Abstract
Investigating the incidence and prevalence of HIV-1 superinfection is challenging due to the complex dynamics of two infecting strains. The superinfecting strain can replace the initial strain, be transiently expressed, or persist along with the initial strain in distinct or in recombined forms. Various selective pressures influence these alternative scenarios in different HIV-1 coding regions. We hypothesized that the potency of the neutralizing antibody (NAb) response to autologous viruses would modulate viral dynamics in env following superinfection in a limited set of superinfection cases. HIV-1 env pyrosequencing data were generated from blood plasma collected from 7 individuals with evidence of superinfection. Viral variants within each patient were screened for recombination, and viral dynamics were evaluated using nucleotide diversity. NAb responses to autologous viruses were evaluated before and after superinfection. In 4 individuals, the superinfecting strain replaced the original strain. In 2 individuals, both initial and superinfecting strains continued to cocirculate. In the final individual, the surviving lineage was the product of interstrain recombination. NAb responses to autologous viruses that were detected within the first 2 years of HIV-1 infection were weak or absent for 6 of the 7 recently infected individuals at the time of and shortly following superinfection. These 6 individuals had detectable on-going viral replication of distinct superinfecting virus in the env coding region. In the remaining case, there was an early and strong autologous NAb response, which was associated with extensive recombination in env between initial and superinfecting strains. This extensive recombination made superinfection more difficult to identify and may explain why the detection of superinfection has typically been associated with low autologous NAb titers.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) superinfection (SI) is the reinfection of a previously infected individual with a distinct heterologous viral strain. This process allows viral recombination to occur between distantly related strains and may facilitate immune evasion (1, 2), development of drug resistance (3), and disease progression (4–6). Moreover, new circulating recombinant forms complicate vaccine development by expanding global viral diversity (7, 8). HIV-1 superinfection appears to occur more often early in infection and is associated with a weaker and immature immune response (9, 10). However, detection of superinfection is difficult and hinges on timing of sampling and molecular evidence of a genetically distinct viral subpopulation. The recent development of more sensitive next-generation sequencing techniques (e.g., ultradeep sequencing [UDS]) facilitates the identification of cases (4, 11, 12) and permits the assessment of intrahost viral subpopulation dynamics.
The role of neutralizing antibodies (NAb) in protection against superinfection has been supported by animal models (13). Analogous to humans, superinfection in animal models has been associated with a preexisting weaker cell-mediated and humoral immune response to autologous and heterologous viruses (6, 9, 14–16). The host NAb response to HIV-1 can exert strong selective pressures that can drive rapid viral adaptation to escape immune recognition in env (15, 17, 18). Nonetheless, factors that modulate intrahost viral evolution after superinfection has occurred have not been well characterized. Here, we investigated the potential role of autologous NAb responses in driving viral evolution of HIV-1 superinfection in seven superinfected individuals monitored longitudinally.
MATERIALS AND METHODS
Population study and design.
Individuals with intrasubtype B HIV-1 superinfection were identified from a previous screen of 118 participants from the San Diego Primary Infection Cohort, enrolled between January 1998 and January 2007 (4). All screened cohort participants deferred antiretroviral therapy for at least 6 months and had at least two plasma samples available for sequencing. Here, we studied seven previously identified individuals with superinfection who had at least four serially sampled time points available (Table 1). All individuals were men who reported having sex with men (MSM) as their primary risk factor for HIV acquisition. CD4 cell counts (LabCorp) and blood plasma HIV-1 RNA levels (Amplicor HIV-1 monitor test; Roche Molecular Systems Inc.) were also longitudinally quantified. Estimated dates of infection (EDI) were determined using standard procedures (19).
Table 1.
Subject baseline characteristics
| Subject ID | Age at enrollment (yr) | Sex | Race | Risk factor | EDI (mo/day/yr) | First visit after EDI (mo) | CD4 T (cells/μl) after first visit | cDNA (log10 copies/ml) after first visit |
|---|---|---|---|---|---|---|---|---|
| D2 | 34 | Male | African-American | MSM | 9/10/97 | 2.5 | 717 | 2.53 |
| G5 | 33 | Male | White | MSM | 6/05/99 | 3.0 | 492 | 6.26 |
| K6 | 30 | Male | White | MSM | 6/23/01 | 0.9 | 711 | 3.38 |
| K9 | 32 | Male | White | MSM | 6/05/01 | 3.3 | 571 | 3.08 |
| P2 | 21 | Male | White | MSM | 4/16/03 | 2.3 | 516 | 4.86 |
| R5 | 51 | Male | White | MSM | 11/23/03 | 5.7 | 471 | 4.77 |
| S1 | 35 | Male | White | MSM | 6/08/04 | 2.3 | 321 | 4.58 |
| Median (IQR) | 33 (31–34.5) | 2.5 (1.7–3.3) | 516 (356.5–675.5) | 4.58 (3.00–6.17) |
RNA extraction and sequencing methods.
