Abstract
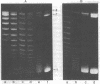
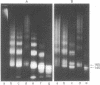
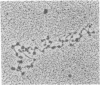
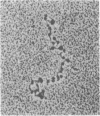
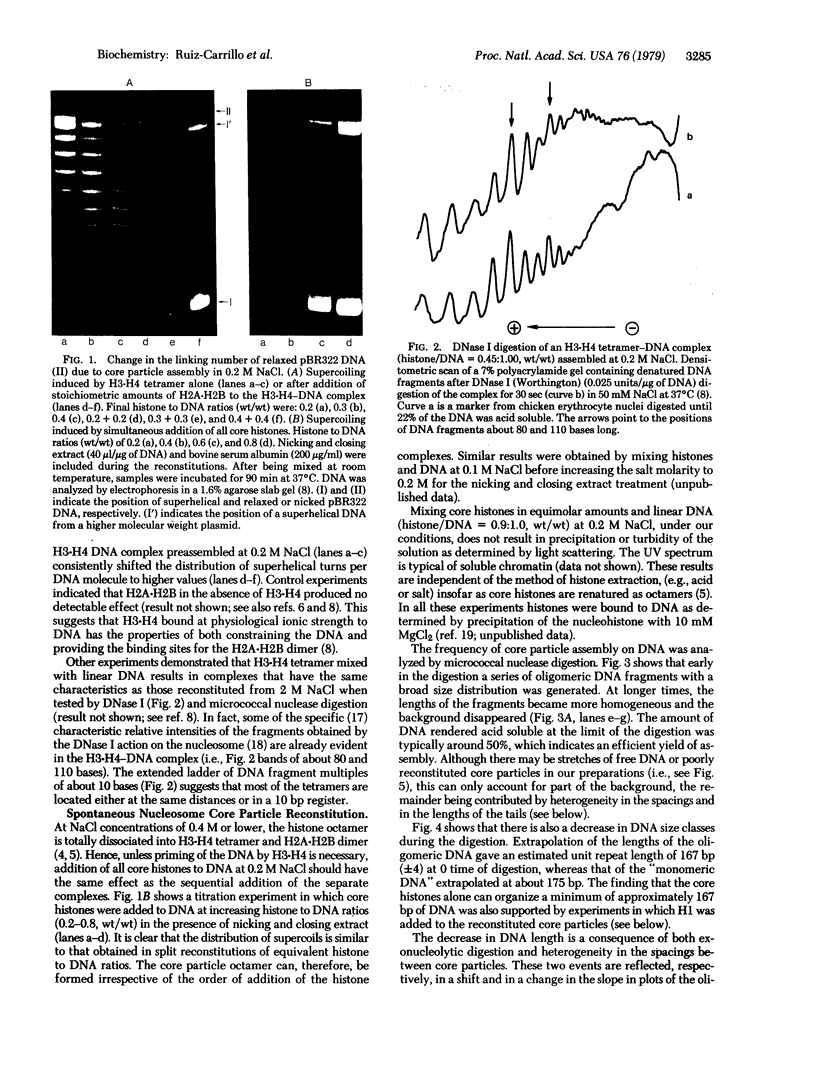
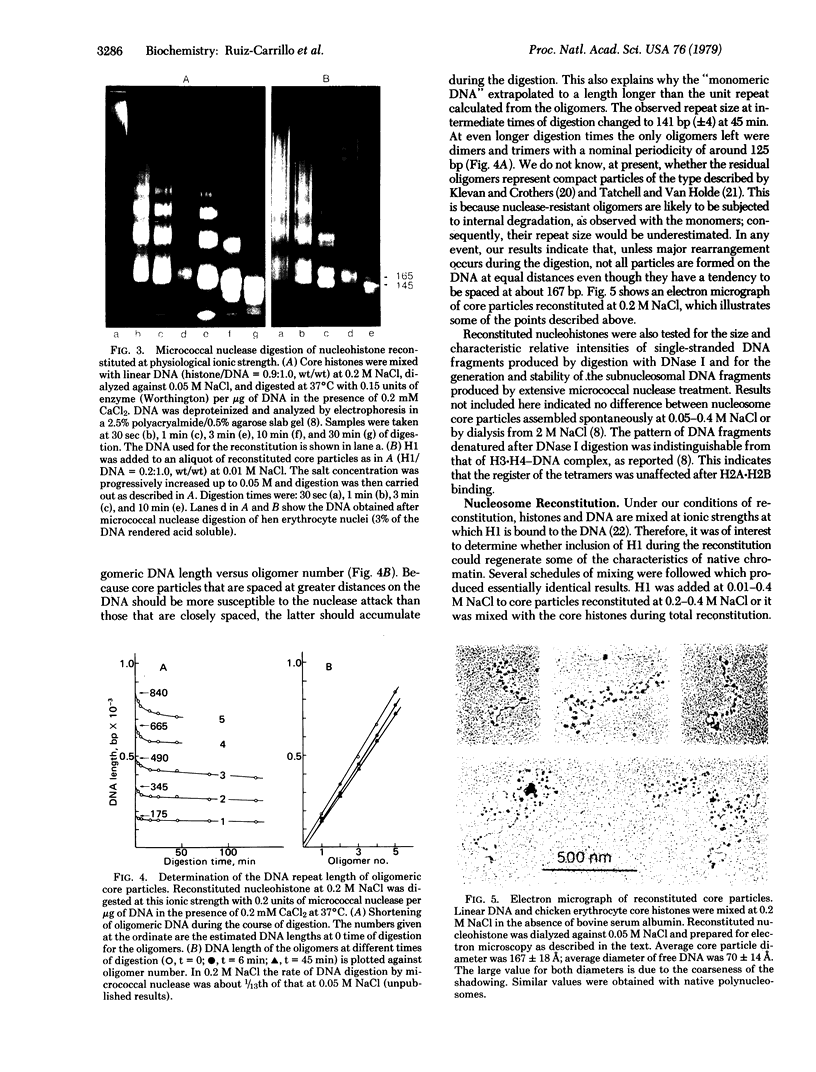
Nucleosome core particles have been efficiently assembled in vitro by direct interaction of histones and DNA at physiological ionic strength, as assayed by digestion with DNases, supercoiling of relaxed circular DNA, and electron microscopy. Reconstitution was achieved either by the simultaneous addition of all core histones, or by the sequential binding of H3 . H4 tetramer and H2A . H2B dimer to DNA. Micrococcal nuclease digestion and electron microscopy studies indicated that there is heterogeneity in the spacings at which core particles are assembly on the DNA. Length measurements of oligomeric DNA produced during the course of the digestion suggest that the core histone octamer can organize 167 (+/- 4) rather than 145 base pairs of DNA, the extra 20 base pairs being quickly digested. Binding of histone H1 to core particles resulted in the protection of about 165 base pairs of DNA from nuclease attack. Because the core histone octamer is fully dissociated into H3 . H4 tetramer and H2A . H2B dimer at physiological ionic strength, our results would suggest that in vivo core particle assembly may also occur by interaction of these two complexes on the nascent DNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldi M. I., Mattoccia E., Tocchini-Valentini G. P. DNA supercoiling by Xenopus laevis oocyte extracts: requirement for a nuclear factor. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4873–4876. doi: 10.1073/pnas.75.10.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boublik M., Bradbury E. M., Crane-Robinson C., Rattle H. W. Proton magnetic resonance ttudies of the interactions of histones F1 and F2B with DNA. Nat New Biol. 1971 Feb 3;229(5):149–150. doi: 10.1038/newbio229149a0. