Abstract
Bunyaviruses are the largest known family of RNA viruses, infecting vertebrates, insects, and plants. Here we isolated three novel bunyaviruses from mosquitoes sampled in Côte d'Ivoire, Ghana, and Uganda. The viruses define a highly diversified monophyletic sister clade to all members of the genus Orthobunyavirus and are virtually equidistant to orthobunyaviruses and tospoviruses. Maximal amino acid identities between homologous putative proteins of the novel group and orthobunyaviruses ranged between 12 and 25%. The type isolates, tentatively named Herbert virus (HEBV), Taï virus (TAIV), and Kibale virus (KIBV), comprised genomes with L, M, and S segments of about 7.4 kb, 2.7 kb, and 1.1 kb, respectively. HEBV, TAIV, and KIBV encode the shortest bunyavirus M segments known and did not seem to encode NSs and NSm proteins but contained an elongated L segment with an ∼500-nucleotide (nt) insertion that shows no identity to other bunyaviruses. The viruses replicated to high titers in insect cells but did not replicate in vertebrate cells. The enveloped virions were 90 to 110 nm in diameter and budded at cellular membranes with morphological features typical of the Golgi complex. Viral RNA recovered from infected cells showed 5′-terminal nontemplated sequences of 9 to 22 nt, suggestive of cap snatching during mRNA synthesis, as described for other bunyaviruses. Northern blotting identified RNA species of full and reduced lengths, suggested upon analogy with other bunyaviruses to constitute antigenomic-sense cRNA and transcript mRNAs, respectively. Functional studies will be necessary to determine if this group of viruses constitutes a novel genus in the bunyavirus family.
INTRODUCTION
The family Bunyaviridae is among the largest and most diversified families of RNA viruses, comprising more than 350 serologically distinct viruses (1). Ninety-six viruses have been formally classified as distinct species by the International Committee on Taxonomy of Viruses (ICTV), and full genome sequences are yet to be determined for the majority of isolates (1). The family comprises five genera whose members can cause pathogenic infections in vertebrates (genera Hantavirus, Nairovirus, Orthobunyavirus, and Phlebovirus) and plants (genus Tospovirus). Several bunyaviruses are considered emerging and reemerging pathogens due to their recent invasion of new habitats and increasing incidence in humans or livestock, such as Crimean-Congo hemorrhagic fever virus (CCHFV), Rift Valley fever virus (RVFV), Sin Nombre virus (SNV), severe fever with thrombocytopenia syndrome virus, and Schmallenberg virus (SBV) (2–7). Orthobunyaviruses, phleboviruses, and nairoviruses are transmitted to their vertebrate hosts by mosquitoes, midges, phlebotomine sandflies, and ticks. The genus Hantavirus is unique in that its members have no arthropod vectors but are transmitted by aerosolized rodent excreta (8).
Bunyaviruses share general features such as their overall virion morphology or their ability to replicate in the cytoplasm and bud into the Golgi cisternae (9–13). Criteria to classify bunyaviruses into genera can be derived from more specific properties such as genome organization, coding strategies, as well as phylogenetic relationships (1). Members of each genus are further subdivided by serology into serogroups and antigenic complexes. Phylogenetic relationships are generally in good agreement with antigenic classification, justifying the use of sequence information as the major criterion for classification of bunyavirus genera (1). Branching inconsistencies within genera have become evident by comparing phylogenetic relationships based on different genes, revealing a potential for bunyaviruses to undergo intrageneric genome segment reassortment (14–16).
The enveloped, spherical bunyavirus virions are ca. 100 nm in diameter and contain segmented, single-stranded, negative-sense RNA genomes implementing negative-sense or ambisense coding strategies (17). The small (S) segment encodes the nucleocapsid (N) protein. The medium (M) segment codes two glycoproteins (Gn and Gc), and the large (L) segment encodes the RNA-dependent RNA polymerase (RdRp). The S and M segments of the genera Orthobunyavirus, Phlebovirus, and Tospovirus encode two additional nonstructural proteins, NSs and NSm, respectively. Orthobunyaviruses encode their N and NSs proteins in overlapping open reading frames (ORFs) translated from one same mRNA that is complementary to the corresponding virion RNA segment (18). Phleboviruses and tospoviruses use an ambisense coding strategy and translate their NSs from a subgenomic mRNA (sg mRNA), which has the same polarity as the virion-sense RNA (vRNA) (19). Recently, it was shown that some hantaviruses also code for an NSs protein in an ORF overlapping the N ORF, with expression enabled by ribosomal leaky scanning (20, 21). Interestingly, accessory proteins are not consistently represented throughout genera, as M segments of tick-transmitted phleboviruses do not encode NSm proteins (7, 22, 23), and viruses in the Anopheles A, Anopheles B, and Tete virus serogroups within the genus Orthobunyavirus do not encode NSs proteins (24). Bunyavirus NSs proteins either inhibit the cellular interferon response in their vertebrate hosts or suppress the RNA interference (RNAi) mechanism in their plant hosts (25–27). Nairoviruses are special regarding their strategy to counteract the antiviral host response, as they code for an ovarian tumor (OTU) domain within their L protein that has been suggested to suppress the host cell inflammatory and antiviral response and thus plays a role as a pathogenicity factor (28–30).
Bunyaviruses are distributed worldwide but appear to have higher diversity and prevalence in tropical and subtropical regions (17). Investigations of bunyaviruses in such regions can yield novel insights into phylogeny and diversity. For instance, Gouléako virus (GOLV) (previously GOUV; the abbreviation was changed as GOUV was already used for Gou virus, a hantavirus isolated from Rattus rattus in China) (31), recently discovered in mosquitoes, is almost equidistant phylogenetically to the five established genera but closest to the genus Phlebovirus (32). Gouléako virus appears to be restricted to arthropod hosts, while all other known phleboviruses can also infect specific vertebrate hosts, suggesting that Gouléako virus represents a new taxonomic entity, potentially a new genus (32).
During a pilot study on mosquito-associated viruses in Côte d'Ivoire, a short reverse transcriptase PCR (RT-PCR) fragment of a putative RdRp gene with a distant relationship to bunyaviruses was encountered (33). The virus was tentatively named Herbert virus (HEBV) (strain F23/CI/2004). Here we provide a full characterization of the virus isolated in cell culture as well as related viruses isolated from mosquitoes in Côte d'Ivoire, Ghana, and Uganda.
MATERIALS AND METHODS
Mosquito collection and species identification.
Mosquitoes were trapped from February to June 2004 in Taï National Park, Côte d'Ivoire (33) and from February to June 2008 in Kibale National Park, Uganda. Habitat types included primary and secondary tropical forests, agricultural plantations, villages, and research camps within primary rainforests. Furthermore, mosquitoes were collected at the botanical garden and at the residential area at the Kwame Nkrumah University of Science and Technology (KNUST) in Kumasi, Ghana. Mosquitoes were trapped with CDC miniature light and gravid traps (John W. Hock Company, USA) and with BG sentinel traps (Biogents, Regensburg, Germany). Traps were baited with octenol, worn socks, Limburger cheese, or simple syrup (1 liter of water mixed with 100 g sugar). Species were identified by morphological criteria (34–37).
Virus isolation, purification, and growth.
Virus isolation from mosquitoes collected in Côte d'Ivoire was done with C6/36 (derived from Aedes albopictus larvae) (38) and Vero E6 (Cercopithecus aethiops kidney) cells as described previously (33, 39). Female mosquitoes from Uganda and Ghana were homogenized individually in 500 μl of L-15 medium without additives by using 3 to 5 ceramic beads and a TissueLyser instrument (Qiagen, Hilden, Germany). Trapped male mosquitoes were pooled (1 to 20 specimens) according to trapping location and genus and homogenized in 1 ml of L-15 medium. Suspensions were cleared from debris by centrifugation at 2,500 rpm for 10 min at 4°C. Pools of female mosquitoes were generated by using 100 μl of supernatant of 10 homogenized mosquito suspensions and used for virus isolation as described previously (39). Virus stocks of the fourth passage of HEBV (isolate C60/CI/2004) and Kibale virus (KIBV) (isolate P07/UG/2008) were generated. Virus titers were determined by 50% tissue culture infective dose (TCID50) titration, and virus-positive wells were identified by real-time PCR. For virus growth kinetics, C6/36 and U4.4 (derived from A. albopictus larvae [40]) cells were infected at multiplicities of infection (MOIs) of 0.1 and 0.01 in duplicate, respectively, as described previously (41). Aliquots of infectious cell culture supernatants were harvested every 24 h for periods of 5 days, and viral genome copy numbers were quantified by real-time RT-PCR (HEBV-F [5′-AGAATGCTTTGTCAGTGG], HEBV-R [5′-AGCAGCAACTTATAAAACAAATC], HEBV-TM [5′–6-carboxyfluorescein {FAM}–TTCTCCGCTAATAAAA–MGB], KIBV-F [5′-TAATTTGAATGGTGAGCCTTTTTCT], KIBV-R [5′-GCTGTCTGAATACCGGATAATCTTG], and KIBV-TM [5′-FAM-ATTCCCTGTCATTGGAGCTTGCTCTTTCTT-TQ2]).
Infection of vertebrate cells.
Green monkey kidney cells (Vero E6), baby hamster kidney cells (BHK-J), mouse embryo fibroblasts (MEFs) from BALB/c MDA5−/− mice, MEFs from BALB/c RIG-I−/− mice, mouse fibroblasts (L929), and porcine stable equine kidney (PSEK) cells were infected with HEBV (fourth passage of isolate F23/CI/2004) at MOIs of 10, 1, 0.5, and 0.1 and incubated at 33°C and 37°C. Cell culture supernatants were passaged in fresh cells every 7 days in 1/10 dilutions for five consecutive passages. Supernatants from identical cell culture types infected at different MOIs were pooled, and all passages were subjected to screening by real-time RT-PCR.
RT-PCR screening.
RNA was extracted from homogenized female and male mosquito pools or from individually homogenized female mosquitoes using 140 μl of the supernatant and a viral RNA kit (Qiagen, Hilden, Germany), and cDNA was synthesized by using SuperScriptII according to the manufacturer's instructions (Invitrogen, Karlsruhe, Germany). Pools were screened by real-time RT-PCR or by nested RT-PCR using primer pair HEBV-F1 (5′-ATGCTGAYATGTCIAAGTGGTSTGC) and HEBV R1 (5′-TGATTGTCATCGSTRTGIACYA) for the first round and primer pair HEBV-F2 (5′-ATGCTGAYATGTCIAAGTGGTSTGC) and HEBV-R2 (5′-TCAARTTVCCTTGGAKCCART) for nested PCR.
Electron microscopy.
For electron microscopy (EM) analyses, viral particles were purified through a 36% sucrose cushion, and the pellet was resuspended in phosphate-buffered saline (PBS) (39, 42). Viral particles were fixed with 2% paraformaldehyde and analyzed by transmission electron microscopy after staining with 1% uranyl acetate (43, 44). For ultrathin sections, infected cells were fixed with 2.5% glutaraldehyde, enclosed in low-melting agar, embedded in resin, and evaluated by transmission EM after ultrathin sectioning (39).
Genome sequencing.
Viral genome fragments from infectious cell culture supernatants of HEBV were generated by random-primed RT-PCR optimized for the detection of encapsidated nucleic acids (so-called “particle-associated nucleic acid PCR” [32, 39]). Briefly, RNA was extracted from ultracentrifuged virus pellets by using the viral RNA kit (Qiagen, Hilden, Germany), and double-strand cDNA was synthesized with random hexamers linked to a defined primer sequence tail by using a double-strand cDNA kit (Promega, Madison, WI, USA). Amplification was performed by using oligonucleotides that bound to the sequence tail and were cloned into the pCR2.1 TOPO vector (Invitrogen, Karlsruhe, Germany). Colonies were analyzed by PCR, and inserts of >500 nucleotides (nt) were sequenced by using dye terminator chemistry (Applied Biosystems, Darmstadt, Germany). Primer sequences were trimmed, and sequences were assembled by using Geneious 6 (106). Consensus sequences were compared at the nucleotide and translated amino acid levels to the GenBank database by applying BLASTn and BLASTx algorithms (http://www.ncbi.nlm.nih.gov/genbank). Fragment-specific primers and generic orthobunyavirus oligonucleotides were used for amplification of sequence gaps. The 3′ and 5′ genome termini were confirmed by rapid amplification of cDNA ends-PCR (RACE-PCR) (Roche, Mannheim, Germany). The complete genome was resequenced for confirmation on both strands by long-range PCR and primer walking techniques. Full-genome sequencing of KIBV was performed by using fragment-specific primers and primers based on the HEBV genome. Full genome sequences of HEBV isolates F33/CI/2004, F45/CI/2005, and F53/CI/2004, as well as that of Taï virus (TAIV) isolate F47/CI/2004, were generated by deep sequencing on the 454 Junior (Roche) and Ion Torrent (Invitrogen) platforms in Bonn, Germany. Reads were identified by reference mapping to HEBV F23/CI/2004 as well as by BLAST comparisons against a local amino acid sequence library containing translations of ORFs detected in the HEBV and KIBV genomes.
