Abstract
Carbapenem-resistant Enterobacter species are emerging nosocomial pathogens. As with most multidrug-resistant Gram-negative pathogens, the polymyxins are often the only therapeutic option. In this study involving clinical isolates of E. cloacae and E. aerogenes, susceptibility testing methods with polymyxin B were analyzed. All isolates underwent testing by the broth microdilution (in duplicate) and agar dilution (in duplicate) methods, and select isolates were examined by the Etest method. Selected isolates were also examined for heteroresistance by population analysis profiling. Using a susceptibility breakpoint of ≤2 μg/ml, categorical agreement by all four dilution tests (two broth microdilution and two agar dilution) was achieved in only 76/114 (67%) of E. cloacae isolates (65 susceptible, 11 resistant). Thirty-eight (33%) had either conflicting or uninterpretable results (multiple skip wells, i.e., wells that exhibit no growth although growth does occur at higher concentrations). Of the 11 consistently resistant isolates, five had susceptible MICs as determined by Etest. Heteroresistant subpopulations were detected in eight of eight isolates tested, with greater percentages in isolates with uninterpretable MICs. For E. aerogenes, categorical agreement between the four dilution tests was obtained in 48/56 (86%), with conflicting and/or uninterpretable results in 8/56 (14%). For polymyxin susceptibility testing of Enterobacter species, close attention must be paid to the presence of multiple skip wells, leading to uninterpretable results. Susceptibility also should not be assumed based on the results of a single test. Until the clinical relevance of skip wells is defined, interpretation of polymyxin susceptibility tests for Enterobacter species should be undertaken with extreme caution.
INTRODUCTION
Enterobacter species are important pathogens in critically ill patients, being the most common Gram-negative pathogen in bloodstream infections and the second most common in patients with pneumonia (1). Many nosocomial isolates are resistant to cephalosporins, quinolones, and aminoglycosides; the acquisition of carbapenemases, including NDM-1 and KPC, further restricts therapeutic options (2, 3). In New York City, approximately 7% of Enterobacter species possess blaKPC (4). In many instances, the polymyxins remain the therapeutic agents of last resort.
The optimal method for polymyxin susceptibility testing remains to be defined. The difficulties in susceptibility testing appear to stem from two sources: (i) the cationic properties of the polymyxins and (ii) the presence of heteroresistance in many Gram-negative pathogens. The poor diffusion of polymyxins in agar, resulting in inaccurate results by disk diffusion assays, has been well chronicled (5). The Etest methodology, compared to dilution susceptibility methods, has resulted in unacceptable major and very major error rates for Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae (6–8). For broth dilution methods, colistin is adsorbed to commonly used labware, including polystyrene, polypropylene, and glass, particularly at low concentrations (9). Although cation-supplemented Mueller-Hinton broth is still the recommended broth medium, the addition of polysorbate 80 appears to reduce the adsorption of colistin to polystyrene wells, resulting in substantially lower MIC values (6).
Second, the presence of resistant subpopulations for several Gram-negative pathogens has contributed to the difficulty with in vitro assessments of polymyxins. “Skip wells” (wells that exhibit no growth although growth does occur at higher concentrations) were identified with Escherichia coli, and the phenomenon was attributed to interference from soap (10). The finding of “cocarde” growth (increased growth near a disk surrounded by a zone of inhibition) has been reported with Serratia marcescens (11). The presence of “skip wells” with polymyxin MIC testing of P. aeruginosa (12) and A. baumannii (13) has also been reported. Heteroresistance, defined by population analysis profiles, has been described with isolates of A. baumannii, especially at concentrations of colistin surrounding the breakpoint of susceptibility (14). Heteroresistance, defined as a small number of persistent colonies on agar plates with increasing concentrations of colistin or growing within the inhibition zone of Etest strips, has been recorded for A. baumannii and E. cloacae (8, 15). Heteroresistant E. cloacae was more readily identified by the Etest method using Isosensitest agar (compared to Mueller-Hinton agar) (15).
In this report, we describe the problems associated with polymyxin MIC testing of clinical isolates of E. cloacae and E. aerogenes.