HIV RNA was extracted from blood plasma (QIAmp viral RNA mini kit; Qiagen, Hilden, Germany), and cDNA was generated (RETROscript kit; Applied Biosystems/Ambion, Austin, TX) from extracted HIV-1 RNA for three or more time points over a minimum of 11 months. Single-genome sequencing (SGS) and UDS of PCR-amplified env C2-V3 (HXB2 coordinates 6928 to 7344), pol reverse transcriptase (RT; HXB2 coordinates 2708 to 3242), and gag p24 (HXB2 coordinates 1366 to 1619) were performed as described previously (4, 11, 20). All UDS and SGS sequences were screened for in-house cross-contamination using BLAST as previously described (21).
Sequence analysis.
UDS sequences were analyzed with the HyPhy software package available on the DataMonkey webserver (Tables 2 and 3) (22, 23). Estimates of sequence diversity were obtained from sliding windows (length, 150 bp; stride, 20 bp) for each of the three sequenced gene regions (4). UDS raw data were then analyzed using ShoRAH (short read assembly into haplotypes) (24), which allows both quantification of genetic diversity and identification of nonredundant sequences. Briefly, this software uses a Bayesian probabilistic clustering method to identify sequence variation. The consensus sequence of each cluster represents a haplotype, and the number of reads within each cluster estimated the prevalence of the haplotype. Hence, the output consists of a list of haplotypes and their relative frequencies (24–27). The viral diversity within each subject's subpopulation and at each time point was further quantified by the mean genetic distance (in number of base substitutions per site) using a Tamura-Nei model (28). Analyses were conducted in MEGA5 (29). For the UDS sequences, we applied the method developed by Poon et al., which consists of the reexpansion of generated viral variants to their relative read counts (30) (Table 4 and Fig. 1).
Table 2.
Sampling time point and env sequences
| Subject ID and env sequence type | Time point (in days after EDI)/no. of partial env sequence haplotypesa |
Total no. of sequences (UDS/SGS) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
| K6 | ||||||||||
| UDS | 219/8 | 95 (8/87) | ||||||||
| SGS | 124/14 | 180/23 | 219/38 | 261/12 | ||||||
| S1 | ||||||||||
| UDS | 85/7 | 167 (7/160) | ||||||||
| SGS | 85/31 | 155/30 | 205/27 | 232/25 | 351/25 | 591/22 | ||||
| D2 | ||||||||||
| UDS | 325/39 | 164 (39/125) | ||||||||
| SGS | 45/22 | 220/24 | 325/26 | 463/24 | 2118/29b | |||||
| K9 | ||||||||||
| UDS | 231/16 | 259/2 | 443/2 | 46 (20/26) | ||||||
| SGS | 136/14 | 315/12 | ||||||||
| P2 | ||||||||||
| UDS | 86/4 | 931/46 | 1015/58 | 277 (108/169) | ||||||
| SGS | 86/22 | 413/25 | 469/9 | 546/25 | 696/24 | 931/28 | 1015/22 | |||
| R5 | ||||||||||
| UDS | 85/5 | 259/5 | 302/2 | 402/2 | 457/16 | 522/5 | 591/11 | 667/19 | 730/11 | 76 (76/0) |
| SGS | ||||||||||
| G5 | ||||||||||
| UDS | 56/14 | 196/11 | 408/2 | 604/9 | 840/1 | 869/9 | 932/26 | 72 (72/0) | ||
| SGS | ||||||||||
Numbers indicate the day of sampling since EDI and the number of sequences available at each time point. For NAb rows, numbers indicate the day of sampling since EDI.
Under antiretroviral therapy. Blank cells indicate that data were unavailable.
Table 3.
Time point of neutralizing antibody assays and mean Nab titers
| Subject ID | Time point (in days after EDI) of NAb assays/ID50a |
No. of time points (UDS and SGS) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
| K6 | 124/43 | 180/21 | 389/15* | 3 | ||||||
| S1 | 85/147 | 155/120 | 351/133 | 497/17 | 4 | |||||
| D2 | 45/42 | 325/18 | 463/24 | 3 | ||||||
| K9 | 94/155 | 231/51 | 315/10 | 443/10 | 4 | |||||
| P2 | 86/585 | 163/737 | 352/406 | 546/291 | 696/262 | 757/146 | 1,015/15 | 7 | ||
| R5 | 85/83 | 259/182 | 730/10* | 3 | ||||||
| G5 | 56/8,513 | 408/1,480 | 604/223 | 840/34 | 869/25 | 93,258 | 6 | |||
Blank cells indicate that data were unavailable. Numbers indicate the day of sampling since EDI and the mean autologous NAb titer at each time point. Boldface indicates time and Nab titers contemporaneous with or following (*) the first evidence of superinfection. For subjects K6 and R5, NAb assays were not available at the time of SI.