Genome and phylogenetic analyses.
Nucleotide and amino acid sequences were compared with other sequences by BLASTn and BLASTx against the GenBank database (http://www.ncbi.nlm.nih.gov/genbank), and protein motifs were identified by a Web-based comparison to the Pfam database (http://pfam.janelia.org/). Identification of cleavage sites of the signal peptide was accomplished by using signalP-NN (http://www.cbs.dtu.dk/services/SignalP/). Prediction of the hydropathy profile was performed with TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/), and N-linked glycosylation sites were identified by using the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/). For phylogenetic analyses, amino acid sequences of the N, Gn, Gc, and RdRp genes were aligned with representative sequences of other bunyaviruses in Geneious by using MAFFT (45). Phylogenetic analyses were conducted by using the maximum likelihood (ML) algorithm with the BLOSUM62 substitution matrix assuming no systematic rate variation across alignment sites, with confidence testing based on 1,000 bootstrap iterations in PhyML (46). Sequence alignments used for phylogenies, including all bunyavirus genera, were 587 amino acids (aa), 140 aa, 622 aa, and 364 aa in length for the N, Gn, Gc, and RdRp proteins, respectively, from which the least conserved columns were removed before analysis. Phylogenetic analyses including HEBV, TAIV, KIBV, all available orthobunyavirus, and tospovirus sequences were based on 3,228 aa, 485 aa, 520 aa, and 331 aa for the RdRp, Gn, Gc, and N proteins, respectively.
mRNA analyses.
C6/36 cells infected with HEBV and KIBV were harvested at 24 h postinfection (hpi). RNA was extracted by using an RNA extraction kit (Qiagen, Hilden) and analyzed by 5′ RACE (Invitrogen, Karlsruhe, Germany) or by Northern blotting as described previously (41, 47). Digoxigenin (DIG)-labeled probes for HEBV and KIBV were generated by using primer pairs HEBV-N-F (5′-TCATCTTATACAGGAGTTCAAAGAAGCGC) and HEBV-N-R (5′-ACATGACTAAACAAGTGTGAGCCTGG), KIBV-N-F (5′-TGGCTTTAAATGGGACCCGGC) and KIBV-N-R (5′-GCTAAACAAGTGAGCACCTGGGG), and KIBV-X1 (5′-CAAGAAGGGCATTGATCTGGTTGTC) and KIBV-X2 (5′-GCACAGGCACACATCCCCTG).
Protein analyses.
Proteins were analyzed as described previously (47). Briefly, viral particles were purified by gradient ultracentrifugation on a continuous gradient of 1 to 2 M sucrose in 0.01 M Tris-HCl–4 mM Na-EDTA at 35,000 rpm (SW40 rotor; Beckman) for 22 h at 4°C. Fractions (0.4 ml each) were tested by real-time PCR, and two fractions with the largest amounts of genome copies were concentrated through a 36% sucrose cushion at 35,000 rpm (SW40 rotor; Beckman) for 2 h at 4°C. The virus pellet was resuspended in 150 μl PBS overnight at 4°C. Proteins were lysed in 4× NuPage LDS sample buffer at 70°C for 10 min and separated by SDS-PAGE on a NuPage Novex 4 to 12% Bis-Tris gel with NuPage MES SDS running buffer (Invitrogen, Darmstadt, Germany). Bands were analyzed by limited tryptic digestion and mass spectrometry using a matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer. RdRp and Gc proteins were additionally analyzed by liquid chromatography mass spectrometry (LC-MS).
Nucleotide sequence accession numbers.
The complete genome sequences of HEBV, TAIV, and KIBV were assigned GenBank accession numbers JQ659256 to JQ659258 and KF590572 to KF590586. Further sequence fragments from HEBV, TAIV, and KIBV strains of over 200 nt were assigned to GenBank accession numbers KF590587 to KF590623.
RESULTS
Detection of a novel clade of mosquito-associated bunyaviruses.
In order to investigate the distribution of HEBV and to detect related viruses, we tested pooled female mosquitoes collected in Taï National Park, Côte d'Ivoire (432 pools consisting of 4,839 mosquitoes); Kibale National Park, Uganda (81 pools consisting of 807 mosquitoes); and Kumasi, Ghana (62 pools consisting of 1,230 mosquitoes) by RT-PCR. HEBV was detected in 39 mosquito pools originating from Côte d'Ivoire and in 6 mosquito pools originating from Ghana, showing nucleotide distances of 94.6 to 98.3% and 94.9 to 99.2% to HEBV (strain F23/CI/2004) within their RdRp genes, respectively (Table 1). Individual mosquitoes from positive pools originating from Ghana were tested for infection with HEBV, resulting in a prevalence of 1.1% (14/1,230). Mosquitoes from positive pools from Côte d'Ivoire could not be tested individually, as in this case, mosquito pools had been homogenized, and no individual mosquitoes were available. Two further distinct viruses with a distant relationship at the nucleotide level to HEBV (72.6 to 72.9%) were obtained from two pools originating from Côte d'Ivoire and from two pools originating from Uganda. At the amino acid level, these viruses had distant relationships to orthobunyaviruses of the Simbu serogroup according to initial BLAST comparisons. The viruses were tentatively named Taï virus (TAIV) and Kibale virus (KIBV). Testing of individual mosquitoes from positive pools from Uganda indicated a prevalence of 0.4% (3/807). Mosquito species and sampling locations are summarized in Table 1.
Table 1.
Mosquito species infected with HEBV, TAIV, or KIBV
| Virus and straina | Mosquito speciesb | No. of mosquitoes | Sampling site | % pairwise identity to HEBV F23/CI/2004 |
|---|---|---|---|---|
| HEBV | 94.8 | |||
| A11/CI/2004 | Culex (Eumelanomyia) spp. | 20 | Camp | |
| A18/CI/2004 | Anopheles spp. | 1 | Camp | 96.3 |
| A26/CI/2004 | C. nebulosus | 10 | Camp | 95.4 |
| A27/CI/2004 | ND | 1 | Camp | 95.2 |
| A28/CI/2004 | C. nebulosus | 22 | Camp | 95.7 |
| A30/CI/2004 | Uranotaenia mashonaensis | 6 | Camp | 95.8 |
| A45/CI/2004 | Culex telesilla | 11 | Camp | 95.8 |
| A52/CI/2004 | ND | 8 | Camp | 96.3 |
| A57/CI/2004 | Culex spp. | 10 | Camp | 96.4 |
| B40/CI/2004 | ND | 2 | Primary forest | 96.1 |
| B42/CI/2004 | Culex spp. | 9 | Primary forest | 95.9 |
| C40/CI/2004 | U. mashonaensis | 20 | Secondary forest | 95.2 |
| C43/CI/2004 | C. nebulosus | 17 | Secondary forest | 96.2 |
| C45/CI/2004 | C. nebulosus | 16 | Secondary forest | 95.8 |
| C57/CI/2004 | Culex decens | 20 | Secondary forest | 95.9 |
| C59/CI/2004 | C. decens | 20 | Secondary forest | 97.1 |
| C60/CI/2004 | C. decens | 9 | Secondary forest | 97.1 |
| C68/CI/2004 | Culex spp. | 21 | Secondary forest | 96.2 |
| C88/CI/2004 | ND | 20 | Secondary forest | 96.3 |
| D24/CI/2004 | Culex spp. | 23 | Plantation | 95.7 |
| D28/CI/2004 | Anopheles spp. | 2 | Plantation | 95.4 |
| D50/CI/2004 | C. nebulosus | 20 | Plantation | 96.5 |
| D60/CI/2004 | ND | 15 | Plantation | 98.3 |
| D61/CI/2004 | ND | 11 | Plantation | 94.6 |
| D62/CI/2004 | Culex spp. | 14 | Plantation | 96.2 |
| F23/CI/2004 | C. nebulosus | 20 | Village | |
| F25/CI/2004 | C. nebulosus | 21 | Village | 95.8 |
| F26/CI/2004 | C. nebulosus | 50 | Village | 95.1 |
| F27/CI/2004 | C. nebulosus | 40 | Village | 96.7 |
| F28/CI/2004 | C. nebulosus | 20 | Village | 96.1 |
| F30/CI/2004 | C. nebulosus | 20 | Village | 96.5 |
| F32/CI/2004 | C. nebulosus | 15 | Village | 96.2 |
| F33/CI/2004 | C. nebulosus | 12 | Village | 96.1 |
| F43/CI/2004 | Culex spp. | 1 | Village | 96.2 |
| F45/CI/2004 | Culex spp. | 26 | Village | 95.8 |
| F47/CI/2004 | Culicidae spp. | 10 | Village | 95.7 |
| F53/CI/2004 | C. quinquefasciatus | 8 | Village | 96.1 |
| F54/CI/2004 | Culex antenatus | 20 | Village | 96.3 |
| F55/CI/2004 | C. antenatus | 9 | Village | 96.1 |
| M257/P13/GH/2011 | C. quinquefasciatus | 1 | Residential area | 95.4 |
| M538/P27/GH/2011 | C. nebulosus | 1 | Botanical garden | 96.7 |
| M540/P27/GH/2011 | C. nebulosus | 1 | Botanical garden | 95.9 |
| M566/P29/GH/2011 | C. nebulosus | 1 | Botanical garden | 100 |
| M569/P29/GH/2011 | C. nebulosus | 1 | Residential area | 95.9 |
| M572/P29/GH/2011 | C. nebulosus | 1 | Residential area | 96.3 |
| M105/P06/GH/2011 | Culex pipiens | 1 | Residential area | 96.6 |
| M120/P06/GH/2011 | C. pipiens | 1 | Residential area | 96.6 |
| M201/P11/GH/2011 | C. quinquefasciatus | 1 | Residential area | 95.4 |
| M206/P11/GH/2011 | C. quinquefasciatus | 1 | Residential area | 97.1 |
| M211/P11/GH/2011 | C. quinquefasciatus | 1 | Botanical garden | 96.2 |
| M213/P11/GH/2011 | C. quinquefasciatus | 1 | Residential area | 97 |
| M219/P11/GH/2011 | C. quinquefasciatus | 1 | Residential area | 97.1 |
| M858/P43/GH/2011 | C. nebulosus | 1 | Botanical garden | 94.9 |
| TAIV | ||||
| C48/CI/2004 | C. nebulosus | ND | Secondary forest | 75.8 |
| F47/CI/2004 | Culicidae spp. | 10 | Village | 76.1 |
| KIBV | ||||
| M15/P05/UG/2008 | C. simpliciforceps | 1 | Forest edge | 72.7 |
| M22/P05/UG/2008 | C. simpliciforceps | 1 | Forest edge | 72.4 |
| M202/P07/UG/2008 | Culex spp. | 1 | Tea plantation | 72.4 |
P, Pool; M, mosquito; CI, Côte d'Ivoire; GH, Ghana; UG, Uganda.
ND, not determined.
Virus isolation, growth, and morphology.
HEBV was successfully isolated from 28 pools of mosquitoes in C6/36 cells. TAIV and KIBV were each isolated from two different mosquito pools, respectively. RT-PCR studies showed that both TAIV-containing cell cultures were coinfected with mesoniviruses (47), and these could not be removed from cell cultures by repeated rounds of endpoint purification. As plaque purification was not possible due to the absence of cytopathic effects (CPE) (see below), TAIV supernatants were not further purified for the purposes of this study, and growth curve studies were done only for HEBV and KIBV, for which pure supernatants were available.
