Abstract
Macrophages play important roles in host immune defense against virus infection. During infection by herpes simplex virus 1 (HSV-1), macrophages acquire enhanced antiviral potential. Restriction of HSV-1 replication and progeny production is important to prevent viral spread, but the cellular mechanisms that inhibit the DNA virus in macrophages are unknown. SAMHD1 was recently identified as a retrovirus restriction factor highly expressed in macrophages. The SAMHD1 protein is expressed in both undifferentiated monocytes and differentiated macrophages, but retroviral restriction is limited to differentiated cells by modulation of SAMHD1 phosphorylation. It is proposed to block reverse transcription of retroviral RNA into DNA by depleting cellular deoxynucleotide triphosphates (dNTPs). Viruses with DNA genomes do not employ reverse transcription during infection, but replication of their viral genomes is also dependent on intracellular dNTP concentrations. Here, we demonstrate that SAMHD1 restricts replication of the HSV-1 DNA genome in differentiated macrophage cell lines. Depleting SAMHD1 in THP-1 cells enhanced HSV-1 replication, while ectopic overexpression of SAMHD1 in U937 cells repressed HSV-1 replication. SAMHD1 did not impact viral gene expression from incoming HSV-1 viral genomes. HSV-1 restriction involved the dNTP triphosphohydrolase activity of SAMHD1 and was partially overcome by addition of exogenous deoxynucleosides. Unlike retroviruses, restriction of HSV-1 was not affected by SAMHD1 phosphorylation status. Our results suggest that SAMHD1 functions broadly to inhibit replication of DNA viruses in nondividing macrophages.
INTRODUCTION
Intrinsic immune mechanisms form a frontline of antiviral host defense and are mediated by constitutively expressed and active cellular proteins (1). Recent years have seen an explosion in the identification of host proteins that act as restriction factors to block specific stages of viral replication cycles in infected cells (2–5). The antiviral activity of restriction factors can be enhanced by interferons that often upregulate their expression. Intrinsic cellular defenses discovered due to their restriction of retroviruses include APOBEC3 proteins (6), tetherin/BST2 (7), and SAMHD1 (8, 9). Interestingly, many of these factors are broad effectors of antiviral defense. In addition to retroviruses, human DNA viruses are also subject to the antiviral activities of interferon-induced cellular restriction factors (10–13). Known intrinsic defenses that function against herpesviruses include the interferon-inducible protein 16 (IFI16) DNA sensor (14, 15), recruitment of DNA damage proteins (16), repression of gene expression by Sp100 and Daxx (11, 13, 17), and blocking virion assembly at late stages of infection by viperin (18). The same strategies used by restriction factors to block retroviruses may also be employed to limit infection of DNA viruses. For example, both the APOBEC3 and tetherin/BST2 proteins have been suggested to inhibit herpesvirus infection (19, 20). The human SAMHD1 protein was recently identified as a retroviruses restriction factor (8, 9). Although demonstrated to limit reverse transcription (RT) for a diverse array of retroviruses (21, 22), it is not yet known the extent to which the same mechanisms might be employed by SAMHD1 to restrict replication of viruses with DNA genomes.
The Herpesviridae are a large family of double-stranded DNA viruses that display distinct tissue tropisms and are responsible for a number of human diseases. Herpes simplex virus 1 (HSV-1) is a ubiquitous human pathogen that causes oral lesions, corneal infections, and encephalitis (23, 24) and persists latently in sensory neurons of the trigeminal ganglia (25). HSV-1 is also a significant contributing factor to genital transmission of human immunodeficiency virus (HIV) in patients who are coinfected. Lytic HSV-1 infection proceeds through immediate early, early, and late phases, leading to production of infectious virions and host cell lysis. DNA replication uses seven essential HSV-encoded proteins but also relies on host factors and nucleotide biosynthetic machinery (26). Control of HSV-1 replication during the initial phase of infection is crucial for the host to limit virus replication and dissemination. Macrophages play a number of important roles in limiting the early stages of HSV-1 infection (27). Resting macrophages are restricted for HSV-1 replication, but the cellular effectors that limit virus replication are unknown.
The SAMHD1 enzyme restricts retrovirus infection in macrophages, dendritic cells, and resting CD4+ T cells (28–30). It was identified through biochemical purification of the accessory factor Vpx encoded by certain lentiviruses (8, 9). The Vpx protein encoded by HIV-2 and closely related simian immunodeficiency virus (SIV) strains is incorporated into viral particles and facilitates infection of myeloid cells through proteasome-mediated degradation of SAMHD1 (8, 9). In contrast, HIV-1 and other retroviruses do not encode Vpx proteins that degrade SAMHD1 (31), rendering them inefficient for replication in myeloid cells. SAMHD1 is implicated as a regulator of the innate immune response, and its expression is induced by interferon (32). The histidine-aspartic acid domain (HD) of the SAMHD1 protein possesses dGTP-regulated deoxynucleotide triphosphohydrolase activity that decreases intracellular deoxynucleotide triphosphate (dNTP) levels (3, 33). This activity is responsible for the very low dNTP concentrations found in terminally differentiated nondividing human monocyte-derived macrophages (34, 35). The SAMHD1 protein also binds to DNA and RNA and is suggested to have exonuclease activity on single-stranded nucleic acids (36). Increasing intracellular dNTP pools by depleting SAMHD1 or treating with dNTP precursors (dNs) rescues HIV-1 infectivity of macrophages and resting CD4+ T cells (37, 38). The Vpx protein of SIV can increase the ability of HIV-1 to infect macrophages and dendritic cells when incorporated into virions or provided in trans (38–40). Since herpesviruses that consist of DNA genomes rely on cellular dNTPs for replication of their DNA genomes in the nucleus, we examined whether SAMHD1 affects infection of macrophages by HSV-1. Here, we demonstrate that SAMHD1 serves as a restriction factor for HSV-1 infection in differentiated human macrophage cell lines. We show that inhibition of HSV-1 by SAMHD1 involves a block in viral DNA replication and has features distinct from that used to restrict HIV-1. Together with recent findings with HSV-1 and vaccinia virus (41), these results suggest that SAMHD1 acts broadly to suppress replication of DNA viruses in nondividing myeloid cells.
