Abstract
The issue of hepatitis B virus (HBV) mutations possibly leading to a gender disparity in the progression of liver diseases has not been explored. We aimed to elucidate the relationships of the novel pre-S1 mutations, W4P/R, with the progression of liver diseases and male predominance in a South Korean chronic cohort by use of a molecular epidemiologic study. We developed a fluorescence resonance energy transfer (FRET)-based real-time PCR (RT-PCR) assay for the detection of the W4P/R mutations and applied it to 292 chronic HBV patients. The pre-S1 mutations from 247 (84.6%) of a total of 292 patients were detected by this assay. W4P/R mutants were found to be significantly related to severe liver diseases (hepatocellular carcinoma [HCC] and liver cirrhosis, 12.4% [19/153] of patients, versus chronic hepatitis and asymptomatic carriage, 1.1% [1/94] of patients) (P < 0.001). All of the W4P/R mutants were found in males only. The novel HBV pre-S1 mutations, W4P/R, may be associated with disease severity in male patients chronically infected with HBV genotype C. The W4P/R mutations may provide in part an explanation for the relatively high ratio of male to female incidence in HCC generation in South Korean chronic HBV patients.
INTRODUCTION
Hepatitis B virus (HBV) infection is a global health problem, and >350 million people are chronic carriers of the virus (1, 2). The infection is associated with a wide spectrum of clinical manifestations, ranging from acute or fulminant hepatitis to various forms of chronic infection, including asymptomatic carriage, chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) (3). South Korea is recognized as an area where HBV infection is endemic; based on the Korean National Health and Nutrition Survey of 2007, the prevalence of HBsAg was 4.2% in men and 3.1% in women at that time (4). Moreover, it was reported that the extraordinary prevalence in South Korea of HBV genotype C2, which is known to be more virulent than HBV genotype B (5), might contribute to the distribution of the characteristic HBV mutation patterns related to the progression of liver diseases (6–14).
HBV produces three envelope proteins, all of which are encoded in the pre-S/S open reading frame. They have been suggested to play a role in virus assembly (15, 16) and attachment to hepatocytes (17). It has also been proposed that large surface proteins (LHBs) with mutations, particularly deletions, in the Pre-S region might contribute to hepatocarcinogenesis through the induction of an endoplasmic reticulum (ER) stress pathway or trans-activating capacity (18–20). HBV pre-S mutations prevail in countries where HBV infection is highly endemic. In general, the Pre-S2 region is known to be more prone to mutations than the Pre-S1 region. Therefore, so far, the mutation patterns related to the progression of liver disease have been described more frequently in the Pre-S2 than in the Pre-S1 region. Recently, we showed a novel pre-S1 deletion type leading to 11 amino acid deletions from the pre-S1 start codon that is related to liver disease progression in HBV genotype C-infected patients (9). However, no other pre-S1 mutations related to HCC, except for this one, have been found so far.
HCC is a common malignancy and a leading cause of cancer-related death worldwide. Risk factors for HCC have been known to be associated with host and viral factors, lifestyle, and superinfection of other viruses (21–24). HCC is more common in males, who have a 3- to 5-times-higher frequency of developing HCC (25–29). In particular, the recent chemically induced model by diethylnitrosamine (DEN) showed that the gender disparity in HCC generation was attributable to a higher production of interleukin 6 (IL-6) in males after toxicant administration (30). However, a study that focuses on HBV viral mutations has not been performed.
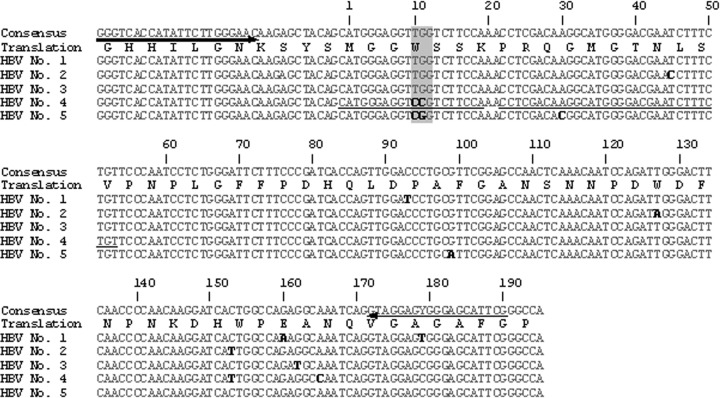
Recently, we identified several characteristic pre-S deletions related to the progression of liver diseases by performing a molecular epidemiology study of South Korean patients with HBV genotype C infections (9). In addition, through further extended sequence analyses of the same patients, we discovered novel Pre-S1 substitutions (W4P/R) related to HCC which change tryptophan to proline or arginine at the 4th codon from the pre-S1 start (Fig. 1). For the current study, we developed a real-time PCR (RT-PCR) based on the fluorescence resonance energy transfer (FRET) technology (31, 32) that facilitates the discrimination of three virus types (wild type and the W4P and W4R variants) at the 4th codon of pre-S1, obtained directly from clinical blood serum DNA samples. The aim of the present study was to prove the relationship between the W4P/R variants and the progression of liver disease and to find out the characteristic clinical factors of patients with W4P/R through a molecular epidemiologic study using an RT-PCR assay we developed for use in South Korean cohorts with diverse clinical statuses.
Fig 1.
Primer and probe position of W4P/R detection for the RT-PCR method. The 3 types of hepatitis B virus (HBV) pre-S1 variants in the 4th codon (wild type [TGG], W4P [CCG], and W4R [CGG]) and primer and probe positions designed for the detection of 3 types of hepatitis B virus pre-S1 variants. Arrows indicate the primer directions. Underlining indicates the probe positions. The numbers indicate the nucleotide position on the pre-S1 gene sequence. Bases in bold type denote bases different from the ones of the consensus sequence. Shading indicates the variants at the 4th codon of the pre-S1 gene. The amino acid sequence is shown using the one-letter amino acid symbols.
