Abstract
Alphaherpesvirinae family members can reactivate from latency following stress. The synthetic corticosteroid dexamethasone induces certain cellular transcription factors in murine and bovine trigeminal ganglionic neurons. Three dexamethasone-induced transcription factors, Krüppel-like factor 15, Slug, and SPDEF, stimulated the herpes simplex virus type 1-infected cell protein 0 (ICP0) promoter more than 150-fold. Conversely, other viral promoters (VP16 and ICP4) were not strongly stimulated, suggesting that the ICP0 promoter is preferentially activated by dexamethasone-simulated stress.
TEXT
The primary site of latency for herpes simplex virus type 1 (HSV-1) is sensory neurons in trigeminal ganglia (TG) (1–3). Abundant viral protein expression and infectious virus are not readily detected during latency, in contrast to the expression of the virus-encoded latency-associated transcript that occurs in latently infected sensory neurons (1–3). The ability of herpes simplex virus 1 (HSV-1) to reactivate from latency is crucial for virus transmission and recurrent disease. Increased stress levels correlate with an increased incidence of reactivation from latency (1–3). Dexamethasone (DEX), a synthetic corticosteroid, increases the incidence of reactivation from latency in TG neuronal cultures prepared from latently infected mice (4) and stimulates reactivation from latency in TG organ cultures latently infected with HSV-1 (5). With respect to reactivation from latency, there are at least two important unresolved issues: (i) which viral genes might be involved in the initiation of reactivation and (ii) whether the cascade of viral gene expression during reactivation is the same as productive infection of cultured cells (i.e., immediate early [IE] to early [E] to late [L]). Recent studies have suggested that VP16, a late transcript, stimulates reactivation from latency (6, 7), presumably because VP16 stimulates IE gene expression. The viral IE protein ICP0 also stimulates reactivation from latency (8–10), whereas others have concluded that ICP0 is not required for reactivation from latency (11–13). Several studies have also proposed that E gene expression and DNA replication occur prior to IE gene expression during reactivation from latency (14–17). Finally, viral gene expression is reported to be initially disorganized during explant-induced reactivation from latency (5, 18). Regardless of whether VP16 or ICP0 is involved or required for reactivation from latency, it is reasonable to predict that stimulus-specific cellular transcription factors may stimulate viral gene expression during the early stages of reactivation.
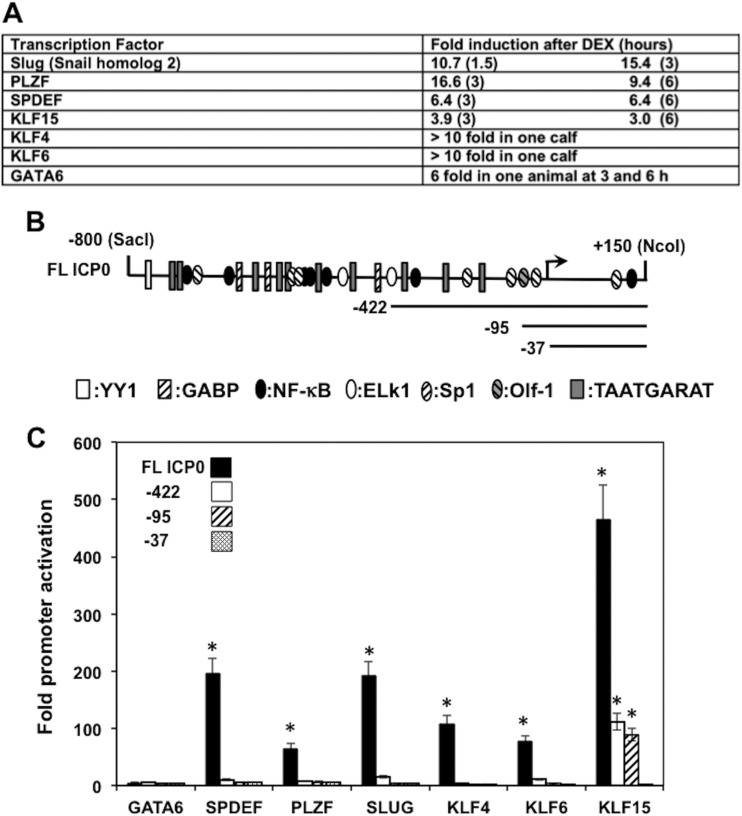
DEX consistently initiates bovine herpesvirus 1 (BHV-1) reactivation from latency in calves and rabbits (1, 2, 19–22), which culminates in lytic cycle viral transcription in neurons within 6 h after treatment (23, 24). We identified DEX-inducible cellular factors in bovine TG neurons during the early phases of reactivation from latency (25). A subset of these cellular genes consists of transcription factors (Fig. 1A gives a summary of the DEX-inducible transcription factors). One of the highly induced transcription factors, promyelocytic leukemia zinc finger (PLZF), stimulated BHV-1 productive infection more than 20-fold. Two other DEX-inducible transcription factors, Krüppel-like factor 15 (KLF15) and KLF4, stimulated BHV-1 productive infection and trans activated the bICP0 E promoter approximately 100-fold. In contrast, PLZF and SPDEF (SAM pointed domain containing Ets transcription factor) stimulated the L BHV-1 gC promoter to a higher degree than the bICP0 E promoter and the IE transcription unit 1 promoter. Members of the KLF family, to which PLZF belongs, can repress or activate transcription, and they regulate cell growth, differentiation, apoptosis, and cancer (reviewed in references 26, 27, and 28). KLF and Sp1 family members may be involved in regulation of HSV-1 transcription, including certain aspects of the latency-reactivation cycle, because these family members bind GC-rich sequences (reviewed in references 27 and 29) and many HSV-1 promoters contain Sp1 binding sites and Sp1 activates IE promoters (30).