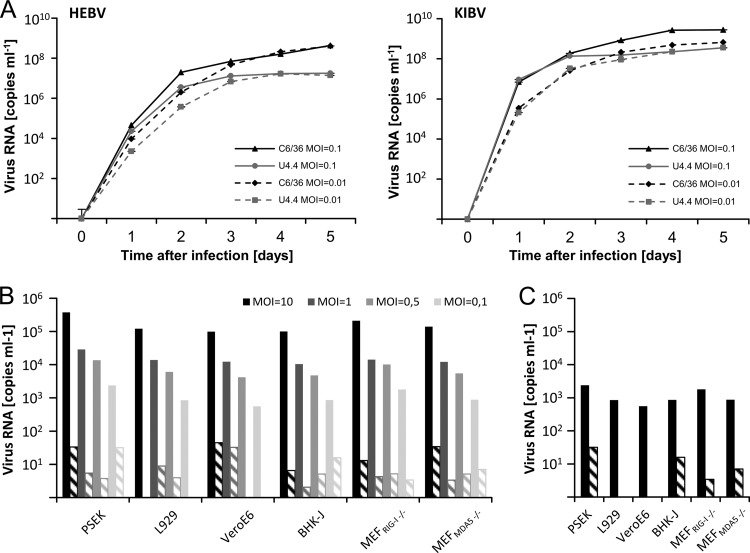
HEBV (isolate C60/CI/2004) and KIBV (isolate P07/UG/2008) reached titers of 3.2 × 109 TCID50/ml and 3.2 × 107 TCID50/ml in infected C6/36 cells, respectively. Growths of HEBV and KIBV in C6/36 and U4.4 cells were compared (Fig. 1A). For both viruses, a 10- to 100-fold-higher level of replication in C6/36 cells than in U4.4 cells was observed by 2 to 3 days postinfection (dpi). Notably, no CPE was observed for both viruses in U4.4 cells, and only weak changes in morphology were detected in C6/36 cells.
Fig 1.
Growth of HEBV and KIBV. (A) C6/36 and U4.4 cells were infected with HEBV and KIBV at MOIs of 0.1 and 0.01, respectively. The genome copy numbers per milliliter in cell culture supernatants were measured by RT-PCR for 5 days. (B) Vertebrate cells were infected with HEBV at the indicated MOIs, and five blind passages at 37°C were performed. The genome copy numbers per milliliter in cell culture supernatants were measured by RT-PCR at 7 days postinfection (solid bars) and at the fifth passage (dashed bars). (C) Cells were infected and passaged as described above for panel B but were incubated at 33°C. Supernatants of the same cell lines infected at different MOIs were pooled, and genome copy numbers were measured by RT-PCR.
In order to get insight in the putative host tropism, growth of HEBV (isolate F23/CI/2004) was investigated using six different vertebrate cell lines. No CPE was observed, and no virus replication was measured by real-time RT-PCR over five blind passages in any of these vertebrate cells (Fig. 1B and C). Additionally, KIBV was inoculated at an MOI of 10 in Vero cells. No virus replication was detected by 7 dpi by real-time RT-PCR.
In order to assess the potential for transovarial or transvenereal transmission, we further tested 269 pools of 1,716 male mosquitoes trapped during the survey in Côte d'Ivoire, 39 pools of 386 male mosquitoes trapped in Ghana, and 11 male mosquitoes trapped in Uganda for infection with HEBV, TAIV, or KIBV. No virus was detected by RT-PCR in any of the male mosquitoes.
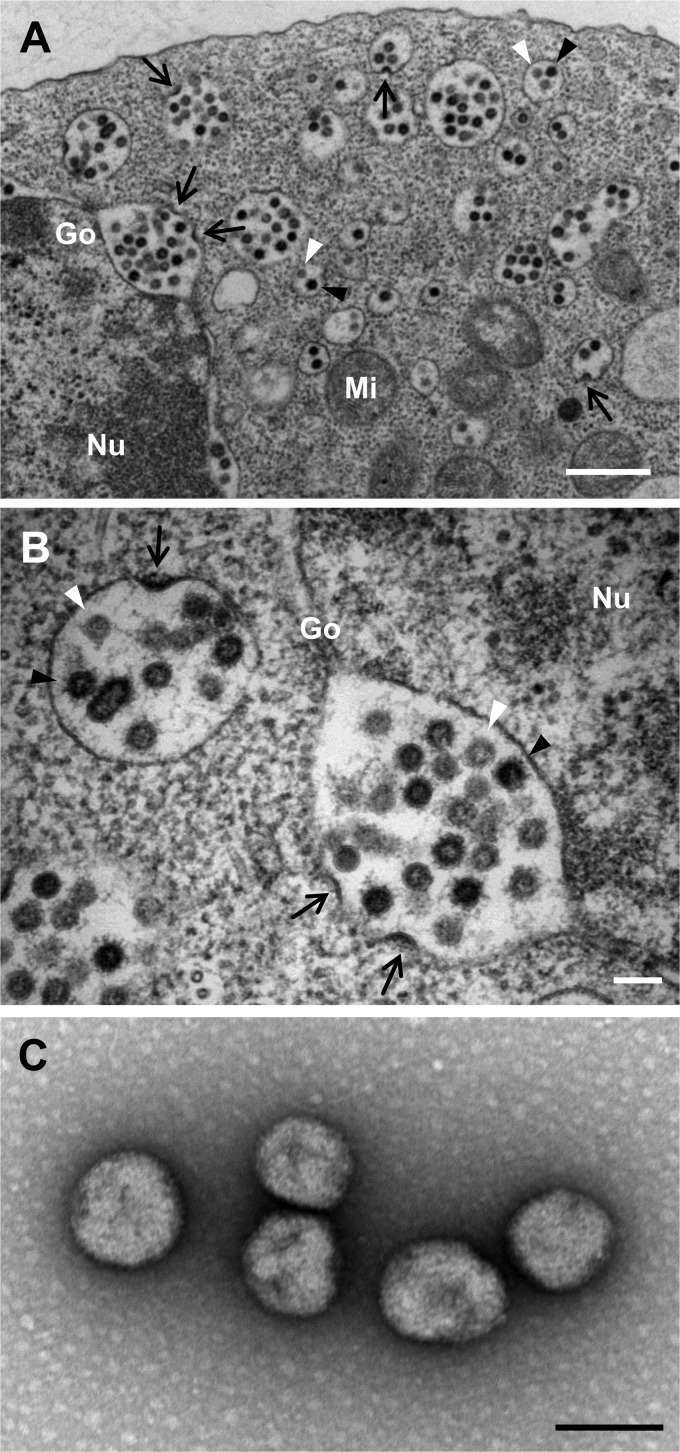
Virus morphology during maturation was studied in ultrathin sections of C6/36 cells infected with HEBV (isolate F23/CI/2004). Two types of spherical viral particles 50 to 60 nm in diameter, of high and low electron densities, respectively, were observed in structures resembling Golgi vesicles (Fig. 2A and B). These were termed intracellular annular viruses (IAV) and intracellular dense viruses (IDV), in agreement with terminology used in studies on Bunyamwera virus (13). Budding or maturation of viral particles at the Golgi membrane was observed in Golgi vesicles filled with IAV and IDV (Fig. 2A and B). Mature spherical, enveloped virions of about 90 to 110 nm in diameter were detected in virus pellets generated by ultracentrifugation of cell culture supernatants infected with HEBV (Fig. 2C).
Fig 2.
Maturation and morphology of HEBV. Shown are ultrathin sections of C6/36 cells infected with HEBV (A and B) and negative-stained ultracentrifuged virions of HEBV (C). Budding arcs are indicated by black arrows, annular spherical particles are indicated by white arrowheads, and dense spherical particles are indicated by black arrowheads. Abbreviations: Nu, nucleus; Mi, mitochondria; Go, Golgi apparatus. Bars = 500 nm (A) and 100 nm (B and C).
Genome sequencing and phylogenetic analyses.
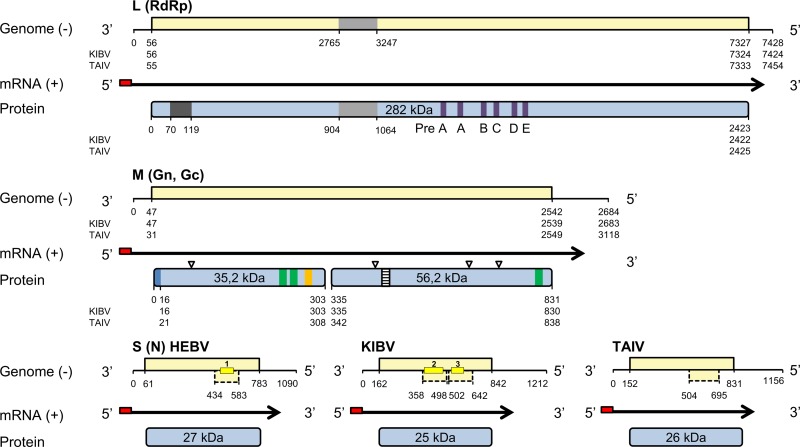
The entire genomes of four different HEBV isolates (isolates F23/CI/2004, F33/CI/2004, F45/CI/2004, and F53/CI/2004), one TAIV isolate (isolate F47/CI/2004), and one KIBV isolate (isolate P05/UG/2008) were sequenced. All genomes were found to comprise three segments (Fig. 3). Seven reverse-complementary terminal nucleotides were found to be conserved between HEBV, TAIV, and KIBV (Table 2). These were identical to terminal sequences in members of the genus Orthobunyavirus, where, however, these conserved sequences are 10 nt in length. The three genomes differed in the lengths of their untranslated regions (UTRs) of S and M segments (Fig. 3). Pairwise nucleotide identities among all HEBV genomes ranged between 96.1 and 99.7%. Nucleotide and amino acid identities of S, M, and L segment ORFs of HEBV, TAIV, and KIBV were >61% (Table 3).
Fig 3.
Schematic view of the genome organization of HEBV, TAIV, and KIBV. Open reading frames are shown as light yellow boxes, mRNAs are indicated by black arrows, and nontemplate sequences at the 5′ terminus are symbolized by red boxes. Predicted proteins are shown as light blue boxes. Northern blot probes are shown as dark yellow boxes, putative transmembrane domains (hydrophobic regions) are marked by green boxes, glycosylation sites are marked by triangles, the unique region in the RdRp gene is indicated by a light gray box, the endonuclease domain is indicated by a dark gray box, the putative signal peptide is indicated by a blue box, the Gn zinc finger motif is indicated by an orange box, and the Gc fusion peptide is indicated by a dashed box. Genome positions and predicted molecular protein masses are indicated.
Table 2.
Genome size and consensus terminal nucleotides of HEBV, TAIV, and KIBV compared to established genera of the family Bunyaviridae
| Genus and virus | Consensus terminal nucleotidesa | Genome size (nt) | Segment size (nt) (GenBank accession no.) |
||
|---|---|---|---|---|---|
| S | M | L | |||
| Hantavirus | |||||
| Hantaan virus | 3′-AUCAUCAUCUG | 11,845 | 1,696 (M14626) | 3,616 (M14627) | 6,533 (X55901) |
| 5′-UAGUAGUAUGC | |||||
| Nairovirus | |||||
| Dugbe virus | 3′-AGAGUUUCU | 18,855 | 1,712 (M25150) | 4,888 (M94133) | 12,255 (U15018) |
| 5′-UCUCAAAGA | |||||
| Tospovirus | |||||
| Tomato spotted wilt virus | 3′-UCUCGUUA | 16,634 | 2,916 (D00645) | 4,821 (S48091) | 8,897 (D10066) |
| 5′-AGAGCAAU | |||||
| Phlebovirus | |||||
| Rift Valley fever virus | 3′-UGUGUUUC | 11,979 | 1,690 (X53771) | 3,885 (M11157) | 6,404 (X56464) |
| 5′-ACACAAAG | |||||
| Unassigned | |||||
| Gouléako virus | 3′-UGUGU | 10,633 | 1,087 (HQ541736) | 3,188 (HQ541737) | 6,358 (HQ541738) |
| 5′-ACACA | |||||
| Orthobunyavirus | |||||
| Bunyamwera virus | 3′-UCAUCACAUG | 12,294 | 961 (D00353) | 4,458 (M11852) | 6,875 (X14383) |
| 5′-AGUAGUGUGC | |||||
| Unassigned | |||||
| Herbert virus | |||||
| S | 3′-UCAUCACACG | 11,202 | 1,090 | 2,684 | 7,428 |
| 5′-AGUAGUGCAC | |||||
| M | 3′-UCAUCACACG | ||||
| 5′-AGUAGUGCAC | |||||
| L | 3′-UCAUCACACG | ||||
| 5′-AGUAGUGUGC | |||||
| Kibale virus | |||||
| S | 3′-UCAUCACACG | 11,322 | 1,212 | 2,683 | 7,427 |
| 5′-AGUAGUGCAC | |||||
| M | 3′-UCAUCACACG | ||||
| 5′-AGUAGUGCAC | |||||
| L | 3′-UCAUCACACG | ||||
| 5′-AGUAGUGCAC | |||||
| Tai virus | |||||
| S | 3′-UCAUCACGUG | 11,728 | 1,156 | 3,118 | 7,454 |
| 5′-AGUAGUGCAC | |||||
| M | 3′-UCAUCACGUG | ||||
| 5′-AGUAGUGCAC | |||||
| L | 3′-UCAUCACGUG | ||||
| 5′-AGUAGUGUGC | |||||
Boldface type indicates conserved terminal nucleotides.