MATERIALS AND METHODS
Unique patient isolates of E. cloacae and E. aerogenes were obtained from a citywide surveillance study conducted in 2009, as previously described (4). Susceptibility testing was performed according to CLSI guidelines (16) using the microdilution method with cation-supplemented Mueller-Hinton broth (Difco Laboratories, Sparks, MD) with polystyrene microwell trays (polysorbate 80 was not added). MICs were defined as the first well displaying no visible growth. As per CLSI guidelines, a single skip well was not counted and did not affect the MIC reading (16). However, tests showing multiple skip wells were considered uninterpretable. Susceptibility testing was also performed by the agar dilution method with Mueller-Hinton agar (Difco Laboratories). MICs were read as the first concentration displaying <2 colonies; a faint haze was not read as growth (16). Twofold concentrations of polymyxin B from 0.12 to 512 μg/ml were used for the dilution assays. Representative isolates demonstrating both consistent and inconsistent/uninterpretable MICs were also tested with the Etest method, using Mueller-Hinton and Isosensitest agar. MICs were read at the intersection of the zone of inhibition with the test strip. Etest MICs were considered uninterpretable when there were isolated colonies within the zone of inhibition. P. aeruginosa ATCC 27853 was used as the control for all susceptibility tests. Although breakpoints have not been defined for Enterobacter species, for this report, isolates with an MIC of >2 μg/ml were considered resistant.
Population analysis profiles (PAPs) were performed, in duplicate, by plating 50 μl of dilutions of an overnight culture (∼ 109 CFU/ml) on Mueller-Hinton agar plates containing polymyxin B at the following concentrations: 0, 0.5, 1, 2, 4, 6, 8, 10, and 64 μg/ml (14). Colonies were counted after overnight incubation at 37°C. Isolates with inconsistent and/or uninterpretable MICs were preferentially selected, along with a single isolate with susceptible MICs, for population analysis profiling.
To begin exploring the mechanism of adaptive resistance, expression of phoP and phoQ was examined in select isolates by real-time reverse transcriptase PCR (RT-PCR) using SYBR green. To determine target sequences for RT-PCR primers, phoP and phoQ were amplified with the following primers (5′ to 3′): phoPf, GCTTTTTCCCCGGTTTACTC, and phoPr, CCCACTTTGCGAGGGTATAA; phoQf, TAATGCTCCAGCTTTACCCG, and phoQr, ATTCCGCAGGCTCTTACTGA. These primers were derived from the published sequences for Enterobacter cloacae ATCC 13047 (GenBank CP001918). For the expression studies, the following primers (5′ to 3′) were used: phosPSYBRfor, GAGGATAACGCATTGCTACG; phosPSYBRrev, TCCACCTGATGTCCCATCTC; phosQSYBRfor, ACCTCTGCGCTGAATAAGGT; phosQSYBRrev, AGCAGGTTACCCATCACCTC. Expression was normalized to that of a ribosomal housekeeping gene using the following primers: rpoDfor, TGCAATCTTCGGTGAGAAG, and rpoDrev, TGCATCTCTTCGATTTCCAG. All primer concentrations were 200 nM in a final volume of 25 μl. Bacteria were grown in cation-supplemented Mueller-Hinton broth; polymyxin-resistant isolates were grown in the presence of 1 μg/ml of polymyxin. RNA was isolated using the RNeasy kit (Qiagen Inc., Germantown, MD) and treated with RNase-free DNase; each RT-PCR well contained 12.5 ng of RNA. Amplification efficiencies were 90 to 110%. Expression studies were performed in triplicate using three unique RNA samples, and each real-time RT-PCR amplification assay was performed in triplicate. Select isolates were fingerprinted by the rep-PCR method using ERIC primers, as previously described (17).
