Abstract
BK polyomavirus (BKV) is an emerging pathogen in immunocompromised individuals. BKV subtype III is rarely identified and has not previously been associated with disease. Here we provide the whole-genome sequence of a subtype III BKV from a pediatric kidney transplant patient with polyomavirus-associated nephropathy.
CASE REPORT
A 16-year-old with a history of posterior urethral valves complicated by obstructive uropathy leading to end-stage renal disease received a kidney transplant from his father (living related donor) in 2000. He was maintained on steroid-free immunosuppression with tacrolimus and mycophenolate mofetil for 8 years. In 2008, the patient was diagnosed with chronic transplant glomerulopathy, eventually requiring dialysis. He received his second renal transplant in 2012 from a 1/6 human leukocyte antigen (HLA)-matched deceased male donor. He had a wide range of anti-HLA class I and II preformed antibodies with percent panel reactive antibody (%PRA) values of 13% class I and 69% class II. For induction immunosuppression, the patient received a total of 7.5 mg/kg (body weight) of rabbit antithymocyte globulin with methylprednisone premedication and a single dose of 500 mg/m2 (body surface area) rituximab. He was maintained on immunosuppression consisting of tacrolimus, mycophenolate mofetil, and a tapering dose of prednisone. A ureteric stent placed routinely at the time of transplant was removed uneventfully at ∼5 weeks posttransplant. BK polyomavirus (BKV) DNA was monitored regularly in plasma and urine specimens by quantitative real-time PCR. Testing was performed using the previously described V3a assay (1), modified for use on a LightCycler 2.0 instrument (see the supplemental material). BKV DNA was first noted in plasma on posttransplant day (PTD) 114. A renal biopsy was performed because of increasing creatinine and plasma BKV levels (293,000 BKV copies/ml on PTD 154, 5 days before the biopsy) despite decreased immunosuppression. The biopsy specimen showed histological evidence of severe tubulointerstitial inflammation, and immunohistochemistry using antibodies against simian virus 40 (SV40) T antigen (dilution, 1:200; EDTA antigen retrieval) (Calbiochem, USA) revealed frequent nuclear staining of tubular epithelial cells, confirming the presence of BKV antigen (Fig. 1). Taken together, these findings were diagnostic for severe BK virus-associated nephropathy (BKVN). BKV levels reached 1.2 million copies/ml in plasma (PTD 210) and >60 million copies/ml in urine (PTD 154). Levels remained at more than 104 copies/ml in serial plasma samples (n = 17) collected up to PTD 308 (results not shown). Throughout the course of infection, the patient received multiple treatments, including leflunomide, intravenous immunoglobulin, ciprofloxacin, and cidofovir.
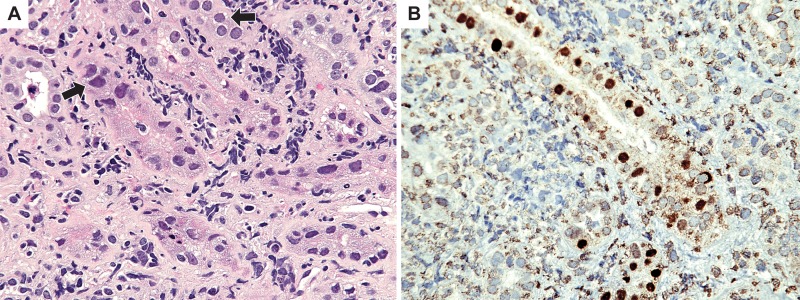
Fig 1.
(A) Hematoxylin-eosin stain showing interstitial edema and inflammation. Several tubular epithelial nuclei are enlarged with ground glass appearance (arrows). (B) Immunohistochemical staining for polyomavirus large T antigen (SV40) highlights multiple tubular epithelial nuclei, confirming the diagnosis of polyomavirus nephropathy. Magnification, ×400 (both panels).