Table 3.
Nucleotide and amino acid pairwise sequence identity values for HEBV, TAIV, KIBV, and OROV as well as pairs of the most distantly related orthobunyaviruses and tospovirusesa
| Gene | Virus | % nucleotide or amino acid sequence identity |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| RdRp | HEBV | TAIV | KIBV | OROV | SIMV | SORV | TZSV | BeNMV | |
| HEBV | 73.9 | 73.7 | 37.8 | 38.9 | 39.0 | 28.4 | 28.0 | ||
| TAIV | 79.2 | 72.1 | 37.4 | 38.5 | 38.8 | 27.7 | 27.8 | ||
| KIBV | 80.0 | 78.5 | 36.8 | 38.0 | 38.1 | 27.9 | 27.5 | ||
| OROV | 24.7 | 24.6 | 24.8 | 60.8 | 56.2 | 27.8 | 27.4 | ||
| SIMV | 24.7 | 24.6 | 24.6 | 58.2 | 55.7 | 27.3 | 27.3 | ||
| SORV | 24.3 | 24.3 | 24.2 | 49.1 | 47.2 | 27.2 | 27.4 | ||
| TZSV | 14.1 | 14.0 | 13.5 | 13.6 | 13.1 | 13.3 | 52.6 | ||
| BeNMV | 13.5 | 14.0 | 13.0 | 14.2 | 12.6 | 12.4 | 41.4 | ||
| GPC | HEBV | TAIV | KIBV | OROV | AKAV | TAHV | MYSV | BeNMV | |
| HEBV | 70.0 | 69.6 | 21.9 | 22.2 | 21.8 | 20.4 | 20.7 | ||
| TAIV | 70.4 | 68.4 | 21.5 | 22.5 | 21.8 | 20.1 | 20.5 | ||
| KIBV | 69.6 | 67.2 | 21.3 | 22.3 | 21.4 | 20.2 | 20.5 | ||
| OROV | 12.3 | 12.6 | 12.0 | 43.2 | 42.6 | 26.5 | 26.5 | ||
| AKAV | 12.0 | 11.5 | 11.7 | 24.7 | 45.0 | 23.6 | 24.3 | ||
| TAHV | 10.7 | 11.4 | 11.8 | 31.8 | 29.3 | 23.1 | 22.5 | ||
| MYSV | 11.6 | 12.5 | 12.4 | 9.8 | 10.9 | 10.3 | 39.3 | ||
| BeNMV | 12.4 | 12.2 | 12.6 | 9.7 | 10.8 | 8.9 | 32.6 | ||
| N | HEBV | TAIV | KIBV | OROV | BMAV | BORV | TZSV | INSV | |
| HEBV | 65.3 | 69.3 | 31.0 | 30.6 | 33.5 | 25.1 | 25.4 | ||
| TAIV | 66.2 | 64.0 | 29.9 | 31.3 | 33.8 | 25.3 | 25.6 | ||
| KIBV | 72.6 | 60.9 | 32.7 | 31.7 | 33.7 | 25.5 | 25.4 | ||
| OROV | 19.8 | 20.2 | 20.2 | 38.2 | 37.3 | 24.4 | 27.1 | ||
| BMAV | 16.5 | 17.4 | 15.9 | 32.1 | 39.7 | 26.9 | 24.3 | ||
| BORV | 17.0 | 16.7 | 18.3 | 31.5 | 24.8 | 27.2 | 25.6 | ||
| TZSV | 10.8 | 11.5 | 12.2 | 10.6 | 9.5 | 11.4 | 24.7 | ||
| INSV | 13.2 | 12.0 | 12.7 | 14.6 | 11.7 | 11.5 | 24.9 | ||
Top right values for each gene indicate nucleotide sequence identity; bottom left values indicate amino acid identity. AKAV, Akabane virus; BeNMV, bean necrotic mosaic virus; BMAV, Batama virus; BORV, Boraceia virus; INSV, impatiens necrotic spot virus; MYSV, melon yellow spot virus; OROV, Oropouche virus; SIMV, Simbu virus; SORV, Sororoca virus; TAHV, Tahyna virus; TZSV, tomato zonate spot virus.
No significant similarity was found between the S, M, and L segment ORFs and ORFs of any other viruses by using nucleotide BLAST. Low but significant levels of identity (ranging from 12 to 25%) with N protein, glycoprotein, and RdRp protein sequences of orthobunyaviruses (the most closely related virus was Oropouche virus) were identified by BLASTx using the deduced amino acid sequences of these ORFs (Table 3).
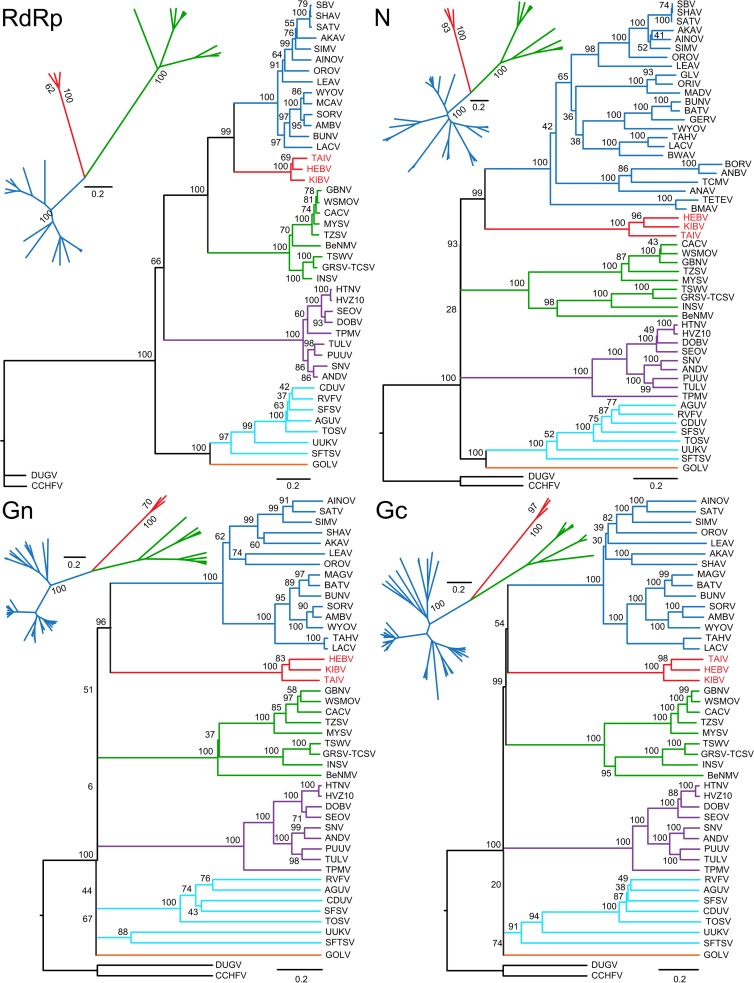
Phylogenetic trees were inferred based on the deduced amino acid sequences of the RdRp, Gn, Gc, and N genes. Analyses of all genes, including representative sequences of established bunyavirus genera, yielded congruent topologies. HEBV, TAIV, and KIBV formed a novel independent monophyletic clade that shared the most recent common ancestor (MRCA) with the genus Orthobunyavirus in all genes (Fig. 4). HEBV, TAIV, and KIBV sequences were almost equidistant to all members of the genera Orthobunyavirus and Tospovirus.
Fig 4.
Phylogenetic relationships of HEBV, TAIV, and KIBV to representative members of the family Bunyaviridae. Phylogenies were investigated for the RdRp, Gn, Gc, and N proteins based on sizes of 364 aa, 140 aa, 622 aa, and 587 aa, respectively. Maximum likelihood (ML) analyses were performed on a gap-free alignment guided by the BLOSUM62 substitution matrix and using MAFFT (E-INS-I algorithm). Confidence testing was performed by 1,000 bootstrap replicates. Bars indicate evolutionary substitutions per position in the alignments. Smaller pictograms represent ML analyses of HEBV, TAIV, KIBV, all available orthobunyavirus, and tospovirus sequences based on sizes of 3,228 aa, 485 aa, 520 aa, and 331 aa for the RdRp, Gn, Gc, and N proteins, respectively. Abbreviations (and GenBank accession numbers for L, M, and S segments, respectively, in parentheses) are as follows: AGUV, Aguacate virus (accession numbers NC_015451, NC_015450, and NC_015452); AINOV, Aino virus (accession numbers NC_018465, NC_018459, and NC_018460); AKAV, Akabane virus (accession numbers NC_009894, NC_009895, and NC_009896); AMBV, Anhembi virus (accession numbers JN572062, JN572063, and JN572064); ANDV, Andes virus (accession numbers NC_003468, NC_003467, and NC_003466); BeNMV, bean necrotic mosaic virus (accession numbers NC_018070, NC_018072, and NC_018071); BUNV, Bunyamwera virus (accession numbers NC_001925, NC_001926, and NC_001927); CACV, Capsicum chlorosis virus (accession numbers NC_008302, NC_008303, and NC_008301); CDUV, Candiru virus (accession numbers NC_015374, NC_015373, and NC_015375); DOBV, Dobrava virus (accession numbers NC_005235, NC_005234, and NC_005233); GBNV, groundnut bud necrosis virus (accession numbers NC_003614, NC_003620, and NC_003619); GOLV, Gouléako virus (accession numbers HQ541738, HQ541737, and HQ541736); GRSV-TCSV, groundnut ringspot and tomato chlorotic spot virus reassortant (accession numbers NC_015469, NC_015468, and NC_015467); HEBV, Herbert virus (accession numbers JQ659256, JQ659257, and JQ659258); HTNV, Hantaan virus (accession numbers NC_005222, NC_005219, and NC_005218); HVZ10, Hantavirus Z10 virus (accession numbers NC_006435, NC_006437, and NC_006433); INSV, Impatiens necrotic spot virus (accession numbers NC_003625, NC_003616, and NC_003624); KIBV, Kibale virus (accession numbers KF590577, KF590576, and KF590575); LACV, La Crosse virus (accession numbers NC_004108, NC_004109, and NC_004110); LEAV, Leanyer virus (accession numbers HM627178, HM627176, and HM627177); MCAV, Macaua virus (accession numbers JN572068, JN572069, and JN572070); MYSV, melon yellow spot virus (accession numbers NC_008306, NC_008307, and NC_008300); OROV, Oropouche virus (accession numbers NC_005776, NC_005775, and NC_005777); PUUV, Puumala virus (accession numbers NC_005225, NC_005223, and NC_005224); RVFV, Rift Valley fever virus (accession numbers NC_014397, NC_014396, and NC_014395); SATV, Sathuperi virus (accession numbers NC_018461, NC_018466, and NC_018462); SBV, Schmallenberg virus (accession numbers JX853179, JX853180, and JX853181); SEOV, Seoul virus (accession numbers NC_005238, NC_005237, and NC_005236); SFSV, sandfly fever Sicilian virus (accession numbers NC_015412, NC_015411, and NC_015413); SFTSV, severe fever with thrombocytopenia syndrome virus (accession numbers NC_018136, NC_018138, and NC_018137); SHAV, Shamonda virus (accession numbers NC_018463, NC_018467, and NC_018464); SIMV, Simbuvirus (accession numbers NC_018476, NC_018478, and NC_018477); SNV, Sin Nombre virus (accession numbers NC_005217, NC_005215, and NC_005216); SORV, Sororoca virus (accession numbers JN572071, JN572072, and JN572073); TAIV, Taï virus (accession numbers KF590574, KF590573, and KF590572); TOSV, Toscana virus (accession numbers X68414, X89628, and X53794); TPMV, Thottapalayam virus (accession numbers NC_010707, NC_010708, and NC_010704); TSWV, tomato spotted wilt virus (accession numbers NC_002052, NC_002050, and NC_002051); TULV, Tula virus (accession numbers NC_005226, NC_005228, and NC_005227); TZSV, tomato zonate spot virus (accession numbers NC_010491, NC_010490, and NC_010489); UUKV, Uukuniemi virus (accession numbers NC_005214, NC_005220, and NC_005221); WSMOV, watermelon silver mottle virus (accession numbers NC_003832, NC_003841, and NC_003843); WYOV, Wyeomyia virus (accession numbers JN572080, JN572081, and JN572082).