Fig 1.
Effect of DEX-inducible transcription factors on ICP0 promoter activity. (A) Cellular transcription factors identified in TG neurons following DEX treatment (25). Fold induction was determined by microarray studies and then confirmed by reverse transcription-PCR (RT-PCR) and immunohistochemistry. KLF4, KLF6, and GATA6 expression was stimulated in 1 of the 3 animals, whereas the other transcription factors were stimulated in 3/3 animals. Plasmids expressing the respective transcription factors were previously described and were shown to express similar levels of protein in transfected Neuro-2A cells (25). (B) The full-length ICP0 promoter (FL ICP0) and the three deletion constructs used in this study. A subset of the known transcription factor binding sites in the FL ICP0 promoter was previously described, and their locations are shown (31). An arrow denotes the start site of ICP0 mRNA. The ICP0 promoter luciferase reporter constructs were obtained from Priscilla Schaffer and were previously described (31). (C) Luciferase activity at 40 h after Neuro-2A cells were cotransfected with the designated ICP0 promoter luciferase reporter construct (1.0 μg DNA) and a DEX-inducible transcription factor (0.5 μg DNA). Transfection of Neuro-2A cells was performed as previously described (25, 48, 49). Luciferase activity was normalized by comparing Renilla luciferase levels, which are regulated by a simple TATA box. The results are the average of data from 5 independent experiments. An asterisk denotes significant differences (P < 0.05) in cells transfected with the designated ICP0 luciferase plasmid and the designated DEX- inducible transcription factor, as determined by the Student t test.
Based on the above observations, we hypothesized that DEX-inducible transcription factors identified in bovine TG neurons (25) might stimulate HSV-1 promoters that would be important for mediating stress-induced reactivation from latency. We initially tested the ICP0 promoter because ICP0 expression stimulates reactivation from latency (8–10), the ICP0 promoter is stimulated by hyperthermic stress (31), and BHV-1 IE and E promoters that drive bICP0 expression are trans activated by certain DEX-inducible transcription factors (25). For these studies, we used a full-length (FL) ICP0 promoter construct that contains sequences spanning −800 to +150 relative to the ICP0 transcription initiation site (31) (Fig. 1B). Numerous transcription factor-binding sites are located in the FL ICP0 promoter, suggesting that more than one cellular transcription factor may regulate its activity, either independently or in a synergistic manner. Three additional deletion mutants, the −422, −95, and −37 mutants, were also examined. Mouse neuroblastoma cells (Neuro-2A) were used for these studies because they are of neuronal origin, they are readily transfected, DEX-inducible transcription factors are not abundantly expressed in Neuro-2A cells, and plasmids expressing the DEX-inducible transcription factors are active in these cells (25). KLF15 stimulated FL ICP0 promoter activity more than 400-fold in Neuro-2A cells (Fig. 1C). The −95 ICP0 construct was activated nearly 100-fold by KLF15, whereas the −37 ICP0 construct was not trans activated. SLUG and SPDEF stimulated the FL ICP0 promoter nearly 200-fold, while PLZF, KLF6, and KLF4 but not GATA6 trans activated the FL ICP0 promoter at least 50-fold. In contrast to KLF15, the ICP0 deletion constructs were not responsive to these factors. Titration studies demonstrated that the concentrations of the respective DEX inducible transcription factors used for this study were optimal for activating the FL ICP0 promoter (data not shown). In summary, these studies demonstrated that DNA sequences located between −800 and −37 were responsive to KLF15 while sequences between −800 and −422 were responsive to SLUG, SPDEF, PLZF, KLF6, and KLF4.