Table 4.
Overall mean viral diversity within env regions across the 7 superinfected subjects
| Subject | Diversitya |
|
|---|---|---|
| Mean | Range (minimum-maximum) | |
| S1 | 0.06 | 0.01–0.14 |
| K6 | 0.03 | 0.02–0.05 |
| K9 | 0.02 | 0.01–0.09 |
| P2 | 0.06 | 0.03–0.13 |
| D2 | 0.04 | 0.02–0.11 |
| R5 | 0.04 | 0.01–0.13 |
| G5 | 0.07 | 0.02–0.14 |
The mean values within each individual and the ranges (minimum and maximum) are indicated. The latest time point for subject D2 (under antiretroviral therapy) was removed.
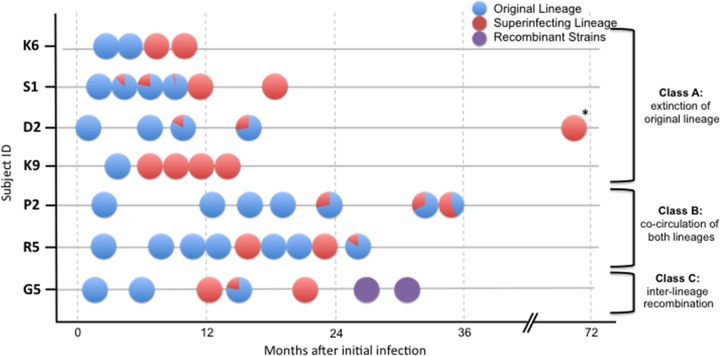
Fig 1.
Relative proportion of original and superinfecting lineages during the follow-up of the 7 subjects. The y axis represents 7 individuals identified as being superinfected, and the x axis is the time from the estimated date of initial infection. Circles represent sampled time points. Blue, initial lineage; red, superinfecting lineages; purple, recombinant strains. Slices represent the relative proportion of each lineage. Classes A and B are associated with a weak autologous NAb response. In the class C individual, there was a strong autologous NAb response toward SI strains. The time point indicated by an asterisk was after initiation of antiretroviral therapy.
Recombination analyses.
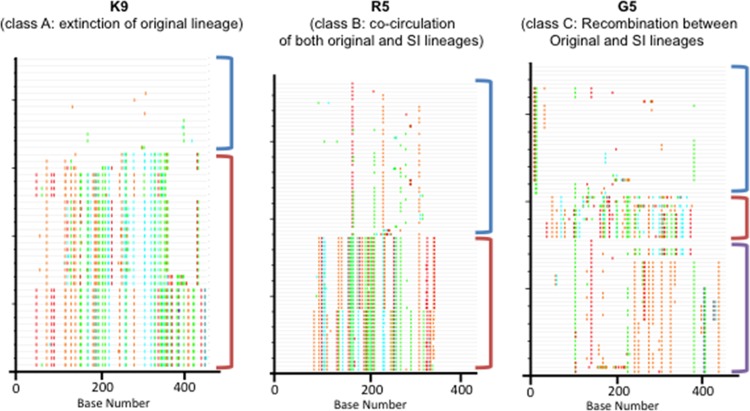
Sequences were also screened for recombinant variants by visual inspection of Highlighter plots using Highlighter from the Los Alamos National Laboratory HIV Sequence Database (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/HIGHLIGHT_XYPLOT/highlighter.html) (31) and a genetic algorithm for identifying recombination breakpoints (GARD) (32) (Fig. 2).
Fig 2.
Highlighter plots of aligned env from three representative individuals with distinct dynamics. In the plots, base differences from the top master sequence are highlighted with colored ticks. Lineages are indicated with colored brackets: original, blue; SI, red; and recombinant strains, purple. K9 is an example of the extinction of the original lineage. R5 shows the cocirculation of both original and superinfected lineages. G5 depicts intense recombination events between the initial lineage and SI lineages associated with escape from contemporaneous autologous NAb response.
Neutralization assays.