For a more detailed assessment, additional phylogenetic analyses were done including only the novel viruses as well as all orthobunyaviruses and tospoviruses, so as to avoid losses of sequence information due to indels (Fig. 4, small pictograms). To investigate whether the novel viruses might fall into the intragenetic distance range of orthobunyaviruses or tospoviruses, pairwise identity rates for viruses the most divergent from each other of both genera were investigated. The three novel viruses showed a similar distance to each pair, indicating a similar distance to all members of both genera (Table 3). HEBV, TAIV, and KIBV showed mean distances of 71 to 79% to orthobunyaviruses and 81 to 86% to tospoviruses in all genes, similar to the distance between orthobunyaviruses and tospoviruses (81 to 86%).
Genome organization of the novel bunyaviruses.
HEBV, TAIV, and KIBV S segments comprised an ORF of 225 to 226 aa in cRNA sense that putatively encoded a 25-kDa to 27-kDa protein, presumably the N protein (Fig. 3). No ORF was present near the N terminus of the N ORF, where an NSs protein of ca. 11 kDa is typically located in all members of the genus Orthobunyavirus. However, additional ORFs of 42 to 63 aa in cRNA sense were identified within the putative N ORFs of HEBV, TAIV, and KIBV (Fig. 3). No similarities to other sequences in GenBank were detected for the smaller ORFs.
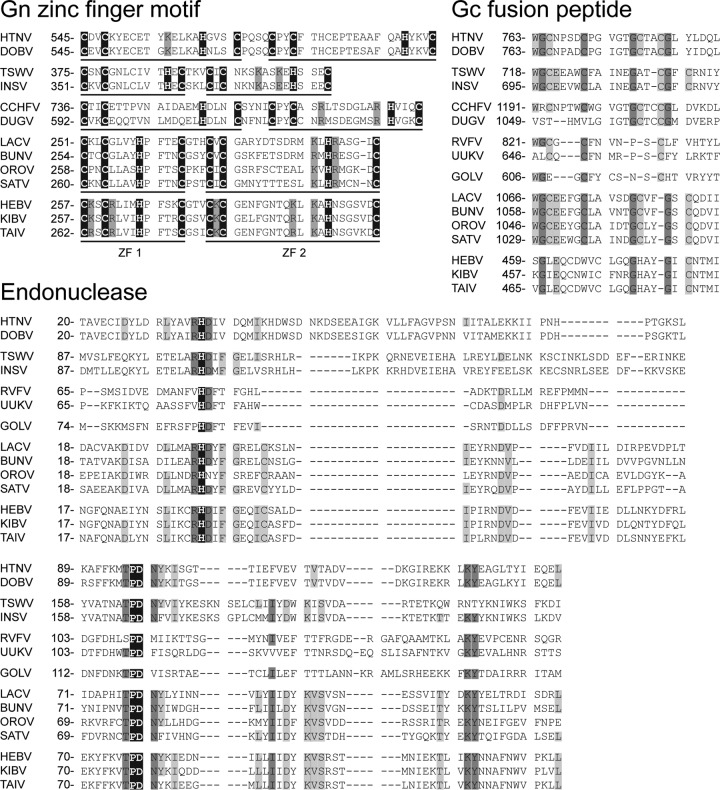
The M segments of HEBV, TAIV, and KIBV were the shortest bunyavirus M segments reported so far, about 1.2 to 1.7 kb shorter than the average size of orthobunyavirus M segments (Table 2). The segments contained a single ORF ranging between 830 aa and 838 aa in length that putatively encoded in cRNA sense the glycoprotein precursor (GPC) polyprotein that is posttranslationally cleaved into the two envelope glycoproteins Gn and Gc (Fig. 3). The GPC polyproteins in HEBV and TAIV had two possible in-frame translation initiation codons (47AUG and 53AUG, and 32AUG and 53AUG, respectively). For KIBV GPC, only one translation initiation codon at 47AUG was found. Signal peptidase cleavage sites, putative transmembrane domains (TMDs), and potential N-linked glycosylation sites of HEBV, TAIV, and KIBV are summarized in Fig. 3. Alignment of the putative GPC ORFs of HEBV, TAIV, and KIBV to the Pfam database and with orthobunyavirus glycoproteins suggested that the Gc proteins of the novel viruses were truncated by 482 aa at their N termini compared to those of orthobunyaviruses and that Gn and Gc have molecular masses of 35 kDa and 56 kDa, respectively (Fig. 3). In contrast to orthobunyaviruses, no coding regions for putative NSm proteins were identified in all three viruses. Putative Gn zinc binding (48) and Gc fusion peptide (49) domains were identified in the predicted Gn and Gc genes of HEBV, TAIV, and KIBV, respectively (Fig. 5).
Fig 5.
Multiple-sequence alignments of conserved domains of HEBV, TAIV, KIBV, and other bunyaviruses. Alignments were performed by using the E-INS-I algorithm in MAFFT and manually edited. Numbers represent genome positions. Amino acids with 100% identity are highlighted in black, those with 75% identity are highlighted in dark gray, and those with 50% identity are highlighted in light gray. Gn zinc finger motifs are highlighted in black, and conserved basic residues are highlighted in dark gray.
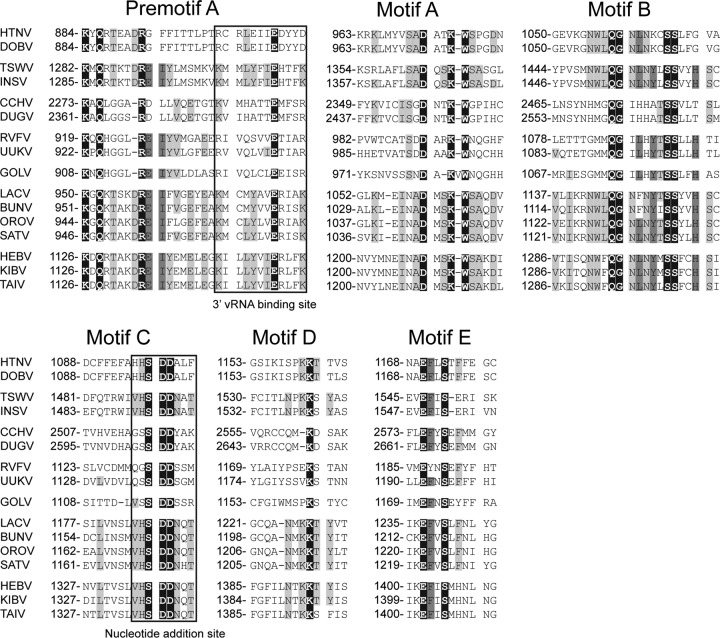
The L segments of the novel viruses were about 500 nt longer than the L segments of orthobunyaviruses due to the insertion of a unique and conserved region from amino acid positions 905LYI to 1064GLY (Fig. 3). No significant similarity to other sequences in GenBank, including those of other bunyaviruses, was identified. A putative endonuclease domain was identified at the N termini of the L proteins in HEBV, TAIV, and KIBV (50, 51) (Fig. 5). HEBV, TAIV, and KIBV were almost identical in the motifs of the third conserved region of the RdRp and exhibited the invariant residues found for bunyaviral RdRp motifs but clearly differed from members of any of the other established genera (Fig. 5).
Transcription mechanism.
To investigate if the novel bunyaviruses contain nontemplated sequences at their 5′ ends, total RNA was analyzed from infected cells by 5′-RACE RT-PCRs with reverse primers placed on all genome segments of HEBV and KIBV. Non-virally templated sequences of 9 to 16 nt and of 10 to 22 nt were detected at the 5′ ends of all HEBV and KIBV segments, respectively, indicating that viral mRNA 5′ ends are formed according to the typical mechanism for bunyaviruses (Fig. 6) (53–56).
Fig 6.

Nontemplated sequences of mRNAs of HEBV and KIBV. Shown are 5′ genome termini of L, M, and S segment mRNAs of HEBV and KIBV. C6/36 cells were infected with HEBV and KIBV, and total RNA was extracted at 1 dpi. Genome termini were amplified by 5′ RACE-PCR, PCR products were cloned, and five random clones were analyzed. Nontemplate sequences (putative transcription primers obtained from host cell mRNAs) are marked by gray boxes. Conserved genome termini of HEBV and KIBV are shown in boldface type.
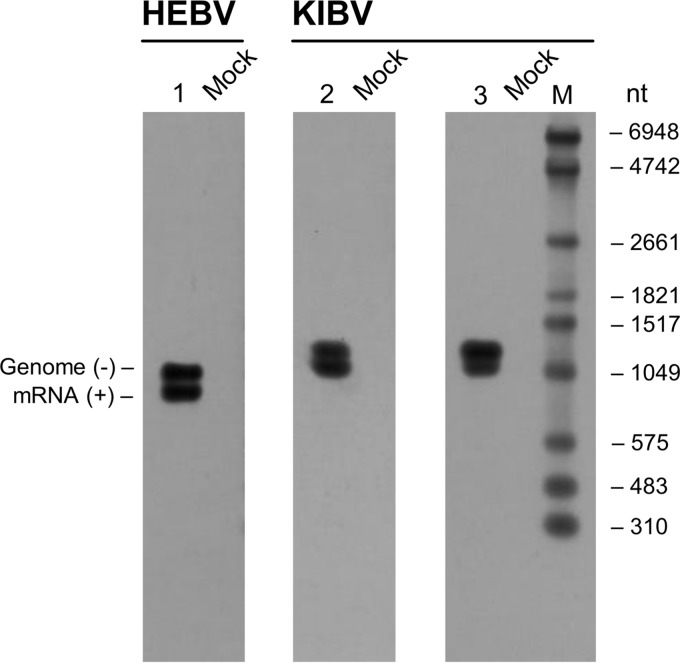
Bunyaviruses generate three different types of RNA for replication and transcription, including negative-sense genomic RNA (vRNA), positive-sense replicative cRNA, and mRNA species that contain 5′-methylated capped nonviral (primer) sequences and truncations at their 3′ ends compared to the vRNA and cRNA (17). We did a preliminary analysis of transcription of the S segments of HEBV and KIBV by Northern blotting. Two bands each were detected for HEBV and KIBV, respectively (Fig. 7). The larger bands likely corresponded to vRNA and cRNA occurring during viral replication, and the smaller bands likely represent viral mRNA transcription products. No shorter RNA transcripts, as would be expected in the case of transcription from hypothetical downstream promoters, were detected (refer to the placement of Northern blot probes shown in Fig. 3).
Fig 7.
S segment replication and transcription products analyzed by Northern blotting. Viral RNA was isolated from HEBV- and KIBV-infected C6/36 cells at 2 dpi. RNA from noninfected C6/36 cells was used as a control. A DIG-labeled RNA was used as a size marker (M), with sizes given in nucleotides at the right. Positions of DIG-PCR probes are shown in Fig. 3.
Major structural proteins.