BKV typing was performed on two plasma specimens, KT-867 (collected on PTD 114, with a viral load of <1,000 copies/ml) and KT-514 (collected on PTD 121, with a viral load of 191,000 copies/ml). Typing was also performed on one urine sample, KT-815.4 (collected on PTD 154, with a viral load of >60 million copies/ml). The 287-bp VP1 typing region, positions 1650 to 1936 relative to BKV Dunlop (GenBank accession no. V01108), was amplified using nested PCR as described previously (2). The PCR amplicons were sequenced by bidirectional dideoxynucleotide termination sequencing, identifying BKV subtype III from all three specimens. Long-range PCR was utilized to amplify the whole genomes of these viruses for sequencing (see the supplemental material). The complete BKV genomes were then acquired by primer walking and were deposited in GenBank (KF055891 to KF055893).
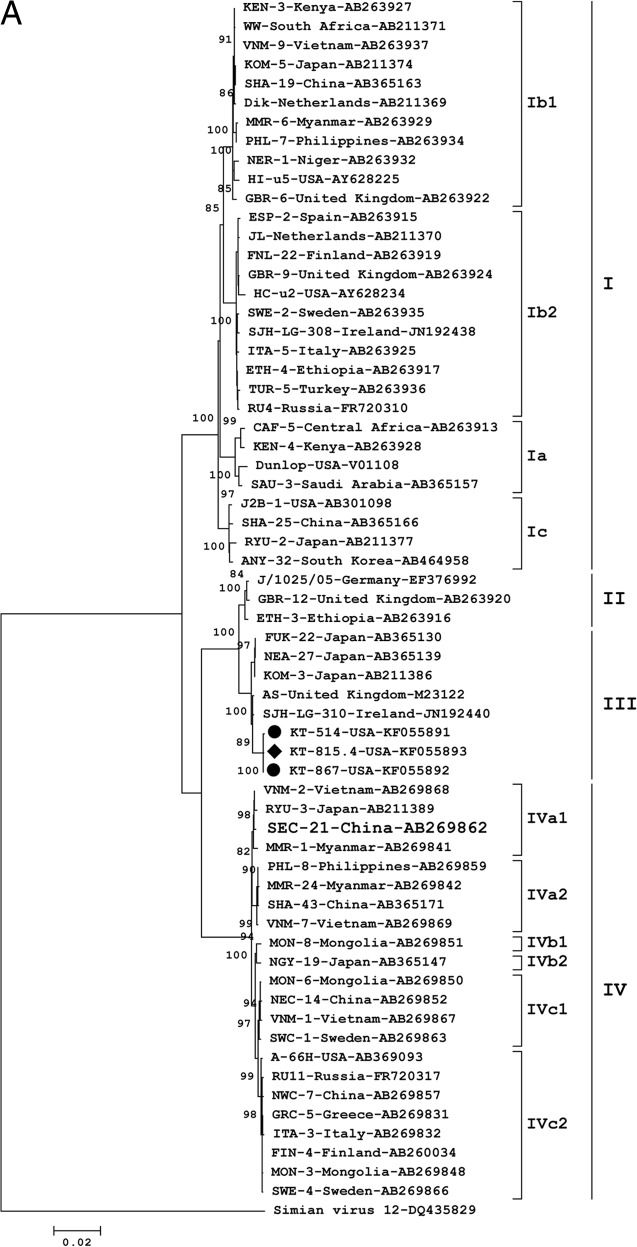
Phylogenetic analysis of the whole genomes showed their close relationship to BKV subtype III reference sequences AS (GenBank accession no. M23122.1), identified from the urine of a pregnant woman in the United Kingdom, and SJH-LG-310 (GenBank accession no. JN192440.1), identified from the urine of a hematopoietic stem cell transplant patient with hemorrhagic cystitis who was born in Africa (Fig. 2A). Only three other BKV subtype III whole genomes have been sequenced (Fig. 2A), two from patients without immune compromise (NEA-27 [GenBank accession no. AB365139.1] and FUK-22 [GenBank accession no. AB365130.1]) and one from a bone marrow transplant recipient (KOM-3 [GenBank accession no. AB211386.1]). There was no evidence of subtype recombination (SimPlot v.3.5.2 [3]; data not shown). However, based on amino acid alignments of VP1 sequences, BKV from this patient was highly divergent from other BK subtype III viruses (Fig. 2B). The reason for this divergence is a substitution of eight amino acids, from MGDPDDNL to DQVSATVI (positions 56 to 63), of the VP1 major capsid protein. These changes affect the BC loop, a surface-exposed region important for viral attachment to host cells (4). No amino acid changes were detected in the other protein-coding regions of the viruses sequenced in this study, except for KT-815.4-USA, in which a Q150H substitution was detected in the pRb domain of the large T antigen.