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. Supercoiling energy and nucleosome formation: the role of the arginine-rich histone kernel. Nucleic Acids Res. 1977;4(5):1159–1181. doi: 10.1093/nar/4.5.1159-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Chattoraj D. K., Gosule L. C., Schellman A. DNA condensation with polyamines. II. Electron microscopic studies. J Mol Biol. 1978 May 25;121(3):327–337. doi: 10.1016/0022-2836(78)90367-4. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Linking numbers and nucleosomes. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2639–2643. doi: 10.1073/pnas.73.8.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Anna J. A., Jr, Isenberg I. A histone cross-complexing pattern. Biochemistry. 1974 Nov 19;13(24):4992–4997. doi: 10.1021/bi00721a019. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Moudrianakis E. N. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978 Nov 14;17(23):4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976 Nov;3(11):3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelm R. P., Kneale G. G., Sauau P., Baldwin J. P., Bradbury E. M., Ibel K. Small angle neutron scattering studies of chromatin subunits in solution. Cell. 1977 Jan;10(1):139–151. doi: 10.1016/0092-8674(77)90148-9. [DOI] [PubMed] [Google Scholar]
- Jorcano J. L., Ruiz-Carrillo A. H3.H4 tetramer directs DNA and core histone octamer assembly in the nucleosome core particle. Biochemistry. 1979 Mar 6;18(5):768–774. doi: 10.1021/bi00572a005. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. I. Isolation of a histone IIb1-IIb2 complex. Biochem Biophys Res Commun. 1973 Oct 15;54(4):1588–1594. doi: 10.1016/0006-291x(73)91168-6. [DOI] [PubMed] [Google Scholar]
- Klevan L., Crothers D. M. Isolation and characterization of a spacerless dinucleosome from H1-deleted chromatin. Nucleic Acids Res. 1977 Dec;4(12):4077–4089. doi: 10.1093/nar/4.12.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevan L., Dattagupta N., Hogan M., Crothers D. M. Physical studies of nucleosome assemble. Biochemistry. 1978 Oct 17;17(21):4533–4540. doi: 10.1021/bi00614a027. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Finch J. T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978 Oct 5;275(5679):416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978 Nov;15(3):979–984. