Abstract
Retroviral integrase (IN) proteins catalyze the permanent integration of proviral genomes into host DNA with the help of cellular cofactors. Lens epithelium-derived growth factor (LEDGF) is a cofactor for lentiviruses, including human immunodeficiency virus type 1 (HIV-1), and targets lentiviral integration toward active transcription units in the host genome. In contrast to lentiviruses, murine leukemia virus (MLV), a gammaretrovirus, tends to integrate near transcription start sites. Here, we show that the bromodomain and extraterminal domain (BET) proteins BRD2, BRD3, and BRD4 interact with gammaretroviral INs and stimulate the catalytic activity of MLV IN in vitro. We mapped the interaction site to a characteristic structural feature within the BET protein extraterminal (ET) domain and to three amino acids in MLV IN. The ET domains of different BET proteins stimulate MLV integration in vitro and, in the case of BRD2, also in vivo. Furthermore, two small-molecule BET inhibitors, JQ1 and I-BET, decrease MLV integration and shift it away from transcription start sites. Our data suggest that BET proteins might act as chromatin-bound acceptors for the MLV preintegration complex. These results could pave a way to redirecting MLV DNA integration as a basis for creating safer retroviral vectors.
INTRODUCTION
Retroviruses depend on the virally encoded IN proteins to facilitate stable insertion of their reverse-transcribed genomes into host cell chromosomes. INs recognize the attachment (att) sites at the ends of long terminal repeats (LTRs) in viral DNA to carry out two sequential enzymatic reactions. In the first reaction, referred to as 3′ processing, IN removes dinucleotides from the 3′ ends of viral DNA to expose the 3′ OH groups attached to the invariant CA dinucleotides. In the second reaction, DNA strand transfer, IN inserts the processed 3′ termini into opposing strands of the host chromosomal DNA via a transesterification mechanism (1, 2). Host cell enzymes complete the process by repairing the single-stranded gaps on both sides of integrated viral DNA. Consequently, the resulting provirus is flanked by short duplications of the target DNA sequences. The duplication size appears to be retroviral genus specific, being 5 bp for human immunodeficiency virus type 1 (HIV-1) and 4 bp for murine leukemia virus (MLV) (3–5). The terminal cleavage and strand transfer steps can be observed in vitro with purified recombinant retroviral IN and DNA substrates, demonstrating that IN alone is sufficient to carry out these reactions (3, 6).
Retroviral IN consists of three structural domains (reviewed in reference 7). The N-terminal domain (NTD) contains the zinc binding HHCC motif, and a highly conserved catalytic core domain (CCD) contains the essential active site Asp, Asp, and Glu (D, D-35-E motif) residues, which are directly involved in the catalytic activities of IN. The C-terminal domain (CTD) is least conserved (8–11). Mounting evidence suggests that IN functions as a tetramer (12–15). Recent crystal structures of the prototype foamy virus (PFV) IN bound to its viral and host DNA substrates revealed that all three IN domains participate in tetramerization and interactions with viral DNA (16, 17).
Retroviral integration into cellular DNA does not occur in a random manner with respect to various genomic features (reviewed in reference 18). HIV-1 and other lentiviruses show a remarkable preference for integration within active transcription units (19). In contrast, MLV, a gammaretrovirus, preferentially integrates near transcription start sites and CpG islands, features that are largely avoided by HIV-1 (20, 21). The remaining retroviral genera show other, albeit far less contrasting, integration patterns (22). Integration site selection of HIV-1 and other lentiviruses was shown to depend on the cellular protein lens epithelium-derived growth factor (LEDGF) (reviewed in reference 23). The IN binding domain (IBD) located within the C-terminal region of LEDGF mediates its interactions with HIV-1 and other lentiviral INs (24–26). LEDGF associates with chromatin via its N-terminal PWWP domain, which selectively binds to nucleosomes containing H3 trimethylated on Lys36 (27, 28), an epigenetic mark associated with bodies of transcription units (29). In cells depleted of LEDGF/p75, HIV-1 integration and replication were significantly affected, while the residual HIV-1 integration sites were less enriched in transcriptional units (30–32). Furthermore, it was possible to retarget HIV-1 integration by chimeric proteins containing the IN binding domain (IBD) of LEDGF/p75 and alternative chromatin binding domains (33–35).
Several cellular proteins, including transcription factors and chromatin and RNA binding proteins, were recently identified as potential interaction partners for MLV IN (36). This diverse group of proteins included BRD2, a member of the bromodomain and extraterminal domain (BET) family of chromatin binding proteins (37). Five mammalian BET family members are known: BRD2/RING3, BRD3/ORFX, BRD4 (includes two splice variants, a short variant termed BRD4/HUNK-1 and a long variant, BRD4/MCAP), and BRD6/BRDT (specifically expressed in testes). BRD2 serves as a transcriptional activator and is ubiquitously expressed in all tissues (38, 39). BRD2 localizes throughout the cell in resting cells, whereas mitogen treatment induces its nuclear localization (40). BRD2, and likely other BET proteins, acts as a scaffold on chromatin to recruit E2F proteins, histone deacetylases (HDACs), histone H4-specific acetyltransferase (HAT), and proteins involved in chromatin remodeling (41–43). BET proteins bind to acetylated histone tails via their bromodomains (44, 45). The structures of BRD2 bromodomains BD1 and BD2 have been solved in association with H4 acetylated on Lys-5 and -12 (46, 47). Recently, small-molecule inhibitors of BET proteins (I-BET and JQ1) have been developed that disrupt the binding interface between the bromodomain and the acetylated lysine groups on chromatin (48–51). In addition to two N-terminal bromodomains, BET proteins also contain a highly conserved C-terminal ET domain. The structure of the ET domain, known to be a protein-protein interaction motif, has been determined (52–54).
Some viruses exploit cellular BET proteins for different aspects of their life cycle (reviewed in reference 55). Thus, human papillomaviruses (HPVs) use BET proteins as cellular adaptors to anchor their genomes to mitotic chromosomes (56). In addition, the HPV E2 protein, required for virus episome maintenance and transcription, interacts with BRD4 to enable both transcriptional activation of E2 target genes (57–59) and repression of oncogenic E6 and E7 genes (60, 61). We and others showed that BRD2, BRD3, and BRD4 interact with Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded latent nuclear antigen 1 (LANA-1) and may contribute to LANA-1-regulated transcription and KSHV episomal maintenance (54, 62–64). Similarly, mutations introduced into the BRD2 and BRD4 binding site on the murine gammaherpesvirus 68 (MHV-68) Orf73 protein, the functional homologue of KSHV LANA-1, compromise promoter transactivation of several cyclin genes such as cyclins D1, D2, and E (65). It has been proposed that Epstein-Barr virus (EBV)-encoded EBV nuclear antigen 1 (EBNA-1), a functional homologue of KSHV LANA-1 that is required for EBV episomal maintenance, transformation, and latency, may also interact with BRD4 (66).
In this study, we show that the ET domains of BRD2/RING3, BRD3/ORFX, and BRD4/HUNK-1 physically and functionally interact with gammaretroviral INs. We mapped the interactions using mutagenesis approaches and found that BRD ET domains stimulate MLV integration in vitro. We also show that BET inhibition by the bromodomain inhibitors I-BET and JQ1 reduces MLV integration in vivo and shifts the MLV integration site preference away from cellular transcription start sites. While the manuscript was being prepared, another group reported similar findings (67). Taken together, these two studies indicate that cellular BET proteins act as cofactors for gammaretroviral integration.