MATERIALS AND METHODS
Patient subjects.
A total of 292 blood serum samples were randomly selected from samples of chronic HBV patients who visited Cheju National University Hospital, Jeju, South Korea, in 2003, the Seoul Veterans Hospital in 2004, or the Seoul National University Hospital in 2005. Among these, 292 serum samples were 162 hepatitis B e antigen (HBeAg) positive (130 HBeAg negative) and 225 samples were from males (67 from females). The clinical diagnoses of the study subjects were as follows: asymptomatic carriage (n = 64), chronic hepatitis (n = 33), liver cirrhosis (n = 69), and HCC (n = 126). All work was approved by the institutional review board of Seoul National University Hospital (institutional review board [IRB] no. C-1007-021-322). The experiment was mainly based on the virion DNA extracted from the isolates; therefore, the research was done without informed consent, and a waiver of informed consent was agreed upon by the IRB.
Primer and probe design.
The primers designed to amplify the HBV S gene fragment and the probes designed to identify the sequence variants at the 4th codon in the Pre-S1 region are shown in Table 1 and Fig. 1. A total of 95 cloned HBV genes were aligned by using the SeqMan II software (DNAStar) (data not shown). A primer pair (preS1F and preS1R) for amplification of the HBV pre-S1 gene fragment was designed by using Oligo version 6.5 (Molecular Biology Insights), producing a 224-bp amplicon. An anchor probe and a sensor probe for the identification of mutants (CCG) were designed by using the LC PDS software (version 2.0) to detect three types of polymorphisms, TGG, W4P variant (CCG), and W4R variant (CGG), at channel 640, using the LightCycler 2.0 RT-PCR system.
Table 1.
Primers and probes developed to identify HBV pre-S1 gene sequence polymorphisms at the 4th amino acid (tryptophan) by nested PCR and real-time PCR
| PCR and primer or probe type | Primer name | Sequence (5′ → 3′) | Tm (°C)a | Location (bp) or target sequence |
|---|---|---|---|---|
| Nested PCR | ||||
| Forward primer | PreS1-Del-F1 | GAAGGCKGGCATTCTATATA | 2762–2781 | |
| Forward primer | Del-PRA-F1 | CTTGGGAACAAGAGCTACAGC | 2827–2847 | |
| Reverse primer | HB2R | CATACTTTCCAATCAATAGG | 970–989 | |
| Real-time PCR | ||||
| Forward primer | HBV_pre-S1_F | GGGTCACCATATTCTTGGGAAC | 61.3 | 224 bp of S gene |
| Reverse primer | HBV_pre-S1_R | CGAATGCTCCCRCTCCTAC | 60.2 or 63 | 224 bp of S gene |
| Anchor probe | CGG_A | ACAGAAAGATTCGTCCCCATGCCTTGTCGAGG-FLb | 72.1 | |
| Sensor probe | CGG_S | LC Red640-TGGAAGACGGACCTCCCATG-PHc | 63.4 | CGG |
Tm, melting temperature, calculated by using LC PDS software version 2.0.
FL, fluorescein.
LC Red640, LightCycler dye Red640, channel 640;. PH, phosphate.
DNA extraction and RT-PCR.
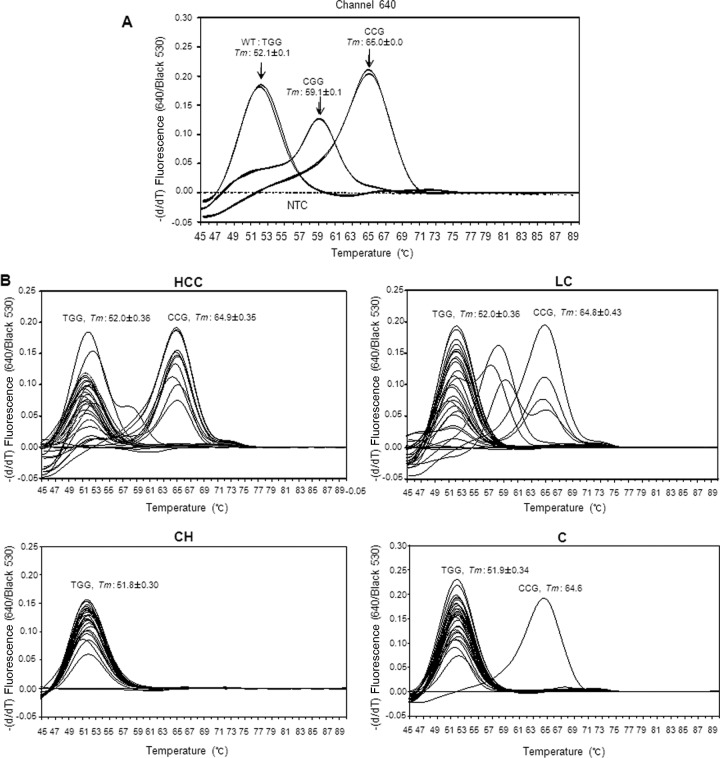
DNA was prepared as previously described (12). A LightCycler 2.0 system was used, and its detection channels were calibrated for color compensation and activated for the experiment. The LightCycler FastStart DNA master HP kit (Roche Diagnostics) was used for the preparation of the master mixture according to the protocol provided with the kit. The cycling conditions were 15 min at 95°C and 50 cycles of 10 s at 95°C, 15 s at 58°C (single acquisition of fluorescence signals), and 20 s at 72°C. The melting curve analysis was followed by cycling for 10 s at 95°C and 30 s at 43°C, and the temperature was then increased from 43°C to 90°C at a temperature transition rate of 0.1°C/s, during which the fluorescence signal was continuously acquired. To determine the precise melting temperatures of the designed probes for the target variants by RT-PCR, triplicate experiments were performed with the cloned DNAs of each known variant, and the average melting temperature (Tm) for each variant was determined (Fig. 2A). DNA extracted from the blood of a total of 292 patients presenting with various degrees of disease severity was subsequently tested for identification of the HBV pre-S1 polymorphisms (Fig. 2B).