RESULTS
E. cloacae.
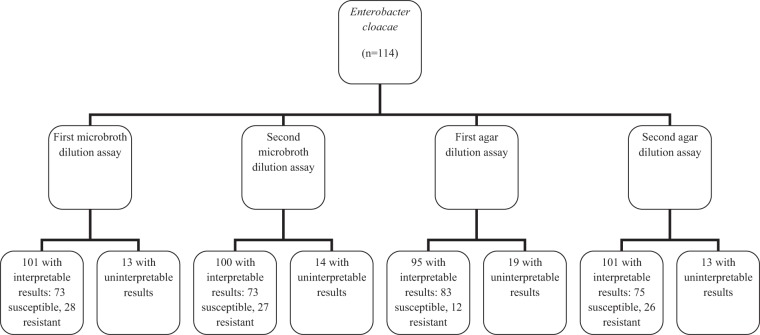
A total of 114 E. cloacae isolates underwent polymyxin B susceptibility testing; 10 were known to carry blaKPC. All isolates underwent four dilution susceptibility tests: twice by the broth microdilution method and twice by the agar dilution method (Fig. 1). For the first broth microdilution assay, of 101 isolates with interpretable results, 7 had a single skip well (most in the range of 0.12 to 0.5 μg/ml). Thirteen isolates had uninterpretable MICs (with all having multiple skip wells in the 0.12 to 8 μg/ml range). For the second broth microdilution assay, of 100 with interpretable results, 15 had a single skip well, mostly at 0.12 to 0.5 μg/ml. The remaining 14 isolates had uninterpretable MICs, with multiple skip wells (in the 0.12 to 1 μg/ml range). For the first agar dilution assay, of 95 isolates with interpretable MICs, four had a single skip concentration. Nineteen isolates had uninterpretable results, with most having skip concentrations involving 0.12 to 4 μg/ml. For the second agar dilution assay, of 101 isolates with interpretable results, seven had a single skip concentration. The remaining 13 isolates had uninterpretable MICs, with 12 involving skip concentrations in the 0.25 to 2 μg/ml range. Of note, similar results were obtained when using colistin sulfate for susceptibility testing; 25 isolates had uninterpretable results by a single microdilution assay and 21 by a single agar dilution assay (data not shown).
Fig 1.
Polymyxin B susceptibility results for Enterobacter cloacae and E. aerogenes by microbroth and agar dilution methods.
When all four polymyxin B susceptibility assays were analyzed, 36 of the 114 isolates were considered resistant at least once (8 once, 9 twice, 8 thrice, and 11 by all four assays). There was categorical agreement for 76/114 (67%) of isolates, with 65 susceptible and 11 resistant in all four tests. Thirty-eight isolates (33%) had either uninterpretable (n = 32) MICs at least once or conflicting (categorical disagreement; n = 6) MICs. Reproducibility was poor even within test methods, with categorical agreement for only 77% by broth microdilution and 77% by agar dilution.
Fifty-seven isolates underwent susceptibility testing by the Etest method. When tested using Mueller-Hinton agar, 39 were considered susceptible (MICs ≤ 2 μg/ml), 7 were considered resistant, and 11 had uninterpretable MICs. When the 39 isolates with susceptible MICs were tested using Isosensitest agar, 14 remained susceptible, 13 were resistant, and results for 12 were uninterpretable. When the seven isolates with resistant MICs with Mueller-Hinton agar were tested using Isosensitest agar, four remained resistant and three had uninterpretable MICs. All 11 isolates with unreadable MICs on Mueller-Hinton agar also had uninterpretable MICs with Isosensitest agar.
For the 39 isolates that were considered susceptible as determined by Etest on Mueller-Hinton agar, there were 156 (four for each isolate) broth and agar dilution susceptibility tests. Of these 156 dilution tests, 70 results were susceptible, 46 were resistant, and 40 were uninterpretable. For the seven isolates considered resistant by Etest on Mueller-Hinton agar, there were 28 broth/agar dilution tests performed. Seven results were susceptible, 13 resistant, and 8 uninterpretable. Of the 44 broth and agar dilution tests performed using the 11 isolates with uninterpretable Etest results, the majority of results were either resistant (34 tests) or uninterpretable (7 tests). Conversely, of 14 isolates with categorical results of susceptibility on all four dilution tests, 13 were found to be susceptible by Etest on Mueller-Hinton agar. Of the 11 isolates with categorical agreement of resistance on all four dilution tests, 5 were susceptible, 2 were resistant, and 4 had uninterpretable MICs by Etest.
