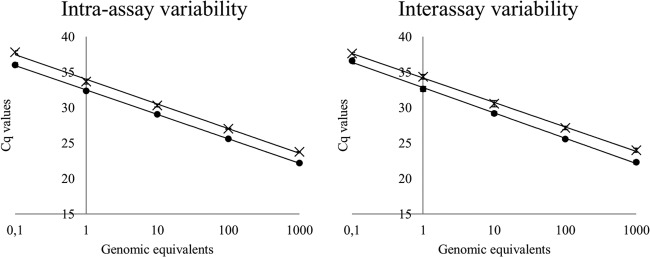
Standard curves for Batrachochytrium dendrobatidis and B. salamandrivorans generated with the duplex real-time PCR. Standard curves for B. dendrobatidis (×) and B. salamandrivorans (●) were generated by assaying triplicates of quantitation standards. Error bars represent the standard deviations of the assayed quantitation standard triplicates.
Assay performance and precision.
Assay performance and precision were evaluated with the described quantitation standards of B. dendrobatidis and B. salamandrivorans. High linear correlation (r2 > 0.995) and amplification efficiency (>94%) values for B. dendrobatidis and B. salamandrivorans in intra-assay and interassay variability experiments, together with low (<1%) intra- and interassay variabilities, demonstrate that the developed duplex real-time PCR has a good performance over the tested quantitation range, with highly reproducible results (Table 2). These traits make the duplex real-time PCR highly suitable for use in screening surveys and disease diagnosis.
Table 2.
Assay precision, efficiencies, and linear correlations of the B. dendrobatidis and B. salamandrivorans real-time duplex PCRs
| Species and variability experiment | Efficiency (%) | Linear correlation (r2) | Coefficient of variation (% [mean ± SD]) |
|---|---|---|---|
| B. dendrobatidis | |||
| Intra-assay | 94.1 | 0.997 | 0.56 ± 0.34 |
| Interassay | 99.4 | 0.996 | 0.99 ± 0.39 |
| B. salamandrivorans | |||
| Intra-assay | 95.7 | 0.999 | 0.39 ± 0.48 |
| Interassay | 96.0 | 0.997 | 0.98 ± 0.27 |
PCR protocol with bovine serum albumin.
Adding BSA to the duplex real-time PCR mixture at a final concentration of 400 ng μl−1 did not significantly affect the generated Cq values (t test, P > 0.05) (Table 3). This allows BSA to be used as an alternative to sample dilution in order to alleviate the influence of PCR inhibitors present in amphibian samples.
Table 3.
Effect of adding BSA (400 ng μl−1) to the duplex real-time PCR mixture on Cq values generated with B. dendrobatidis and B. salamandrivorans quantitation standardsa
| Genomic equivalent(s) |
Cq (mean ± SD) for: |
|||
|---|---|---|---|---|
|
B. dendrobatidis |
B. salamandrivorans |
|||
| With BSA | Without BSA | With BSA | Without BSA | |
| 1,000 | 24.95 ± 0.02 | 23.96 ± 0.04 | 23.00 ± 0.04 | 22.90 ± 0.10 |
| 100 | 27.22 ± 0.03 | 27.21 ± 0.07 | 26.32 ± 0.06 | 26.34 ± 0.17 |
| 10 | 30.48 ± 0.10 | 30.55 ± 0.11 | 29.69 ± 0.15 | 29.67 ± 0.17 |
| 1 | 33.53 ± 0.10 | 33.67 ± 0.32 | 33.32 ± 0.62 | 32.99 ± 0.19 |
| 0.1 | 37.33 ± 0.17 | 37.43 ± 0.23 | 36.18 ± 0.23 | 36.18 ± 0.16 |
Four replicates with and four replicates without added BSA were assayed in order to evaluate the effect on standard dilutions of B. dendrobatidis and B. salamandrivorans DNA. No significant difference was found between the two conditions for any of the dilutions for either pathogen (t test, P > 0.05).
Amphibian samples.