Fig 2.
Real-time PCR melting curve analysis and its application. (A) Real-time PCR melting curve analysis to identify the melting temperatures (Tms) of hepatitis B virus (HBV) pre-S1 gene sequence variants (TGG, CCG, or CGG) at the 4th amino acid (tryptophan) of the Pre-S1 region with cloned positive-control sequences and nontemplate controls (NTC). The Tms were determined by duplicate runs and are shown as the average and standard deviation values. The y axis is the negative differential of fluorescence over temperature at the detection channel and normalized by background fluorescence at channel 530. (B) Application of RT-PCR melting curve analysis into blood serum DNAs of the patients with diverse liver diseases. Thirty samples of each disease status, including a nontemplate control, were arbitrarily chosen to provide an exemplary view of the melting temperatures to detect the mutants in each channel. HCC, hepatocellular carcinoma; LC, liver cirrhosis; CH, chronic hepatitis; C, nonsymptomatic carrier. The y axis is the negative differential of fluorescence over the temperature at the detection channel and normalized by background fluorescence at channel 530.
Sequencing analysis.
To confirm the RT-PCR results, direct sequencing of the pre-S1 genomic regions was applied to the same 224-bp amplicon as RT-PCR by preS1F and preS1R primers for all 247 subjects, which, using RT-PCR melting curve analysis, proved to be positive. During direct sequencing analysis, we used purified PCR amplicons (224 bp) from 30 subjects as the template and PreS1R, a reverse primer, as a direct sequencing primer. Sequences of more than ∼220 bp were obtained. The electropherogram profiles of direct sequencing were compared with the results obtained by RT-PCR.
To address issues of whether the W4P/R variants originated from a mutation of the preexisting wild type or de novo horizontal infections, a quasispecies analysis from five patients, in which the coexistence of the wild type and W4P/R variants had already been confirmed, were performed. For PCR amplification that included the entire LHB region (1,378 bp), a nested PCR method was used. The first round of PCR was carried out using the PreS1-Del-F1 and HB2R primers and the second-round amplification was performed using the Del-PRA-F1 and HB2R (antisense) primers. The 1,378 bp of PCR products was cloned into the Topo TA cloning kit (Invitrogen Co., Carlsbad, CA, USA). A total of 108 clones from five patients (>21 clones from one patient) with W4P/R mutations and a total of 63 clones from four patients (>15 clones from one patient) with wild-type infection (chronic hepatitis and asymptomatic carriage) for a comparison were analyzed. Sequencing was conducted using the Applied Biosystems model 377 DNA automatic sequencer (PerkinElmer Applied Biosystems, Warrington, United Kingdom), known as the automatic sequencing system. Phylogenetic analyses of 23 selected clones based on 1,378-bp sequences were performed using the neighbor-joining method of MEGA version 4.1 (33).
Statistical analysis.
All of the detection steps in this study were repeated at least three times, and the results were expressed as percentages, means ± standard deviation (SD), or medians (range). The differences between categorical variables were analyzed using Fisher's exact test or the chi-square test. For continuous variables, the Student t test was used when the data were normally distributed, and the Mann-Whitney U test was used when the data were not normally distributed. The SPSS version 18.0 software (Professional Statistic, Chicago, IL) was used for the performance of all statistical analyses, and a P value of <0.05 (two-tailed) was considered to be statistically significant.
Nucleotide sequence accession numbers.
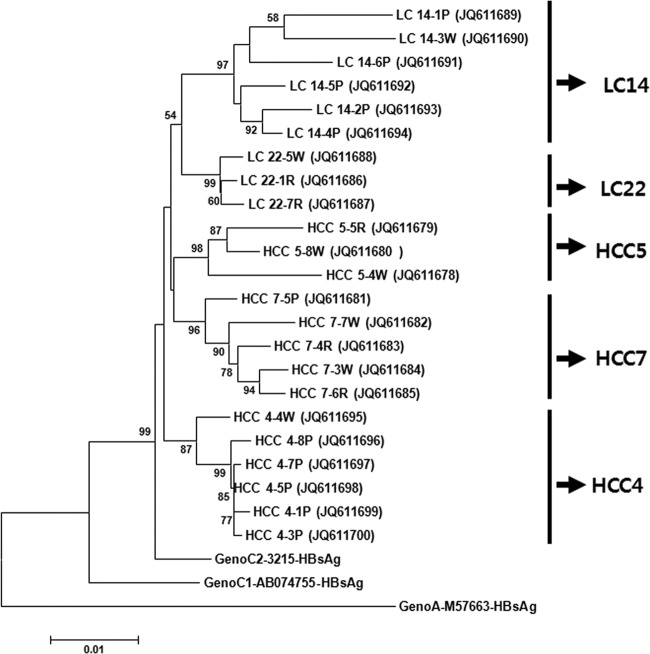
The clinical isolates characterized in this study have been deposited in GenBank under accession numbers JQ611678 through JQ611700. For further details, see Fig. 4.
Fig 4.