MATERIALS AND METHODS
Cells and reagents.
Cells were obtained from the American Type Culture Collection and were grown in a 5% CO2 humidified incubator at 37°C. THP-1 and U937 cells were grown in RPMI 1640 with GlutaMAX, while 293T and Vero cells were gown in Dulbecco's modified Eagle's medium (DMEM). The cell growth medium also contained 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin. THP-1 and U937 cells were differentiated for 48 h with 50 ng/ml phorbol myristate acetate (PMA). Deoxynucleosides (dNs; 2′-deoxyadenosine monohydrate, 2′-deoxycytidine, 2′-deoxyguanosine monohydrate, and thymidine), PMA, and phosphonoacetic acid (PAA) were purchased from Sigma. dN solutions were prepared and treated as described previously (38). PAA was used at a final concentration of 200 μg/ml.
Viruses and infections.
All experiments used either HSV-1 strain 17syn+ or a green fluorescent protein (GFP)-expressing 17syn+ strain containing a cytomegalovirus (CMV)-GFP cassette in the US5 region. Viruses were propagated and titrated on Vero cells. Infections were performed in serum-free medium. After 1 h at 37°C, cells were washed twice with PBS, and fresh medium with serum was added. For HSV-1 plaque assays, Vero cells in 24-well plates were infected with 10-fold serial dilutions of viruses. After virus adsorption for 1 h, the cells were overlaid with medium containing 0.5% carboxymethylcellulose. Plaques were stained with crystal violet at 3 days postinfection.
Retroviral transduction and VLP generation.
Lentiviral vectors were used to generate SAMHD1-depleted cells. The pLKO.1-expressing short hairpin RNAs (shRNAs) for luciferase (TRCN0000072243) and SAMHD1 (TRCN0000343807 and TRCN0000343808) were purchased from Open Biosystems. Lentiviruses were prepared by cotransfecting 293T cells with lentiviral vectors and the packaging plasmids pCMV-ΔR8.74 and pMD.G using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Cell supernatants were harvested at 48 h after transfection and filtered at 0.45 μm. THP-1 cells were transduced with lentiviruses in the presence of Polybrene (10 μg/ml) and then selected with puromycin (1 μg/ml). The selected cells were maintained in medium containing 0.5 μg/ml of puromycin. Retroviral transduced U937 cells expressing SAMHD1 wild type and mutants were described previously (22, 42). For viruslike particle (VLP)-hemagglutinin (HA)-Vpx production, pcHA-Vpx, pPBj.pack, and pMD.G were cotransfected in 293T cells. Cell supernatants were harvested at 48 h after transfection and filtered. After 2 h of incubation with VLP, differentiated THP-1 cells were infected with HSV-1 at a multiplicity of infection (MOI) of 0.1.
Antibodies.
The antibody used to detect ICP0 was Mab11060 (43), and the antibody to ICP4 was generated from 58S, an ATCC hybridoma cell line. Primary antibodies were purchased from Abcam (SAMHD1, ab117908; β-actin, ab8227), Santa Cruz (GAPDH, sc-25778; α-tubulin, sc-69969), Covance (HA, MMS-101R), and Sigma (Flag, F3165). All secondary antibodies were from Jackson Laboratories.
Immunoblot analysis and fluorescence-activated cell sorting (FACS).
Cells were washed with phosphate-buffered saline (PBS), and total cell extracts were prepared by boiling the cell pellets in sodium dodecyl sulfate (SDS) loading buffer. Proteins were separated via SDS-PAGE, and immunoblotting was carried out by standard protocols. Proteins were visualized using a luminol/enhancer solution (Thermo), ECL+Plus (GE Healthcare), and G:Box imaging system (Syngene). For counting GFP-positive THP-1 cells after 24 h of HSV-1–GFP infection, trypsinized cells were washed with PBS, fixed with 4% paraformaldehyde in PBS, and analyzed on a BD FACSCanto II flow cytometer.
RT-PCR and quantitative PCR.
Cells were infected with HSV-1 at an MOI of 0.1 and harvested at the indicated time points. Total DNA was extracted using the PureLink genomic DNA minikit (Invitrogen). Viral DNA was quantified by using the ICP27-specific primers GCATCCTTCGTGTTTGTCATT (forward) and GCATCTCTCCGACCCCG (reverse) and normalized to the endogenous RPLP0 gene, which was amplified using primers CTGGAAGTCCAACTACTTCC (forward) and TGCTGCATCTGCTTGGAGCC (reverse). For RT-PCR, total RNA was isolated (RNeasy minikit; Qiagen) and was reverse transcribed using a high-capacity RNA-to-cDNA kit (Applied Biosystems). Amplifying sequences were detected using Power SYBR green (Applied Biosystems) PCR reporter dye in a ViiA7 real-time PCR system (Applied Biosystems).
RESULTS
SAMHD1 depletion increases HSV-1 replication in differentiated THP-1 cells.