To validate the developed duplex real-time PCR, amphibian skin swabs and skin tissue were assayed (Table 4). The samples included known negative and positive B. dendrobatidis and B. salamandrivorans samples. All tested samples were run in the simplex and duplex real-time PCR assays to compare the variability in Cq values between the simplex and duplex runs (Fig. 3). Very little variation was found between the results of the duplex real-time PCR and both simplex real-time PCRs, as indicated by a high degree of correlation (r2 > 0.995) for the Cq values from the simplex and duplex runs for B. dendrobatidis and B. salamandrivorans. In the setup used in this study, the lowest detectable number of GEs per swab was 20, which corresponds to 0.1 GE per PCR. Dilution occurring in the process of DNA extraction and in the prevention of PCR inhibition accounts for this difference in detection limits between swabs and reactions. All samples that tested negative in the B. dendrobatidis and B. salamandrivorans simplex PCR assays also tested negative in the duplex PCR assay. The samples that tested positive in the B. dendrobatidis or B. salamandrivorans simplex PCR assay also tested positive for the corresponding pathogen in the duplex PCR assay. The noninvasive sampling technique (skin swabbing) resulted in overall lower GE numbers than the invasive technique (skin tissue collection) (Table 4). In the S. salamandra samples taken from the declining Dutch population, skin swabs were collected from live and apparently healthy animals, while skin tissues were collected from animals found dead on site. A possible explanation for the higher number of GEs found in the skin tissue samples could therefore be a more advanced disease state of the animals, accompanied by increased infection intensity. The smaller difference between GE numbers in the skin swab and skin tissue samples from the B. salamandrivorans-positive fire salamanders (S. salamandra) in the B. salamandrivorans infection experiment could be explained by the short period of time between swabbing and the animals dying due to infection with B. salamandrivorans. For B. dendrobatidis, a threshold in infection intensity (mean of >10,000 GEs per swab) predicts whether an amphibian population will decline due to B. dendrobatidis infection (16). In the B. salamandrivorans infection experiment, the mean value for B. salamandrivorans GEs per swab was comparable to this threshold (mean of 6,920 GEs per swab), indicating that this could also be the case for B. salamandrivorans.
Fig 3.
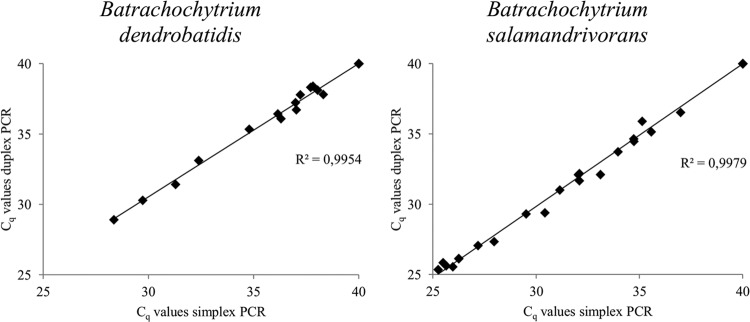
Cq values generated by assaying amphibian samples with the Batrachochytrium dendrobatidis and B. salamandrivorans duplex real-time PCR assay and both simplex real-time PCR assays. An overview of all assayed amphibian samples is presented in Table 1. Samples that did not generate a signal were assigned a Cq value of 40, corresponding to the maximum number of cycles run in this real-time PCR setup.
Conclusion.
The described B. dendrobatidis and B. salamandrivorans duplex real-time PCR can be used to accurately and reliably detect these two pathogens in amphibian samples. The real-time PCR can be used to aid in chytridiomycosis disease diagnosis and in mapping of the worldwide distribution of B. dendrobatidis and B. salamandrivorans.
ACKNOWLEDGMENT
This study was funded by a Dehousse research grant provided by the Royal Zoological Society of Antwerp (RZSA).