Eight isolates were examined for the presence of heteroresistance by population analysis profiles. Two isolates (MAEL4 and KCEL1) had either susceptible or uninterpretable MICs when tested by broth/agar methods; at 2 to 6 μg/ml polymyxin B, approximately 0.00017 to 0.00076% of the inoculum demonstrated heteroresistance (Fig. 2A). Neither isolate had detectable heteroresistant populations at 64 μg/ml. Two isolates (KCEL23 and KCEL27) had MICs that varied by dilution testing. The percentage of heteroresistant colonies varied widely between these two isolates, being 0.005% and 34% at concentrations of 2 to 6 μg/ml (Fig. 2A). Three isolates (BVEL5, KCEL2, and CIEL2) had consistently uninterpretable MICs when tested by broth/agar dilution methods; at 2 to 6 μg/ml, 0.0006 to 0.29% of the original inoculum demonstrated heteroresistance (Fig. 2B). Finally, one isolate (WOEL3) had either highly resistant or uninterpretable MICs by dilution testing. Heteroresistance was detected in ∼0.1% of the original inoculum (Fig. 2B). It is noteworthy that for several of the isolates with repeatedly uninterpretable MICs in dilution testing, the number of heteroresistant colonies was greater when grown in 4 to 8 μg/ml of polymyxin B compared to 0.5 to 2 μg/ml.
Fig 2.
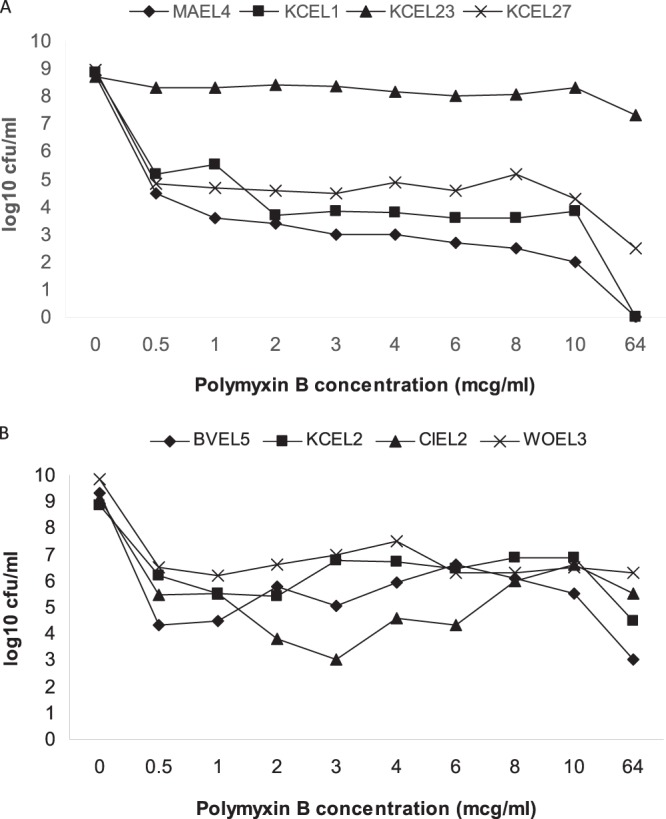
Population analysis profiles of clinical isolates of Enterobacter cloacae. (A) The isolates MAEL4 and KCEL1 had susceptible and/or uninterpretable MICs when tested by broth/agar methods; the isolates KCEL23 and KCEL27 had conflicting and/or uninterpretable MICs by dilution testing. (B) The isolates BVEL5, KCEL2, and CIEL2 had consistently uninterpretable MICs, and the isolate WOEL3 had either highly resistant or uninterpretable MICs. mcg, micrograms.