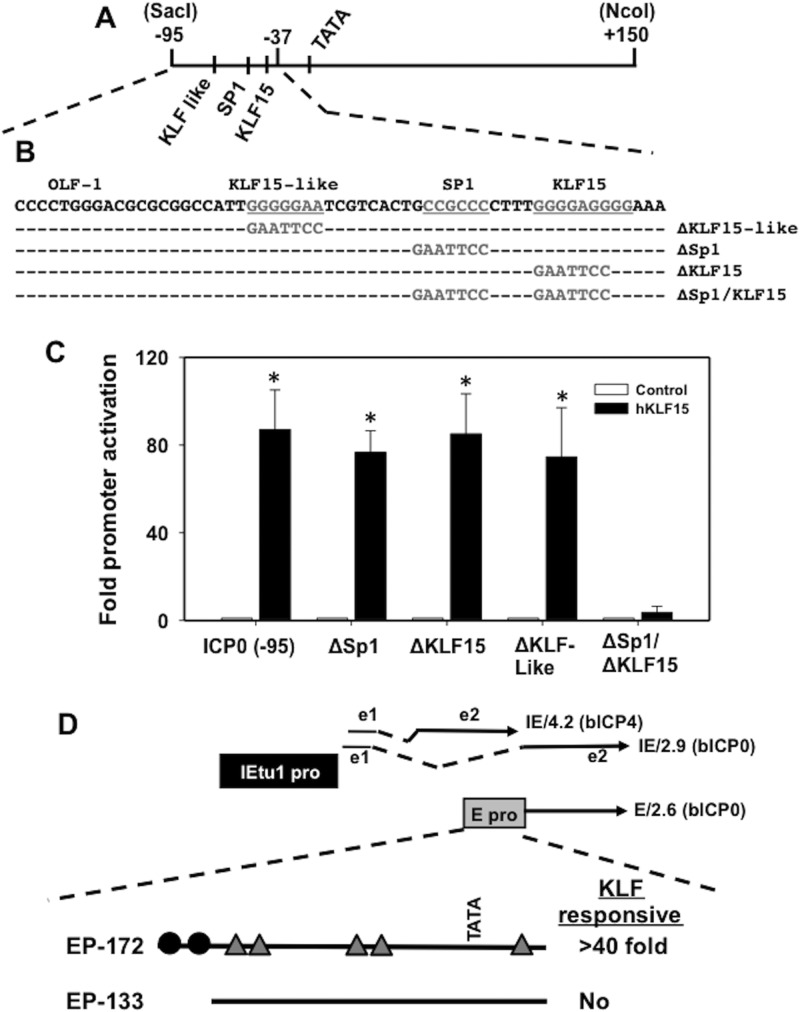
The ICP0 promoter appeared to contain 2 separate KLF15 response elements: the first between −800 and −422 and the second between −95 and −37. Deleting DNA sequences between −800 and −422 reduced promoter activity 4- to 5-fold, whereas deletion of DNA sequences between −95 and −37 reduced promoter activity 80-fold. An ICP0 promoter deletion mutant virus that lacks −70 to −420 (32) does not reactivate from latency following hyperthermic stress (13), suggesting that KLF15-mediated trans activation of the ICP0 promoter via sequences in the −95 ICP0 construct are important for stress-induced reactivation from latency. These observations led us to hypothesize that KLF15-mediated trans activation of sequences in the −95 ICP0 construct are important for certain aspects of reactivation from latency. DNA sequences between −37 and −95 contain three distinct motifs for KLF15-induced trans activation (Fig. 2A). These include a KLF binding site analogous to that located in the promoter of the gene encoding an interphotoreceptor binding protein (33), a KLF-like binding site, and a consensus Sp1 binding site. To identify DNA sequences between −95 and −37 that are responsive to KLF15, the core sequence of each potential KLF15-responsive motif was mutated and an EcoRI linker inserted (see Fig. 2B for a schematic of the respective mutants). In several independent experiments, the ΔSp1/KLF15 mutant was not trans activated by KLF15 (Fig. 2C). As expected, the wild-type (wt) −95 ICP0 construct was trans activated by KLF15 at least 80-fold. KLF15 also trans activated the ΔKLF15-like, ΔSp1, and ΔKLF15 promoter constructs approximately 80-fold. It is interesting to note that KLF15 and Sp1 have been shown to synergistically activate cellular promoters (34, 35), and synergistic activation correlates with the ability of KLF15 to stably interact with Sp1 (35). Since the KLF15 binding site in the ICP0 promoter is 4 bp from the 3′ end of the Sp1 binding site, it is possible that an interaction between SP1 and KLF15 is important for trans activation of the ICP0 promoter.
Fig 2.