MATERIALS AND METHODS
DNA constructs for expression in human cells.
Plasmids for the expression of pEGFP-BRD2 and pEGFP-BRD4 were described previously (64). Primer sequences used in this work are listed in Table 1. The following site-directed mutations were introduced into pEGFP-BRD2 using the QuikChange procedure (Stratagene): E682A/E683A/E685A, D687A/E689A, R648A/N655A/L662E, L676A/S679A, R648A/N655A/L676A/S679A, R648A, N655A, L662E, D687A, E689A, F688Y, S651E, S651N, and S651K. Synthetic genes encoding various retroviral INs, codon optimized for mammalian expression, were synthetized by GeneArt, Regensburg, Germany. To generate murine leukemia virus (MLV), feline leukemia virus (FeLV), Mason-Pfizer monkey virus (MPMV), and prototype foamy virus (PFV) IN expression constructs, the corresponding open reading frames (ORFs) including C-terminal FLAG tags were PCR amplified using the following primer sets (Table 1): pQMLV-INS-FLAG, PC594 and PC595; pQFeLV-INS-FLAG, PC596 and PC595; pQMPMV-INS-FLAG, PC654 and PC655; pQPFV-INS-FLAG, PC656 and PC655. The PCR products were digested with NotI and EcoRI (pQMLV-INS and pQFeLV-INS) or NotI and BamHI (pQMPMV-INS and pQPFV-INS) and then ligated into a similarly digested retroviral vector, pQCXIP (Clontech). To construct pQHTLV-1-INS-FLAG, a PCR fragment was obtained using primers PC632 and PC633 and reamplified using primers PC632 and PC595, digested with NotI and EcoRI, and ligated into the NotI and EcoRI sites of pQCXIP. To construct pQBLV-INS-FLAG, the same procedure was carried out using primers PC630 and PC631 followed by a second PCR using primers PC630 and PC595. The construct pCEP-INSalaFLAG was used to express FLAG-tagged HIV-1 IN (68).
Table 1.
Primer sequences used to construct plasmids described in Materials and Methodsa
| Primer name | Sequence (5′ OH-) | Description |
|---|---|---|
| PC524 | GGGATAGAAAAATTCATCACCCTACACC | Sense (S) MLV IN to recreate SmaI site |
| PC525 | GCGTGGATCCTTAGGGGGCCTCGCGGGTTAACCTTATTTTTAAGG | Antisense (AS) MLV IN-BamHI |
| PC594 | GCCAGCGGCCGCAGACACCATGGCGATCGAGAACAGCAGCCCCTACAC | (S) NotI-Kozak-MetAlaMLVsynthIN |
| PC595 | GCCAGAATTCATCACTTGTCGTCGTCGTCCTTGTAGTC | (AS) Stop-Stop-EcoRI-FLAG-synth |
| PC596 | GCCAGCGGCCGCAGACACCATGGCGCCACAGAGCTGATCGAGGG | (S) NotI-Kozak-MetAlaFeLVsynthIN |
| PC654 | GCCAGCGGCCGCAGACACCATGGCGAGCAACATCAACACCAACCTGGAAAGC | (S) NotI-Kozak-MetAlaMPMVsynthIN |
| PC655 | GCGTGGATCCTCACTTGTCGTCGTCGTCCTTGTAGTC | (AS) Stop-BamHI-FLAG-synth |
| PC656 | GCCAGCGGCCGCAGACACCATGGCGTGCAACACCAAGAAGCCCAACCTGGAC | (S) NotI-Kozak-MetAlaPFVsynthIN |
| PC632 | GCCAGCGGCCGCAGACACCATGGCGCAGCTGAGCCCTGCCGACCTGC | (S) NotI-Kozak- MetAlaHTLV-1synthIN |
| PC633 | GTCGTCGTCGTCCTTGTAGTCGCCGTGGTGCTGGTGGTCC | (AS) HTLV-1 IN with part of FLAG tag |
| PC630 | GCCAGCGGCCGCAGACACCATGGCGCTGGAAACCCCGAGCAGTGG | (S) NotI-Kozak-MetAlaBLVsynthIN |
| PC631 | GTCGTCGTCCTTGTAGTCGCCGTCGCTGCTTCTGTTGTGG | (AS) BLV IN with part of FLAG tag |
| SG1 | GCCAGCGGCCGCAGACACCATGGCGCAGGGCACAAGAGTGCGGGGCC | (S) NotI-Kozak-MetAlaMLVIN dNTD |
| SG2 | GCCAGCGGCCGCAGACACCATGGCGACCGCCAAGGTGGTGACCAAG | (S) NotI-Kozak-MetAlaMLVIN dCCD |
| SG3 | GCCAGAATTCTCACTTGTCGTCGTCGTCCTTGTAGTCGCCTCTGTCCAGCTGTTCCTGGTAGGC | (AS) Stop-EcoRI FLAG-MLV IN dCTD |
| SG4 | GCCAGAATTCTCACTTGTCGTCGTCGTCCTTGTAGTCGCCAGGGGTGGTCAGCAGCACGGTGTAAGGTCC | (AS) Stop-EcoRI FLAG-MLV IN dCCD+CTD |
| SG5 | ACAACCGGTAGACACCATGGCGATCGAGAACAGCAGCCCCTACACC | (S) AgeI-Kozak-MetAlaMLVIN |
| SG6 | ACAGGTACCGCCAGGGGCCTCTCTGGTCAG | (AS) KpnI-MLV IN |
| SG7 | GCATGGATCCAGGCCCATGAGTTACGATGAGAAGCGGCAG | (S) BamHI-BRD2(640–801) |
| SG8 | GCCAGAATTCGCCTGAGTCTGAATCACTGGTGTCTGAAGACGA | (AS) EcoRI-BRD2(640–801) |
| SG9 | GATTGACTACCCGTCAGCGGGGGTCTTTCA | |
| SG10 | AATGAAAGACCCCCGCTGACGGGTAGTCAATC | |
| GM36 | GAGGGATCCCCCATGAGTTACGATG | (S) BamHI-BRD2(641–710) |
| GM37 | GGCCCTCGAGTCATTTCTTACGTAGGCAGG | (AS) XhoI-BRD2(641–710) |
| TM1 | GCCCATGGTGTACCCATACGACGTACCAGATTACGCTCACC | (S) NcoI-HA |
| TM2 | ATGCGGCCGCTCAGTGGTGGTGGTGAGCGTAATCTGGTACGT | (AS) NotI-Stop-HA |
| TM3 | GAGGAGTTGTGGCCCGTTGT | (S) PRE |
| TM4 | TGACAGGGTGGTGGCAATGCC | (AS) PRE |
| TM5 | GTCTCCATTCCCTATGTTCATGC | (S) PTBP2 |
| TM6 | GTTCCCGCAGAATGGTGAGGT | (AS) PTBP2 |
| MG1 | CCTCGATTGACTGAGTCGC | (S) MLV-SF11 |
| MG2 | GAGACCCTCCCAAGGATCAG | (AS) MLV-SF11 |
| MG3 | CAAATCTCGGTGGAACCTCCA | (AS) MLV-PBS |
| TM7 | GAACCCACTGCTTAAGCCTCA | LV-SIN-LTR-I |
| TM8 | CGCTAGCGATATCGAATTCAC | GV-SIN-LTR-I |
| TM9 | AGCTTGCCTTGAGTGCTTCA | LV-SIN-LTR-II |
| TM10 | AGCGATATCGAATTCACAACC | GV-SIN-LTR-II |
| TM11 | GACCCGGGAGATCTGAATTC | OC1 |
| TM12 | CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXXXXXAGTAGTGTGTGCCCGTCTGT | LV-SIN-BC-LTR-III |
| TM13 | CCATCTCATCCCTGCGTGTCTCCGACTCAGATCGCGTACCCAATAAAGCCTCTTGCTGT | GV-SIN-BC-LTR-III |
| TM14 | CCTATCCCCTGTGTGCCTTGGCAGTCTCAGAGTGGCACAGCAGTTAGG | OC2-TitaniumB |
Nucleotides in boldface type represent barcode sequences.