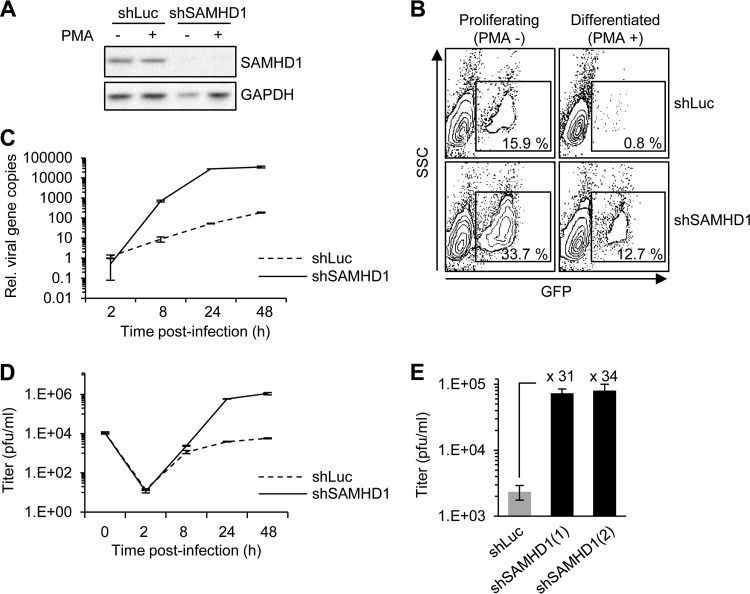
We examined HSV-1 infection in the THP-1 cells, a human cell line derived from peripheral blood of an acute monocytic leukemia patient that can be differentiated with phorbol esters. THP-1 cells were infected in the undifferentiated state or after differentiation by the phorbol ester PMA. We generated THP-1 cells in which SAMHD1 expression was knocked down using a lentivirus vector encoding shRNA (shSAMHD1) and compared their viral transduction to that of cells containing control shRNA to the luciferase gene (shLuc). The efficiency of SAMHD1 knockdown was confirmed by immunoblotting of endogenous protein levels (Fig. 1A). Cells were infected with HSV-1 expressing a GFP reporter at an MOI of 0.1 and analyzed by flow cytometry at 24 h postinfection (hpi). In proliferating THP-1 cells, knockdown of SAMHD1 had a marginal effect (2.1-fold increase) on the number of GFP-positive cells (Fig. 1B). In contrast, infection was severely restricted in PMA-treated differentiated cells (19.9-fold decrease), and depleting SAMHD1 had a significant rescue of virus replication (15.9-fold increase). These results show that differentiation of THP-1 cells leads to restriction of HSV-1 replication and that SAMHD1 is a major restriction factor in these cells. This conclusion was supported by measuring the accumulation of viral DNA using quantitative PCR (qPCR) and the amount of progeny virus generated by plaque assay (Fig. 1C and D). HSV-1 replication in differentiated THP-1 cells was severely attenuated, but depletion of SAMHD1 resulted in a 538-fold increase in viral DNA and a 150-fold increase in progeny production at 24 hpi. Equivalent results were obtained with two different shRNAs to SAMHD1 (Fig. 1E).
Fig 1.
Replication of HSV-1 is enhanced in SAMHD1-depleted THP-1. (A) THP-1 cells were stably transduced with lentiviruses containing luciferase shRNA (shLuc) or SAMHD1 shRNA (shSAMHD1). SAMHD1 protein levels were analyzed by immunoblotting in lysates from untreated or PMA-treated THP-1 cells. GAPDH served as a loading control. (B) Untreated and PMA-treated differentiated THP-1 cells were infected at an MOI of 0.1 with HSV-1 (strain 17syn+) expressing GFP as a reporter. Cells were analyzed by flow cytometry at 24 hpi. (C) DNA was extracted at the indicated time points from differentiated THP-1 cells infected with HSV-1 at an MOI of 0.1. The relative number of viral genomes was determined by quantitative PCR using primers to the ICP27 gene. Error bars represent the standard deviations from triplicate samples. (D) Cell culture supernatants from differentiated THP-1 cells infected with HSV-1 at an MOI of 0.1 were analyzed at the indicated time points for progeny virus titers using plaque assays in Vero cells. (E) Two independent shRNAs to SAMHD1 (labeled 1 and 2) achieved equivalent levels of HSV-1 enhancement. Cell culture supernatants from cells infected with HSV-1 at an MOI of 0.1 were analyzed at 24 hpi for progeny virus titers using plaque assays in Vero cells. Experiments were performed in triplicate, and the fold increase in shSAMHD1 cells is indicated.