Identification of KLF15-responsive regions in the −95 ICP0 promoter construct. (A) Schematic of the −95 ICP0 promoter deletion and location of potential cis-acting motifs between −37 and −95 that might be important for KLF15 mediated trans activation. (B) ICP0 promoter DNA sequences that are located between −37 and −95. Locations of KLF15-like motif, Sp1, and KLF15 sites are denoted by underlined gray nucleotides. The respective mutant constructs contain an EcoRI linker located in the KLF15 site or the KLF15-like site. In addition, a construct containing EcoRI linker insertions in the Sp1 and KLF15 site was synthesized. Integrated DNA Technology (Iowa) synthesized the respective mutant −95 ICP0 promoters. The respective ICP0 mutant −95 promoter constructs were then cloned into the promoterless luciferase reporter construct (pGL3-Basic; Promega) using unique SacI and NcoI restriction enzyme sites. The ΔSp1 binding site mutant was previously described (31). (C) Neuro-2A cells were cotransfected with the designated −95 ICP0 mutant constructs and KLF15 or a control empty vector. At 40 h after transfection, luciferase activity was measured. Luciferase activity was normalized by comparing Renilla luciferase levels, which are regulated by a simple TATA box. The results are presented as fold induction relative to results for the empty vector control and are the averages of data from 3 independent experiments. An asterisk denotes significant differences (P < 0.05) in cells transfected with the designated ICP0 luciferase plasmid and the DEX-inducible transcription factor KLF15, as determined by the Student t test. (D) The positions of BHV-1 transcripts that encode bICP4 and bICP0 are shown. The immediate early transcription unit 1 (IEtu1) encodes bICP4 (IE/4.2) and bICP0 (IE/2.9) (50, 51). The IEtu1 promoter (denoted by the black rectangle) activates IE expression of IE/4.2 and IE/2.9. E/2.6 is the early transcript that encodes bICP0, and an early promoter (denoted by the gray rectangle) activates expression of the early bICP0 transcript (E/2.6) (36). Exon 2 (e2) of bICP0 contains all of the protein coding sequences of bICP0. The dashed lines are intron sequences. The ability of KLF4 and KLF15 to trans activate the bICP0 promoter constructs (EP-172 and EP-133) is also included and was summarized from a previous study (25).
bICP0 RNA expression is regulated by an IE promoter (IEtu1) and a separate E promoter (36) (Fig. 2D). The IEtu1 and E bICP0 promoters were trans activated approximately 100-fold by KLF4 and KLF15 (25). In contrast to the ICP0 promoter, KLF4 trans activated both bICP0 promoters more efficiently than KLF15 (25). Although the KLF-responsive region of the IEtu1 promoter was not precisely localized, there are numerous SP1 binding sites and potential KLF binding sites in this promoter. The KLF15-responsive region present in the −95 ICP0 promoter is not present in the smallest IETu1 promoter trans activated by KLF4 or KLF15 (data not shown). Comparison of the sequences of the E bICP0 promoter construct that was not responsive to KLF4 and KLF15 (EP-133) to the sequence of the minimal bICP0 promoter construct responsive to KLF4 and KLF15 (EP-172) revealed that there is no Sp1 binding site or consensus KLF15 binding site present in the −95 ICP0 promoter (Fig. 2D) (25). However, there are several potential KLF binding sites (Fig. 2D, closed circles), and two Sp1 binding sites are 13 bp from the 5′ terminus of the EP133 promoter (denoted by triangles). Since KLF family members bind to multiple GC-rich and related CACCC sequences in DNA, reviewed in reference 29, it is not surprising that the KLF15-responsive sequences in the HSV-1 ICP0 promoter are not identical to KLF-responsive sequences in the BHV-1 ICP0 promoters.
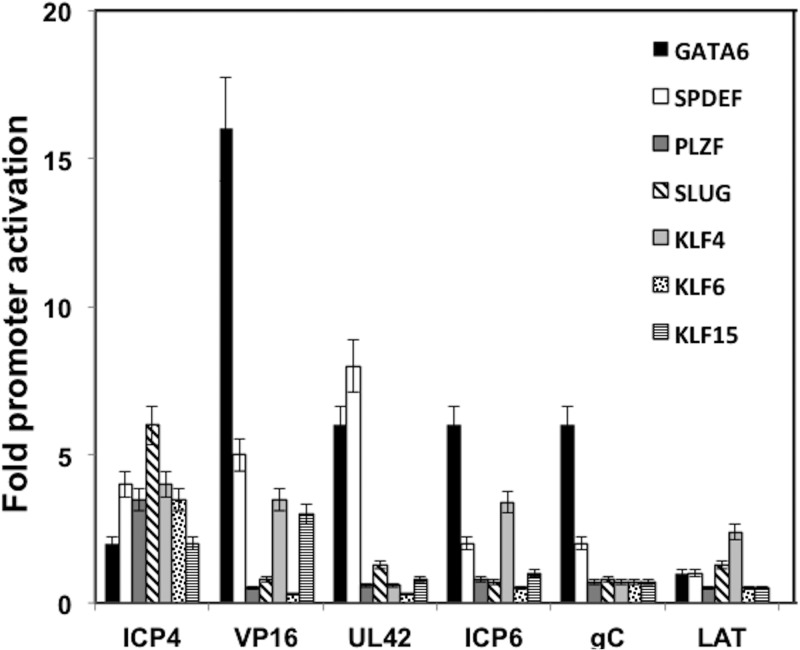
The ability of the DEX-inducible transcription factors to trans activate additional HSV-1 promoter reporter constructs was also examined. These promoter constructs contain representative IE, E, or L promoters. The VP16 promoter (L) was stimulated by GATA6 (>15-fold), SPDEF (∼5-fold), and KLF4 or KLF15 (4-fold) (Fig. 3). GATA6 also stimulated the UL42 (E), ICP6 (E), and gC (L) promoters (∼5-fold). SLUG was the only transcription factor that stimulated the ICP4 (IE) promoter more than 5-fold. In contrast, LAT promoter activity was not trans activated by any of the DEX-inducible transcription factors more than 3-fold. Compared to the ICP0 promoter, the other HSV-1 promoters examined in this study were not efficiently trans activated by the DEX-inducible transcription factors.