Phylogenetic analysis based on 1,378 bp covering an LHB region, including pre-S1, pre-S2, and the HBsAg region of three references (genotypes A, C1, and C2), and the 23 HBV strains from five patients (two liver cirrhosis patients [LC14 and LC22] and three HCC patients [HCC4, HCC5, and HCC7]) with coexisting wild-type and W4P/R mutations. Genetic distances were estimated using the Kimura two-parameter matrix, and the phylogenetic tree was constructed using the neighbor-joining method. The percentages indicated at nodes represent bootstrap levels supported by 1,000 resampled data sets. Bootstrap values of <50% are not shown. The numbers in parentheses indicate GenBank accession numbers. P, R, and W indicate the polymorphisms of W4P, W4R, and the wild type, respectively.
RESULTS
Determination of melting temperatures of RT-PCR for W4P/R detection.
The application of the RT-PCR method we developed to three types of control plasmid DNA demonstrated that not only was separation between the wild-type infection and variants possible, but separation between two different types of variants (CGG [W4R] and CCG [W4P]) was also possible, as they showed distinct melting temperatures according to their respective probe. While a clone of the wild type (TGG) formed a distinct melting peak at 52.0 ± 0.15°C, a CGG variant (W4R) and a CCG variant clone (W4P) formed distinct melting peaks at 59.1 ± 0.10°C and at 65.0 ± 0.02°C, respectively, at channel 640 (Fig. 2A).
Application of RT-PCR to DNA from clinical blood samples.
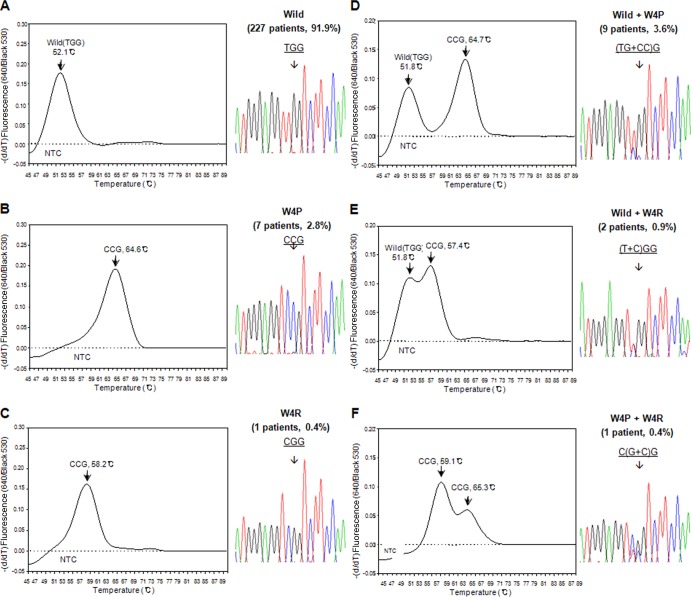
The application of this method to the 292 DNA samples from the clinical blood samples taken from South Korean chronic liver infection patients resulted in the successful differentiation in a total of 247 patients (84.6%). The clinical details of the 247 patients are presented in Table S1 in the supplemental material. Details of the sensitivity of the RT-PCR according to the respective disease stage are presented in Table S2 in the supplemental material. All the 247 detected samples were clearly separated into 3 genotypes, wild type, W4R, and W4P, by the newly developed RT-PCR protocol, showing a difference of <l°C compared with each standard melting temperature (Fig. 2B). We detected W4P/R variants in 20 (8.1%) of the 247 patients studied. Among 20 patients with W4P/R mutations, 16 (6.5%), 3 (1.2%), and 1 (0.4%) had W4P, W4R, and both mutations, respectively (Table 2). Overall, a total of 6 types of infection polymorphisms in the melting temperature curve analysis were found (Fig. 3): the wild type alone (227 patients [91.9%]), W4P alone (7 patients [2.8%]), W4R alone (1 patient [0.4%]), and three types of mixed infection, wild type plus W4P (9 patients [3.6%]), wild type plus W4R (2 patients [0.4%]), and W4P plus W4R (1 patient [0.4%]). The results obtained by the RT-PCR assay of all 247 samples were completely concordant with those obtained by PCR direct sequencing (data not shown), suggesting the feasibility of the RT-PCR assay we developed as a diagnostic or epidemiologic tool for the detection of W4P polymorphisms.
Table 2.
Prevalence of W4P/R variants between patients with diverse chronic clinical statuses determined by real-time PCR
| Clinical status (n)a | No. (%) with each type or variant |
P value |
|||||
|---|---|---|---|---|---|---|---|
| Wild type | W4P | W4R | W4P/R | W4P | W4P/R | HCC+LC vs CH+C | |
| HCC (96) | 83 (86.5) | 12 (12.5) | 1 (1.0) | 13 (13.5) | 0.036b | 0.028b | 0.004f |
| LC (57) | 51 (89.5) | 4 (7.0) | 3 (5.3) | 6 (10.5) | 0.016c | 0.009c | <0.001g |
| CH (32) | 32 (100) | 0 (0) | 0 (0) | 0 (0) | 0.057d | ||
| C (62) | 61 (98.4) | 1 (1.6) | 0 (0) | 1 (1.6) | 0.039e | ||
| Total (247) | 227 | 17 | 4h | 20 | |||
HCC, hepatocellular carcinoma; LC, liver cirrhosis; CH, chronic hepatitis; C, carrier.
HCC versus CH.
HCC versus C.
LC versus CH.
LC versus C.
W4P type.
W4P/R type.
Three are only W4R type and one is combined mutations of W4P and W4R from an LC patient.
Fig 3.
Representative views of the melting temperatures for all six polymorphisms of the 4th codon of HBV pre-S1 detected in the present study, the corresponding direct sequencing data, and their prevalences in 247 chronic patients. All 247 samples were clearly separated into six genotypes using RT-PCR melting curve analysis: wild-type (TGG) (A), W4P (CCG) alone (B), W4R (CGG) alone (C), and 3 types of mixed infection, wild type plus W4P (D), wild type plus W4R (E), and W4P plus W4R (F). The y axis is the negative differential of the fluorescence over temperature at the detection channel and is normalized by the background fluorescence at channel 530.