To clone FLAG-tagged deletion constructs for MLV IN, IN fragments were PCR amplified from pQMLV-INS-FLAG plasmid using the following primer pairs: IN(110-409) (ΔNTD), SG1 and PC595; IN(160-409) (δCCD), SG2 and PC595; IN(1-278) (ΔCTD), PC594 and SG3; IN(160-359) (δCCD + δCTD), SG2 and SG4. The resulting PCR products were then digested with NotI and EcoRI and ligated into NotI- and EcoRI-digested pQCXIP. To construct pcDNA-MLV-INS-FLAG, the insert was removed from pQMLV-INS-FLAG by digestion with NotI and EcoRI and ligated into NotI and EcoRI sites of pcDNA3.1 vector (Invitrogen). The following alanine mutations were generated in pcDNA-MLV-INS-FLAG using the QuikChange mutagenesis procedure: S239A/R240A/D241A, G261A/L262A/T263A, P264A/Y265A/E266A, I267A/L268A/Y269A, S239A, R240A, D241A, G261A, L262A, T263A, P264A, Y265A, E266A, I267A, L268A, and Y269. To construct p189-MLV-IN for the expression of MLV IN with an mCherry tag at the C terminus, a PCR fragment obtained using primers SG5 and SG6 was digested with AgeI and KpnI and cloned into the similarly digested lentiviral vector p189 (a kind gift from Benno Woelk, MHH). The retroviral vector pQFLAG-puro was described previously (69). For cloning of the MLVg/p-IN-HA construct, an NcoI-HA-4×His-Stop-NotI-PCR product was generated with primers TM1 and TM2, digested with NcoI/NotI, and ligated into the pcDNA3.MLVg/p-INGFP SfiI/NotI- and SfiI/NcoI-digested backbone via a three-fragment ligation, thereby replacing the green fluorescent protein (GFP)-encoding sequence at the end of the integrase with the hemagglutinin (HA) tag. The sequence of the HA tag was verified by sequencing. The construct pcDNA3.MLVg/p-INGFP has been described before (70).
DNA constructs for bacterial protein expression.
To produce MLV IN with a removable N-terminal hexahistidine (His6) tag, MLV IN, amplified using primers PC524 and PC525, was digested with BamHI and ligated into pCPH6P-BIV-IN (24) digested with SmaI and BamHI, replacing the original insert. The plasmid pKB-IN6H used for the bacterial expression of HIV-1 IN with a C-terminal His6 tag and the plasmids for the production of C-terminally His6-tagged INs (HIV-2, bovine immunodeficiency virus [BIV], equine infectious anemia virus [EIAV], FeLV, MPMV, human T-lymphotropic virus type 1 [HTLV-1], human spumaretrovirus type 2 [HSRV-2], feline immunodeficiency virus [FIV], visna virus, and simian immunodeficiency virus [SIV]) have been described previously (24, 26). To generate pGST-BRD2(641–710), the fragment was PCR amplified from the pEGFP-BRD2 plasmid using primers GM36 and GM37. The resulting PCR product was digested with BamHI and XhoI and ligated into BamHI/XhoI-digested pGEX-6P3. The following point mutations were generated in pGST-BRD2(641–710) using the QuikChange mutagenesis protocol: D687A/E689A, L662E, F688Y, and S651E. The construct GST-BRD4(607–722) (64), as well as the pGST-LEDGF/p75(347–471) fusion construct, have been described previously (25). Unmodified pGEX-6P3 was used to produce glutathione S-transferase (GST) as a control for pulldown experiments.
Cell transfection and coimmunoprecipitation.
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal calf serum (FCS) (Gibco), 50 IU/ml penicillin, and 50 μg/ml streptomycin (Cytogen) at 37°C in 5% CO2 and a humidified atmosphere. Cells grown to 80% confluence in six-well plates were transfected with 1 μg of plasmid expressing FLAG-tagged avian sarcoma leukosis virus (ASLV) IN, MPMV IN, MLV IN, FeLV IN, BIV IN, HTLV IN, HIV-1 IN, and MLV IN deletion or point mutants as described above, along with pEGFP-BRD2, pEGFP-BRD4, or pEGFP-BRD2 point mutants using FuGENE 6 according to the instructions of the manufacturer (Promega). Cells were lysed 48 h after transfection in 300 μl of 150 mM NaCl, 25 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 10 mM dithiothreitol (DTT), 0.5% (vol/vol) NP-40 in the presence of protease inhibitors. For immunoprecipitation, lysates (250 μl) precleared with 15 μl of protein A Sepharose beads (GE Healthcare) were incubated with 20 μl rabbit polyclonal anti-GFP antibody (Clontech) bound to protein A Sepharose beads. After 24 h of rocking at 4°C, the beads were washed eight times in extraction buffer and bound proteins were eluted in 20 μl of SDS-PAGE sample buffer. Cell extracts and immunoprecipitates were analyzed by Western blotting using monoclonal anti-GFP J1 (Clontech) or anti-FLAG M2 (Sigma-Aldrich).
Generation of stable cell lines expressing BRD2 C-terminal domain.
To express a FLAG-tagged BRD2 fragment, BRD2(640–801) was PCR amplified from pEGFP-BRD2 using primers SG7 and SG8, digested with BamHI and EcoRI, and ligated with similarly digested pQFLAG-puro. Stably expressing cell lines were made by transient transfection of HEK293T cells using FuGENE 6 (Promega) with retroviral vectors pQ-BRD2(640-801)-FLAG-puro and pQFLAG-puro. The cells were selected with 2.5 μg/ml puromycin, and individual clones were selected on the basis of expression by Western blotting using anti-FLAG M2 antibody (Sigma-Aldrich).
Purification of GST-tagged proteins.
