Abstract
Two novel protocols for inactivation and extraction were developed and used to identify 107 Mycobacterium clinical isolates, including Mycobacterium tuberculosis complex, from solid cultures using Vitek matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry. The protocol using heat inactivation with sonication and cell disruption with glass beads resulted in 82.2% and 88.8% species and genus level identifications, respectively.
TEXT
The Mycobacterium genus consists of over 100 species of rapidly growing and slow-growing acid-fast bacilli (AFB) (1–4). Rapid and accurate diagnosis of mycobacteria infections is important to patient care and public health (4). Inappropriate treatment may lead to unnecessary exposure to toxic drugs or drug resistance (5). Rapid identification (ID) of mycobacteria has proven difficult due in part to their fastidious growth requirements and low growth rate (3, 6). Molecular probes and DNA hybridization are relatively fast and simple but are available only for a limited number of clinically common species (2, 3, 5, 7, 8). High-performance liquid chromatography (HPLC) (2, 3, 9) and electrospray ionization-tandem mass spectrometry analysis (2) have recently been used to analyze mycolic acid but are labor-intensive and require technical expertise (1, 2).
Recent studies have shown that matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is an accurate and rapid method for identifying bacteria and yeast from solid culture media (6, 10–13). MALDI-TOF MS has also recently been adapted for the identification of mycobacteria (1–5, 8, 9, 14), mostly using a Bruker Daltonics Flex system (1, 2, 4, 5, 8). We previously evaluated one inactivation procedure, described in a 2010 training manual from bioMérieux, that suspended mycobacteria in trifluoroacetic acid (TFA) for 30 min (15) and found that both the procedure and the database were ineffective (data not shown). There is no standard inactivation procedure currently available for identifying mycobacteria by using MALDI-TOF MS for either the Bruker or bioMérieux system.
This study evaluates two novel inactivation and extraction protocols used to identify clinical mycobacterial isolates, including Mycobacterium tuberculosis complex (MTC), from solid culture media. In addition, this study evaluates an improved database for mycobacteria by using Vitek MALDI-TOF MS RUO (Vitek MS) (bioMérieux, Durham, NC) with a reference database developed in-house.
(Parts of these results were presented at the 113th General Meeting of the American Society for Microbiology, 18 to 21 May 2013.)
Clinical mycobacterial isolates from a culture positive for AFB were identified by either a DNA probe or HPLC. Organisms grown on BBL Middlebrook 7H11 solid agar media (Becton, Dickinson [BD], Sparks, MD) and incubated at 37°C in 4.0 to 8.0% CO2 were used for identification by Vitek MS.
Due to the lack of a mycobacterial superspectrum in the RUO database at the time, we developed a reference database consisting of 50 relevant clinical isolates of 18 Mycobacterium species by importing the spectrum into Vitek MS with SARAMIS (spectral archive and microbial identification system). The imported spectra had at least 80 peaks from isolates that were inactivated using protocol A and identified by DNA probe or HPLC. For Vitek MS results showing more than one species ID under the Mycobacterium genus, we concluded the results to be correct to the genus level. When a spectrum was obtained but the database was unable to identify the spectrum, the result was categorized as no identification.
The first protocol, protocol A, using heat inactivation in ethanol followed by sonication, was independently developed in our laboratory. A 1-μl disposable inoculation loop was used to transfer a colony of mycobacteria from 7H11 agar medium into a microcentrifuge tube containing 500 ml of 70% ethanol. The microcentrifuge tube was heated for 30 min on a heat block at 95°C ± 5°C. The tube was then centrifuged at 18,000 × g for 2 min. The resulting pellet was washed and dispersed with sterile water. Centrifugation and washing were repeated two more times. After the 3rd wash, the vial was placed in a sonication bath for 15 min. It was then centrifuged at 18,000 × g for 10 min, and the supernatant was removed. The pellet was resuspended in 5 μl of 85% formic acid and centrifuged at 15,000 × g for 1 min. Five microliters of acetonitrile was added before the tube was again centrifuged at 15,000 × g for 1 min. One microliter of supernatant was added to a spot on a disposable MALDI target plate (Shimadzu Biotech; catalog no. 220-99999-FM1). The spot was allowed to dry completely and covered with 1 μl of an α-cyano-4-hydroxycinnamic acid (CHCA) matrix (bioMérieux; catalog no. 411071). A flow chart of protocol A is shown in Fig. 1.
Fig 1.