To identify the major structural proteins, HEBV particles were purified by gradient ultracentrifugation, and viral proteins were separated by SDS-PAGE before staining with Coomassie brilliant blue. Four distinct proteins, of about 280 kDa, 60 kDa, 36 kDa, and 27 kDa, were identified (Fig. 8). MALDI-TOF mass spectroscopy confirmed two bands, corresponding to the Gn and N proteins (Fig. 8). The RdRp and Gc proteins were identified by LC-MS because MALDI-TOF analysis yielded no conclusive results for these proteins (Fig. 8). While migrations of the L and N proteins corresponded well with their predicted molecular masses, the bands corresponding to Gc and Gn proteins migrated at higher-molecular-mass equivalents than predicted based upon their amino acid sequences, which would be compatible with N-linked glycosylation at the sites described above (Fig. 3).
Fig 8.
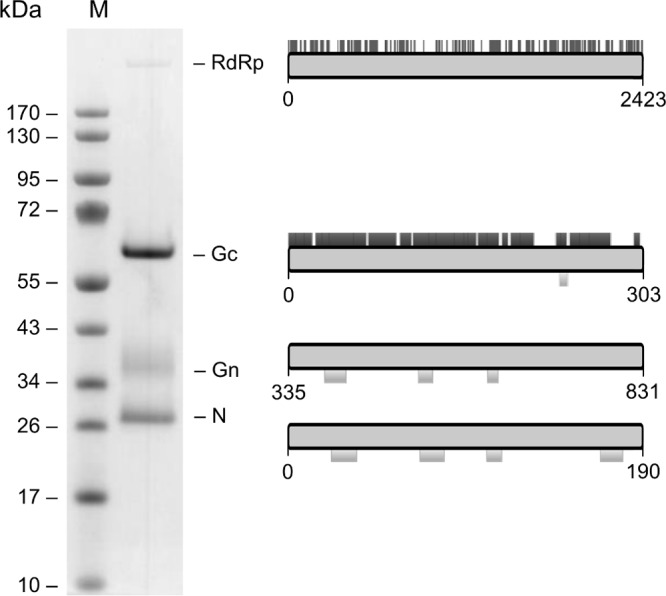
SDS-PAGE analysis of HEBV major structural proteins. Particles were purified from cell culture supernatants of infected C6/36 cells by gradient ultracentrifugation. Proteins were stained with Coomassie blue R-250. Obtained MALDI-TOF data are shown below and LC-MS data are shown above the schematic view of proteins to the right.
DISCUSSION
In this study, we discovered and characterized three novel bunyaviruses detected in mosquitoes from Côte d'Ivoire, Ghana, and Uganda. The data showed that HEBV, TAIV, and KIBV represent three novel bunyaviruses that do not group with any of the established bunyavirus genera. Although formal classification criteria for bunyavirus genera are not established, inferred tree topologies showed that the novel viruses form a novel phylogenetic sister group to orthobunyaviruses. Phylogenetic distances and comparisons of sequence similarity suggested these viruses to be sufficiently related to each other to classify them into one genus. In contrast, they were collectively about as distant from the established bunyavirus genera as the latter were from each other. This suggests that the novel viruses might form a separate genus. In order to generate auxiliary classification criteria, we investigated host range, viral growth and morphology, genome organization, as well as features of genome replication and gene expression.
HEBV, TAIV, and KIBV were detected in mosquitoes of three different genera (mainly Culex nebulosus, Culex quinquefasciatus, and Culex simpliforceps) and replicated well in RNA interference (RNAi)-competent U4.4 cells (57, 58) and in C6/36 cells that have impaired Dicer 2-based RNAi responses (59–61), indicating no growth restrictions in insect cells with an intact antiviral RNAi system. The growth phenotype in insect cells involving no or very little CPE and the inability to replicate in a large range of vertebrate cells was unexpected. Insect-restricted viruses normally cause clear CPE in insect cells. The absence of CPE in insect cells is rather typical for viruses that can additionally infect vertebrate hosts (1), which in turn could not be confirmed here by cell culture experiments. Notably, for the maintenance of insect-restricted viruses in nature, insect cycles involving horizontal (transvenereal) and vertical (transovarial) transmission are necessary. For instance, transovarial and transvenereal transmission to up to 30% of arthropod offspring has been described for bunyaviruses (62–64). Some viruses can be maintained in overwintering vectors or during time periods with a low density of amplifying hosts (65, 66). In contrast, in this study, we have gained no evidence for infection of any of the novel viruses in male mosquitoes, which is a hallmark of transovarial or transvenereal transmission. Further infection studies with a larger range of vertebrate cell lines as well as ecological investigations of insects and potential amplificatory vertebrate hosts will be necessary to clarify whether the novel viruses constitute arboviruses. Critically, proof of their insect restriction would constitute a criterion to delineate the novel viruses from the genus Orthobunyavirus, a classical group of arboviruses employing vertebrate-based amplification.
Species within the genus Orthobunyavirus are classically defined by serological criteria (1). The genetic distance between established orthobunyavirus serogroups ranges between 27 and 53% based on glycoprotein and nucleocapsid protein amino acids. Serogroups will not serologically cross-react with each other (67–70). Because the amino acid distance between the novel viruses and any orthobunyavirus ranged from 88 to 89%, and similar distances existed between orthobunyaviruses and tospoviruses, we could not expect the new viruses to yield any meaningful cross-reactivities using any animal serum directed against orthobunyaviruses or tospoviruses. Serological cross-comparisons were therefore not attempted.
Various pathogenicity- and tropism-related functions of orthobunyavirus and phlebovirus NSs proteins have been found in mammalian cells, including the suppression of host protein synthesis (26, 71–73), the inhibition of the host cell antiviral interferon response (25, 71, 74–78), as well as the inhibition of RNA polymerase II-mediated transcription (73, 79, 80). The inability of the novel bunyaviruses to replicate in vertebrate cells might be due to the putative absence of an NSs protein. Putative NSs proteins similar in sequence or position to those in orthobunyaviruses, tospoviruses, and phleboviruses were not identified in HEBV, TAIV, and KIBV. The smaller ORFs located in the C-terminal half of the N ORF of the novel bunyaviruses may encode proteins of only 5 to 7 kDa, which are significantly smaller than NSs proteins of other bunyaviruses. Moreover, no mRNAs corresponding in size to the smaller ORFs were detected by Northern blotting.
Interestingly, viruses of the Anopheles A, Anopheles B, and Tete serogroups were able to replicate in newborn mice and Vero cells, although these viruses were shown not to encode NSs proteins and were not able to counteract the antiviral interferon response (24). Another group of viruses within the genus Orthobunyavirus, the Wyeomyia group viruses, have truncated NSs sequences that may not code for functional proteins (81). However, antibodies were detected in humans, and the viruses are associated with febrile illness (52, 82, 83). Determination of whether the inability of HEBV, TAIV, and KIBV to replicate in vertebrate cells is due to the absence of an NSs protein or is encoded within another genome region needs further in-depth studies.
The only other known nonstructural protein in bunyaviruses, the NSm protein, which was shown to play a role in the pathogenesis of Rift Valley fever virus (84), was also not present in the three novel viruses. The NSm protein is encoded within orthobunyaviruses between the Gn and Gc proteins. The three proteins are expressed as a polyprotein from the M segment ORF and posttranslationally cleaved. So far, no orthobunyavirus (or tospovirus) without an NSm protein has been reported, providing an additional indication of the uniqueness of the novel viruses as a separate taxonomic entity.
There is little information on the role of NSs and NSm proteins in mosquitoes. It has been shown that the BUNV NSs protein is essential for replication in U4.4 and Aedes aegypti (Ae) cells and is required for replication and spread in Aedes aegypti mosquitoes (85). In contrast, no specific function of the La Crosse virus NSs protein and of the Rift Valley fever virus NSs protein was found in mosquito cells and mosquitoes, respectively (86–88). However, the NSm protein seems to be essential for replication of Rift Valley fever virus in mosquitoes (87). Rift Valley fever virus NSm was also found to inhibit apoptosis in mammalian cells (89). In contrast, viruses of the California serogroup (genus Orthobunyavirus) seem to induce apoptosis triggered by the NSs protein (90), a function homologous to Reaper, a Drosophila melanogaster protein that induces apoptosis (91, 92). Interestingly, sequence similarities to the Trp/GH3 motif of Reaper and the corresponding Reaper-like regions in the NSs of California serogroup viruses were identified in the L proteins of HEBV, TAIV, and KIBV (283WRILESKLLET293, 283WKDLETKLTST293, and 283WKMLEEKLEK293, respectively [conserved sequences among Reaper and HEBV, TAIV, and KIBV are underlined]). The Trp/GH3 motif is conserved among Reaper and two other Drosophila proteins, Grim and Sickle, which have crucial functions in programmed cell death (93–95). Whether this Trp/GH3-like motif in HEBV, TAIV, and KIBV may have homologous functions needs to be studied.
The absence of any NS protein ORFs conserved across the clade comprising tospoviruses, orthobunyaviruses, and the novel viruses suggests that the most recent common ancestor of all of these viruses would not have encoded any of these genes. Rather, the different coding strategies for NS proteins suggest independent acquisitions during the formation of generic viral lineages. In particular, NSs and NSm proteins might have been acquired during the evolution of orthobunyaviruses in the course of acquiring replicative capability in vertebrate hosts.
A unique insertion of about 500 nt was identified in the RdRp genes of HEBV, TAIV, and KIBV. This additional region, not found in any other bunyaviruses, might represent a putative accessory protein domain. The presence of an accessory domain in the L protein is not unprecedented. For example, the CCHFV L protein contains an OTU-like cysteine protease that has been suggested to suppress the host cell inflammatory and antiviral response (28). The L proteins of orthobunyaviruses, tospoviruses, hantaviruses, and nairoviruses contain an N-terminal endonuclease domain (50, 51, 96). However, no sequence similarities of the unique region in HEBV, TAIV, and KIBV to any other viral proteins were found. We further specifically searched for GW/WG motifs found to be conserved within viral RNA silencing suppressor proteins encoded by many insect-restricted viruses (97). No such motifs were detected in all translated HEBV, TAIV, and KIBV ORFs. Determination of whether HEBV, TAIV, and KIBV express any accessory proteins at all will therefore require further experimental studies.
While the ORFs were well conserved among HEBV, TAIV, and KIBV, the high level of variability of the UTRs and the extended length of up to 569 nt in the TAIV M segment 5′ UTR were surprising. The UTRs have many different functions and play a role during replication, transcription, encapsidation, and packaging of the viral genome (98–101). 3′ and 5′ UTR lengths of the three genome segments are generally well conserved among different orthobunyaviruses, with M and L segment 3′ and 5′ UTRs of about 50 to 100 nt and S segment 3′ and 5′ UTRs of about 80 to 200 nt. It will be interesting to study the functions of these highly different UTRs. Interestingly, the terminal nucleotides of the UTRs are strictly conserved among bunyaviruses of the same genus, serving as a criterion for genus classification (1). HEBV, TAIV, and KIBV contained unique terminal nucleotides that were truncated compared to orthobunyaviruses, precluding their grouping into the genus Orthobunyavirus and providing further support that the viruses constitute a separate taxonomic entity.
Segmented negative-strand RNA viruses of the families Orthomyxoviridae, Bunyaviridae, and Arenaviridae use capped RNA primers that are cleaved from the 5′ termini of host cell mRNAs in order to initiate their transcription (53, 54, 56, 102–104). The lengths of reported capped primers vary from 10 to 20 nt (53, 54, 56, 102–104). We found nontemplated sequences of 9 to 16 nt and 10 to 22 nt at the 5′ termini of HEBV and KIBV mRNAs, respectively. Primer sequences containing a 3′ U residue were found preferentially, suggesting that the 3′ U residue might be able to undergo base pairing with the terminal 5′ A residue of the vRNA during transcription initiation. This would be in good agreement with previous observations in orthobunyaviruses and hantaviruses, where capped primers preferentially terminate at G residues, potentially facilitating RNA primer binding to the terminal 5′ C residue (53). As observed for orthobunyaviruses, a number of primer sequences contained 3′ GU or 3′ AGU residues (105).
Analyses of RNA products in infected cells indicated that HEBV and KIBV generate truncated mRNAs, similar to what has been described for other bunyaviruses such as snowshoe hare virus, an orthobunyavirus whose S segment mRNA is about 85 nt shorter than the vRNA species (18).