NAb assays against autologous pseudoviruses were performed by Monogram Biosciences Inc. (15). Briefly, autologous viral envelope populations were used to produce pseudotyped virus stocks for each of the subjects at each available time point. The plasmid preparations used in the neutralization experiments contained the HIV-1 Env quasispecies derived as a population from the patient plasma, so for the neutralization assays a representative sampling of all of the Env variants present in the subject's plasma were also present in pseudotyped viral stocks. Each individual's plasma sample was tested in the neutralization assay with each of the longitudinal pseudovirus stocks as previously described (15). These pseudovirions were incubated for 1 h with serial 4-fold dilutions of autologous plasma. Virus infectivity was determined 72 h postinoculation by measuring the amount of luciferase activity expressed in infected cells. Neutralizing activity is displayed as the percent inhibition of viral replication (luciferase activity) at each antibody (Ab) dilution compared to an antibody-negative control: percent inhibition = [1 − (luciferase + Ab/luciferase − Ab]) × 100. The titer was defined as the reciprocal of the dilution of plasma that produces 50% inhibition of virus replication (ID50). A threshold ID50 titer of 20 was used to identify significant NAb responses (Table 5) (9, 15).
Table 5.
Autologous NAb titers to contemporaneous viruses at the time of first evidence of superinfection
| Subject | Estimated time of SI (in days since EDI) | ANAb ID50 |
||
|---|---|---|---|---|
| Minimum | Mean | SD | ||
| K6 | 219 | 10 | 16 | 9.1 |
| R5 | 457 | 10 | 139 | 258 |
| 667 | 10 | 10 | 10 | |
| G5 | 408 | 10 | 1,480 | 1,209 |
| K9 | 231 | 10 | 51 | 33.1 |
| S1 | 155 | 10 | 120 | 182.7 |
| P2 | 546 | 10 | 291 | 118.2 |
| D2 | 325 | 10 | 18 | 13.9 |
The means, medians, and standard deviations (SD) of autologous NAb (ANAb) titer at the time of the superinfecting event within each individual are denoted. For subject R5, two possible superinfecting events were identified. ID50 titers below the limit of detection (<20) were assigned to half the limit of detection (i.e., 10).
Phylogenetic reconstruction.
Multiple alignments of the partial HIV-1 env, gag, and RT regions were constructed with MAFFT (33) and then manually edited using BioEdit (34). Sequences that could not be unambiguously aligned were removed.
Demographic and evolutionary parameters of intrahost dynamics of initial and superinfecting viral lineages were also estimated by a Bayesian Markov chain Monte Carlo (BMCMC) inference implemented in BEAST v1.7.4 (35) using partial env sequences. Briefly, BMCMC runs of 50 to 100 million generations were performed for each analysis with a GTR+Γ4 substitution model under an exponential coalescent model tree prior. All analyses were performed using an uncorrelated log-normal relaxed molecular clock (35). Maximum clade credibility trees were selected with the software TreeAnnotator v1.7.4, with the first 10% of generations discarded as burn-in. Trees were visualized in FigTree v.1.4.0, and Tracer v.1.5 (http://beast.bio.ed.ac.uk/Tracer) was used to check for convergence.
RESULTS
Population and sequence data.
Seven HIV-1 intrasubtype B superinfection cases were analyzed, with a mean duration of follow-up of 18.2 months (range, 4.1 to 31.0). All were men who reported sex with other men as their main HIV risk factor. Their mean age at enrollment was 33.7 years (range, 21 to 51), and their mean estimated duration of infection at enrollment was 85.4 days (range, 26 to 170) (Table 1).
HIV-1 env sequences were generated from multiple time point samples for each individual with a mean of 6.3 time points (range, 5 to 9 time points) (Tables 2 and 3 and Fig. 1). The numbers of UDS haplotypes and SGS sequences generated at each time point are summarized in Table 2. The mean sequence length was 192 bp (range, 50 to 471 bp). Altogether, the mean number of partial env sequences available for each individual was 132.4 sequences (range, 46 to 277 sequences) (Table 2). Overall, superinfection was detected within the first 2 years after EDI (mean date of SI of 330 days after EDI [range, 155 to 546 days]) (Fig. 1).
Intrahost viral dynamics and evolution.
As expected, phylogenetic analysis of the combined data sets showed well-supported clades for each individual (data not shown), and original and superinfecting lineages were observed in each individual. In subject G5, we found evidence for a recombination event (breakpoint at position 227 [GARD; P = 0.01]) between original and superinfecting viral populations. Therefore, we performed separate Bayesian phylogenetic analyses for each G5 env nonrecombinant region. In each individual, the relative proportion of the original infecting lineage, superinfecting lineage, and recombinant lineage varied over time (Fig. 1). Highlighter plot analysis confirmed the presence of distinct lineages, consistent with superinfection (Fig. 2).