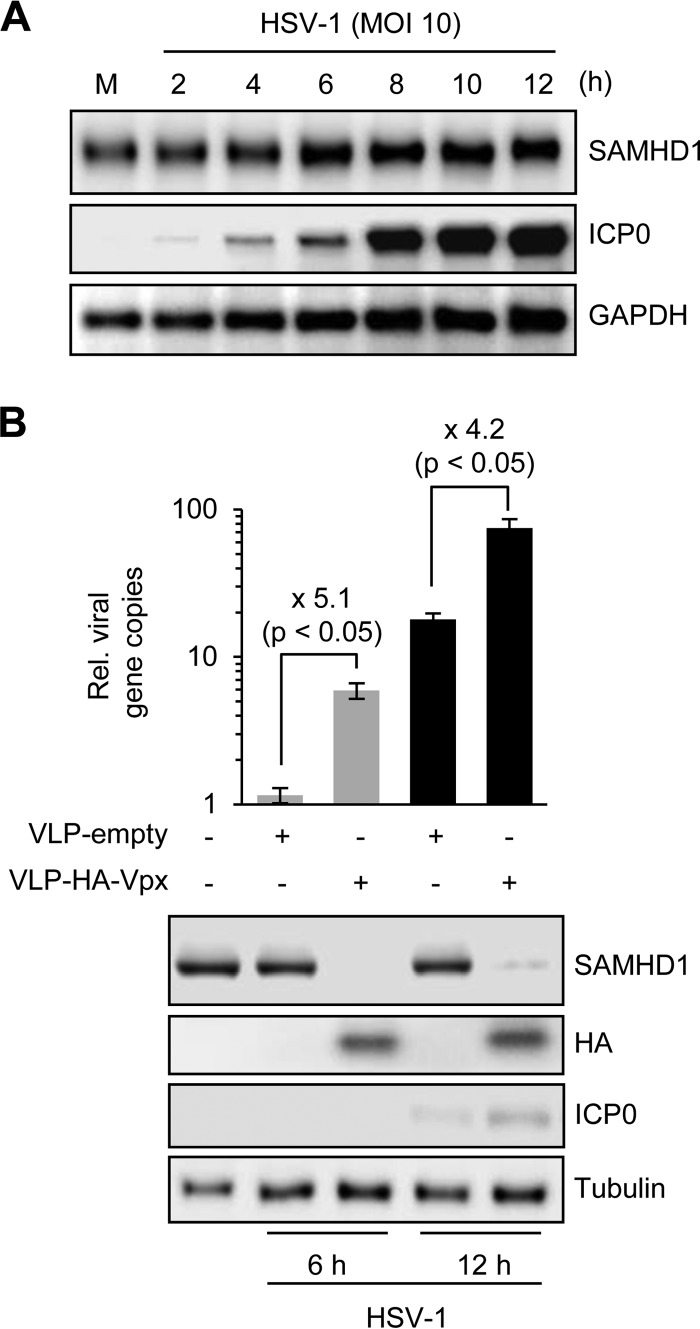
Since the Vpx protein of HIV-2 depletes SAMHD1 (8, 9), we examined protein levels during HSV-1 infection and also tested the impact of Vpx on HSV-1 infection (Fig. 2). In contrast to the function of the Vpx protein, we found that HSV-1 infection did not decrease the level of cellular SAMHD1 protein (Fig. 2A). Over the course of HSV-1 infection, there was a slight increase in total levels of SAMHD1, which may reflect upregulation by the virus-induced interferon response. Since knockdown of SAMHD1 in THP-1 cells enhances HSV-1 infection, we reasoned that Vpx-mediated depletion would also promote HSV-1 replication. Viruslike particles (VLPs) were generated with and without SIV Vpx, and PMA-differentiated THP-1 cells were incubated with VLPs for 2 h prior to infection with HSV-1 (Fig. 2B). Immunoblotting confirmed the presence of Vpx and the resulting reduction of endogenous SAMHD1. When replication of HSV-1 was assessed at 6 and 12 hpi, cells incubated with VLPs containing Vpx showed a 4- to 5-fold increase in HSV-1 DNA accumulation compared to control VLPs. Enhancement of HSV-1 replication was less effective with VLP-Vpx compared to that with shRNA knockdown, possibly reflecting incomplete depletion. Together, these results demonstrate that HSV-1 replication is enhanced by depletion of endogenous SAMHD1 in differentiated macrophage cell lines.
Fig 2.
HSV-1 does not alter SAMHD1 levels but is enhanced by Vpx-mediated downregulation of endogenous SAMHD1. (A) The levels of SAMHD1 were examined over a time course of HSV-1 infection in differentiated THP-1 cells. An antibody to ICP0 was included as a marker of virus infection, and GAPDH served as a loading control. (B) SAMHD1 depletion by Vpx enhances HSV-1 infection. PMA-treated THP-1 cells were incubated with VLPs that did or did not contain HA-tagged Vpx protein. After 2 h, cells were infected with HSV-1 at an MOI of 0.1. At 6 and 12 hpi, DNA was extracted for qPCR and proteins were harvested for immunoblotting. Experiments were performed in triplicate, and P values were calculated by a Student t test. Cells incubated with VLPs containing Vpx (detected with HA antibody) showed depleted SAMHD1 and enhanced HSV-1 replication. Expression of viral protein ICP0 confirmed increased virus replication, and tubulin served as a loading control.
Ectopic expression of SAMHD1 inhibits HSV-1 replication in differentiated U937 cells.
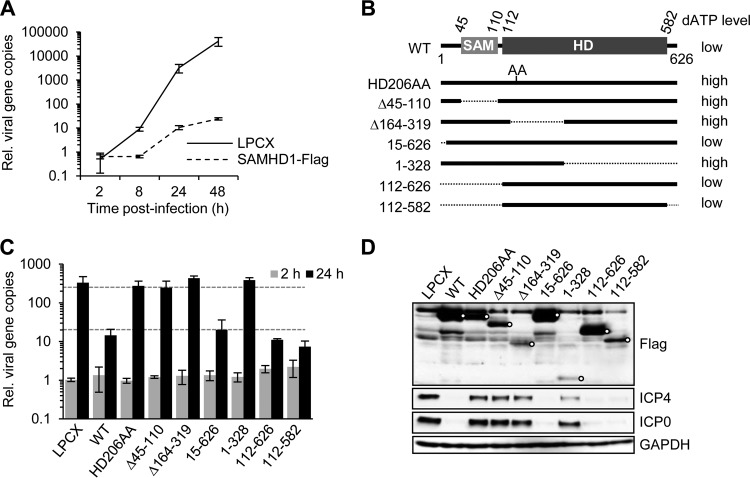
We next examined whether ectopic expression of SAMHD1 would restrict HSV-1 infection using the human monocyte cell line U937 that does not express endogenous SAMHD1 protein. We compared infection of HSV-1 at an MOI of 0.1 in PMA-treated U937 cells transduced with a retrovirus expressing wild-type full-length SAMHD1 or an empty vector LPCX (22) and measured viral DNA accumulation (Fig. 3A). HSV-1 replication was severely attenuated in cells expressing wild-type SAMHD1, consistent with results from depletion of endogenous protein.
Fig 3.