Comparison of clinical factors between patients with and without W4P/R mutations.
The prevalence of patients with W4P/R mutations was significantly higher in HCC patients than in patients with asymptomatic carriage or chronic hepatitis infection (HCC, 13/96 patients [13.5%], versus carrier, 1/62 patients [1.6%] [P = 0.028]; HCC versus chronic hepatitis, 0/32 patients [0%] [P = 0.009]). Similar trends were observed in comparisons between patients with liver cirrhosis and a mild presentation of disease (chronic hepatitis and asymptomatic carriage) (cirrhosis, 6/57 patients [10.5%], versus carrier, 1/62 patients [1.6%] [P = 0.057]; cirrhosis versus chronic hepatitis, 0/32 patients [0%] [P = 0.039]). In general, the prevalence of W4P/R variants was significantly higher in patients with severe forms of liver disease (HCC and liver cirrhosis) than in patients with mild forms of disease (chronic hepatitis and asymptomatic carriage) (HCC and liver cirrhosis, 12.4% [19/153 patients], versus chronic hepatitis and carrier, 1.1% [1/94 patients]; [P < 0.001]) (Table 2).
A comparison of the clinical factors between patients with W4P/R mutations (20 patients) and those with the wild type (227 patients) showed no significant differences between the two groups with regard to age, HBeAg positivity, alanine aminotransferase (ALT) levels, and HBV DNA levels. However, significant differences in gender distribution were observed. Compared with the wild type, W4P/R mutations were only detected in male patients (100% versus 74%) (P = 0.010), suggesting that W4P/R mutations occur predominantly in males (Table 3).
Table 3.
Comparison of clinical profiles between patients with wild-type HBV and W4P/R variants
| Patient characteristics | Wild type (n = 227) | W4P/R (n = 20) | P |
|---|---|---|---|
| Age (mean ± SD) (yr) | 44.8 ± 16.4 | 49.4 ± 11.3 | 0.221 |
| Male (no. [%])a | 169 (74.4) | 20 (100) | 0.010 |
| HBeAg-positive (no. [%])b | 127 (55.9) | 15 (75) | 0.155 |
| Liver disease (no.) C:CH:LC:HCCa | 61:32:51:83 | 1:0:6:13 | |
| ALT level (mean ± SD) (IU/liter) | 83.2 ± 171.1 | 61.7 ± 35.8 | 0.643 |
| HBV-DNA level (median [range]) (pg/ml) | 1,066.7 (0 to ∼6,000) | 2.9 (0 to ∼8) | 0.145 |
C, carrier; CH, chronic hepatitis; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
ALT, alanine aminotransferase.
Quasispecies analysis of five patients showing the coexistence of wild type and W4P/R mutations.
To address issues of whether W4P/R variants originated from mutations of the preexisting wild-type or de novo horizontal infections, quasispecies analysis from 108 clones of five patients (three patients with HCC and two patients with liver cirrhosis [LC]), in which the coexistence of wild type and W4P/R variants had already been confirmed by RT-PCR assay, were performed. The results obtained by quasispecies analysis showed that the wild type and mutants coexisted in all five patients. In two patients, LC14 and HCC4, the coexistence of the W4P mutation and the wild type was observed. In one patient, LC22, the coexistence of the W4R mutation and the wild type was observed. In two patients, HCC5 and HCC7, who had been confirmed as having a mixed infection of the wild type and W4R mutant and both the wild type and the W4P mutation by RT-PCR, respectively, three types of polymorphisms, the wild type and the W4P and W4R mutants, were observed in the nested PCR cloning-based quasispecies analysis (Table 4). This disparity between the results obtained by RT-PCR and quasispecies analysis may be due to the difference in sensitivity between the two protocols, conventional-based and nested-based PCRs. Quasispecies analysis from 63 clones from 4 patients identified as only wild types by RT-PCR proved that no W4P/R mutations were found from their serum samples (Table 4). The phylogenetic analysis based on the entire LHB region (1,378 bp) indicated that all of the 23 W4P/R variants from the five patients may be from respective wild types rather than from a superinfection of these variants into someone already infected with HBV. Moreover, quasispecies analysis from the HCC5 and HCC7 patients in whom three types, the wild type and the W4P and W4R mutants, coexist suggested that the W4R mutation (CGG, where the mutated part of the sequence is underlined) with its one substitution may be an intermediate type in the conversion from the wild type (TGG) into W4P (CCG) with its double mutation (Fig. 4).
Table 4.
Quasispecies distribution between W4P/R variants and wild types
| Serum sample (no. of subclones) | RT-PCR detection results | Diagnosisa | No. (%) of samples with: |
Other amino acid substitution (no. of subclones) | |||
|---|---|---|---|---|---|---|---|
| Wild type | W4P | W4R | W4P/R | ||||
| HCC4 (22) | W4P+WT | HCC | 9 | 13 | 0 | 13 (59.1) | |
| HCC5 (21) | W4R+WT | HCC | 7 | 6 | 8 | 14 (66.7) | |
| HCC7 (21) | W4P+WT | HCC | 7 | 11 | 2 | 13 (61.9) | W4Stop (1) |
| LC14 (22) | W4P+WT | LC | 6 | 16 | 0 | 16 (72.7) | |
| LC22 (22) | W4R+WT | LC | 15 | 0 | 6 | 6 (27.3) | W4Q (1) |
| CH31 (16) | WT | CH | 16 | 0 | 0 | 0 | |
| CH32 (16) | WT | CH | 15 | 0 | 0 | 0 | W4Stop (1) |
| C33 (16) | WT | C | 16 | 0 | 0 | 0 | |
| C34 (15) | WT | C | 15 | 0 | 0 | 0 | |
HCC, hepatocellular carcinoma; LC, liver cirrhosis; CH, chronic hepatitis; C, carrier.