Fig 2.
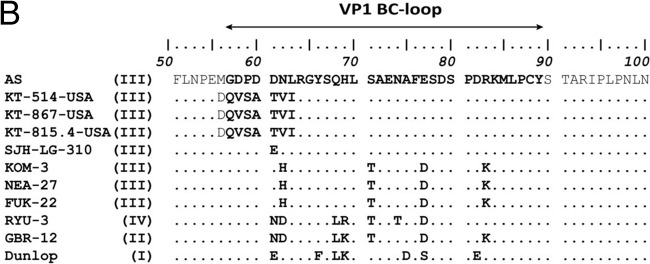
(A) Maximum likelihood phylogenetic tree constructed from complete BKV genomes. The newly described subtype III viruses are labeled as KT-867-USA, KT-514-USA, and KT-815.4-USA. Circle, plasma; diamond, urine. The tree includes the five subtype III whole genomes available in GenBank, as well as one representative whole genome per country, per subtype for subtype II and per country, per subgroup for subtypes I and IV. SV12 (GenBank accession no. AY614708) was used as an outgroup. The numbers at nodes give the bootstrap confidence levels (percentages) obtained for 1,000 replicates (only values of ≥50% are shown for major nodes). The scale bar indicates nucleotide substitutions per site. (B) Amino acid identity plot of the BC loop domain of the VP1 protein.
Here we provide the whole-genome sequence of a subtype III BKV from a case of BKVN in a pediatric kidney transplant patient. To our knowledge, this study describes the first whole genome of BKV subtype III identified in the United States and represents the first reported case of BKVN associated with BKV subtype III worldwide.
BKV belongs to the family Polyomaviridae, of small, nonenveloped DNA viruses with an icosahedral capsid of 45 nm in diameter that contains a circular, double-stranded genome of approximately 5 kb (5). The BKV genome contains a noncoding control region (NCCR), which regulates the expression of small t and large T antigens, capsid proteins (VP1, VP2, and VP3), and the agnoprotein (6). BKV strains can be classified based on polymorphisms in the VP1 region (subtypes I to IV) (2, 7).
BKV is transmitted via the respiratory or oral/enteric route, infecting children asymptomatically and then persisting in the kidney, where it can cause nephropathy in renal transplant patients (8). BKVN occurs in up to 10% of kidney transplants and is a significant cause of allograft dysfunction and graft loss. Treatment of BKVN is currently based on reduction of immunosuppression. There is no established antiviral therapy for BKVN, although cidofovir and leflunomide may be considered (8).
BKV subtype I is the most commonly identified subtype, accounting for 80% of BKVs and demonstrating a global geographic distribution (7, 9). BKV subtype IV makes up 15% and is found primarily in East Asia and Europe. Subtypes II and III are rarely identified, though primer and/or probe subtype-specific mismatches in certain BKV molecular diagnostic assays may contribute to underdetection (1, 10). Interestingly, BKV subtype III demonstrates reduced replicative capacity in vitro (11). This may correspond to limited pathogenicity in vivo and contribute to the low prevalence of BKV subtype III worldwide. Only once has BKV subtype III been reported in a patient with BKVN previously, and in that case it was identified in a mixed infection with subtype I and another variant that could not be conclusively subtyped; thus, the pathology could not be definitively attributed to subtype III (12).
The development of BKVN in kidney transplant recipients is a process involving the complex interplay of host and viral factors. The patient described herein was at very high risk of BKVN, given that he received a second transplant with only 1/6 HLA matching. Furthermore, he was highly sensitized, requiring potent induction immunosuppression.
While these immunologic elements likely contributed to the development of disease, evaluation of the BKV genomes from this patient's plasma and urine specimens suggests that intrinsic viral factors may also play a role. We identified viral amino acid changes that are unique among BKV sequences, most of which affected the BC loop of VP1. It has been proposed that amino acid changes within the BC loop may be associated with an increase in pathogenic potential and therefore may contribute to the development of BKVN (11, 12). In addition, phosphorylation has been hypothesized to influence VP1 function, and two of the amino acid changes (P59S and N61T) create potential new phosphorylation sites for cellular kinases (13). Future experiments will be required to determine the effects that these VP1 changes have on virion assembly, replication, and target cell binding.