Fig 3.
Effect of DEX-inducible transcription factors on additional HSV-1 promoters. The ICP4, VP16, UL42, ICP6, gC, and LAT promoter luciferase reporter constructs were previously described (31). Neuro-2A cells were cotransfected with 1 μg of the designated reporter plasmid and 0.5 μg of a plasmid expressing the designated DEX-inducible transcription factor. Levels of DNA were made the same in each transfection by adding the pcDNA3.1 empty vector. Transfection of Neuro-2A cells was performed as previously described (25, 48, 49). At 40 h after transfection, luciferase activity was measured. The numbers represent fold induction relative to results for the empty vector control. The results are the average of data from 4 independent experiments.
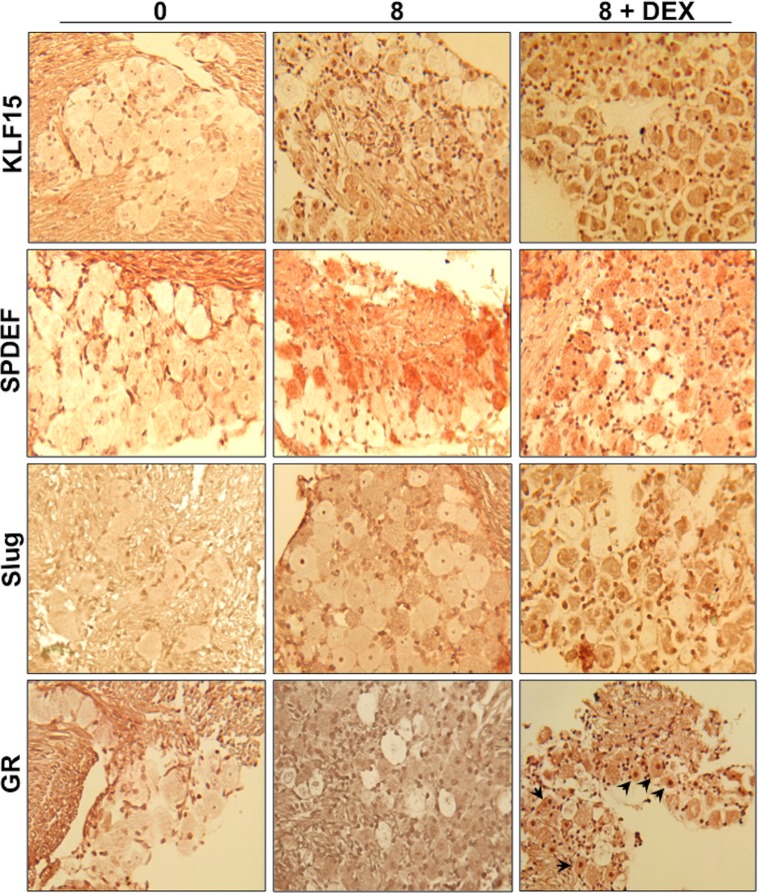
Immunohistochemistry (IHC) studies were performed to determine whether explanted mouse TG expressed any of the transcription factors that strongly trans activated the ICP0 promoter (KLF15, Slug, and SPDEF) and whether DEX stimulated their expression. TG were excised from adult female Swiss Webster mice, and each TG was minced into 4 pieces (37, 38). At 8 h after explantation, more neurons were KLF15+ and SPDEF+ than at time zero, where explanted TG were immediately formalin fixed (Fig. 4). Many TG neurons were KLF15+ and SPDEF+ when explanted for 8 h in the presence of 100 μM DEX. Slug+ neurons were not readily detected at time zero or following explant for 8 h: however, Slug+ neurons were readily detected in TG after explant for 8 h in the presence of DEX (Fig. 4). At 2 and 4 h after explant, lower numbers of KLF15+, SPDEF+, and Slug+ neurons were detected even when incubated with DEX (data not shown). As expected, more neurons were glucocorticoid receptor positive (GR+) at 8 h after explant, and many of these neurons exhibited nuclear GR staining when explanted TG were incubated with medium containing DEX (Fig. 4, GR panel, arrows). This result is expected because DEX activates the GR, resulting in nuclear localization (39, 40). A subset of TG neurons were weakly GR+ at time zero, which was not surprising because approximately 30% of rat TG neurons express the GR (41). KLF15 expression is induced by DEX in human airway smooth muscle cells (42), suggesting that KLF15 expression is stimulated by corticosteroids in multiple tissues. In summary, these studies were consistent with the findings observed in bovine TG (25). However, it took longer for DEX to induce expression of these transcription factors in sensory neurons of explanted mouse TG.