Production of GST-tagged LEDGF/p75 has been described previously (25). To isolate GST-tagged BET proteins, Escherichia coli PC2 cells (24) containing the expression plasmid pGST-BRD2(641–710), pGST-BRD4(607–722), or pGST-BRD2 point mutants (L662E, D687A E689A, S651E, and F688Y) were grown to an A600 of ∼1.0 in the presence of 120 μg/ml ampicillin and induced with 0.15 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 25°C for 5 h. Cells were lysed by sonication in core buffer (25 mM Tris-HCl, pH 7.4, 0.5 M NaCl) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF). The fusion protein was captured on glutathione Sepharose (GE Healthcare Biosciences, Sweden) and, following extensive washing, was eluted in 25 mM Tris-HCl, pH 7.4, 0.5 M NaCl, 20 mM glutathione (Sigma-Aldrich), 10 mM 1,4-dithiothreitol (DTT). The protein was then dialyzed against excess 0.5 M NaCl, 25 mM Tris-HCl, pH 7.4, 5 mM DTT; supplemented with 10% glycerol; and flash-frozen in liquid nitrogen.
GST tag pulldown assay.
GST-tagged proteins [GST, GST-BRD2(641–710), or GST-tagged LEDGF/p75(347–471)] were bound to glutathione Sepharose slurry (GE Healthcare Biosciences) at a concentration of 1 μg/μl settled bead volume. Five microliters of settled beads was used per pulldown experiment, to which the following were added: 600 μl of binding buffer (150 mM NaCl, 2 mM MgCl2, 0.1% NP-40, 50 mM Tris, pH 7.4, 1 mM DTT), 10 μg of bovine serum albumin (BSA), and 5 μg of IN. Binding was allowed to occur for 2 h by incubation at 4°C with gentle rocking. The beads were then washed four times with 1 ml of binding buffer. The entire remaining buffer was removed from the beads, and 50 μl of 3× Laemmli buffer was added to each tube and boiled for 5 min. The samples (10 μl) were separated by SDS-PAGE and detected by staining in Coomassie R-250 (Serva). A similar protocol was followed for other GST pulldowns. Briefly, 60 μl glutathione Sepharose beads was incubated for 3 h at 4°C with 50 μg GST-BRD2, GST-BRD4, GST-BRD2 mutants (L662E, D687A E689A, S651E, and F688Y), or GST in 100 μl of 0.25 M NaCl, 25 mM Tris-HCl, pH 7.4, 10 mM DTT. Beads collected by gentle centrifugation were washed with several changes of ice-cold pulldown buffer (PDB; 0.5 M NaCl, 0.1% NP-40, 25 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 10 mM DTT) supplemented with complete EDTA-free protease inhibitor cocktail (Roche). Forty microliters of glutathione Sepharose preloaded with GST-BRD2, GST-BRD4, GST-BRD2 mutants, or GST was then incubated with 10 μg MLV IN and 10 μg BSA in 650 μl PDB. Reaction mixtures were rocked at 4°C for 4 to 5 h. After gentle centrifugation, the beads were washed with several changes of ice-cold PDB. Bound proteins were eluted in 15 μl of 2× Laemmli sample buffer supplemented with 25 mM DTT, separated in 12% SDS-PAGE gels, and detected by staining with Coomassie R-250.
Colocalization studies.
HeLa cells were plated onto 20- by 20-mm glass coverslips in a six-well plate prior to transfection with plasmids pEGFP-BRD2 and p189-MLV-IN (as described above). Forty-eight hours after transfection, cells were fixed by incubation with 4% paraformaldehyde in PBS for 10 min, washed three times with PBS, quenched with 50 mM NH4Cl in PBS for 10 min, and permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature and the nuclear DNA was stained with 1 μM TO-PRO-3 (Invitrogen) in PBS for 20 min. Finally, the cells were washed three times with PBS and once with distilled water and the coverslips were mounted in 30 μl Mowiol containing 25 mg/ml DABCO [1,4-diazabicyclo(2,2,2)octane]. Images were acquired with an Axio Observer microscope (Carl Zeiss).
Production of recombinant retroviral IN proteins.
Isolation of His6-tagged retroviral IN (HIV-1, HIV-2, BIV, EIAV, FeLV, MPMV, HTLV-1, HSRV-2, FIV, visna virus, and SIV) proteins was carried out as described before (24, 26). To prepare MLV IN, Escherichia coli PC2 cells transformed with pCPH6P-MLVIN were grown in Lennox LB broth in shaker flasks at 30°C to an A600 of ∼1.0 in the presence of 120 μg/ml ampicillin prior to the addition of 0.25 mM IPTG. Following induction for 5 h at 25°C, bacteria were harvested and stored at −80°C. For purification, thawed bacterial paste was lysed by sonication in buffer 1 (0.5 M NaCl, 50 mM Tris-HCl, pH 7.4, 7.5 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate [CHAPS]) in the presence of 0.5 mM PMSF. The lysate, precleared by centrifugation at 20,000 × g for 30 min and supplemented with 20 mM imidazole, pH 7.5, was incubated with nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) at 4°C for 1 h. The resin was washed in buffer 1 containing 20 mM imidazole, and bound proteins were eluted with 200 mM imidazole in buffer 1 supplemented with 10 mM DTT. Following removal of the His6 tag by overnight digestion with human rhinovirus 14 protease 3C at 4°C, the protein was diluted with 5 volumes of ice-cold buffer 1 (50 mM Tris-HCl, pH 7.4, 7.5 mM CHAPS) and loaded onto a 5-ml HiTrap heparin column (GE Healthcare). The IN proteins were eluted with a linear gradient of 0.2 to 1.0 M NaCl in buffer 1, and peak fractions were pooled, supplemented with 10 mM DTT, concentrated to 4 to 13 mg/ml, supplemented with 10% (vol/vol) glycerol, flash-frozen in liquid nitrogen, and stored at −80°C. Protein concentration was determined using the Bradford assay with a BSA standard.
In vitro integration assay and sequence analysis of integration products.
Donor DNA substrates were obtained by annealing pairs of high-pressure liquid chromatography (HPLC)-purified synthetic oligonucleotides. Preprocessed 32-bp U5 donor was prepared using SG9 and SG10 (Thermo Scientific). The donor oligonucleotides correspond to the end of the U5 region of the MLV LTR. The final strand transfer reaction mixture contained 20 mM Tris-HCl, pH 7.4, 5 mM DTT, 5 mM MgCl2, 125 mM NaCl, 5 μM ZnCl2, 300 ng supercoiled pGEM3Zf(+) target DNA, 3 μM donor DNA substrate, 8 μM IN, and 24 μM BET proteins (final volume of 20 μl). Reactions were initiated by addition of IN and allowed to proceed for 90 min at 37°C. To stop the reaction, 0.5% (wt/vol) SDS and 25 mM EDTA were added. Reaction products, deproteinized by digestion with 20 μg proteinase K for 1 h at 37°C and precipitated with ethanol, were separated on 1.5% agarose gels and visualized by staining with Gel Red (Biotium). Sequencing of concerted integration products was done according to the method of Valkov et al. (71).
Production of MLV and HIV-1 retroviral stocks.