Taken together, our findings suggest that HEBV, TAIV, and KIBV cannot be assigned to any existing bunyavirus genus, while they share common features with each other sufficient to classify them as one genus. Although they are somewhat more closely related to orthobunyaviruses than to other bunyavirus genera, their genome organization and phylogenetic relationships separate them from other genera. Further studies, particularly on their host restriction and antigenic properties, will be necessary to support their putative classification into a separate novel genus.
ACKNOWLEDGMENTS
We thank the Uganda Wildlife Authority and Uganda National Council of Science and Technology, the Ivorian Ministry of Environment and Forest and the Ministry of Research, as well as the directorship of the Taï National Park and Makerere University Biological Field Station for permission to conduct this research. We thank Clement Nyikiriza and Charles Kyalisima for assistance in the field. We are grateful to Fabian Leendertz for assistance with fieldwork research design and logistic support. We are grateful to Annika Branting and Priyal de Zoysa for performing Illumina next-generation sequencing.
Funding for fieldwork in Uganda was provided by the Emory University Global Health Institute. The project was supported by the National Institutes of Health (AI057158 Northeast Biodefense Center—Lipkin); by the United States Department of Defense and the United States Agency for International Development (USAID) Emerging Pandemic Threats (EPT) Program, PREDICT project, under the terms of cooperative agreement GHN-A-OO-09-00010-00; by European Union DG Research through the program ANTIGONE (grant agreement no. 278976); and by the Deutsche Forschungsgemeinschaft (grant agreement no. DR 772/3-1).
Footnotes
Published ahead of print 25 September 2013
REFERENCES
- 1.Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist Å, Schmaljohn CS, Tesh RB. 2012. Family Bunyaviridae, p 725–741 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus Taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 2.Beer M, Conraths FJ, van der Poel WH. 2013. ‘Schmallenberg virus'—a novel orthobunyavirus emerging in Europe. Epidemiol. Infect. 141:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird BH, Nichol ST. 2012. Breaking the chain: Rift Valley fever virus control via livestock vaccination. Curr. Opin. Virol. 2:315–323 [DOI] [PubMed] [Google Scholar]
- 4.Ergonul O. 2012. Crimean-Congo hemorrhagic fever virus: new outbreaks, new discoveries. Curr. Opin. Virol. 2:215–220 [DOI] [PubMed] [Google Scholar]
- 5.Soldan SS, Gonzalez-Scarano F. 2005. Emerging infectious diseases: the Bunyaviridae. J. Neurovirol. 11:412–423 [DOI] [PubMed] [Google Scholar]
- 6.Watson DC, Sargianou M, Papa A, Chra P, Starakis I, Panos G. Epidemiology of hantavirus infections in humans: a comprehensive, global overview. Crit. Rev. Microbiol., in press. 10.3109/1040841X.2013.783555 [DOI] [PubMed] [Google Scholar]
- 7.Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, Zhang L, Zhang QF, Popov VL, Li C, Qu J, Li Q, Zhang YP, Hai R, Wu W, Wang Q, Zhan FX, Wang XJ, Kan B, Wang SW, Wan KL, Jing HQ, Lu JX, Yin WW, Zhou H, Guan XH, Liu JF, Bi ZQ, Liu GH, Ren J, Wang H, Zhao Z, Song JD, He JR, Wan T, Zhang JS, Fu XP, Sun LN, Dong XP, Feng ZJ, Yang WZ, Hong T, Zhang Y, Walker DH, Wang Y, Li DX. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 364:1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai TF. 1987. Hemorrhagic fever with renal syndrome: mode of transmission to humans. Lab. Anim. Sci. 37:428–430 [PubMed] [Google Scholar]
- 9.Kuismanen E, Bang B, Hurme M, Pettersson RF. 1984. Uukuniemi virus maturation: immunofluorescence microscopy with monoclonal glycoprotein-specific antibodies. J. Virol. 51:137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuismanen E, Hedman K, Saraste J, Pettersson RF. 1982. Uukuniemi virus maturation: accumulation of virus particles and viral antigens in the Golgi complex. Mol. Cell. Biol. 2:1444–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy FA, Harrison AK, Whitfield SG. 1973. Bunyaviridae: morphologic and morphogenetic similarities of Bunyamwera serologic supergroup viruses and several other arthropod-borne viruses. Intervirology 1:297–316 [DOI] [PubMed] [Google Scholar]
- 12.Novoa RR, Calderita G, Cabezas P, Elliott RM, Risco C. 2005. Key Golgi factors for structural and functional maturation of bunyamwera virus. J. Virol. 79:10852–10863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salanueva IJ, Novoa RR, Cabezas P, Lopez-Iglesias C, Carrascosa JL, Elliott RM, Risco C. 2003. Polymorphism and structural maturation of bunyamwera virus in Golgi and post-Golgi compartments. J. Virol. 77:1368–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briese T, Kapoor V, Lipkin WI. 2007. Natural M-segment reassortment in Potosi and Main Drain viruses: implications for the evolution of orthobunyaviruses. Arch. Virol. 152:2237–2247 [DOI] [PubMed] [Google Scholar]
- 15.Yanase T, Aizawa M, Kato T, Yamakawa M, Shirafuji H, Tsuda T. 2010. Genetic characterization of Aino and Peaton virus field isolates reveals a genetic reassortment between these viruses in nature. Virus Res. 153:1–7 [DOI] [PubMed] [Google Scholar]
- 16.Yanase T, Kato T, Aizawa M, Shuto Y, Shirafuji H, Yamakawa M, Tsuda T. 2012. Genetic reassortment between Sathuperi and Shamonda viruses of the genus Orthobunyavirus in nature: implications for their genetic relationship to Schmallenberg virus. Arch. Virol. 157:1611–1616 [DOI] [PubMed] [Google Scholar]
- 17.Schmaljohn C, Nichol ST. 2007. Bunyaviridae, p 1741–1789 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, ]?> 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 18.Eshita Y, Ericson B, Romanowski V, Bishop DH. 1985. Analyses of the mRNA transcription processes of snowshoe hare bunyavirus S and M RNA species. J. Virol. 55:681–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikegami T, Peters CJ, Makino S. 2005. Rift Valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J. Virol. 79:5606–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaaskelainen KM, Kaukinen P, Minskaya ES, Plyusnina A, Vapalahti O, Elliott RM, Weber F, Vaheri A, Plyusnin A. 2007. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J. Med. Virol. 79:1527–1536 [DOI] [PubMed] [Google Scholar]
- 21.Vera-Otarola J, Solis L, Soto-Rifo R, Ricci EP, Pino K, Tischler ND, Ohlmann T, Darlix JL, Lopez-Lastra M. 2012. The Andes hantavirus NSs protein is expressed from the viral small mRNA by a leaky scanning mechanism. J. Virol. 86:2176–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palacios G, Savji N, Travassos da Rosa A, Guzman H, Yu X, Desai A, Rosen GE, Hutchison S, Lipkin WI, Tesh R. 2013. Characterization of the Uukuniemi virus group (Phlebovirus: Bunyaviridae): evidence for seven distinct species. J. Virol. 87:3187–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rönnholm R, Pettersson RF. 1987. Complete nucleotide sequence of the M RNA segment of Uukuniemi virus encoding the membrane glycoproteins G1 and G2. Virology 160:191–202 [DOI] [PubMed] [Google Scholar]
- 24.Mohamed M, McLees A, Elliott RM. 2009. Viruses in the Anopheles A, Anopheles B, and Tete serogroups in the Orthobunyavirus genus (family Bunyaviridae) do not encode an NSs protein. J. Virol. 83:7612–7618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, Haller O. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 75:1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 98:664–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeda A, Sugiyama K, Nagano H, Mori M, Kaido M, Mise K, Tsuda S, Okuno T. 2002. Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 532:75–79 [DOI] [PubMed] [Google Scholar]
- 28.Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, Garcia-Sastre A, Virgin HW., IV 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honig JE, Osborne JC, Nichol ST. 2004. Crimean-Congo hemorrhagic fever virus genome L RNA segment and encoded protein. Virology 321:29–35 [DOI] [PubMed] [Google Scholar]
- 30.Kinsella E, Martin SG, Grolla A, Czub M, Feldmann H, Flick R. 2004. Sequence determination of the Crimean-Congo hemorrhagic fever virus L segment. Virology 321:23–28 [DOI] [PubMed] [Google Scholar]
- 31.Plyusnina A, Ibrahim IN, Plyusnin A. 2009. A newly recognized hantavirus in the Asian house rat (Rattus tanezumi) in Indonesia. J. Gen. Virol. 90:205–209 [DOI] [PubMed] [Google Scholar]
- 32.Marklewitz M, Handrick S, Grasse W, Kurth A, Lukashev A, Drosten C, Ellerbrok H, Leendertz FH, Pauli G, Junglen S. 2011. Gouleako virus isolated from West African mosquitoes constitutes a proposed novel genus in the family Bunyaviridae. J. Virol. 85:9227–9234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Junglen S, Kurth A, Kuehl H, Quan PL, Ellerbrok H, Pauli G, Nitsche A, Nunn C, Rich SM, Lipkin WI, Briese T, Leendertz FH. 2009. Examining landscape factors influencing relative distribution of mosquito genera and frequency of virus infection. EcoHealth 6:239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards FW. 1941. Mosquitoes of the Ethiopian region. III. Culicine adults and pupae, vol 3. Oxford University Press, London, United Kingdom [Google Scholar]
- 35.Gaffigan TV, Wilkerson RC, Pecor JE, Stoffer JA, Anderson T. 2013. Systematic catalog of culicidae. Walter Reed Biosystematics Unit, Division of Entomology, Walter Reed Army Institute of Research, Silver Spring, MD: http://www.mosquitocatalog.org [Google Scholar]
- 36.Gillies MT, De Meillon B. 1968. The Anophelinae of Africa south of the Sahara (Ethiopian Zoogeographical Region), 2nd ed, vol 54 South Africa Institute for Medical Research, Johannesburg, South Africa [Google Scholar]
- 37.Jupp PG. 1996. Mosquitoes of South Africa—Culicinae and Toxorhynchitinae. Ekogilde Publishers, Hartebeespoort, Republic of South Africa [Google Scholar]
- 38.Igarashi A. 1978. Isolation of a Singh's Aedes albopictus cell clone sensitive to dengue and Chikungunya viruses. J. Gen. Virol. 40:531–544 [DOI] [PubMed] [Google Scholar]
- 39.Junglen S, Kopp A, Kurth A, Pauli G, Ellerbrok H, Leendertz FH. 2009. A new flavivirus and a new vector: characterization of a novel flavivirus isolated from Uranotaenia mosquitoes from a tropical rain forest. J. Virol. 83:4462–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh KR. 1967. Cell cultures derived from larvae of Aedes albopictus (Skuse) and Aedes aegypti (L). Curr. Sci. 36:506–508 [Google Scholar]
- 41.Zirkel F, Kurth A, Quan PL, Briese T, Ellerbrok H, Pauli G, Leendertz FH, Lipkin WI, Ziebuhr J, Drosten C, Junglen S. 2011. An insect nidovirus emerging from a primary tropical rainforest. mBio 2(3):e00077–11. 10.1128/mBio.00077-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quan PL, Junglen S, Tashmukhamedova A, Conlan S, Hutchison SK, Kurth A, Ellerbrok H, Egholm M, Briese T, Leendertz FH, Lipkin WI. 2010. Moussa virus: a new member of the Rhabdoviridae family isolated from Culex decens mosquitoes in Cote d'Ivoire. Virus Res. 147:17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biel SS, Gelderblom HR. 1999. Diagnostic electron microscopy is still a timely and rewarding method. J. Clin. Virol. 13:105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayat MA. 2000. Principles and techniques of electron microscopy: biological applications. Macmillan Press Houndmills, London, United Kingdom [Google Scholar]
- 45.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 47.Zirkel F, Roth H, Kurth A, Drosten C, Ziebuhr J, Junglen S. 2013. Identification and characterization of genetically divergent members of the newly established family Mesoniviridae. J. Virol. 87:6346–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Estrada DF, De Guzman RN. 2011. Structural characterization of the Crimean-Congo hemorrhagic fever virus Gn tail provides insight into virus assembly. J. Biol. Chem. 286:21678–21686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plassmeyer ML, Soldan SS, Stachelek KM, Roth SM, Martin-Garcia J, Gonzalez-Scarano F. 2007. Mutagenesis of the La Crosse virus glycoprotein supports a role for Gc (1066-1087) as the fusion peptide. Virology 358:273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Y, Wang W, Ji W, Deng M, Sun Y, Zhou H, Yang C, Deng F, Wang H, Hu Z, Lou Z, Rao Z. 2012. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc. Natl. Acad. Sci. U. S. A. 109:5046–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reguera J, Weber F, Cusack S. 2010. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 6:e1001101. 10.1371/journal.ppat.1001101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aitken TH, Spence L, Jonkers AH, Anderson CR. 1968. Wyeomyla-virus isolations in Trinidad, West Indies. Am. J. Trop. Med. Hyg. 17:886–888 [DOI] [PubMed] [Google Scholar]