Fig 4.
Examination of transcription factors that trans activated the ICP0 promoter in explanted mouse TG. TG from adult female Swiss Webster mice (Charles River Labs) were minced into four pieces/TG and either explanted in Eagle minimal essential medium (EMEM) containing 2% charcoal-stripped fetal bovine serum (FBS) or immediately formalin fixed. Normal FBS but not charcoal-stripped FBS contains glucocorticoids that activate the GR, which masks the effect of glucocorticoids (52). As indicated, certain TG samples were also incubated with DEX (100 μM) for 8 h. Samples marked “0” were TG extracted from mice and then immediately fixed (no explant). All samples were fixed in neutral buffered formalin, paraffin embedded, and thin section prepared. IHC was performed as previously described (25, 48, 52). The goat anti-KLF15 antibody (sc-34826), rabbit anti-Slug antibody (sc-67022), and rabbit anti-SPDEF antibody (sc-67022) were purchased from Santa Cruz Biotechnology. The rabbit anti-GR antibody (36605) was purchased from Cell Signaling. All antibodies were diluted 1:1,000. Vectastain ABC kits containing biotinylated goat anti-rabbit IgG (PK-6101; Vector Laboratories) or biotinylated rabbit anti-goat IgG (PK-6105; Vector Laboratories) were incubated with sections to allow visualization of TG neurons that were recognized by the respective antibody. Magnification, ×200.
Exogenous expression of ICP0, independent of other viral gene products, can initiate HSV-1 (8) or HSV-2 (43) reactivation from latency using an in vitro neuronal culture system. Furthermore, in the absence of VP16, ICP0 enhances the ability of transfected viral DNA to initiate productive infection in cell culture (44). These observations suggest that reactivation stimuli activate the ICP0 promoter as an important event during reactivation from latency. Consequently, it is not surprising that ICP0 promoter activity is stimulated by hyperthermic stress (31). In transgenic mice, the ICP0 promoter but not the ICP4 promoter is active in a subset of neurons at certain times after birth (45, 46), providing additional evidence that specific neuronal signaling pathways regulate ICP0 promoter activity. VP16, the trans activator of IE promoters (47), has also been proposed to initiate reactivation from latency (6, 7). Although the VP16 promoter was trans activated by GATA6 15-fold, stimulation by DEX-inducible transcription factors was in general minimal relative to findings for the ICP0 promoter. It remains possible that other specific cellular factors may trans activate the VP16 promoter during reactivation from latency or that low levels of VP16 in certain “permissive” neurons may promote reactivation from latency. It is also possible that certain reactivation stimuli, DEX for example, directly stimulate ICP0, leading to induction of viral gene expression, including that of VP16, and further amplification of viral gene expression by this viral activator. Irrespective of this, it seems unlikely that successful reactivation from latency (shedding of infectious virus following a reactivation stimulus) occurs in the absence of ICP0. In summary, our studies provide evidence that certain cellular transcription factors induced by DEX in bovine or mouse TG neurons, including KLF15, preferentially activate the ICP0 promoter and mediate certain aspects of the HSV-1 latency-reactivation cycle.
ACKNOWLEDGMENTS
We thank the late Priscilla Schaffer for the FL, −420, −95, and −37 ICP0 promoter constructs as well as the other luciferase constructs used in this study.
This research was supported in part by grants from the USDA, National Institute of Food Animals (09-01653 and 2013-01041). A grant to the Nebraska Center for Virology (1P20RR15635) also supported certain aspects of these studies. Aspen Workman and Devis Sinani were partially supported by a fellowship from a Ruth L. Kirschstein National Research Service Award, 1 T32 AIO60547 (National Institute of Allergy and Infectious Diseases).