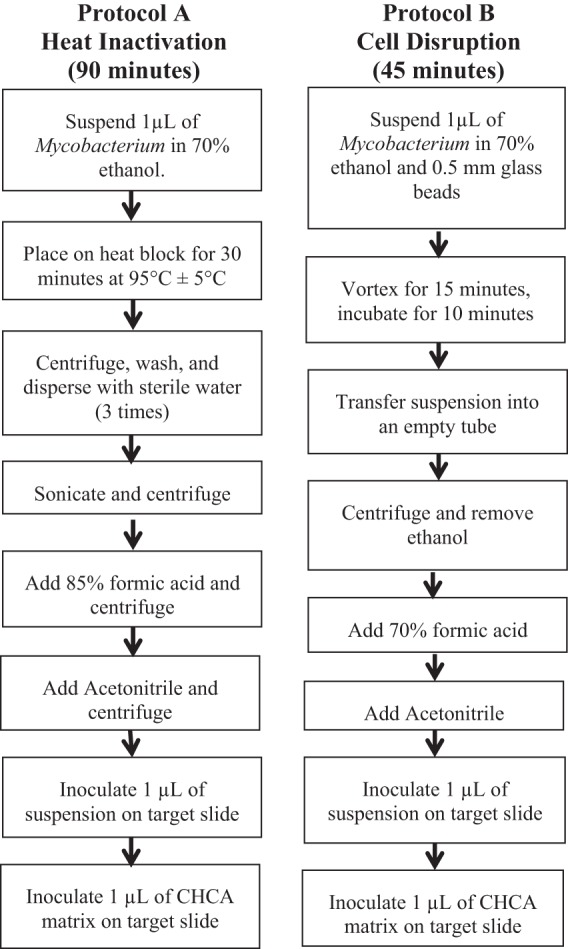
Flow chart of inactivation protocols used prior to identification (ID) by Vitek MALDI-TOF MS RUO (Vitek MS). Note that the suspension spot must dry completely before adding the CHCA matrix.
The second protocol, protocol B, using cell disruption with glass bead in ethanol, was developed by bioMérieux and provided for research use for this study. A 1-μl loop was used to transfer a colony of mycobacteria from 7H11 solid medium into a microcentrifuge tube containing 500 ml of 70% ethanol and 200 ml of 0.5-mm glass beads (Sartorius Stedim; catalog no. 14-559-084). The microcentrifuge tube was vortexed for 15 min using a vortex adaptor (MoBio; catalog no. BMX13000-V1-24) and allowed to incubate at room temperature for another 10 min. The contents of the tube were vortexed for 5 to 10 s to suspend the mycobacteria. The suspension was transferred to an empty microcentrifuge tube with care to avoid the transfer of any bead. The tube was centrifuged at 10,000 × g for 2 min to create a sufficient pellet. The pellet was resuspended in 10 μl of 70% formic acid and allowed to incubate for 2 to 5 min at room temperature. Ten microliters of acetonitrile was added to the suspension. The tube was then centrifuged at 10,000 × g for 2 min. One microliter of supernatant was added to a spot on a disposable target plate. The spot was allowed to dry completely and covered with 1 μl of CHCA matrix. A flow chart of protocol B is shown in Fig. 1.
The remaining supernatant of each run was inoculated onto H711 solid medium to ensure successful inactivation. No mycobacterial growth was seen after 6 weeks of incubation at 37°C in 4.0 to 8.0% CO2. This is particularly important for MTC inactivation and identification. Thus, the inactivated isolates can be used for MALDI-TOF MS in the clinical setting.
One hundred seven clinically relevant isolates, consisting of 14 species of mycobacteria, were included in the study (Table 1). After inactivation by protocol A, 88/107 (82.2%) mycobacterial isolates were correctly identified to the species or genus level. After inactivation by protocol B, 95/107 (88.8%) mycobacterial isolates were correctly identified to the species or genus level. Inactivation by protocol A resulted in a higher percentage of correct identifications at species level for M. avium complex (MAC) and M. kansasii isolates. It could be argued that protocol A had an unfair advantage, because the database used to identify isolates was built using spectral fingerprints of isolates inactivated with the same protocol. Inactivation by protocol B resulted in a higher percentage of correct identifications at species level. Protocol B requires less processing time and fewer steps than protocol A. The higher percentage of correct identifications at species level may be a result of fewer protocol steps, resulting in fewer chances to decrease the sample recovery yield. One more MTC isolate was correctly identified by using protocol B than by using protocol A.
Table 1.