Replication of HSV-1 in U937 cells expressing ectopic SAMHD1. The human monocytic cell line U937 does not express SAMHD1 and was used to assess the impact of SAMHD1 ectopic expression and contribution of different protein domains. (A) HSV-1 replication is restricted by wild-type SAMHD1. U937 cells with empty vector (LPCX) or expressing wild-type SAMHD1 were treated with PMA and then infected with HSV-1 (MOI of 0.1) and harvested at the indicated time points. The relative number of viral genomes was determined by qPCR using primers to the ICP27 gene. Error bars represent the standard deviations from triplicate samples. (B) Wild-type and mutant SAMHD1 proteins were expressed in U937 cells by stable retrovirus vector transduction. The sterile alpha motif (SAM) and histidine-aspartic acid (HD) protein domains are indicated along with the numbers of amino acid residues at the boundaries. At the right are indicated the previously determined cellular dATP levels (22). (C) The contribution of protein domains to HSV-1 restriction was determined by assessing the relative number of viral genomes by qPCR at 2 and 24 hpi. (D) Accumulation of viral proteins and SAMHD1-Flag proteins was assessed by immunoblotting at 24 hpi. Open circles indicate SAMHD1-Flag proteins.
The HD of SAMHD1 is required for inhibition of HSV-1 replication.
Functional domains within the SAMHD1 protein are the sterile alpha motif (SAM) domain and histidine-aspartic acid domain (HD). The SAM domain mediates protein interactions, while the HD confers the phosphohydrolase activity (33). To assess the contribution of different domains of SAMHD1 to HSV-1 restriction, we examined a series of deletion mutants (Fig. 3B) stably expressed in the human monocyte cell line U937, which lacks SAMHD1 expression (22). The U937 cells stably expressing SAMHD1 variants were induced to differentiation by PMA treatment and then infected with HSV-1 (Fig. 3C). Deletion constructs 112-626 and 112-582, which lack the SAM domain but retain an intact HD, inhibited HSV-1 to the same extent as wild-type SAMHD1. This suggests that the SAM domain is dispensable for restriction of HSV-1. Variants with mutations of the histidine and aspartic acid residues (the catalytically inactive HD206AA mutant) or larger deletions within the HD had lost HSV-1 restriction (Fig. 3C). Immunoblotting confirmed expression of all SAMHD1 variants (Fig. 3D). Together, these experiments suggest that the HD of SAMHD1 is sufficient to achieve inhibition of HSV-1 replication in differentiated U937 cells.
Inhibition of HSV-1 is distinct from restriction of retroviruses.
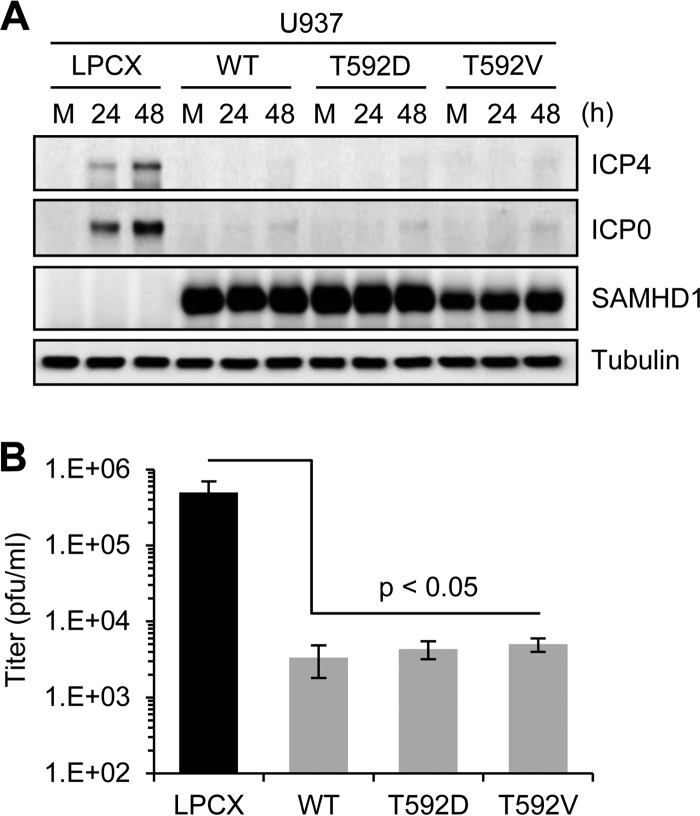
The antiviral activity of SAMHD1 against retroviruses is limited to noncycling cells. Two recent reports demonstrated that the ability of SAMHD1 to restrict retroviruses is regulated by phosphorylation (42, 44). SAMHD1 in cycling cells is phosphorylated on residue T592, which renders it unable to block retroviral infection. SAMHD1 containing a phosphomimetic residue at this site (T592D) loses the ability to block retrovirus restriction in noncycling differentiated U937 cells (42). This modification does not affect the ability of SAMHD1 to decrease the cellular dNTP levels (42). To determine whether SAMHD1 phosphorylation also impacts inhibition of HSV-1, we compared infection in differentiated U937 cells expressing SAMHD1 variants with T592 replaced by a phosphomimetic (T592D) or unphosphorylatable (T592V) residue (42). Cells infected with HSV-1 (MOI of 0.1) were analyzed for accumulation of viral early proteins and production of viral progeny (Fig. 4). Immunoblotting of lysates harvested at 24 hpi and 48 hpi showed an increase in viral proteins for infections of U937 cells transduced with the empty vector LPCX, but this was prevented in cells expressing ectopic wild-type SAMHD1 (Fig. 4A). Infections of cells expressing both T592D and T592V demonstrated similar inhibition of HSV-1 as the wild-type SAMHD1. Analysis of progeny production by plaque assay at 48 hpi also demonstrated that T592D and T592V variants both inhibited HSV-1 replication to the same extent as wild-type SAMHD1 (Fig. 4B). These data suggest that unlike restriction of HIV-1, inhibition of HSV-1 is not affected by the T592 phosphorylation status of SAMHD1.
Fig 4.