Footnotes
Published ahead of print 11 September 2013
REFERENCES
- 1.Jones C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 51:81–133 [DOI] [PubMed] [Google Scholar]
- 2.Jones C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16:79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perng G-C, Jones C. 2010. Towards an understanding of the herpes simplex virus type 1 latency-reactivation cycle. Interdiscip. Perspect. Infect. Dis. 2010:262415. 10.1155/2010/262415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halford WP, Gebhardt BM, Carr DJ. 1996. Persistent cytokine expression in trigeminal ganglion latently infected with herpes simplex virus type 1. J. Immunol. 157:3542–3549 [PubMed] [Google Scholar]
- 5.Du T, Zhou G, Roizman B. 2012. Induction of apoptosis accelerates reactivation from latent HSV-1 in ganglionic organ cultures and replication in cell cultures. Proc. Natl. Acad. Sci. U. S. A. 109:14616–14621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JY, Mandarino A, Chao MV, Mohr I, Wilson AC. 2012. Transient reversal of episome silencing precedes VP16-dependent transcription during reactivation of HSV-1 in neurons. PLoS Pathog. 8:e1002540. 10.1371/journal.ppat.1002540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson RL, Preston CM, Sawtell NM. 2009. De novo synthesis of VP16 coordinates the exit from HSV latency in vivo. PLoS Pathog. 5:e1000352. 10.1371/journal.ppat.1000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halford WP, Kemp CD, Isler JA, Davido DJ, Schaffer PA. 2001. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J. Virol. 75:6143–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halford WP, Schaffer PA. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75:3240–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMahan L, Schaffer PA. 1990. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J. Virol. 64:3471–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller CS, Danaher RJ, Jacob RJ. 2006. ICP0 is not required for efficient stress-induced reactivation of herpes simplex virus type 1 from cultured quiescently infected neuronal cells. J. Virol. 80:3360–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Preston CM. 2007. Reactivation of expression from quiescent herpes simplex virus type 1 genomes in the absence of immediate-early protein ICP0. J. Virol. 81:11781–11789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson RL, Sawtell NM. 2006. Evidence that the herpes simplex virus type 1 ICP0 protein does not initiate reactivation from latency in vivo. J. Virol. 80:10919–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosz-Vnenchak JJ, Coen DM, Knipe DM. 1993. Evidence for a novel regulatory pathway for herpes simplex virus gene expression in trigeminal ganglion neurons. J. Virol. 67:5383–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nichol PF, Chang JY, Johnson EM, Jr, Olivo PD. 1996. Herpes simplex virus gene expression in neurons: viral DNA synthesis is a critical regulatory event in the branch point between lytic and latent pathways. J. Virol. 70:5476–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pesola JM, Zhu J, Knipe DM, Coen DM. 2005. Herpes simplex virus 1 immediate-early and early gene expression during reactivation from latency under conditions that prevent infectious virus production. J. Virol. 79:14516–14525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tal-Singer R, Lasner TM, Podrzucki W, Skokotas A, Leary JJ, Berger SL, Frazer NW. 1997. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J. Virol. 71:5268–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du T, Zhou G, Roizman B. 2011. HSV-1 gene expression from reactivted ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc. Natl. Acad. Sci. U. S. A. 108:18820–18824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inman M, Lovato L, Doster A, Jones C. 2002. A mutation in the latency related gene of bovine herpesvirus 1 interferes with the latency-reactivation cycle of latency in calves. J. Virol. 76:6771–6779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones C, Newby TJ, Holt T, Doster A, Stone M, Ciacci-Zanella J, Webster CJ, Jackwood MW. 2000. Analysis of latency in cattle after inoculation with a temperature sensitive mutant of bovine herpesvirus 1 (RLB106). Vaccine 18:3185–3195 [DOI] [PubMed] [Google Scholar]
- 21.Jones C, Geiser V, Henderson G, Jiang Y, Meyer F, Perez S, Zhang Y. 2006. Functional analysis of bovine herpesvirus 1 (BHV-1) genes expressed during latency. Vet. Microbiol. 113:199–210 [DOI] [PubMed] [Google Scholar]
- 22.Rock D, Lokensgard J, Lewis T, Kutish G. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 66:2484–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler MT, Doster A, Sur JH, Jones C. 2002. Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet. Microbiol. 86:139–155 [DOI] [PubMed] [Google Scholar]
- 24.Winkler MTC, Doster A, Jones C. 2000. Persistence and reactivation of bovine herpesvirus 1 in the tonsil of latently infected calves. J. Virol. 74:5337–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Workman A, Eudy J, Smith L, Frizzo da Silva L, Sinani D, Bricker H, Cook E, Doster A, Jones C. 2012. Cellular transcription factors induced in trigeminal ganglia during dexamethasone-induced reactivation from latency stimulate bovine herpesvirus 1 productive infection and certain viral promoters. J. Virol. 86:2459–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bieker JJ. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 276:34355–34358 [DOI] [PubMed] [Google Scholar]