In conclusion, we describe the identification and whole-genome sequencing of subtype III BKV in a pediatric kidney transplant recipient with nephropathy. Findings such as those reported here may contribute to a better understanding of the pathogenic potential of BKV.
Nucleotide sequence accession numbers.
The BKV genomes determined here were deposited in GenBank under accession numbers KF055891 to KF055893.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the Pediatric Kidney Transplant Clinic at Lucile Packard Children's Hospital and the staff of the Stanford Clinical Virology Laboratory for their continued diligent work and dedication to patient care.
This research was supported by Beta Sigma Phi and the Transplantation and Tissue Engineering Center of Excellence Program Endowment Fund, Lucile Packard Children's Hospital. We declare no conflicts of interest in the publication of this research.
Footnotes
Published ahead of print 18 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01801-13.
REFERENCES
- 1.Hoffman NG, Cook L, Atienza EE, Limaye AP, Jerome KR. 2008. Marked variability of BK virus load measurement using quantitative real-time PCR among commonly used assays. J. Clin. Microbiol. 46:2671–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin L, Gibson PE, Knowles WA, Clewley JP. 1993. BK virus antigenic variants: sequence analysis within the capsid VP1 epitope. J. Med. Virol. 39:50–56 [DOI] [PubMed] [Google Scholar]
- 3.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dugan AS, Gasparovic ML, Tsomaia N, Mierke DF, O'Hara BA, Manley K, Atwood WJ. 2007. Identification of amino acid residues in BK virus VP1 that are critical for viability and growth. J. Virol. 81:11798–11808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett SM, Broekema NM, Imperiale MJ. 2012. BK polyomavirus: emerging pathogen. Microbes Infect. 14:672–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gosert R, Rinaldo CH, Funk GA, Egli A, Ramos E, Drachenberg CB, Hirsch HH. 2008. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J. Exp. Med. 205:841–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng HY, Nishimoto Y, Chen Q, Hasegawa M, Zhong S, Ikegaya H, Ohno N, Sugimoto C, Takasaka T, Kitamura T, Yogo Y. 2007. Relationships between BK virus lineages and human populations. Microbes Infect. 9:204–213 [DOI] [PubMed] [Google Scholar]
- 8.Hirsch HH, Randhawa P. 2013. BK polyomavirus in solid organ transplantation. Am. J. Transplant. 13(Suppl 4):179–188 [DOI] [PubMed] [Google Scholar]
- 9.Zhong S, Randhawa PS, Ikegaya H, Chen Q, Zheng HY, Suzuki M, Takeuchi T, Shibuya A, Kitamura T, Yogo Y. 2009. Distribution patterns of BK polyomavirus (BKV) subtypes and subgroups in American, European and Asian populations suggest co-migration of BKV and the human race. J. Gen. Virol. 90:144–152 [DOI] [PubMed] [Google Scholar]
- 10.Luo C, Bueno M, Kant J, Randhawa P. 2008. Biologic diversity of polyomavirus BK genomic sequences: implications for molecular diagnostic laboratories. J. Med. Virol. 80:1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tremolada S, Delbue S, Larocca S, Carloni C, Elia F, Khalili K, Gordon J, Ferrante P. 2010. Polymorphisms of the BK virus subtypes and their influence on viral in vitro growth efficiency. Virus Res. 149:190–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tremolada S, Akan S, Otte J, Khalili K, Ferrante P, Chaudhury PR, Woodle ES, Trofe-Clark J, White MK, Gordon J. 2010. Rare subtypes of BK virus are viable and frequently detected in renal transplant recipients with BK virus-associated nephropathy. Virology 404:312–318 [DOI] [PubMed] [Google Scholar]
- 13.Li M, Garcea RL. 1994. Identification of the threonine phosphorylation sites on the polyomavirus major capsid protein VP1: relationship to the activity of middle T antigen. J. Virol. 68:320–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.