DISCUSSION
Notably, the gender dimorphism in HCC generation differs between HBV- and hepatitis C virus (HCV)-related cases. The predominance of males affected by HBV-related HCC is more pronounced than that of males affected with HCV-related HCC, with a ratio of 5:1 to 11:1 versus 2:1 to 3:1 males to females, respectively (34–36). Recently, the HBV X protein (HBx) has been reported to play a very pivotal role in the predominance of males affected by the HBV-related HCC via enhancement of the transcriptional activity of the androgen receptor (AR) (37). However, the above-mentioned factor cannot provide a likely explanation for the geographical difference in the predominance of males with regards to HCC. Other factors reflecting the geographic background, such as HBV genotype or HBV mutation type, need to be considered. The mortality from HCC in South Korea is the highest among the Organisation for Economic Co-operation and Development (OECD) nations (38). Furthermore, HCC in South Korea is approximately 4.2 to 5.9 times more prevalent in men than in women (4). This male-to-female ratio of HCC incidence is higher than the average 2.9:1 male-to-female ratio found worldwide (39), suggesting that there might be crucial factors in South Korean chronic HBV patients that make them distinct from patients in other areas that favor the predominance of HCC in males. Therefore, we tried to search for the specific HBV mutation type that is related to liver disease progression in male South Korean chronic patients. The W4P/R mutations introduced in the present study were two of the putative male-specific mutations.
In this report, we describe the establishment of an RT-PCR assay for the detection of W4P/R variants in clinical samples. The test is based on the application of FRET probes that provide a precise genotypic characterization of three polymorphisms in the 4th codon of Pre-S1. Three genotypes (wild type, W4P, and W4R) were discriminated reliably in our cohort. There are some findings worthy of note regarding our FRET-based RT-PCR assay used for epidemiologic or diagnostic purposes. First, W4P/R variants were detected with a relatively high rate from our male cohort with severe liver diseases (19/127 HCC or liver cirrhosis patients [15.0%]), meaning that these variants may be promising as a diagnostic marker for disease progression in male HBV patients. Second, characteristic mutation patterns, such as the substitution of two consecutive nucleotides between the wild type and W4P variant (TGG→CCG) and a transversion substitution between the wild type and W4R variant (TGG→CGG), might provide the rationale to guarantee the reliable and reproducible data in RT-PCR melting curve analysis. Actually, differences of >5°C in the melting temperatures between three polymorphisms, wild type (52.0°C ± 0.1°C), W4R (59.0°C ± 0.1°C), and W4P (65.0°C ± 0.0°C), were observed, which enabled the simultaneous identification of three variants not only in the case of single infection but also in the mixed infection, while using only one pair of probes in one channel instead of using multiprobe sets in multichannels. Third, since the HBV sequences flanking the W4P/R mutations are highly conserved among the HBV genotype C strains, as shown in Fig. 1, these mutations are very appropriate as a target for the molecular method. Actually, in the melting curve analysis of the 247 detected cases, this method proved to produce reliable results from all samples without producing ambiguous melting temperatures due to variations (Fig. 2B). Fourth, despite the direct application of this assay to serum DNA samples, instead of using a nested PCR strategy, a high level of sensitivity (84.6% [247/292 patients] was obtained. This means that this assay might minimize the potential risk of the cross contamination that can be found with the nested-PCR-based diagnostic method. It should be noted that the sensitivity level gradually decreased with increasing clinical severity. While a sensitivity of 96.9% (94/97 patients) was found for patients with a mild type of HBV disease (chronic hepatitis and asymptomatic carriage), a lower level of sensitivity of 78.9% (153/194 patients) was found for patients with a severe type of HBV disease (liver cirrhosis and HCC) (see Table S2 in the supplemental material). This may be due to the lower level of HBV DNA in the severe forms of liver diseases than in the mild forms. Collectively, our FRET-based RT-PCR assay may have promise as a diagnostic method for the disease progression in male patients with chronic HBV infection.
In the current study, we have provided intriguing data to support the relationships between the novel pre-S1 mutations, W4P/R, and the disease progression in male chronic HBV patients through a molecular epidemiologic study using an RT-PCR assay we developed. Notably, the W4P/R mutations were detected exclusively in males (20 male patients). To the best of our knowledge, this may be the first viral single (SNP) or double nucleotide polymorphism related to male HCC generation. A collective consideration of our epidemiologic (higher prevalence of the W4R mutation in liver cirrhosis patients [5.3% {3/57 patients}] than in HCC [1.0% {1/96 patients}]) and quasispecies (coexistence of wild type and W4R and W4P mutations in an HCC7 patient) data suggests that the occurrence of a mutation in the 4th codon of pre-S1 in a sequential manner (wild type [TGG]→W4R [CGG]→W4P [CCG]) might be associated with liver disease progression during the natural course of HBV infection (mild type [asymptomatic carriage and chronic hepatitis] → liver cirrhosis → HCC). The predominance of W4P over the intermediated type, W4R, in HCC patients was observed (12.5% [12/96 patients] versus 1.0% [1/96 patients]), suggesting that the former has advantages over the latter in HBV survival on the transformed hepatocytes.
Interestingly, the W4P/R mutations are located in the 11-amino-acid region of the pre-S1 5′ end, which is reported to be absent in HBV genotype D. Although the function of this region is still unknown, its absence in genotype D strains suggests that it may be dispensable in the HBV life cycle. However, together with the previous report that deletions leading to 11 amino acids in the pre-S1 5′ region were observed more frequently in HCC patients (9), our data also support the significance of this region in the progression of liver diseases. Furthermore, the functional significance of this region is also indirectly proved by the sequence conservation between the HBV genotype C isolates from our HBV sequence databases of 95 chronic patients (data not shown), which provide the rationale for the development of the hybridization-based RT-PCR method of this study.