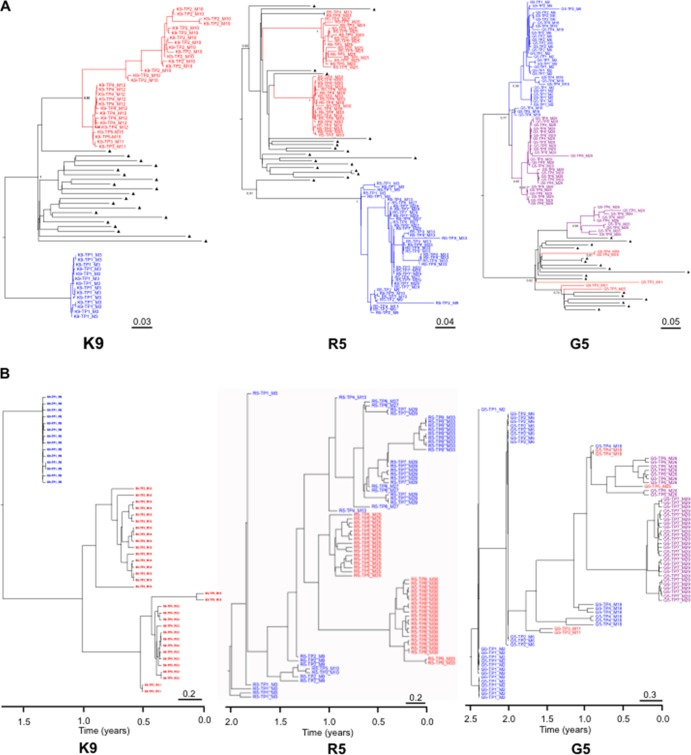
We classified viral evolution profiles according to the persistence or extinction of original and superinfecting lineages at the subsequent time points. Altogether, these results allowed us to characterize three different virologic profiles after superinfection: (i) extinction of the original lineage (individuals K6, S1, D2, and K9) (Fig. 1 to 3); (ii) cocirculation of both original and superinfecting lineages (individuals P2 and R5; Fig. 1 to 3); and (iii) recombination between original and superinfecting lineages for individual G5. These phylogenetic patterns within the first env region derived from either the original or the superinfecting lineage were highly supported (posterior probability P of 1) (Fig. 3). Interestingly, the tree topology for subject R5 (Fig. 3A) demonstrated distinct superinfecting lineages at the 5th and the 8th sampled time points with high posterior probability (P = 0.95 to 0.99). More precisely, it showed the absence of ongoing replication of the first SI subpopulation (time point 5) at time points 6 and 7, which could illustrate the extinction of this first SI subpopulation (i.e., SI failure or SI extinction), and a second potential SI event at time point 8, with no close phylogenetic relationship either with initial strains or with the first potential SI strains.
Fig 3.
(A) BMCMC tree of partial env sequences for individuals K9, R5, and G5. The sequences from the original lineage are depicted in blue. The sequences from the SI lineage are depicted in red, while those in purple represent potential recombinant sequences between original and SI strains. Background sequences are labeled with black triangles. Posterior probably of main lineages are indicated at the root. Tip labels indicate time points of sampling (TP) and month after infection (M). Scale bars represent genetic distances in substitutions/site. (B) Time-scaled BMCMC phylogenetic trees of partial env sequences for individuals K9, R5, and G5. The sequences from the original lineage are depicted in blue. The sequences from the SI lineage are depicted in red, while those in purple represent potential recombinant sequences between original and SI strains. Tip labels indicated time points of sampling (TP) and month after infection (M). The trees represent the ancestral relationships of sequences belonging to each lineage. The x axis represents time in years before the latest sampling date. Scale bars represent time in years.
Similar analyses of viral dynamics were performed for both RT (6 individuals) and gag (4 individuals) UDS data. Recombination events within gag or RT coding regions were not detected, including for subject G5. Viral dynamics of the HIV-1 RT region within each subject were consistent with env analyses. Superinfection with extinction of the original lineage or cocirculation of both the original and superinfecting lineages occurred for all subjects except for K9 and K6, for whom sufficient RT sequences were not available (see Table S1 in the supplemental material). The limited numbers of gag sequences available for most of the individuals did not permit inference in this coding region.
Viral dynamics and autologous neutralizing antibody response.