Two isolates with uninterpretable MICs were studied in further detail. Isolate KCEL6 had skip wells involving 0.25, 0.5, 1 and 2 μg/ml. Isolate CIEL1 had skip wells involving 0.12, 0.25, 0.5, 1, 2, and 4 μg/ml. Bacteria were recovered from polymyxin B concentrations above those of the skip wells (at 4 μg/ml for KCEL6, yielding isolate KCEL6pxn4, and at 8 μg/ml for CIEL1, yielding isolate CIEL1pxn8). DNA fingerprinting confirmed that the isolates recovered in the polymyxin wells were identical to the parents (Fig. 3). Compared to results for the respective parents, only polymyxin B MICs increased for KCEL6pxn4 and CIEL1pxn8 (Table 1). Population analysis profiles for these isolates revealed a marked increase in the percentage of heteroresistant colonies for the isolates collected above the skip wells. For the parent isolates KCEL6 and CIEL1, at concentrations of polymyxin B from 0.5 to 10 μg/ml, the percentages of heteroresistant colonies ranged from 0.0015 to 0.0082% and 0.005 to 7.3%, respectively. In comparison, for KCEL6pxn4 and CIEL1pxn8, the percentages of heteroresistant colonies ranged from 26 to 100% and 40 to 78%, respectively. At 64 μg/ml, the percentages of heteroresistant colonies for KCEL6 and CIEL1 were 0.0005% and 0.0001%, respectively; for KCEL6pnx4 and CIEL1pnx8, the percentages increased to 8.3% and 12.3%, respectively. For KCEL6, relative expression of phoP (1.3 ± 0.86) and phoQ (1.1 ± 0.79) was similar to that for E. cloacae ATCC 13047. However, relative expression increased approximately 10-fold for both phoP (9.1 ± 8.6) and phoQ (12.6 ± 4.2) for KCEL6pxn4. Compared to the parent isolate, there were no mutations affecting phoP in KCELpxn4; however, there were mutations leading to amino acid changes in PhoQ (L25S, L110P, and N158K). For CIEL1, compared to E. cloacae ATCC 13047 (and KCEL6), there were multiple mutations (including one in the RT-PCR primer region), leading to six amino acid changes in PhoP. The isolate from the skip well (CIEL1Pxn8) had two additional mutations affecting PhoP (R25H and F153L). For both CIEL1 and CIEL1Pxn8, the gene for PhoQ was not amplifiable with the primers used, suggesting mutations in this operon. Because of these mutations, the RT-PCR results were considered invalid for this isolate.
Fig 3.
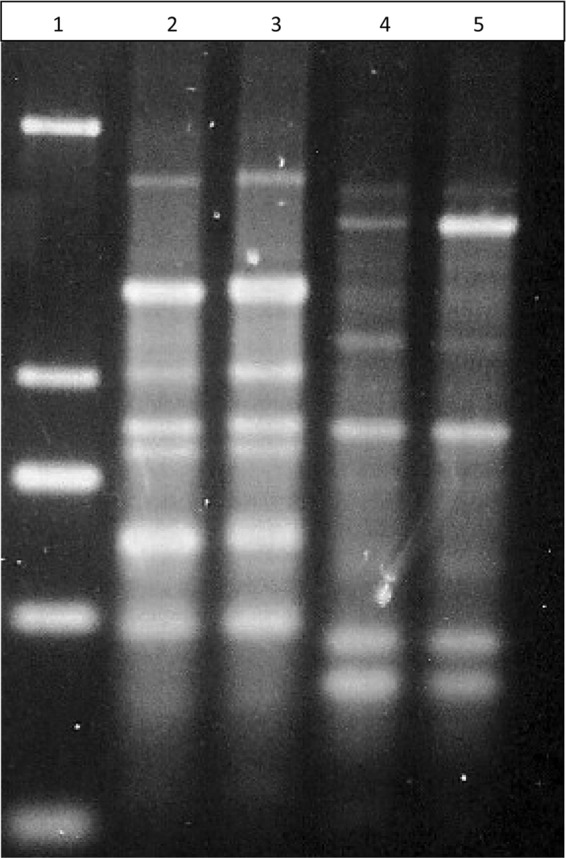
rep-PCR patterns of isolates demonstrating skip well phenomenon. Lane 1, molecular weight marker; lane 2, isolate CIEL1; lane 3, isolate CIEL1pxn8 (recovered from a well containing 8 μg/ml of polymyxin B); lane 4, isolate KCEL6; lane 5, isolate KCEL6pxn4 (recovered from a well containing 4 μg/ml of polymyxin B).
Table 1.
Susceptibility tests, using the Etest method, of isolates demonstrating growth in polymyxin B above skip well concentrations