- 27.Black AR, Black JD, Azizkhan-Clifford J. 2001. Sp1 and Kruppel-like transcription factor family of transcription factors in cell growth and cancer. J. Cell Physiol. 188:143–160 [DOI] [PubMed] [Google Scholar]
- 28.McConnell BB, Yang VW. 2010. Mammalian Kruppel-like factors in health and diseases. Physiol. Rev. 90:1337–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaczynski J, Cook T, Urrutia R. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol. 4:206.201–206.208. 10.1186/gb-2003-4-2-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones KA, Tjian R. 1985. Sp1 binds to promoter sequences and activates herpes simples virus ‘immediate-early' gene transcription in vitro. Nature 317:179–182 [DOI] [PubMed] [Google Scholar]
- 31.Kushnir AS, Davido DJ, Schaffer PA. 2010. Role of nuclear factor Y in stress-induced activation of the herpes simplex virus type 1 ICP0 promoter. J. Virol. 84:188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davido DJ, Lieb DA. 1996. Role of cis-acting sequences of the ICP0 promoter of herpes simplex virus type 1 in viral pathogenesis, latency, and reactivation. J. Gen. Virol. 77:1853–1863 [DOI] [PubMed] [Google Scholar]
- 33.Otteson DC, Lai H, Liu Y, Zack DJ. 2005. Zinc-finger domains of the transcriptional repressor KLF15 binds multiple sites in rhodopsin and IRBP promoters including the CRS-1 and G-rich elements. BMC Mol. Biol. 6:15. 10.1186/1471-2199-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Yang Y, Jiang B, Zhang X, Zhou Y, Gong Y. 2010. SP1 and KLF15 regulate basal transcription of the human LRP5 gene. BMC Genet. 11:12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto J, Ikeda Y, Iguchi H, Fujino T, Tanaka T, Asaba H, Iwasaki S, Ioka RX, Kaneko IW, Magoori K, Takahashi S, Mori T, Sakaue H, Kodama T, Yanagisawa M, Yamamoto TT, Ito S, Sakai J. 2004. A Kruppel-like factor KLF15 contributes fasting-induced transcriptional activation of mitochondrial acetyl-CoA synthetase gene AceCS2. J. Biol. Chem. 279:16954–16962 [DOI] [PubMed] [Google Scholar]
- 36.Wirth UV, Fraefel C, Vogt B, Vlcek C, Paces V, Schwyzer M. 1992. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 66:2763–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perng G-C, Slanina S, Ghiasi H, Nesburn AB, Wechsler SL. 2001. The effect of latency-associated transcript on the herpes simplex virus type 1 latency-reactivation phenotype is mouse strain-dependent. J. Gen. Virol. 82:1117–1122 [DOI] [PubMed] [Google Scholar]
- 38.Whitlow ZW, Kristie TM. 2009. Recruitment of the transcriptional coactivator HCF-1 to viral immediate-early promoters during initiation of reactivation from latency of herpes simplex virus type 1. J. Virol. 83:9591–9595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Funder JW. 1997. Glucocorticoids and mineralocorticoid receptors: biology and clinical relevance. Annu. Rev. Med. 48:231–240 [DOI] [PubMed] [Google Scholar]
- 40.Pratt WB, Toft DO. 1997. Steroid receptor interactions with heat shock protein and immunophillin chaperones. Endocr. Rev. 18:306–360 [DOI] [PubMed] [Google Scholar]
- 41.DeLeon M, Covenas R, Chadi G, Narvaez JA, Fuxe K, Cintra A. 1994. Subpopulations of primary sensory neurons show coexistence of neuropeptides and glucocorticoid receptors in the rat spinal and trigeminal ganglia. Brain Res. 14:338–342 [DOI] [PubMed] [Google Scholar]
- 42.Masuno K, Haldar SM, Jeyaraji D, Mailoux C, Huang X, Panettieri RA, Jain MK, Gerber AN. 2011. Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. Am. J. Respir. Cell Mol. Biol. 45:642–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu XX, Chen JX, Young CS, Silverstein S. 1990. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J. Virol. 64:4489–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai WZ, Schaffer PA. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loiacono CM, Myers R, Mitchell WJ. 2002. Neurons differentially activate the herpes simplex virus type 1 immediate-early gene ICP0 and ICP27 promoters in transgenic mice. J. Virol. 76:2449–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taus NS, Mitchell WJ. 2001. The transgenic ICP4 promoter is activated in Schwann cells in trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J. Virol. 75:10401–10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Hare P. 1993. The virion transactivator of herpes simplex virus. Semin. Virol. 4:145–155 [Google Scholar]
- 48.Sinani D, Frizzo da Silva L, Jones C. 2013. A bovine herpesvirus 1 protein expressed in latently infected neurons (ORF2) promotes neurite sprouting in the presence of activated Notch1 or Notch3. J. Virol. 87:1183–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinani D, Jones C. 2011. Localization of sequences in a protein encoded by the latency related gene of bovine herpesvirus 1 (ORF2) that inhibits apoptosis and interferes with Notch1 mediated trans-activation of the bICP0 promoter. J. Virol. 85:12124–12133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirth UV, Gunkel K, Engels M, Schwyzer M. 1989. Spatial and temporal distribution of bovine herpesvirus 1 transcripts. J. Virol. 63:4882–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wirth UV, Vogt B, Schwyzer M. 1991. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J. Virol. 65:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frizzo da Silva L, Kook I, Doster A, Jones C. 2013. Bovine herpesvirus 1 regulatory proteins bICP0 and VP16 are readily detected in trigeminal ganglionic neurons expressing the glucocorticoid receptor during the early stages of reactivation from latency. J. Virol. 87:11214–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]