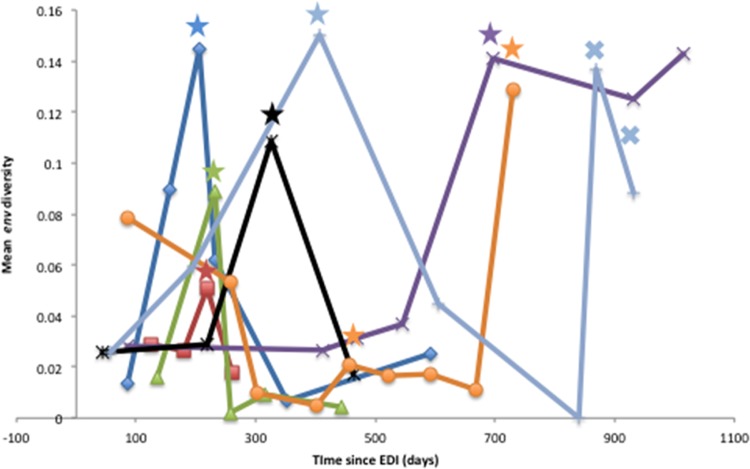
We next considered the relationship between viral evolution and autologous NAb development during the period of observation. The mean env nucleotide diversity of the sampled HIV RNA population was measured at each time point sampled and within each subject. The global mean viral diversity within each subject varied from 0.02 to 0.07 (Table 4). Neutralizing activity of antibody in each individual's plasma sample against autologous contemporaneous pseudoviruses was then evaluated, and NAb ID50 titers were generated for 3 to 7 time points per participant (Tables 3 and 5). Consistent with the detection of superinfection, maximum viral diversity for each participant was observed at the time of superinfection and at the time of recombination for subject G5 (Fig. 4). Interestingly, mean NAb titers to autologous viruses at the time of and shortly following superinfection were barely detectable for six of the seven subjects. Specifically at the time of superinfection, the ID50 titers for participants K6 and D2 were the lowest, at 15 and 18, respectively, while for K9, P2, S1, and R5, ID50 titers ranged from 51 to 291. However, a strong NAb response to the autologous contemporaneous viral population was observed at the time of superinfection for G5 (mean ID50, 1,480) (Table 5). Participant G5 also displayed high viral diversity within env at the two latest time points available (with mean diversities of 0.14 and 0.09, respectively) where recombination events were observed. Taken together, these associated virologic and antibody data suggest that higher levels of autologous NAb drives viral recombination after superinfection has occurred.
Fig 4.
Trend of mean viral diversification within env regions across the 7 superinfected subjects. Mean env viral diversity at each time point is plotted relative to the time since the estimated date of infection (EDI). Participant S1 is shown as the dark blue line, K6 as the red line, K9 as the green line, P2 as the purple line, D2 as the black line, R5 as the orange line, and G5 as the light blue line. Colored stars indicate the earliest time point with evidence of superinfection for each individual. Crosses indicate times of recombinant events identified in subject G5. The highest diversity within each subject corresponded to the time of superinfecting event(s) and, for G5, to the time when recombination was evident.
DISCUSSION
Understanding the immunological and virological dynamics of HIV-1 superinfection may be important in the development of efficient vaccine strategies, since superinfection can be considered a vaccine failure. Therefore, correlates of susceptibility to superinfection may reflect correlates of protection with vaccines. In this longitudinal study of seven superinfected individuals, we provided unique insights into the postsuperinfection evolution of viral lineages under the selective pressure of the NAb response. We observed three different patterns of viral subpopulation dynamics. In four of seven individuals, there was evidence of extinction of the original lineage, highlighting the ability of the superinfecting lineage to overcome the preexisting immune response within these hosts. Distinct original and superinfecting lineages cocirculated in two individuals, and the two strains recombined in one individual. This recombination between original and superinfecting lineages likely facilitated evasion of host immune responses, since viral replication continued at high levels (5.23 log10 HIV-1 RNA copies/ml) despite preexisting NAb.
Immune correlates of protection against superinfection have been investigated previously. Hence, a broad CD8+ T-cell immune response does not seem to prevent HIV-1 superinfection (6) but could drive selection of new recombinant variants after superinfection (36). Similarly, humoral immune responses around superinfection events have been previously studied, and a lack of NAb was associated with superinfection events (9, 37). However, it was previously unknown if NAb responses could also drive recombination between original and superinfecting strains. In this study, we observed significantly increased viral diversity and recombination between original and superinfecting lineages in the one individual who developed the highest autologous NAb response.
Next-generation sequencing is a promising approach to better estimate the genetic diversity of viral population and by detecting low-frequency variants (25, 26, 38). However, the large number of possible haplotypes in a set of reads generated by UDS raises computational and analytical problems. Using computational and statistical procedures for haplotype reconstruction and haplotype frequency, our study confirms the potential benefits of UDS for the analysis of the genetic diversity of viral populations. With these methods, we observed that the highest amount of env diversity was associated with the identification of superinfecting events for all of the individuals. Interestingly, it also confirmed the possibility that two distinct superinfections with two different strains occurred within subject R5, as suggested by both highlighter plots (Fig. 2) and tree topology (Fig. 3A). For subject G5 we were able to identify a putative recombination event within env arising at the latest time points, corresponding with an increase in observed viral diversity. Despite the limited size of this study, these observations demonstrate that this computational strategy can be used to evaluate the viral dynamics occurring during superinfection.