- 53.Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D. 1995. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J. Virol. 69:5754–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bishop DH, Gay ME, Matsuoko Y. 1983. Nonviral heterogeneous sequences are present at the 5′ ends of one species of snowshoe hare bunyavirus S complementary RNA. Nucleic Acids Res. 11:6409–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin H, Elliott RM. 1993. Characterization of Bunyamwera virus S RNA that is transcribed and replicated by the L protein expressed from recombinant vaccinia virus. J. Virol. 67:1396–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simons JF, Pettersson RF. 1991. Host-derived 5′ ends and overlapping complementary 3′ ends of the two mRNAs transcribed from the ambisense S segment of Uukuniemi virus. J. Virol. 65:4741–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Attarzadeh-Yazdi G, Fragkoudis R, Chi Y, Siu RW, Ulper L, Barry G, Rodriguez-Andres J, Nash AA, Bouloy M, Merits A, Fazakerley JK, Kohl A. 2009. Cell-to-cell spread of the RNA interference response suppresses Semliki Forest virus (SFV) infection of mosquito cell cultures and cannot be antagonized by SFV. J. Virol. 83:5735–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. 2012. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 8:e1002470. 10.1371/journal.ppat.1002470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, Schilkey FD, Mudge J, Wilusz J, Olson KE, Blair CD, Ebel GD. 2010. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl. Trop. Dis. 4:e856. 10.1371/journal.pntd.0000856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scott JC, Brackney DE, Campbell CL, Bondu-Hawkins V, Hjelle B, Ebel GD, Olson KE, Blair CD. 2010. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl. Trop. Dis. 4:e848. 10.1371/journal.pntd.0000848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vodovar N, Bronkhorst AW, van Cleef KW, Miesen P, Blanc H, van Rij RP, Saleh MC. 2012. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One 7:e30861. 10.1371/journal.pone.0030861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schopen S, Labuda M, Beaty B. 1991. Vertical and venereal transmission of California group viruses by Aedes triseriatus and Culiseta inornata mosquitoes. Acta Virol. 35:373–382 [PubMed] [Google Scholar]
- 63.Thompson WH, Beaty BJ. 1977. Venereal transmission of La Crosse (California encephalitis) arbovirus in Aedes triseriatus mosquitoes. Science 196:530–531 [DOI] [PubMed] [Google Scholar]
- 64.Turell MJ, Reeves WC, Hardy JL. 1982. Evaluation of the efficiency of transovarial transmission of California encephalitis viral strains in Aedes dorsalis and Aedes melanimon. Am. J. Trop. Med. Hyg. 31:382–388 [DOI] [PubMed] [Google Scholar]
- 65.McGaw MM, Chandler LJ, Wasieloski LP, Blair CD, Beaty BJ. 1998. Effect of La Crosse virus infection on overwintering of Aedes triseriatus. Am. J. Trop. Med. Hyg. 58:168–175 [DOI] [PubMed] [Google Scholar]
- 66.Tesh RB, Lubroth J, Guzman H. 1992. Simulation of arbovirus overwintering: survival of Toscana virus (Bunyaviridae: Phlebovirus) in its natural sand fly vector Phlebotomus perniciosus. Am. J. Trop. Med. Hyg. 47:574–581 [DOI] [PubMed] [Google Scholar]
- 67.Calisher CH. 1996. History, classification, and taxonomy of viruses in the family Bunyaviridae, p 1–17 In Elliott RM. (ed), The Bunyaviridae. Plenum Press, New York, NY [Google Scholar]
- 68.de Brito Magalhaes CL, Drumond BP, Novaes RF, Quinan BR, de Magalhaes JC, dos Santos JR, do Amaral Pinto C, Assis MT, Bonjardim CA, Kroon EG, Ferreira PC. 2011. Identification of a phylogenetically distinct orthobunyavirus from group C. Arch. Virol. 156:1173–1184 [DOI] [PubMed] [Google Scholar]
- 69.Saeed MF, Li L, Wang H, Weaver SC, Barrett AD. 2001. Phylogeny of the Simbu serogroup of the genus Bunyavirus. J. Gen. Virol. 82:2173–2181 [DOI] [PubMed] [Google Scholar]
- 70.Savji N, Palacios G, Travassos da Rosa A, Hutchison S, Celone C, Hui J, Briese T, Calisher CH, Tesh RB, Lipkin WI. 2011. Genomic and phylogenetic characterization of Leanyer virus, a novel orthobunyavirus isolated in northern Australia. J. Gen. Virol. 92:1676–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798–9806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hart TJ, Kohl A, Elliott RM. 2009. Role of the NSs protein in the zoonotic capacity of orthobunyaviruses. Zoonoses Public Health 56:285–296 [DOI] [PubMed] [Google Scholar]
- 73.Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, Egly JM. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550 [DOI] [PubMed] [Google Scholar]
- 74.Kohl A, Clayton RF, Weber F, Bridgen A, Randall RE, Elliott RM. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 77:7999–8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 4:e13. 10.1371/journal.ppat.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Streitenfeld H, Boyd A, Fazakerley JK, Bridgen A, Elliott RM, Weber F. 2003. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J. Virol. 77:5507–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Knippenberg I, Carlton-Smith C, Elliott RM. 2010. The N-terminus of Bunyamwera orthobunyavirus NSs protein is essential for interferon antagonism. J. Gen. Virol. 91:2002–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weber F, Bridgen A, Fazakerley JK, Streitenfeld H, Kessler N, Randall RE, Elliott RM. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. 2009. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 5:e1000287. 10.1371/journal.ppat.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas D, Blakqori G, Wagner V, Banholzer M, Kessler N, Elliott RM, Haller O, Weber F. 2004. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 279:31471–31477 [DOI] [PubMed] [Google Scholar]
- 81.Chowdhary R, Street C, Travassos da Rosa A, Nunes MR, Tee KK, Hutchison SK, Vasconcelos PF, Tesh RB, Lipkin WI, Briese T. 2012. Genetic characterization of the Wyeomyia group of orthobunyaviruses and their phylogenetic relationships. J. Gen. Virol. 93:1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Souza Lopes O, de Abreu Sacchetta L, Fonseca IE, Lacerda JP. 1975. Bertioga (Guama group) and Anhembi (Bunyamwera group), two new arboviruses isolated in Sao Paulo, Brazil. Am. J. Trop. Med. Hyg. 24:131–134 [DOI] [PubMed] [Google Scholar]
- 83.Sirhongse S, Johnson CM. 1965. Wyeomyia subgroup of arbovirus: isolation from man. Science 149:863–864 [DOI] [PubMed] [Google Scholar]
- 84.Bird BH, Albarino CG, Hartman AL, Erickson BR, Ksiazek TG, Nichol ST. 2008. Rift Valley fever virus lacking the NSs and NSm genes is highly attenuated, confers protective immunity from virulent virus challenge, and allows for differential identification of infected and vaccinated animals. J. Virol. 82:2681–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Szemiel AM, Failloux AB, Elliott RM. 2012. Role of Bunyamwera orthobunyavirus NSs protein in infection of mosquito cells. PLoS Negl. Trop. Dis. 6:e1823. 10.1371/journal.pntd.0001823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blakqori G, Delhaye S, Habjan M, Blair CD, Sanchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, Weiss S, Michiels T, Staeheli P, Weber F. 2007. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J. Virol. 81:4991–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Crabtree MB, Kent Crockett RJ, Bird BH, Nichol ST, Erickson BR, Biggerstaff BJ, Horiuchi K, Miller BR. 2012. Infection and transmission of Rift Valley fever viruses lacking the NSs and/or NSm genes in mosquitoes: potential role for NSm in mosquito infection. PLoS Negl. Trop. Dis. 6:e1639. 10.1371/journal.pntd.0001639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moutailler S, Krida G, Madec Y, Bouloy M, Failloux AB. 2010. Replication of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, in Aedes and Culex mosquitoes. Vector Borne Zoonotic Dis. 10:681–688 [DOI] [PubMed] [Google Scholar]
- 89.Won S, Ikegami T, Peters CJ, Makino S. 2007. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J. Virol. 81:13335–13345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Colon-Ramos DA, Irusta PM, Gan EC, Olson MR, Song J, Morimoto RI, Elliott RM, Lombard M, Hollingsworth R, Hardwick JM, Smith GK, Kornbluth S. 2003. Inhibition of translation and induction of apoptosis by bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol. Biol. Cell 14:4162–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. 2000. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19:589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. 2002. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat. Cell Biol. 4:439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Christich A, Kauppila S, Chen P, Sogame N, Ho SI, Abrams JM. 2002. The damage-responsive Drosophila gene sickle encodes a novel IAP binding protein similar to but distinct from reaper, grim, and hid. Curr. Biol. 12:137–140 [DOI] [PubMed] [Google Scholar]
- 94.Claveria C, Caminero E, Martinez AC, Campuzano S, Torres M. 2002. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. EMBO J. 21:3327–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wing JP, Karres JS, Ogdahl JL, Zhou L, Schwartz LM, Nambu JR. 2002. Drosophila sickle is a novel grim-reaper cell death activator. Curr. Biol. 12:131–135 [DOI] [PubMed] [Google Scholar]
- 96.Heinemann P, Schmidt-Chanasit J, Günther S. 2013. The N terminus of Andes virus L protein suppresses mRNA and protein expression in mammalian cells. J. Virol. 87:6975–6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bivalkar-Mehla S, Vakharia J, Mehla R, Abreha M, Kanwar JR, Tikoo A, Chauhan A. 2011. Viral RNA silencing suppressors (RSS): novel strategy of viruses to ablate the host RNA interference (RNAi) defense system. Virus Res. 155:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barr JN, Wertz GW. 2005. Role of the conserved nucleotide mismatch within 3′- and 5′-terminal regions of Bunyamwera virus in signaling transcription. J. Virol. 79:3586–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kohl A, Dunn EF, Lowen AC, Elliott RM. 2004. Complementarity, sequence and structural elements within the 3′ and 5′ non-coding regions of the Bunyamwera orthobunyavirus S segment determine promoter strength. J. Gen. Virol. 85:3269–3278 [DOI] [PubMed] [Google Scholar]
- 100.Kohl A, Lowen AC, Leonard VH, Elliott RM. 2006. Genetic elements regulating packaging of the Bunyamwera orthobunyavirus genome. J. Gen. Virol. 87:177–187 [DOI] [PubMed] [Google Scholar]
- 101.Osborne JC, Elliott RM. 2000. RNA binding properties of bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. J. Virol. 74:9946–9952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Caton AJ, Robertson JS. 1980. Structure of the host-derived sequences present at the 5′ ends of influenza virus mRNA. Nucleic Acids Res. 8:2591–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jin H, Elliott RM. 1993. Non-viral sequences at the 5′ ends of Dugbe nairovirus S mRNAs. J. Gen. Virol. 74(Part 10):2293–2297 [DOI] [PubMed] [Google Scholar]
- 104.Raju R, Raju L, Hacker D, Garcin D, Compans R, Kolakofsky D. 1990. Nontemplated bases at the 5′ ends of Tacaribe virus mRNAs. Virology 174:53–59 [DOI] [PubMed] [Google Scholar]
- 105.Bouloy M, Pardigon N, Vialat P, Gerbaud S, Girard M. 1990. Characterization of the 5′ and 3′ ends of viral messenger RNAs isolated from BHK21 cells infected with Germiston virus (Bunyavirus). Virology 175:50–58 [DOI] [PubMed] [Google Scholar]
- 106.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]