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Wright E. B., Olins D. E. Chromatin nu bodies: isolation, subfractionation and physical characterization. Nucleic Acids Res. 1976 Dec;3(12):3271–3291. doi: 10.1093/nar/3.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardon J. F., Worcester D. L., Wooley J. C., Cotter R. I., Lilley D. M., Richards R. M. The structure of the chromatin core particle in solution. Nucleic Acids Res. 1977 Sep;4(9):3199–3214. doi: 10.1093/nar/4.9.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roark D. E., Geoghegan T. E., Keller G. H. A two-subunit histone complex from calf thymus. Biochem Biophys Res Commun. 1974 Jul 24;59(2):542–547. doi: 10.1016/s0006-291x(74)80014-8. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Jorcano J. L. An octamer of core histones in solution: central role of the H3-H4 tetramer in the self-assembly. Biochemistry. 1979 Mar 6;18(5):760–768. doi: 10.1021/bi00572a004. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Jorcano J. L. Nucleohistone assembly: sequential binding of histone H3-H4 tetramer and histone H2A-H2B dimer to DNA. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):165–170. doi: 10.1101/sqb.1978.042.01.018. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Wangh L. J., Allfrey V. G. Processing of newly synthesized histone molecules. Science. 1975 Oct 10;190(4210):117–128. doi: 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Streeck R. E., Zachau H. G. Closely spaced nucleosome cores in reconstituted histone.DNA complexes and histone-H1-depleted chromatin. Eur J Biochem. 1978 Feb;83(2):615–628. doi: 10.1111/j.1432-1033.1978.tb12131.x. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Compact oligomers and nucleosome phasing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3583–3587. doi: 10.1073/pnas.75.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Butler P. J. Characterization of the octamer of histones free in solution. J Mol Biol. 1977 Nov;116(4):769–781. doi: 10.1016/0022-2836(77)90270-4. [DOI] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M. L. Assembly of newly replicated chromatin. Cell. 1978 Nov;15(3):969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]