In summary, the humoral response can exert very strong selective pressures on HIV-1 and can drive rapid viral escape (15, 18). In this study, we confirmed that humoral immune selection pressure is also associated with viral recombination and likely NAb escape, similar to previous reports on cytotoxic T lymphocyte pressure (36). Such immune-driven viral recombination may also make superinfection harder to detect by homogenizing the circulating viral population, and this observation may explain why the detection of superinfection typically has been associated with low autologous NAb titers (9).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Caroline Ignacio for her wonderful technical support and Demetrius Dela Cruz for his administrative assistance, as well as to all the participants in the San Diego Primary Infection Cohort.
This work was supported by the Department of Veterans Affairs and grants from the National Institutes of Health (AI090970, AI100665, MH097520, DA034978, AI43629, AI36214, AI74621, and AI47745), the International AIDS Vaccine Initiative, the National Science Foundation (DMS0714991), and the James B. Pendleton Charitable Trust.
Footnotes
Published ahead of print 18 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02260-13.
REFERENCES
- 1.Ramos A, Hu DJ, Nguyen L, Phan K-O, Vanichseni S, Promadej N, Choopanya K, Callahan M, Young NL, McNicholl J, Mastro TD, Folks TM, Subbarao S. 2002. Intersubtype human immunodeficiency virus type 1 superinfection following seroconversion to primary infection in two injection drug users. J. Virol. 76:7444–7452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jost S, Bernard MC, Kaiser L, Yerly S, Hirschel B, Samri A, Autran B, Goh LE, Perrin L. 2002. A patient with HIV-1 superinfection. N. Engl. J. Med. 347:731–736 [DOI] [PubMed] [Google Scholar]
- 3.Smith DM, Wong JK, Hightower GK, Ignacio CC, Koelsch KK, Petropoulos CJ, Richman DD, Little SJ. 2005. HIV drug resistance acquired through superinfection. AIDS 19:1251–1256 [DOI] [PubMed] [Google Scholar]
- 4.Pacold ME, Pond SLK, Wagner GA, Delport W, Bourque DL, Richman DD, Little SJ, Smith DM. 2012. Clinical, virologic, and immunologic correlates of HIV-1 intraclade B dual infection among men who have sex with men. AIDS 26:157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottlieb GS, Nickle DC, Jensen MA, Wong KG, Grobler J, Li F, Liu S-L, Rademeyer C, Learn GH, Karim SSA, Williamson C, Corey L, Margolick JB, Mullins JI. 2004. Dual HIV-1 infection associated with rapid disease progression. Lancet 363:619–622 [DOI] [PubMed] [Google Scholar]
- 6.Altfeld M, Allen TM, Yu XG, Johnston MN, Agrawal D, Korber BT, Montefiori DC, O'Connor DH, Davis BT, Lee PK, Maier EL, Harlow J, Goulder PJ, Brander C, Rosenberg ES, Walker BD. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420:434–439 [DOI] [PubMed] [Google Scholar]
- 7.Kijak GH, McCutchan FE. 2005. HIV diversity, molecular epidemiology, and the role of recombination. Curr. Infect. Dis. Rep. 7:480–488 [DOI] [PubMed] [Google Scholar]
- 8.Yang C, Li M, Shi Y-P, Winter J, van Eijk AM, Ayisi J, Hu DJ, Steketee R, Nahlen BL, Lal RB. 2004. Genetic diversity and high proportion of intersubtype recombinants among HIV type 1-infected pregnant women in Kisumu, western Kenya. AIDS Res. Hum. Retrovir. 20:565–574 [DOI] [PubMed] [Google Scholar]
- 9.Smith DM, Strain MC, Frost SDW, Pillai SK, Wong JK, Wrin T, Liu Y, Petropolous CJ, Daar ES, Little SJ, Richman DD. 2006. Lack of neutralizing antibody response to HIV-1 predisposes to superinfection. Virology 355:1–5 [DOI] [PubMed] [Google Scholar]
- 10.Basu D, Kraft CS, Murphy MK, Campbell PJ, Yu T, Hraber PT, Irene C, Pinter A, Chomba E, Mulenga J, Kilembe W, Allen SA, Derdeyn CA, Hunter E. 2012. HIV-1 subtype C superinfected individuals mount low autologous neutralizing antibody responses prior to intrasubtype superinfection. Retrovirology 9:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pacold M, Smith D, Little S, Cheng PM, Jordan P, Ignacio C, Richman D, Pond SK. 2010. Comparison of methods to detect HIV dual infection. AIDS Res. Hum. Retrovir. 26:1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cousins MM, Ou S-S, Wawer MJ, Munshaw S, Swan D, Magaret CA, Mullis CE, Serwadda D, Porcella SF, Gray RH, Quinn TC, Donnell D, Eshleman SH, Redd AD. 2012. Comparison of a high-resolution melting assay to next-generation sequencing for analysis of HIV diversity. J. Clin. Microbiol. 