Despite the predominance of W4P/R mutations in South Korean patients, the possibility that their prevalence might be increased in the HBV-infected populations of other areas, particularly areas where genotype C is endemic, such as China and east Asia, cannot be excluded. The predominance of the W4P/R mutations in South Korean patients might be explained by the distinct molecular epidemiologic trait of genotype C, the only genotype which infects South Korean patients at a higher mutation rate. In actuality, recent papers (6–14) proving the presence of several unique HBV mutation patterns in South Korean patients support the above hypothesis.
Recently, it was reported that HBV pre-S/S variants led to a significant reduction in HBsAg secretion due to the retention of envelope proteins in the endoplasmic reticulum (ER), leading to liver cell damage (40). The issue that the W4P/R mutations can also lead to ER stress by affecting HBsAg secretion capacity should be addressed in the future.
The most important drawbacks of our study are as follows. First, a relatively lower number of female patients and only one ethnic group were analyzed, which led to the biased conclusion on the gender disparity of the W4P/R mutations. Second, a longitudinal study following quasispecies evolution is necessary to achieve the goals of the study. Therefore, a longitudinal study using larger populations, particularly one including more women and other ethnic groups, should be done in the future. In addition, molecular mechanisms supporting the idea that the W4P/R mutations play a pivotal role in liver disease progression in male chronic HBV patients need to be elucidated.
In conclusion, the novel HBV pre-S1 mutations, W4P/R, may be associated with disease severity in male patients chronically infected with HBV genotype C. The W4P/R mutations may in part provide a likely explanation as to the relatively high ratio of male to female incidence in HCC generation in South Korean chronic HBV patients. In addition, the RT-PCR assay developed for the detection of W4P/R mutations holds promise for the prognosis of male chronic patients with HCC.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (grant no. 2013005810).
Footnotes
Published ahead of print 11 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01505-13.
REFERENCES
- 1.Kao JH, Chen PJ, Lai MY, Chen DS. 2002. Genotypes and clinical phenotypes of hepatitis B virus in patients with chronic hepatitis B virus infection. J. Clin. Microbiol. 40:1207–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavanchy D. 2005. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J. Clin. Virol. 34(Suppl 1):S1–S3 [DOI] [PubMed] [Google Scholar]
- 3.Chen DS. 1993. From hepatitis to hepatoma: lessons from type B viral hepatitis. Science 262:369–370 [DOI] [PubMed] [Google Scholar]
- 4.KCDC 2007. National health and nutrition survey. Korea Centers for Disease Control and Prevention, Chungcheongbuk-do, South Korea [Google Scholar]
- 5.Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, Okanoue T, Yotsuyanagi H, Iino S. 2001. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology 33:218–223 [DOI] [PubMed] [Google Scholar]
- 6.Song BC, Kim SH, Kim H, Ying YH, Kim HJ, Kim YJ, Yoon JH, Lee HS, Cha CY, Kook YH, Kim BJ. 2005. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J. Med. Virol. 76:194–202 [DOI] [PubMed] [Google Scholar]
- 7.Kim H, Jee YM, Song BC, Shin JW, Yang SH, Mun HS, Kim HJ, Oh EJ, Yoon JH, Kim YJ, Lee HS, Hwang ES, Cha CY, Kook YH, Kim BJ. 2007. Molecular epidemiology of hepatitis B virus (HBV) genotypes and serotypes in patients with chronic HBV infection in Korea. Intervirology 50:52–57 [DOI] [PubMed] [Google Scholar]
- 8.Kim HJ, Park JH, Jee Y, Lee SA, Kim H, Song BC, Yang S, Lee M, Yoon JH, Kim YJ, Lee HS, Hwang ES, Kook YH, Kim BJ. 2008. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J. Med. Virol. 80:1337–1343 [DOI] [PubMed] [Google Scholar]
- 9.Mun HS, Lee SA, Jee Y, Kim H, Park JH, Song BC, Yoon JH, Kim YJ, Lee HS, Hyun JW, Hwang ES, Kook YH, Kim BJ. 2008. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J. Med. Virol. 80:1189–1194 [DOI] [PubMed] [Google Scholar]
- 10.Lee SA, Mun HS, Kim H, Lee HK, Kim BJ, Hwang ES, Kook YH, Kim BJ. 2011. Naturally occurring hepatitis B virus X deletions and insertions among Korean chronic patients. J. Med. Virol. 83:65–70 [DOI] [PubMed] [Google Scholar]
- 11.Mun HS, Lee SA, Kim H, Hwang ES, Kook YH, Kim BJ. 2011. Novel F141L preS2 mutation in hepatitis B virus increases the risk of hepatocellular carcinoma in patients with chronic genotype C infections. J. Virol. 85:123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SA, Kim K, Kim H, Kim BJ. 2012. Nucleotide change of codon 182 in the surface gene of hepatitis B virus genotype C leading to truncated surface protein is associated with progression of liver diseases. J. Hepatol. 56:63–69 [DOI] [PubMed] [Google Scholar]
- 13.Kim DW, Lee SA, Hwang ES, Kook YH, Kim BJ. 2012. Naturally occurring precore/core region mutations of hepatitis B virus genotype C related to hepatocellular carcinoma. PLoS One 7:e47372. 10.1371/journal.pone.0047372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H, Lee SA, Kim DW, Lee SH, Kim BJ. 2013. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS One 8:e54486. 10.1371/journal.pone.0054486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganem D. 1991. Assembly of hepadnaviral virions and subviral particles. Curr. Top. Microbiol. Immunol. 168:61–83 [DOI] [PubMed] [Google Scholar]