Results of Mycobacterium ID by Vitek MS after inactivation by either protocol A or protocol B
| Organism | Total no. of isolates | No. (%) of isolates after inactivation by protocol: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| A with: |
B with: |
||||||||
| Species level ID | Genus level ID | No ID | Incorrect ID | Species level ID | Genus level ID | No ID | Incorrect ID | ||
| M. avium complex | 64 | 42 | 10 | 10 | 2a | 54 | 2 | 8 | 0 |
| M. tuberculosis complex | 18 | 16 | 0 | 2 | 0 | 15 | 0 | 3 | 0 |
| M. kansasii | 9 | 5 | 1 | 3 | 0 | 9 | 0 | 0 | 0 |
| M. fortuitum | 4 | 4 | 0 | 0 | 0 | 3 | 1 | 0 | 0 |
| M. abscessus | 3 | 2 | 0 | 1 | 0 | 3 | 0 | 0 | 0 |
| M. gordonae | 2 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| M. kubicae | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| M. lentiflavum | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| M. mucogenicum | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| M. scrofulaceum | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| M. simiae | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| M. szulgai | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| M. triplex | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Total | 107 | 74 (69.2) | 14 (13.1) | 17 (15.9) | 2 (1.9) | 88 (82.2) | 7 (6.5) | 12 (11.2) | 0 (0.0) |
One M. avium complex isolate was incorrectly identified as a Microbacterium species and the other as Trichophyton violaceum.
Failure to obtain an ID can most likely be attributed to an insufficient protein signal or an absence of an adequate reference spectrum in the database (16–18). Protocol A yielded an average of 119 mass spectrum peaks. This was not statistically different from protocol B, which yielded an average of 128 peaks (P = 0.1426). Spectral fingerprints of representative MAC and MTC isolates can be found in Fig. 2. The median percent match for protocol A was 61% with a range of 40 to 88%. The median percent match for protocol B was 50% with a range of 42 to 76%.
Fig 2.
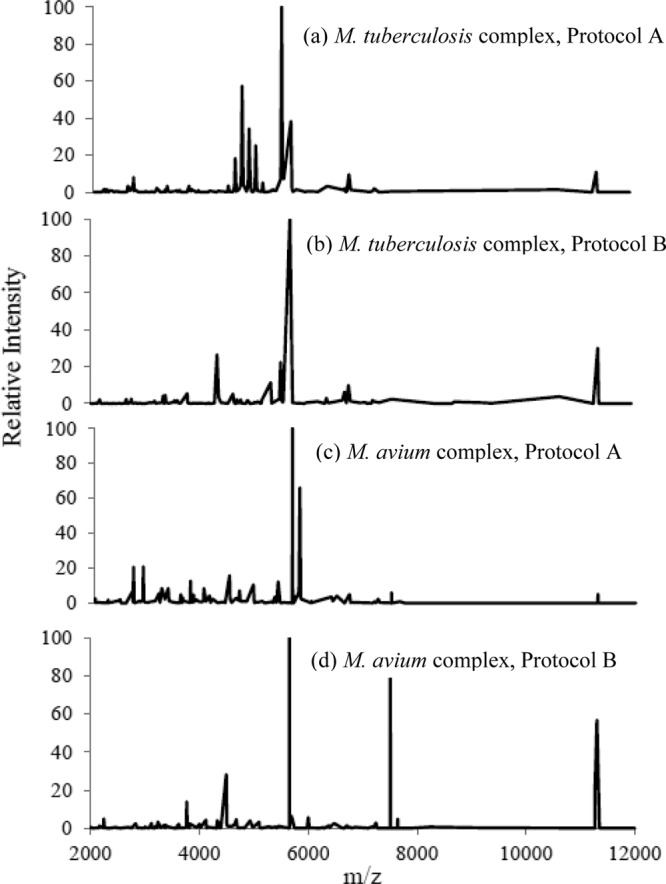
Spectral fingerprints obtained by use of Vitek MS after inactivation. m/z, mass-to-charge ratio.
Protocols A and B are significantly simpler and safer than previously published protocols (1, 2, 5, 8, 9). Protocol A does not require washes or centrifugation before heating. Protocol B does not require any washes; processing time is under 1 h.
One limitation of this study is the relatively small database, which resulted in lower percent matches and a lack of identifications below the complex level for MTC and MAC. Though it seems that there is a lack of diversity of mycobacterial isolates in this study, MAC isolates are the most common mycobacteria isolated in our clinical setting, partially due to the high number of HIV-positive patients. Though it is important to study the clean, pure colony from solid media, future studies should adapt these protocols for liquid culture.
In summary, this study demonstrates the effectiveness of a novel heat inactivation protocol as well as a novel cell disruption inactivation protocol used for identification of clinically relevant mycobacterial isolates, including M. tuberculosis complex, from solid culture media by using Vitek MALDI-TOF (RUO) MS.