| Drug | MIC (μg/ml) for isolate: |
|||
|---|---|---|---|---|
| CIEL1 | CIEL1pxn8 | KCEL6 | KCEL6pxn4 | |
| Polymyxin B | 3 | 96 | 0.5 | 128 |
| Cefepime | 0.032 | 0.047 | 0.125 | 0.094 |
| Meropenem | 0.125 | 0.094 | 0.125 | 0.094 |
| Gentamicin | 1.5 | 1.5 | 3 | 1.5 |
| Amikacin | 2 | 1.5 | 2 | 1.5 |
| Ciprofloxacin | 0.006 | 0.012 | 0.023 | 0.023 |
| Tigecycline | 0.75 | 0.5 | 1.5 | 1 |
E. aerogenes.
A total of 56 isolates underwent polymyxin B susceptibility testing (Fig. 1), including four known to harbor blaKPC. For the first broth microdilution test, 51 isolates had interpretable results, with 3 having a single skip well (all at 0.12 μg/ml). Five isolates had uninterpretable MICs (most having multiple skip wells in the 0.12 to 0.5 μg/ml range). For the second broth microdilution assay, 54 had interpretable results (2 with a single skip well at 0.12 μg/ml). The remaining two isolates had uninterpretable MICs, with multiple skip wells involving 0.12 to 2 μg/ml). For the first agar dilution assay, all 56 isolates had interpretable MICs (1 with a single skip concentration at 32 μg/ml). For the second agar dilution assay, all isolates had interpretable results, including one with a single skip concentration (at 0.5 μg/ml).
Analysis of all four susceptibility assays demonstrated that three isolates were considered resistant at least once (one twice, one thrice, and one by all four assays). Categorical agreement of susceptibility was achieved for 47 isolates with all four assays and of resistance for 1 isolate. Eight of the 56 isolates (14%) had either conflicting categorical or uninterpretable results.
Eight isolates underwent susceptibility testing by the Etest method. When tested using Mueller-Hinton agar, five were considered susceptible, two were considered resistant, and one had an uninterpretable MIC. When the five isolates with susceptible MICs were tested using Isosensitest agar, three remained susceptible and two were resistant. Both of the resistant isolates on Mueller-Hinton remained resistant when Isosensitest agar was used, and results for the single isolate with an uninterpretable MIC on Mueller-Hinton agar remained unreadable with Isosensitest agar.
All five isolates with susceptible MICs by Etest on Mueller-Hinton agar had consistently susceptible MICs by broth microdilution or agar dilution. For the two isolates considered resistant by Etest on Muller Hinton agar, of the eight broth/agar dilution tests, six demonstrated resistant MICs and two resulted in susceptible MICs. For the one isolate with an uninterpretable Etest result, the dilution MICs were either resistant (three times) or uninterpretable (once).
DISCUSSION
Despite decades of clinical use, the optimal methodology for susceptibility testing of polymyxins remains undefined. With the emergence of multidrug-resistant Gram-negative bacilli, accurate determination of susceptibility to these agents is essential. Unacceptable errors have been identified with the disk diffusion and Etest methods due to the poor diffusion of these compounds in agar (5–8). Dilution methods for susceptibility testing are preferred, but problems with antibiotic adsorption to labware and adaptive resistance confound this methodology (9, 18). Our results with E. cloacae and E. aerogenes again highlight the difficulties associated with susceptibility testing with these agents. For E. cloacae in particular, MIC determinations by dilution methods lack precision and often lead to uninterpretable results. Only two-thirds of the tested isolates had categorical agreement of susceptible or resistant when four dilution tests were performed. Conflicting MICs or uninterpretable results, due to multiple skip wells, were commonplace. It does not appear that polymyxin adsorption to the polystyrene trays accounted for these findings, since most skip wells were at the lower concentrations, where adsorption is the greatest (9). Rather, it appears that Enterobacter species possess intrinsic mechanisms of resistance that can become apparent during susceptibility testing.
All of our tested isolates of E. cloacae demonstrated heteroresistance when assessed by population analysis profiling. Isolates that consistently had low (susceptible) MICs tended to have a lower percentage of heteroresistant subpopulations, and heteroresistance was not detected at 64 μg/ml. Isolates with consistently indeterminate MICs had greater degrees of heteroresistance, including at 64 μg/ml. It was noteworthy that for several isolates, the percentage of resistant subpopulations actually increased at concentrations of 2 to 8 μg/ml of polymyxin B. This correlates with the frequent finding of skip wells at lower concentrations and the emergence of growth in concentrations around 4 μg/ml. It appears that critical concentrations propel the mechanisms leading to polymyxin resistance.