The details of gammaretroviral constructs can be found in a previous publication (72). To prepare MLV vector particles, 293T cells were cotransfected using the calcium phosphate transfection method with three plasmids: the vesicular stomatitis virus G-producing plasmid (pMD.G), the MLV backbone (SRS11.eGFP.pre), and a packaging construct (pcDNA3.MLVgag-pol or pcDNA3.MLVgag-pol-HA). Lentiviral supernatants were produced as previously described (73). Briefly, 293T cells were cotransfected using the calcium phosphate transfection method using the following plasmids: HIV-1 vector backbone (pRRL.PPT.SF.GFP.pre), HIV-1 packaging construct (pcDNA3.LVgag-pol.4XCTE), pRSV-Rev, and pMD.G. Culture supernatants were harvested 36 h and 48 h posttransfection. All vector stocks were treated with 1 μg/ml of DNase I (Roche Applied Science) for 1 h at room temperature to remove contaminating plasmid DNA. Subsequently, cell debris from the supernatant was cleared by filtration through 0.45-μm filters and then concentrated by ultracentrifugation at 10,000 × g at 4°C for 16 to 18 h using an SW-28 rotor in a Beckman ultracentrifuge, resuspended in fresh medium, aliquoted, and stored at −80°C.
Retroviral transduction.
HEK293T cells (200,000) were transduced with MLV-based vector supernatants (see above) in a 12-well plate, at a multiplicity of infection (MOI) of 1, in the presence of protamine sulfate. To increase transduction efficiency, the virus-cell mixtures were subjected to centrifugation for 1 h at 450 × g at room temperature. Viral vector supernatant-containing medium was replaced after 12 h with fresh medium.
For inhibitor studies, HEK293T cells seeded in 12-well plates were transduced with wild-type (WT) MLV or HIV-1 vector supernatants at an MOI of 0.1, in the presence of a bromodomain inhibitor, I-BET 151 (ChemieTek), JQ1 (BioVision), or dimethyl sulfoxide (DMSO) (Roth). After 12 h, the virus-containing medium was replaced with fresh medium containing inhibitors or DMSO, and then after 24 h, inhibitors were removed and replaced with fresh medium. Transduced cells were harvested at 3 h and 8 h for the quantification of minus-strand strong-stop (MSSS) intermediates and late reverse transcripts (RTs), respectively.
After 3 days, 90% of the cells were harvested for fluorescence-activated cell sorting (FACS) analysis, while the remaining 10%, to be used to quantify integrated vector copy numbers, were subcultured for at least 20 days before processing to eliminate nonintegrated viral DNA, and genomic DNA was purified with the DNeasy blood and tissue kit (Qiagen). The percentage of GFP-positive cells was determined by FACS on a Beckman Coulter instrument. Data were analyzed using Kaluza 1.2 (Beckman Coulter). All experiments were performed independently at least three times.
qPCR.
Quantitative real-time PCRs (qPCRs) were performed by using the Step One Plus real-time PCR system (Applied Biosystems) and the QuantiFast SYBR green PCR kit (Qiagen, Hilden, Germany) according to the manufacturers' instructions. Primers for quantification of minus-strand strong-stop extension intermediate (MSSE) products were MG1 and MG2, while primers MG1 and MG3 were used for the quantification of late reverse transcript (RT) products. Primers specific for genomic PTBP2 (polypyrimidine tract binding protein 2) genes (TM5 and TM6) were used for an internal control to normalize for input DNA. Vector integrations were detected by posttranscriptional regulatory element (PRE)-specific primers (TM3 and TM4), and results were normalized to a genomic PTBP2 gene fragment. PCR efficiencies were determined on a dilution series of a standard plasmid containing PRE and PTBP2 fragments, and subsequent vector copy number calculations were performed according to the method of Pfaffl (74).
LM-PCR and 454 pyrosequencing.
Infections in the presence of 0.2 μM JQ1 or 0.01% DMSO were performed as described above. Linker-mediated PCRs (LM-PCRs) were performed as previously described with minor modifications (73, 75). Two hundred nanograms of genomic DNA was digested with Tsp509I/MluCI (HIV-1 samples; New England BioLabs) or MseI (MLV samples; Thermo Scientific) prior to primer extension with biotinylated LV-SIN-LTR-I (TM7) or GV-SIN-LTR-I (TM8) primer. After ligation of the polylinker, subsequent PCRs were performed with primer LV-SIN-LTR-II (TM9) or GV-SIN-LTR-II (TM10) in combination with OC-1 (TM11). The resulting PCR products were digested with SacI (lentiviral; New England BioLabs) or BglII (gammaretroviral; Thermo Scientific) for 2 h at 37°C to prevent amplification of an internal control band during subsequent nested PCR with barcoded primers (LV-SIN-BC-LTR-III [TM12] or GV-SIN-BC-LTR-III [TM13] and OC2-TitaniumB [TM14]), each of which was equipped with adaptor sequences for 454 high-throughput sequencing. Sequencing was performed in the Institute for Medical Microbiology and Hospital Epidemiology at Hannover Medical School, Germany, using Lib-L chemistry.
RESULTS
BET proteins specifically bind to gammaretroviral IN proteins.
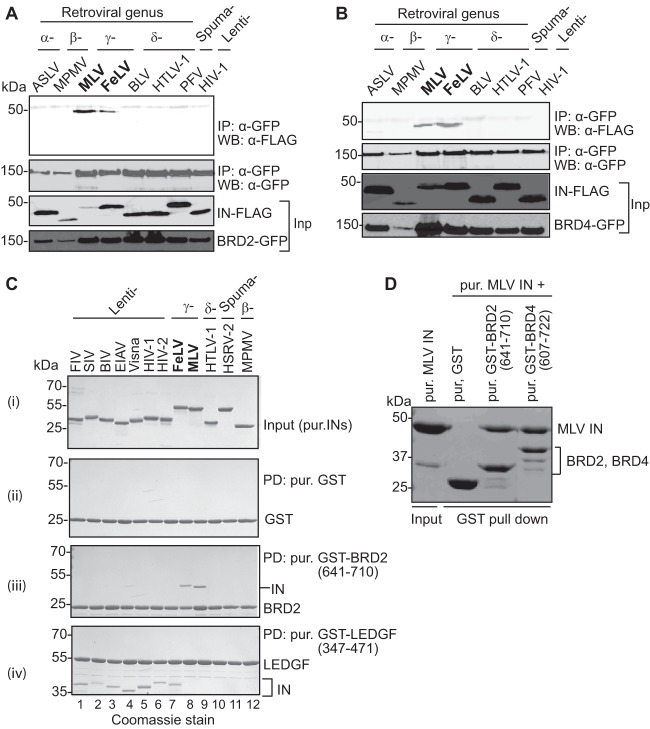
Following up on an earlier report (36) on the isolation of BRD2 in a yeast two-hybrid screen using MLV IN as bait, we investigated the interaction of different retroviral INs with members of the BET family. A panel of divergent INs from several retroviral genera were tested for the interaction with human BET proteins, BRD2 and BRD4, in transfected cells. FLAG-tagged MLV and FeLV gammaretroviral INs were efficiently recovered through coimmunoprecipitation with GFP-tagged BRD2 (Fig. 1A) and BRD4 (Fig. 1B). In contrast, IN proteins from other retroviral genera, including alpharetroviral (ASLV), betaretroviral (MPMV), deltaretroviral (HTLV-1), spumaretroviral (PFV), and lentiviral (HIV-1) INs failed to coimmunoprecipitate with BRD2 (Fig. 1A) or BRD4 (Fig. 1B). The interaction of BRD2 with gammaretroviral but not other retroviral integrases could also be shown using purified recombinant proteins. In these experiments, a C-terminal portion of BRD2, spanning its ET domain (residues 641 to 710), was sufficient for a specific interaction with MLV and FeLV INs in GST pulldown assays (Fig. 1C, lanes 8 and 9). In accord with the results of the coimmunoprecipitation experiments, the interaction was specific to gammaretroviral INs, and GST-BRD2(641–710) failed to pull down lentiviral, betaretroviral, deltaretroviral, or spumaviral INs (Fig. 1C, lanes 1 to 7 and 10 to 12). BRD4 also robustly interacted with MLV IN (Fig. 1D). In contrast, a GST-LEDGF(347–471) fusion, containing the lentivirus-specific IBD (24), specifically and exclusively interacted with the lentiviral INs (Fig. 1C, lanes 1 to 7). These results suggested that gammaretroviral INs evolved to engage the ET domains of BET proteins. Since ET domains are highly conserved between different BET family members, we focused most of our subsequent studies on BRD2.