SAMHD1 phosphorylation status does not affect HSV-1 restriction. PMA-treated U937 cells transduced with empty vector control (LPCX), wild-type SAMHD1 (WT), or phosphorylation variants (T592D and T592V) were infected with HSV-1 at an MOI of 0.1. (A) Proteins were harvested from uninfected mock (M) and infected cells at 24 and 48 hpi. Levels of viral proteins ICP4 and ICP0, and the Flag-tagged SAMHD1 proteins, were detected by immunoblotting. Tubulin served as a loading control. (B) Cell culture supernatants were analyzed at 48 hpi for progeny virus titers by using plaque assays in Vero cells. Experiments were performed in triplicate, and P values were calculated by a Student t test.
SAMHD1 inhibits HSV-1 replication but not gene expression.
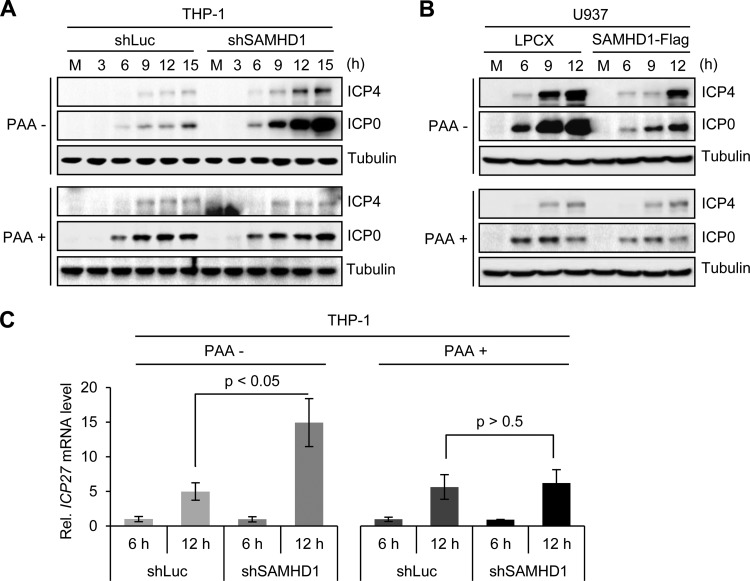
To determine whether SAMHD1 affects HSV-1 at the level of gene expression or DNA replication, we examined infections in cells treated with the drug phosphonoacetic acid (PAA), a specific inhibitor of the viral polymerase. THP-1 cells expressing shSAMHD1 or the control shLuc were differentiated with PMA and then infected with HSV-1 (MOI of 3) in the presence or absence of PAA (Fig. 5A). We measured accumulation of the early viral proteins ICP0 and ICP4 over a time course of infection. Although the levels of viral proteins were higher in shSAMHD1 cells in the absence of PAA than in the presence, inhibition of DNA replication by PAA resulted in similar levels of viral proteins in the shSAMHD1 and shLuc cell lines (Fig. 5A). This result suggests that the increase observed in the absence of the drug reflects increased protein expression due to the greater number of viral genomes able to express viral proteins. A similar experiment was performed in differentiated U937 cells expressing ectopic SAMHD1 (Fig. 5B). In the absence of PAA, viral protein accumulation was reduced in cells expressing SAMHD1 compared to that of the empty vector control, but in the presence of the drug, the two cell lines were equivalent. Our conclusion from these experiments is that the impact of SAMHD1 on HSV-1 infection is not due to an effect on early viral gene expression from incoming viral genomes. To confirm this conclusion, we examined accumulation of mRNA from the viral ICP27 gene (Fig. 5C). THP-1 cells with shLuc or shSAMHD1 were infected with HSV-1 (MOI of 3), and RNA harvested at 6 and 12 hpi was reverse transcribed and analyzed by qPCR with primers targeted to the viral ICP27 transcript. In the absence of PAA, we observed that the ICP27 transcripts increased by 12 hpi in the SAMHD1-depleted cells, but in the presence of PAA, there was no difference between the two cell lines. Together, these results demonstrate that depletion of SAMHD1 increases HSV-1 replication but not through effects on expression of early HSV-1 genes.
Fig 5.
SAMHD1 affects HSV-1 replication but not gene expression. (A) PMA-treated control and SAMHD1-depleted THP-1 cells were infected with HSV-1 at an MOI of 3 in the absence or presence of 200 μg/ml phosphonoacetic acid (PAA). Accumulation of viral proteins ICP0 and ICP4 was analyzed by immunoblotting at the indicated time points. Tubulin served as a loading control. (B) Infections were performed in PMA-treated control (LPCX) and SAMHD1-Flag-expressing U937 cells in the absence or presence of PAA. (C) The relative number of mRNA transcripts for the viral ICP27 gene was determined by RT-PCR analysis of mRNA extracted under infection conditions described for panel A. Error bars represent the standard deviations from three independent experimental samples, and P values were calculated by a Student t test.
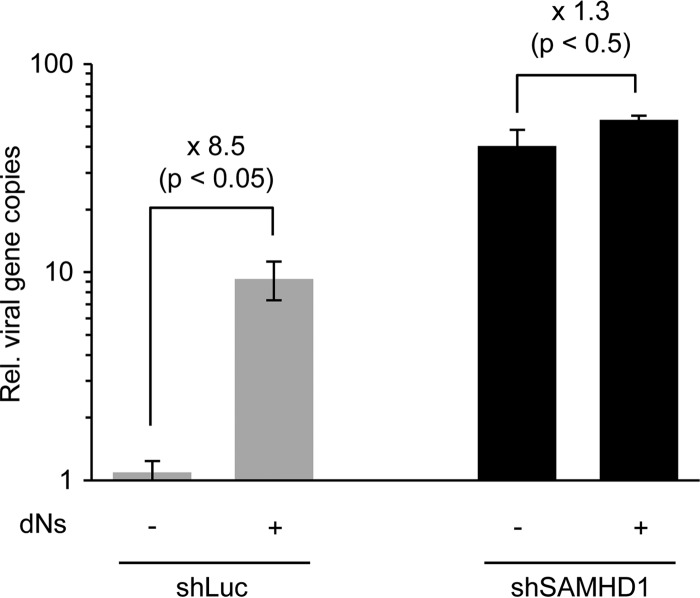
It has been proposed that SAMHD1 restricts retrovirus infection by hydrolyzing intracellular dNTPs to levels below the concentration required for synthesis of viral DNA by reverse transcriptase (35, 37, 38). The restriction of HSV-1 that we observed (Fig. 3 and 5) correlates with low dNTP concentrations previously reported for the cell lines that we used (22, 42). We therefore examined whether providing a source of dNTPs could relieve the restriction of HSV-1 (Fig. 6). Infections were performed in PMA-treated THP-1 cells exposed to deoxynucleosides (dNs), and DNA accumulation was assessed at 8 hpi by qPCR analysis of extracted DNA. Incubating cells with dNs led to a significant enhancement (8.5-fold) of HSV-1 DNA accumulation in differentiated THP-1 control cells. In cells depleted of endogenous SAMHD1, there was no significant additional effect of the dNs (Fig. 6), supporting the conclusion that SAMHD1 restricts HSV-1 in differentiated macrophages by limiting the intracellular pool of dNTPs.