Resistance to polymyxins is related to alterations in the lipopolysaccharide structure, reducing the potential binding sites. Most of our understanding regarding mechanisms contributing to polymyxin resistance comes from studies involving P. aeruginosa. Derepression of the arn operon is associated with the addition of 4-amino-l-arabinose to lipid A, thereby reducing the number of binding sites for polymyxins. Several two-component regulatory systems that control expression of the arn operon have been identified. Under the pressure of low-Mg conditions (19) or exposure to polymyxins (12, 18, 20, 21), PhoPQ has been demonstrated to regulate expression of the arn operon. Similarly, PmrAB can also affect expression of the arn operon under low-cation conditions (22) or following exposure to polymyxins (12, 18, 22, 23). Polymyxin resistance has been associated with altered expression of these regulators (12, 24), including upregulation of pmrA (18), pmrB (12, 23), and phoQ (12) and the disabling of phoQ (21). The “skip well” isolates of P. aeruginosa have been found to have increased expression of pmrAB, phoQ, and the arn operon (12). A third regulator, ParRS, has also been shown to be involved in adaptive resistance to polymyxins (18). More recently, two additional regulators (ColRS and CprRS) that are essential for PhoPQ-mediated peptide resistance have been identified (20, 25). Adding to the complexity, ColRS and CprRS also appear to regulate polymyxin resistance outside of 4-amino-l-arabinose modification of lipid A.
Similar observations concerning mechanisms contributing to polymyxin resistance have been observed in other genera of bacteria. Mutations in the pmrCAB operon, leading to increased production of the PmrAB system and modification of lipid A, have been identified in isolates of polymyxin-resistant A. baumannii (26, 27). In addition, absence of lipid A in the cellular membrane, due to alterations in the lpx operon, has been found in laboratory-derived and clinical isolates of polymyxin-resistant A. baumannii (28, 29). Corresponding findings have also been obtained for members of the Enterobacteriaceae. Alterations in lipopolysaccharide have been associated with polymyxin resistance for K. pneumoniae (30), Escherichia coli (31, 32), and Salmonella enterica serovar Typhimurium (32). In S. Typhimurium, the phoPQ operon regulates pmrAB expression, affecting polymyxin susceptibility (33). For one isolate of E. cloacae tested in our work, increased expression of phoP and phoQ was evident in bacteria growing in 4 μg/ml of polymyxin B. For a second isolate, mutations involving phoP were evident in bacteria growing in 8 μg/ml. While it appears that alterations in these regulators correlate with adaptive resistance, further studies are needed to further define the mechanisms of resistance.
Based on our results, several recommendations can be made regarding susceptibility testing of polymyxins with Enterobacter species and in particular E. cloacae. First, Etests may not reliably detect resistant isolates; of 11 isolates found to be consistently resistant by dilution tests, 5 were found to be susceptible by Etest. Therefore, isolates found to be susceptible by Etest should have MICs confirmed by a dilution method. Second, multiple dilution tests should be performed, with close attention to the presence of skip wells. Isolates should be considered susceptible only if interpretable and repeatedly susceptible results are obtained. Isolates demonstrating more than one skip well in any of the MIC determinations should be considered to have an uninterpretable polymyxin MIC. Third, a wide range of concentrations should be used; using only a few concentrations around 4 μg/ml (a frequently employed breakpoint) may not detect isolates demonstrating adaptive resistance. Even with these recommendations, until the clinical relevance of skip wells is defined and the methodology of susceptibility testing is refined, interpretation of polymyxin susceptibility tests for Enterobacter species should be undertaken with extreme caution.