ACKNOWLEDGMENTS
We thank Tim Drake, Mahin (May) Park, Eileen M. Burd, and Colleen Kraft for help. We are grateful to the medical technologists for their help in obtaining patient samples necessary for this study.
No financial support was provided for performance of the study.
Footnotes
Published ahead of print 25 September 2013
REFERENCES
- 1.El Khéchine A, Couderc C, Flaudrops C, Raoult D, Drancourt M. 2011. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of mycobacteria in routine clinical practice. PLoS One 6:e24720. 10.1371/journal.pone.0024720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lotz A, Ferroni A, Beretti JL, Dauphin B, Carbonnelle E, Guet-Revillet H, Nicolas Veziris N, Heym B, Jarlier V, Gaillard JL, Pierre-Audigier C, Frapy E, Berche P, Nassif X, Bille E. 2010. Rapid identification of mycobacterial whole cells in solid and liquid culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:4481–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pignone M, Greth KM, Cooper J, Emerson D, Tang J. 2006. Identification of mycobacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 44:1963–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shitikov E, Ilina E, Chernousova L, Borovskaya A, Rukin I, Afanas'ev MT, Smirnovab A, Vorobyevab E, Larionovab S, Andreevskayab M, Kostrzewac Govorun V. 2012. Mass spectrometry based methods for the discrimination and typing of mycobacteria. Infect. Genet. Evol. 12:838–845 [DOI] [PubMed] [Google Scholar]
- 5.Saleeb PG, Drake SK, Murray PR, Zelazny AM. 2011. Identification of mycobacteria in solid culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:1790–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel R. 2013. Matrix-assisted laser desorption ionization–time of flight mass spectrometry in clinical microbiology. Clin. Infect. Dis. 57:564–572 [DOI] [PubMed] [Google Scholar]
- 7.Schweickert B, Moter A, Lefmann M, Göbel UB. 2004. Let them fly or light them up: matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry and fluorescence in situ hybridization (FISH). APMIS 112:856–885 [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Chen WF, Li QX. 2012. Rapid identification and classification of Mycobacterium spp. using whole-cell protein barcodes with matrix assisted laser desorption ionization time of flight mass spectrometry in comparison with multigene phylogenetic analysis. Anal Chim. Acta 716:133–137 [DOI] [PubMed] [Google Scholar]
- 9.Hettick JM, Kashon ML, Simpson JP, Siegel PD, Mazurek GH, Weissman DN. 2004. Proteomic profiling of intact mycobacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 76:5769–5776 [DOI] [PubMed] [Google Scholar]
- 10.Christner M, Rohde H, Wolters M, Sobottka I, Wegscheider K, Aepfelbacher M. 2010. Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry fingerprinting. J. Clin. Microbiol. 48:1584–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drancourt M. 2010. Detection of microorganisms in blood specimens using matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a review. Clin. Microbiol. Infect. 16:1620–1625 [DOI] [PubMed] [Google Scholar]
- 12.Neville S, LeCordier A, Ziochos H, Chater M, Gosbell I, Maley M, Van Hal S. 2011. Utility of matrix-assisted laser desorption ionization–time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 49:2980–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez K, Olsen R, Musick W, Cernoch PL, Davis JR, Land GA, Peterson LE, Musser JM. 2013. Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch. Pathol. Lab. Med. 137:1247–1254 [DOI] [PubMed] [Google Scholar]
- 14.Hettick JM, Kashon ML, Slaven JE, Ma Y, Simpson JP, Siegel PD, Mazurek GN, Weissman DN. 2006. Discrimination of intact mycobacteria at the strain level: a combined MALD-TOF MS and biostatistical analysis. Proteomics 6:6416–6425 [DOI] [PubMed] [Google Scholar]
- 15.bioMerieux 2010. Customer training manual. bioMérieux, Durham, NC [Google Scholar]
- 16.Bizzini A, Jaton K, Romo D, Bille J, Prod'hom G, Greub G. 2011. Matrix-assisted laser desorption ionization–time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J. Clin. Microbiol. 49:693–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wieser A, Schneider L, Jung J, Schuber S. 2012. MALDI-TOF MS in microbiological diagnostics—identification of microorganisms and beyond. Appl. Microbiol. Biotechnol. 93:965–974 [DOI] [PubMed] [Google Scholar]
- 18.Alatoom AA, Cunningham SA, Ihde SM, Mandrekar J, Patel R. 2011. Comparison of direct colony method versus extraction method for identification of Gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]