Fig 6.
Restriction of HSV-1 can be overcome by deoxynucleoside (dN) treatment. PMA-treated THP-1 cells were infected with HSV-1 at an MOI of 0.1 for 8 h in cells incubated with 2 mM dNs. The relative number of viral genomes was determined by qPCR using primers to the ICP27 gene. Error bars represent the standard deviations from triplicate samples, and P values were calculated by a Student t test.
DISCUSSION
We have demonstrated that SAMHD1 is a restriction factor for HSV-1 replication in human macrophage cell lines. The major mechanism appears to be through depletion of intracellular dNTPs that are required for replication of the HSV-1 DNA genome. SAMHD1 is phosphorylated at T592 in cycling cells but is unphosphorylated at this residue in noncycling myeloid cells where HIV-1 is restricted (42). Since the phosphomimetic amino acid replacement at residue T592 completely abrogated HIV-1 restriction by SAMHD1 (42) but had no effect on HSV-1 restriction, our data suggest that the mechanism employed to inhibit the two viruses are not completely analogous. Decreasing cellular dNTP levels by SAMHD1 appears to be sufficient to achieve inhibition of HSV-1 replication, but an additional function or cofactor may be required for retrovirus restriction (42). Further understanding of the role for SAMHD1 phosphorylation may help to explain the mechanistic differences between inhibition of HIV-1 and HSV-1. The HD might also contribute to viral inhibition via interaction with RNA (22, 45), its exonuclease activity (36), or other roles in regulating innate immunity (32). Since viruses with DNA genomes such as HSV-1 are particularly susceptible to the intracellular concentration of dNTPs, it will be interesting to determine whether SAMHD1 has additional restrictive effects against HSV-1 in other noncycling cell types. Hollenbaugh et al. recently reported similar findings to our observations in differentiated THP-1 cells and also demonstrated that Vpx can promote HSV-1 replication in human primary dendritic cells and monocyte-derived macrophages (41). Although overexpression of SAMHD1 did not significantly affect infection by HIV-1 and other viruses (46), this has not been examined for HSV-1 in different cells under a range of proliferation conditions. We saw a small but reproducible effect of SAMHD1 depletion on HSV-1 replication in untreated proliferating THP-1 cells (Fig. 1B). Future studies will examine the effects of depleting or overexpressing SAMHD1 in additional cell types.
Viruses have evolved distinct means to counteract the hurdles they encounter during infection (47). It is possible that viruses unable to counteract SAMHD1 do not require infection of myeloid cells as a crucial step during natural infection in vivo. Since HIV-1 cannot counteract SAMHD1, overcoming its antiviral function must not be necessary for efficient infection and spread in the host organism. In this study, we did not uncover ways that HSV-1 counteracts the effects of SAMHD1, at least not in macrophages. As previously reported, we observed SAMHD1 localized in the nucleus (32, 48), and it was unaltered with respect to abundance or localization during HSV-1 infection. The HSV-1 genome encodes proteins involved in nucleotide biosynthesis and DNA metabolism (thymidine kinase, ribonucleotide reductase, deoxyuridine triphosphatase, and uracil-DNA-glycosylase) (26), and these may overcome limitations induced by SAMHD1 in some cellular settings. Future work will determine whether other herpesviruses are also susceptible to SAMHD1 depletion of dNTP concentrations. Since the human cytomegalovirus (HCMV) also infects myeloid cells (49), it will be interesting to determine whether SAMHD1 plays a role in HCMV latency or is counteracted during reactivation. Our results raise the intriguing idea that degradation of SAMHD1 by retroviral Vpx proteins could also enhance replication of opportunistic herpesviruses in coinfected patients.
ACKNOWLEDGMENTS
We thank members of the Weitzman Lab for insightful discussions and input. We are grateful to R. Everett and R. Koenig for generous gifts of reagents.
E.T.K. was supported by the Basic Science Research Program of the National Research Foundation (NRF) of Korea through the Ministry of Education, Science and Technology of the Republic of Korea (357-2011-1-C00092). Work in the Weitzman lab was supported by funds from the Children's Hospital of Philadelphia. Work in the Diaz-Griffero lab was supported in part by NIH grant RO1 AI087390 to F.D.-G.