Footnotes
Published ahead of print 9 October 2013
REFERENCES
- 1.Stuart SN, Chanson JS, Cox NA, Young BE, Rodrigues ASL, Fischman DL, Waller RW. 2004. Status and trends of amphibian declines and extinctions worldwide. Science 306:1783–1786 [DOI] [PubMed] [Google Scholar]
- 2.Daszak P, Berger L, Cunningham AA, Hyatt AD, Green DE, Speare R. 1999. Emerging infectious diseases and amphibian population declines. Emerg. Infect. Dis. 5:735–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skerratt LF, Berger L, Speare R, Cashins S, McDonald KR, Phillott AD, Hines HB, Kenyon N. 2007. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth 4:125–134 [Google Scholar]
- 4.Longcore JE, Pessier AP, Nichols DK. 1999. Batrachochytrium dendrobatidis gen. et sp. nov., a chytrid pathogenic to amphibians. Mycologia 91:219–227 [Google Scholar]
- 5.Spitzen-van der Sluijs A, Spikmans F, Bosman W, De Zeeuw M, van der Meij T, Goverse E, Kik M, Pasmans F, Martel A. 2013. Rapid enigmatic decline drives the fire salamander (Salamandra salamandra) to the edge of extinction in the Netherlands. Amphibia-Reptilia 34:233–239 [Google Scholar]
- 6.Martel A, Spitzen-van der Sluijs A, Blooi M, Bert W, Ducatelle R, Fisher MC, Woeltjes A, Bosman W, Chiers K, Bossuyt F, Pasmans F. 2013. Batrachochytrium salamandrivorans sp. nov. causes lethal chytridiomycosis in amphibians. Proc. Natl. Acad. Sci. U. S. A. 110:15325–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Annis SL, Dastoor FP, Ziel H, Daszak P, Longcore JE. 2004. A DNA-based assay identifies Batrachochytrium dendrobatidis in amphibians. J. Wildl. Dis. 40:420–428 [DOI] [PubMed] [Google Scholar]
- 8.Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD. 2004. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis. Aquat. Organ. 60:141–148 [DOI] [PubMed] [Google Scholar]
- 9.Hyatt AD, Boyle DG, Olsen V, Boyle DB, Berger L, Obendorf D, Dalton A, Kriger K, Hero M, Hines H, Phillott R, Campbell R, Marantelli G, Gleason F, Colling A. 2007. Diagnostic assays and sampling protocols for the detection of Batrachochytrium dendrobatidis. Dis. Aquat. Organ. 73:175–192 [DOI] [PubMed] [Google Scholar]
- 10.Garland S, Baker A, Phillott AD, Skerratt LF. 2010. BSA reduces inhibition in a TaqMan (R) assay for the detection of Batrachochytrium dendrobatidis. Dis. Aquat. Organ. 92:113–116 [DOI] [PubMed] [Google Scholar]
- 11.Kreader CA. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Rooij P, Martel A, Nerz J, Voitel S, Van Immerseel F, Haesebrouck F, Pasmans F. 2011. Detection of Batrachochytrium dendrobatidis in Mexican bolitoglossine salamanders using an optimal sampling protocol. EcoHealth 8:237–243 [DOI] [PubMed] [Google Scholar]
- 13.Bandi C, Damiani G, Magrassi L, Grigolo A, Fani R, Sacchi L. 1994. Flavobacteria as intracellular symbionts in cockroaches. Proc. Biol. Sci. 257:43–48 [DOI] [PubMed] [Google Scholar]
- 14.Kirshtein JD, Anderson CW, Wood JS, Longcore JE, Voytek MA. 2007. Quantitative PCR detection of Batrachochytrium dendrobatidis DNA from sediments and water. Dis. Aquat. Organ. 77:11–15 [DOI] [PubMed] [Google Scholar]
- 15.Longo AV, Rodriguez D, da Silva Leite D, Toledo LF, Mendoza Almeralla C, Burrowes PA, Zamudio KR. 2013. ITS1 copy number varies among Batrachochytrium dendrobatidis strains: implications for qPCR estimates of infection intensity from field-collected amphibian skin swabs. PLoS One 8(3):e59499. 10.1371/journal.pone.0059499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vredenburg VT, Knapp RA, Tunstall TS, Briggs CJ. 2010. Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proc. Natl. Acad. Sci. U. S. A. 107:9689–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]