50:3054–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibata R, Siemon C, Cho MW, Arthur LO, Nigida SM, Jr, Matthews T, Sawyer LA, Schultz A, Murthy KK, Israel Z, Javadian A, Frost P, Kennedy RC, Lane HC, Martin MA. 1996. Resistance of previously infected chimpanzees to successive challenges with a heterologous intraclade B strain of human immunodeficiency virus type 1. J. Virol. 70:4361–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232–13252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortiz GM, Wellons M, Brancato J, Vo HT, Zinn RL, Clarkson DE, Van Loon K, Bonhoeffer S, Miralles GD, Montefiori D, Bartlett JA, Nixon DF. 2001. Structured antiretroviral treatment interruptions in chronically HIV-1-infected subjects. Proc. Natl. Acad. Sci. U. S. A. 98:13288–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 18.Frost SDW, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, Petropoulos CJ, Little SJ, Richman DD. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. U. S. A. 102:18514–18519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le T, Wright EJ, Smith DM, He W, Catano G, Okulicz JF, Young JA, Clark RA, Richman DD, Little SJ, Ahuja SK. 2013. Enhanced CD4+ T-cell recovery with earlier HIV-1 antiretroviral therapy. N. Engl. J. Med. 368:218–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gianella S, Delport W, Pacold ME, Young JA, Choi JY, Little SJ, Richman DD, Kosakovsky Pond SL, Smith DM. 2011. Detection of minority resistance during early HIV-1 infection: natural variation and spurious detection rather than transmission and evolution of multiple viral variants. J. Virol. 85:8359–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith D, Delport W, Butler D, Little S, Richman D, Pond SK. 2010. Response to comment on “The origins of sexually transmitted HIV among men who have sex with men.” Sci. Transl. Med. 2:501r1. 10.1126/scitranslmed.3001473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pond SLK, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679 [DOI] [PubMed] [Google Scholar]
- 24.Zagordi O, Klein R, Däumer M, Beerenwinkel N. 2010. Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies. Nucleic Acids Res. 38:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zagordi O, Bhattacharya A, Eriksson N, Beerenwinkel N. 2011. ShoRAH: estimating the genetic diversity of a mixed sample from next-generation sequencing data. BMC Bioinformatics 12:119. 10.1186/1471-2105-12-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eriksson N, Pachter L, Mitsuya Y, Rhee S-Y, Wang C, Gharizadeh B, Ronaghi M, Shafer RW, Beerenwinkel N. 2008. Viral population estimation using pyrosequencing. PLoS Comput. Biol. 4:e1000074. 10.1371/journal.pcbi.1000074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beerenwinkel N, Zagordi O. 2011. Ultra-deep sequencing for the analysis of viral populations. Curr. Opin. Virol. 1:413–418 [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512–526 [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony. Methods Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon AFY, McGovern RA, Mo T, Knapp DJHF, Brenner B, Routy J-P, Wainberg MA, Harrigan PR. 2011. Dates of HIV infection can be estimated for seroprevalent patients by coalescent analysis of serial next-generation sequencing data. AIDS 25:2019–2026 [DOI] [PubMed] [Google Scholar]
- 31.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping L-H, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098 [DOI] [PubMed] [Google Scholar]
- 33.Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537:39–64 [DOI] [PubMed] [Google Scholar]
- 34.Hall TA. 1999. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 35.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Streeck H, Li B, Poon AFY, Schneidewind A, Gladden AD, Power KA, Daskalakis D, Bazner S, Zuniga R, Brander C, Rosenberg ES, Frost SDW, Altfeld M, Allen TM. 2008. Immune-driven recombination and loss of control after HIV superinfection. J. Exp. Med. 205:1789–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blish CA, Dogan OC, Derby NR, Nguyen MA, Chohan B, Richardson BA, Overbaugh J. 2008. Human immunodeficiency virus type 1 superinfection occurs despite relatively robust neutralizing antibody responses. J. Virol. 82:12094–12103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zagordi O, Geyrhofer L, Roth V, Beerenwinkel N. 2010. Deep sequencing of a genetically heterogeneous sample: local haplotype reconstruction and read error correction. J. Comput. Biol. J. Comput. Mol. Cell. Biol. 17:417–428 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.