Footnotes
Published ahead of print 2 October 2013
REFERENCES
- 1.Fridkin SK. 2001. Increasing prevalence of antimicrobial resistance in intensive care units. Crit. Care Med. 29:N64–N68 [DOI] [PubMed] [Google Scholar]
- 2.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob. Agents Chemother. 55:1274–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchaim D, Navon-Venezia S, Schwaber MJ, Carmeli Y. 2008. Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob. Agents Chemother. 52:1413–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landman D, Bratu S, Kochar S, Panwar M, Trehan M, Doymaz M, Quale J. 2007. Evolution of antimicrobial resistance among Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae in Brooklyn, NY. J. Antimicrob. Chemother. 60:78–82 [DOI] [PubMed] [Google Scholar]
- 5.Gales AC, Reis AO, Jones RN. 2001. Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J. Clin. Microbiol. 39:183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hindler JA, Humphries RM. 2013. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J. Clin. Microbiol. 51:1678–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan TY, Ng SY. 2007. Comparison of Etest, Vitek, and agar dilution for susceptibility testing of colistin. Clin. Microbiol. Infect. 13:541–544 [DOI] [PubMed] [Google Scholar]
- 8.Turnidge JD, Bell JM, Jones RN. 2007. Emergence of colistin-resistant Klebsiella spp. and Enterobacter spp. in the Asia-Pacific (APAC) region: a SENTRY antimicrobial surveillance program report (2006), abstr C2-2054 Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC [Google Scholar]
- 9.Karvanen M, Malmber C, Moamed A, Lagerback P, Friberg LE, Cars O. 2011. Colistin is extensively lost during normal experimental conditions, abstr D-690 Abstr. 51st Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC [Google Scholar]
- 10.Bliss EA, Chandler CA, Schoenbach EB. 1949. In vitro studies of polymyxin. Ann. N. Y. Acad. Sci. 51:944–951 [DOI] [PubMed] [Google Scholar]
- 11.Traub WH. 1982. Polymyxin B-induced cocarde growth phenomenon of Serratia marcescens due to cationic detergent-like activity of polymyxin B. Chemotherapy 28:363–368 [DOI] [PubMed] [Google Scholar]
- 12.Schurek KN, Sampaio JLM, Kiffer CRV, Sinto S, Mendes CMF, Hancock REW. 2009. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:4345–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawley JS, Murray CK, Griffith ME, McElmeel ML, Fulcher LC, Hospenthal DR, Jorgensen JH. 2007. Susceptibility of Acinetobacter strains isolated from deployed U.S. military personnel. Antimicrob. Agents Chemother. 51:376–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:2946–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo-Ten-Foe JR, de Smet AMGA, Diederen BMW, Kluytmans JAJW, van Keulen PHJ. 2007. Comparative evaluation of the VITEK 2, disk diffusion, Etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob. Agents Chemother. 51:3726–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 9th ed, vol 32, no. 2 Approved standard M7-A9. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 17.Landman D, Butnariu M, Bratu S, Quale J. 2009. Genetic relatedness of multidrug-resistant Acinetobacter baumannii endemic to New York City. Epidemiol. Infect. 137:174–180 [DOI] [PubMed] [Google Scholar]
- 18.Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock REW. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 54:3372–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macfarlane ELA, Kwasnicka A, Ochs MM, Hancock REW. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305–316 [DOI] [PubMed] [Google Scholar]
- 20.Gutu AD, Sgambati N, Strasbourger P, Brannon MK, Jacobs MA, Haugen E, Kaul RK, Johansen HK, Hoiby N, Moskowitz SM. 2013. Polymyxin resistance of Pseudomonas aeruginosa phoQ mutants is dependent on additional two-component regulatory systems. Antimicrob. Agents Chemother. 57:2204–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Hoiby N, Moskowitz SM. 2011. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 55:5761–5769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205–217 [DOI] [PubMed] [Google Scholar]
- 23.Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrow K, Kwon DH. 2009. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:5150–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez L, Jenssen H, Bains M, Wiegand I, Gooderham WJ, Hancock REW. 2012. The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrob. Agents Chemother. 56:6212–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53:3628–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock REW. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 55:3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moffatt JH, Harper M, Adler B, Nation RL, Li J, Boyce JD. 2011. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:3022–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffatt JH, Harper M, Harrison P, Hale JDF, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54:4971–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helander IM, Kato Y, Kilpelainen I, Kostianen R, Lindner B, Nummila K, Sugiyama T, Yokochi T. 1996. Characterization of lipopolysaccharides of polymyxin-resistant and polymyxin-sensitive Klebsiella pneumoniae O3. Eur. J. Biochem. 237:272–278 [DOI] [PubMed] [Google Scholar]
- 31.Nummila K, Kilpelainen I, Zahringer U, Vaara M, Helander IM. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-amino-ethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16:271–278 [DOI] [PubMed] [Google Scholar]
- 32.Peterson AA, Fesik SW, McGroarty EJ. 1987. Decreased binding of antibiotics to lipopolysaccharides from polymyxin-resistant strains of Escherichia coli and Salmonella typhimurium. Antimicrob. Agents Chemother. 31:230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunn JS. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16:284–290 [DOI] [PubMed] [Google Scholar]