- 16.Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. 1998. Role of the Pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J. Virol. 72:5573–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh Y, Takai E, Ohnuma H, Kitajima K, Tsuda F, Machida A, Mishiro S, Nakamura T, Miyakawa Y, Mayumi M. 1986. A synthetic peptide vaccine involving the product of the pre-S(2) region of hepatitis B virus DNA: protective efficacy in chimpanzees. Proc. Natl. Acad. Sci. U. S. A. 83:9174–9178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HC, Huang W, Lai MD, Su IJ. 2006. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 97:683–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh YH, Su IJ, Wang HC, Chang WW, Lei HY, Lai MD, Chang WT, Huang W. 2004. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis 25:2023–2032 [DOI] [PubMed] [Google Scholar]
- 20.Caselmann W, Meyer M, Kekulé A, Lauer U, Hofschneider P, Koshy R. 1990. A trans-activator function is generated by integration of hepatitis B virus preS/S sequences in human hepatocellular carcinoma DNA. Proc. Natl. Acad. Sci. U. S. A. 87:2970–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lok AS, McMahon BJ. 2007. Chronic hepatitis B. Hepatology 45:507–539 [DOI] [PubMed] [Google Scholar]
- 22.McMahon BJ. 2009. The influence of hepatitis B virus genotype and subgenotype on the natural history of chronic hepatitis B. Hepatol. Int. 3:334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukuma H, Hiyama T, Tanaka S, Nakao M, Yabuuchi T, Kitamura T, Nakanishi K, Fujimoto I, Inoue A, Yamazaki H, Kawashima T. 1993. Risk factors for hepatocellular carcinoma among patients with chronic liver disease. N. Engl. J. Med. 328:1797–1801 [DOI] [PubMed] [Google Scholar]
- 24.Fattovich G, Stroffolini T, Zagni I, Donato F. 2004. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology 127:S35–S50 [DOI] [PubMed] [Google Scholar]
- 25.Yeh SH, Chen PJ. 2010. Gender disparity of hepatocellular carcinoma: the roles of sex hormones. Oncology 78:172–179 [DOI] [PubMed] [Google Scholar]
- 26.Jepsen P, Vilstrup H, Tarone RE, Friis S, Sørensen HT. 2007. Incidence rates of hepatocellular carcinoma in the U.S. and Denmark: recent trends. Int. J. Cancer 121:1624–1626 [DOI] [PubMed] [Google Scholar]
- 27.Ruggieri A, Barbati C, Malorni W. 2010. Cellular and molecular mechanisms involved in hepatocellular carcinoma gender disparity. Int. J. Cancer 127:499–504 [DOI] [PubMed] [Google Scholar]
- 28.El-Serag HB, Rudolph KL. 2007. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132:2557–2576 [DOI] [PubMed] [Google Scholar]
- 29.Wang SH, Yeh SH, Lin WH, Wang HY, Chen DS, Chen PJ. 2009. Identification of androgen response elements in the enhancer I of hepatitis B virus: a mechanism for sex disparity in chronic hepatitis B. Hepatology 50:1392–1402 [DOI] [PubMed] [Google Scholar]
- 30.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. 2007. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 317:121–124 [DOI] [PubMed] [Google Scholar]
- 31.Lay MJ, Wittwer CT. 1997. Real-time fluorescence genotyping of factor V Leiden during rapid-cycle PCR. Clin. Chem. 43:2262–2267 [PubMed] [Google Scholar]
- 32.Selvin PR. 2000. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 7:730–734 [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 34.Lee CM, Lu SN, Changchien CS, Yeh CT, Hsu TT, Tang JH, Wang JH, Lin DY, Chen CL, Chen WJ. 1999. Age, gender, and local geographic variations of viral etiology of hepatocellular carcinoma in a hyperendemic area for hepatitis B virus infection. Cancer 86:1143–1150 [DOI] [PubMed] [Google Scholar]
- 35.Shiratori Y, Shiina S, Imamura M, Kato N, Kanai F, Okudaira T, Teratani T, Tohgo G, Toda N, Ohashi M, Ogura K, Niwa Y, Kawabe T, Omata M. 1995. Characteristic difference of hepatocellular carcinoma between hepatitis B- and C-viral infection in Japan. Hepatology 22:1027–1033 [DOI] [PubMed] [Google Scholar]
- 36.Sun Z, Lu P, Gail MH, Pee D, Zhang Q, Ming L, Wang J, Wu Y, Liu G, Wu Y, Zhu Y. 1999. Increased risk of hepatocellular carcinoma in male hepatitis B surface antigen carriers with chronic hepatitis who have detectable urinary aflatoxin metabolite M1. Hepatology 30:379–383 [DOI] [PubMed] [Google Scholar]
- 37.Yang WJ, Chang CJ, Yeh SH, Lin WH, Wang SH, Tsai TF, Chen DS, Chen PJ. 2009. Hepatitis B virus X protein enhances the transcriptional activity of the androgen receptor through c-Src and glycogen synthase kinase-3β kinase pathways. Hepatology 49:1515–1524 [DOI] [PubMed] [Google Scholar]
- 38.Bréchot C. 2004. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology 127(5 Suppl 1):S56–S61 [DOI] [PubMed] [Google Scholar]
- 39.KNSO 2007. Annual report on the cause of the death statistics. Korea National Statistical Office, Daejeon, South Korea [Google Scholar]
- 40.Pollicino T, Amaddeo G, Restuccia A, Raffa G, Alibrandi A, Cutroneo G, Favaloro A, Maimone S, Squadrito G, Raimondo G. 2012. Impact of hepatitis B virus (HBV) preS/S genomic variability on HBV surface antigen and HBV DNA serum levels. Hepatology 56:434–443 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.