Footnotes
Published ahead of print 25 September 2013
REFERENCES
- 1.Bieniasz PD. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109–1115 [DOI] [PubMed] [Google Scholar]
- 2.Hatziioannou T, Bieniasz PD. 2011. Antiretroviral restriction factors. Curr. Opin. Virol. 1:526–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco-Melo D, Venkatesh S, Bieniasz PD. 2012. Intrinsic cellular defenses against human immunodeficiency viruses. Immunity 37:399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris RS, Hultquist JF, Evans DT. 2012. The restriction factors of human immunodeficiency virus. J. Biol. Chem. 287:40875–40883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf D, Goff SP. 2008. Host restriction factors blocking retroviral replication. Annu. Rev. Genet. 42:143–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 7.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 8.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossini G, Cerboni C, Santoni A, Landini MP, Landolfo S, Gatti D, Gribaudo G, Varani S. 2012. Interplay between human cytomegalovirus and intrinsic/innate host responses: a complex bidirectional relationship. Mediators Inflamm. 2012:607276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tavalai N, Stamminger T. 2011. Intrinsic cellular defense mechanisms targeting human cytomegalovirus. Virus Res. 157:128–133 [DOI] [PubMed] [Google Scholar]
- 12.Weitzman MD, Ornelles DA. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24:7686–7696 [DOI] [PubMed] [Google Scholar]
- 13.Everett RD, Chelbi-Alix MK. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830 [DOI] [PubMed] [Google Scholar]
- 14.Orzalli MH, DeLuca NA, Knipe DM. 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. U. S. A. 109:E3008–E3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lilley CE, Chaurushiya MS, Boutell C, Everett RD, Weitzman MD. 2011. The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS Pathog. 7:e1002084. 10.1371/journal.ppat.1002084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geoffroy MC, Chelbi-Alix MK. 2011. Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res. 31:145–158 [DOI] [PubMed] [Google Scholar]
- 18.Chin KC, Cresswell P. 2001. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 98:15125–15130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suspene R, Aynaud MM, Koch S, Pasdeloup D, Labetoulle M, Gaertner B, Vartanian JP, Meyerhans A, Wain-Hobson S. 2011. Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo. J. Virol. 85:7594–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardieu C, Vigan R, Wilson SJ, Calvi A, Zang T, Bieniasz P, Kellam P, Towers GJ, Neil SJ. 2010. The RING-CH ligase K5 antagonizes restriction of KSHV and HIV-1 particle release by mediating ubiquitin-dependent endosomal degradation of tetherin. PLoS Pathog. 6:e1000843. 10.1371/journal.ppat.1000843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gramberg T, Kahle T, Bloch N, Wittmann S, Mullers E, Daddacha W, Hofmann H, Kim B, Lindemann D, Landau NR. 2013. Restriction of diverse retroviruses by SAMHD1. Retrovirology 10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen L, Kim B, Brojatsch J, Diaz-Griffero F. 2013. Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436:81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arduino PG, Porter SR. 2008. Herpes simplex virus type 1 infection: overview on relevant clinico-pathological features. J. Oral Pathol. Med. 37:107–121 [DOI] [PubMed] [Google Scholar]
- 24.Kennedy PG, Steiner I. 2013. Recent issues in herpes simplex encephalitis. J. Neurovirol. 19:346–350 [DOI] [PubMed] [Google Scholar]
- 25.Held K, Derfuss T. 2011. Control of HSV-1 latency in human trigeminal ganglia—current overview. J. Neurovirol. 17:518–527 [DOI] [PubMed] [Google Scholar]
- 26.Weller SK, Coen DM. 2012. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb. Perspect. Biol. 4:a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellermann-Eriksen S. 2005. Macrophages and cytokines in the early defence against herpes simplex virus. Virol. J. 2:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laguette N, Benkirane M. 2012. How SAMHD1 changes our view of viral restriction. Trends Immunol. 33:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luban J. 2012. Innate immune sensing of HIV-1 by dendritic cells. Cell Host Microbe 12:408–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan X, Baldauf HM, Keppler OT, Fackler OT. 2013. Restrictions to HIV-1 replication in resting CD4 T lymphocytes. Cell Res. 23:876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M. 2012. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11:194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. 2009. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41:829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480:379–382 [DOI] [PubMed] [Google Scholar]
- 34.Amie SM, Noble E, Kim B. 2013. Intracellular nucleotide levels and the control of retroviral infections. Virology 436:247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. 2012. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287:21570–21574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF. 2013. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J. Biol. Chem. 288:8101–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13:223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. 2012. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18:1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, Mulder LC, Fernandez-Sesma A, Rutsch F, Simon V, Konig R, Flory E. 2011. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7:e1002425. 10.1371/journal.ppat.1002425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sunseri N, O'Brien M, Bhardwaj N, Landau NR. 2011. Human immunodeficiency virus type 1 modified to package simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J. Virol. 85:6263–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollenbaugh JA, Gee P, Baker J, Daly MB, Amie SM, Tate J, Kasai N, Kanemura Y, Kim DH, Ward BM, Koyanagi Y, Kim B. 2013. Host factor SAMHD1 restricts DNA viruses in nondividing myeloid cells. PLoS Pathog. 9:e1003481. 10.1371/journal.ppat.1003481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. 2013. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13:441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Everett RD, Cross A, Orr A. 1993. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell type dependent manner. Virology 197:751–756 [DOI] [PubMed] [Google Scholar]
- 44.Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. 2013. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3:1036–1043 [DOI] [PubMed] [Google Scholar]
- 45.Goncalves A, Karayel E, Rice GI, Bennett KL, Crow YJ, Superti-Furga G, Burckstummer T. 2012. SAMHD1 is a nucleic-acid binding protein that is mislocalized due to Aicardi-Goutieres syndrome-associated mutations. Hum. Mutat. 33:1116–1122 [DOI] [PubMed] [Google Scholar]
- 46.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirchhoff F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8:55–67 [DOI] [PubMed] [Google Scholar]
- 48.Brandariz-Nunez A, Valle-Casuso JC, White TE, Laguette N, Benkirane M, Brojatsch J, Diaz-Griffero F. 2012. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodrum F, Caviness K, Zagallo P. 2012. Human cytomegalovirus persistence. Cell Microbiol. 14:644–655 [DOI] [PMC free article] [PubMed] [Google Scholar]