Fig 1.
Interaction with BET proteins, BRD2 and BRD4, is limited to and conserved among gammaretroviral INs. (A and B) Extracts of HEK293T cells expressing IN-FLAG along with GFP-tagged BRD2 (A) or BRD4 (B) were immunoprecipitated (IP) with a GFP affinity matrix, and immunoprecipitates were analyzed by Western blotting (WB) using mouse anti-FLAG or mouse anti-GFP antibody as indicated. Migration positions of molecular mass standards (kDa) are shown alongside the gel. (C) GST pulldown experiments using GST alone (ii), GST-tagged BRD2(641–710) (iii), or LEDGF(347–471) (control) (iv). Proteins recovered on glutathione Sepharose beads were separated by SDS-PAGE and detected with Coomassie R-250. (i) Input samples of retroviral IN proteins; lanes 1 to 7, FIV, SIV, BIV, EIAV, visna virus, HIV-1, and HIV-2 (lentiviral IN proteins), respectively; lanes 8 and 9, FeLV and MLV (gammaretroviral IN), respectively; lane 10, HTLV-1 IN (δ-retroviral IN); lane 11, HSRV-2 (spumaretroviral IN); lane 12, MPMV (β-retroviral IN). (ii) GST pulldown using GST as negative control. (iii) GST pulldown with GST-BRD2(641–710); only IN proteins belonging to the gammaretroviral genus are specifically coprecipitated with the GST-tagged BRD2 fragment. (iv) GST pulldown using the C-terminal domain of LEDGF as bait; lentiviral IN specifically binds to LEDGF. Positions of the migration of GST-tagged proteins and the molecular mass markers are indicated on the right or left side of the gel. (D) GST pulldown assay. Purified GST-tagged BRD2(641–710) and BRD4(607–722) were incubated with purified MLV IN and glutathione Sepharose beads. Proteins bound to beads were separated by 12% denaturing PAGE and detected by staining with Coomassie blue. Inp, input; PD, pulldown; pur., purified.
The IN binding site in the BRD2 ET domain.
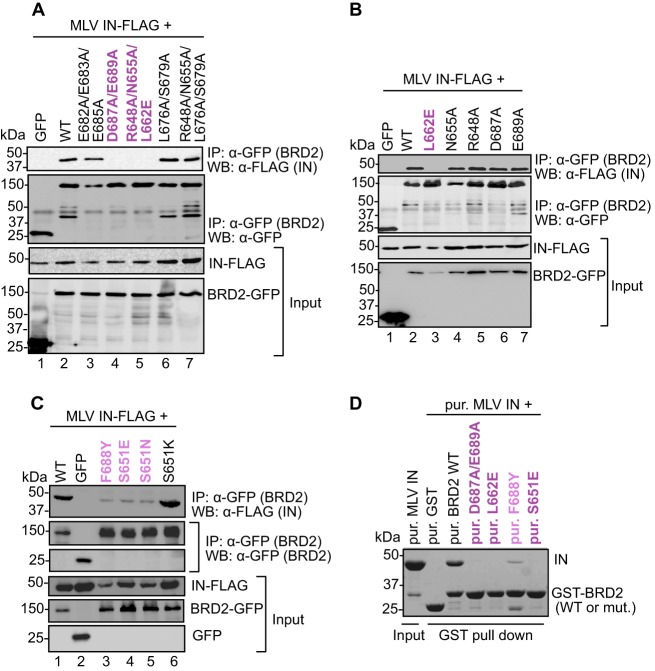
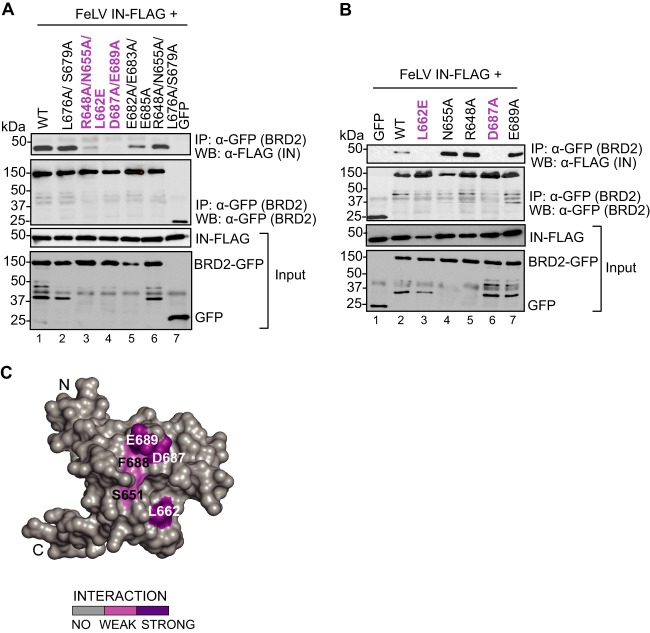
To confirm the involvement of the BRD2 ET domain in the binding to MLV IN and to identify residues participating in this process, we employed point mutants, which we had previously used when studying the interaction of BRD2 with KSHV LANA (76). We targeted, in total, 14 amino acids, located on an acidic ridge or in a neighboring pocket of the ET domain, structural elements previously identified by nuclear magnetic resonance (NMR) (53). The GFP-tagged BRD2 mutants were tested for binding to FLAG-tagged MLV IN by coimmunoprecipitation from transfected HEK293T cells. Alteration of BRD2 residue Asp687 in combination with Glu689 (D687A/E689A; Fig. 2A, lane 4) and Leu662 alone (L662E; Fig. 2B, lane 3) abrogated binding to MLV IN, whereas alteration of BRD2 residue Phe688 substantially reduced binding to IN (F688Y; Fig. 2C, lane 3). Furthermore, mutations of Ser651 to a glutamate (S651E) or an asparagine (S651N) also reduced binding to IN, whereas a mutation to a lysine (S651K) showed binding similar to that of WT BRD2 (Fig. 2C, lanes 4 to 6). The results obtained with all other BRD2 mutants are summarized in Table 2. The same set of mutants, in the context of GST-BRD2(641-710), was expressed in bacteria, purified, and tested for binding to MLV IN in a GST pulldown assay. In agreement with the results of the coimmunoprecipitation assays, GST-BRD2(641-710) mutants L662E, D687A/E689A, and S651E failed to pull down MLV IN, while F688Y was greatly reduced in its capacity to do so (Fig. 2D). These results narrow down the location of the IN binding surface in the ET domain to two main contact points: residues Glu689 and Asp687 located at the edge of the negative ridge and Leu662 positioned in a neighboring pocket (Fig. 3C). Moreover, the point mutations L662E and D687A in the BRD2 ET domain also ablated the interaction with FeLV IN, another gammaretroviral IN (Fig. 3A, lanes 3 and 4, and B, lanes 3 and 6), strongly suggesting that different gammaretroviral INs share the same binding site in the ET domain.
Fig 2.
Mapping the IN binding interface in the BRD2 ET domain. (A to C) Coimmunoprecipitation of FLAG-tagged MLV IN with GFP-tagged WT BRD2 or its point mutants. Proteins recovered after coimmunoprecipitation with a GFP affinity matrix were analyzed by Western blotting using mouse anti-GFP or anti-FLAG antibodies as indicated. (D) GST pulldown assay. GST-tagged BRD2(641–710) or its point mutants were incubated with MLV IN and glutathione Sepharose beads. Proteins bound to beads were separated by 12% denaturing PAGE and detected by staining with Coomassie blue. Residues whose mutations abrogate binding are colored dark purple; residues whose mutations displayed weaker binding to MLV IN are shown in light purple.
Table 2.
Summary of interaction studies with MLV IN and the BRD2 ET domain
| No. | BRD2 protein | Interaction with MLV INa |
|---|---|---|
| 1 | Wild type | + |
| 2 | E682A/E683A/E685A | + |
| 3 | D687A/E689A | − |
| 4 | R648A/N655A/L662E | − |
| 5 | L676A/S679A | + |
| 6 | R648A/N655A/L676A/S679A | + |
| 7 | L662E | − |
| 8 | N655A | + |
| 9 | R648A | + |
| 10 | D687A | + |
| 11 | E689A | + |
| 12 | F688Y | +/− |
| 13 | S651E | +/− |
| 14 | S651N | +/− |
| 15 | S651K | + |
+, strong; +/−, weak; −, none.
Fig 3.
Mapping the FeLV IN binding interface on BRD2. (A and B) Coimmunoprecipitation of FLAG-tagged FeLV IN with GFP-tagged BRD2 wild type (WT) or GFP-tagged BRD2 point mutants. Proteins recovered after coimmunoprecipitation with a GFP affinity matrix were analyzed by Western blotting using mouse anti-GFP or anti-FLAG antibodies as indicated. (C) Homology model of BRD2 ET domain based on Protein Data Bank accession no. 2JNS. Homology model of the BRD2 ET domain based on Protein Data Bank accession no. 2JNS (76). The coimmunoprecipitation results are mapped on the surface of the BRD2 ET domain. Residues whose mutations abrogate binding are colored dark purple; residues whose mutations displayed weaker binding to MLV IN are shown in light purple.
To explore a possible colocalization of BRD2 and MLV IN in cells, we transfected HeLa cells with enhanced GFP (EGFP)-BRD2 and/or MLV IN-mCherry expression constructs. Expressed separately, MLV IN-mCherry and EGFP-BRD2 (WT or L662E) showed a diffuse cytoplasmic (Fig. 4D and F) and nuclear (Fig. 4B, C, N, and O) localization, respectively. Strikingly, when coexpressed with EGFP-BRD2, MLV IN-mCherry changed localization pattern from diffuse cytoplasmic to punctate nuclear (Fig. 4G to I). In contrast, EGFP-BRD2 carrying the L662E mutation, which disrupts the interaction with MLV IN (Fig. 2), failed to redirect the viral protein to the nucleus (Fig. 4J to L). In the presence of MLV IN, we could also detect a weak BRD2 signal in the cytoplasm (Fig. 4H). Although this subcellular redistribution of MLV IN in the presence of BRD2 was obtained with overexpressed proteins, these results strongly corroborate a specific interaction between BRD2 and MLV IN as well as the importance of Leu662 in the BRD2 ET domain for this interaction.
Fig 4.
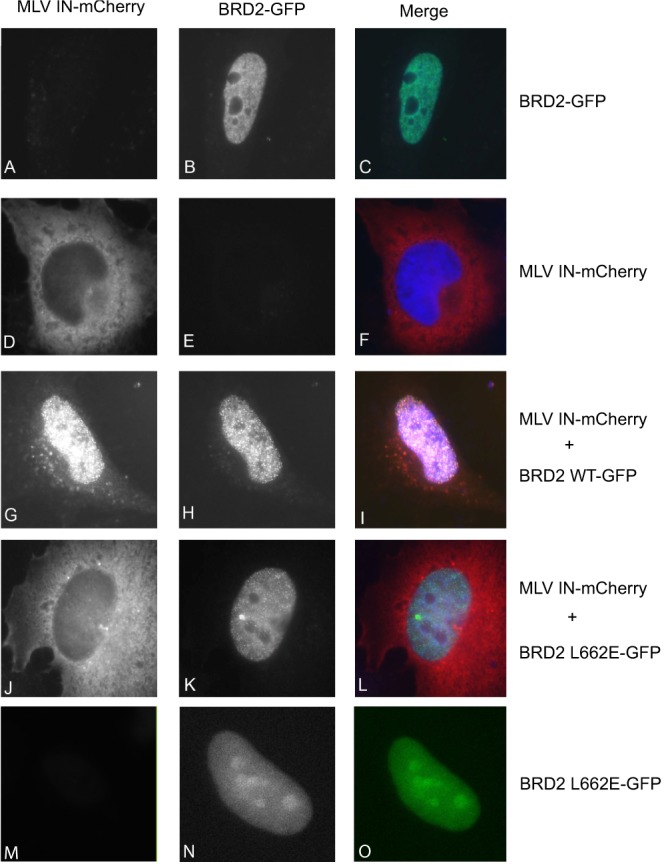
Colocalization of MLV IN with BRD2. HeLa cells were transfected with EGFP-BRD2, EGFP-BRD2 L662E, or MLV IN-mCherry or the indicated combinations. At 48 h posttransfection, cells were fixed and the localization of the fluorescent fusion proteins was documented. The mCherry and EGFP signals and their overlays are shown in the left, middle, and right images of each panel, respectively. DNA was stained with TO-PRO-3. The merged images contain signals of IN-mCherry (red), GFP-BRD2 (green), and DNA (blue). Images were taken at ×63 magnification.
BRD2 interacts with MLV IN in infected cells.
We next asked whether IN interacts with BRD2 during MLV infection. HEK293T cells were transfected with a plasmid encoding the N-terminally EGFP-tagged BRD2, BRD4, or LEDGF protein. Twenty-four hours posttransfection, cells were infected with an MLV vector produced using a packaging construct expressing C-terminally HA-tagged POL (pcDNA3.MLVgag-pol-HA) at an MOI of 1. Extracts from infected cells were incubated with anti-GFP antibody bound to protein A Sepharose beads, and immunoprecipitated proteins were detected by Western blotting (Fig. 5). BRD2 and BRD4 pulled down detectable amounts of HA-tagged MLV IN (Fig. 5, lanes 1 and 7). In contrast, BRD2 mutants L662E and D687A/E689A were defective for this interaction (Fig. 5, lanes 2 and 3). As expected, LEDGF failed to interact with the gammaretroviral IN (Fig. 5, lane 4). These results suggest that the BET proteins BRD2 and BRD4 can interact with MLV IN during infection.
Fig 5.
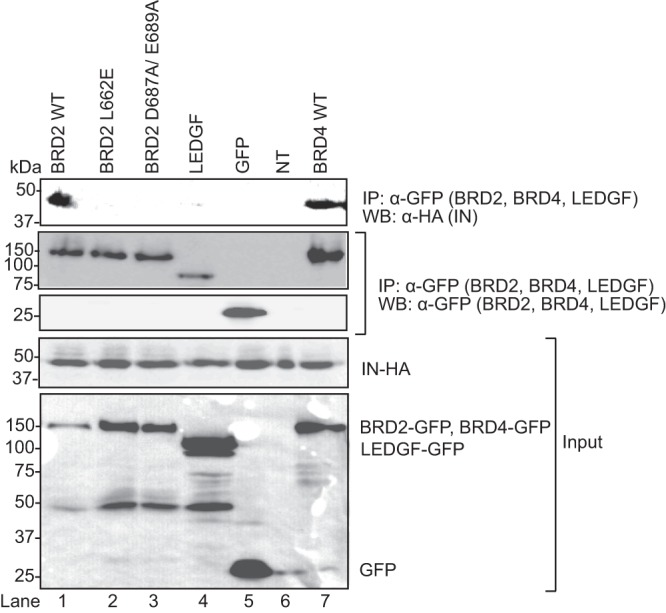
BRD2 mutants, L662E and D687A/E689A, fail to interact with MLV IN in vivo. HEK293T cells were transfected with the plasmids encoding the N-terminally GFP-tagged BRD2 WT (wild type) or BRD2 mutant L662E, D687A/E689A, or LEDGF or BRD4 and infected (MOI of 1) 24 h posttransfection with an MLV containing an HA-tagged IN. Extracts from the infected cells were immunoprecipitated with anti-GFP antibody coupled to protein A Sepharose. GFP-BRD2 WT, mutants, LEDGF, and BRD4 were detected by Western blotting using mouse monoclonal anti-GFP antibody, whereas MLV IN was detected with a rat anti-HA antibody.
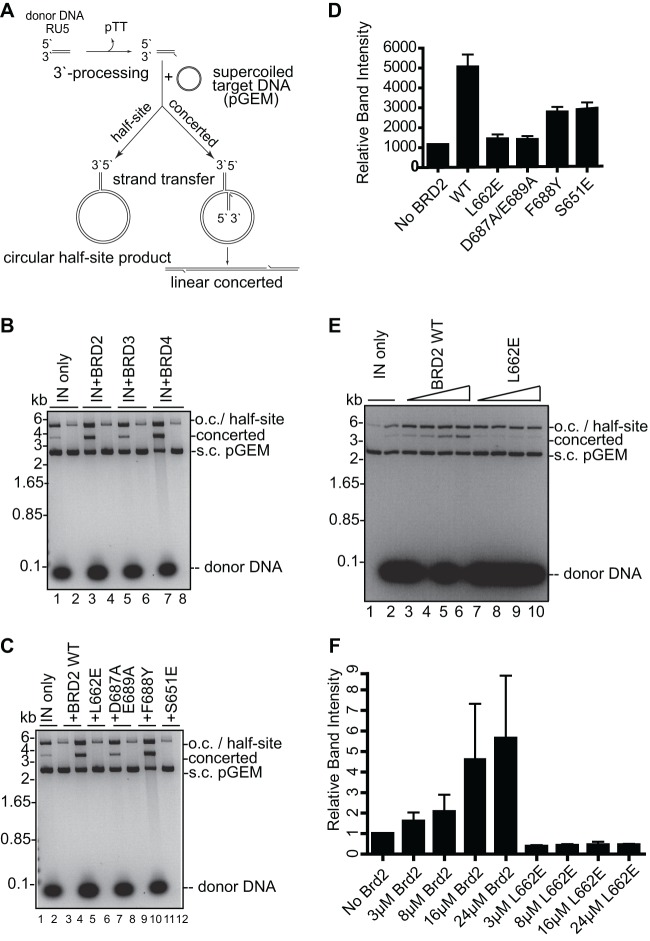
The ET domain of BET proteins augments the integration activity of MLV IN in vitro.
To investigate if binding of the ET domain to MLV IN could have an impact on its ability to mediate the integration of retroviral DNA into host DNA, we employed an in vitro integration assay. This assay uses short mimics of viral DNA ends (referred to as donor DNA substrates) in the presence of supercoiled target DNA and results in the formation of half-site and concerted integration products (71, 77, 78). The circular half-site results from the integration of a single LTR into one strand of the target plasmid, whereas linear concerted products reflect the integration of pairs of LTRs into opposing strands of the target DNA (Fig. 6A). Incubation of the preprocessed 32-bp donor DNA with 8 μM MLV IN in the presence of supercoiled plasmid DNA (pGEM) resulted in the accumulation of a product migrating at ∼3,900 bp, which corresponds to linear concerted integration reaction products, whereas the half-site reaction product comigrates with the open circular (o.c.) form of pGEM at ∼5,000 bp (Fig. 6B, lane1). Importantly, no linear products were formed when donor DNA was omitted from the reaction mixture (lanes 2, 4, 6, and 8), ruling out contamination of recombinant protein preparations with an endonuclease activity. Addition of purified ET domains of BRD2, BRD3, and BRD4 stimulated both half-site and concerted integration activity of MLV IN (Fig. 6B, lanes 3, 5, and 7). To confirm the structure of the concerted integration product, we treated gel-purified material with phi29 DNA polymerase and ligated it to a blunt-ended cassette encoding kanamycin resistance (71). Sequencing of a subset of kanamycin-resistant clones (52 and 91 for the condition without and with 24 μM BRD2, respectively) confirmed that the majority of them (>85%) contained pairs of donor DNA molecules, of which most were integrated with the expected 4-bp duplication (76% and 60%, for the conditions without and with BRD2, respectively).
Fig 6.
BET proteins stimulate strand transfer activity of recombinant MLV IN in vitro. (A) Schematic of strand transfer reactions using a circular DNA target. Concerted integration results in a gapped linear product whereas the half-site integration yields tailed relaxed circles. (B) Concerted integration activity of MLV IN in the presence of BRD2, BRD3, and BRD4. The enzyme (8 μM) was incubated with supercoiled pGEM target DNA in the absence (lanes 2, 4, 6, and 8) or presence (lanes 1, 3, 5, and 7) of 3 μM preprocessed donor DNA and in the absence (lane 1) or presence of 24 μM BET proteins BRD2 ET (lane 3), BRD3 ET (lane 5), and BRD4 ET (lane 7). Donor DNA (32 bp) mimics the U5 MLV cDNA terminus. Deproteinized reaction products were separated in 1.5% agarose gels and detected by staining with ethidium bromide. Migration positions of concerted and half-site reaction products, supercoiled (s.c.) pGEM target DNA forms, donor DNA, and a DNA size ladder are indicated. (C) Comparison of strand transfer activities of MLV IN in the presence of WT and mutant BRD2 ET domain. The donor DNA was omitted in lanes 2, 4, 6, 8, 10, and 12. (D) Quantification using ImageJ software of the band intensities of the concerted integration products in the presence of WT or mutant BRD2 ET domain as shown in panel C. (E) Comparison of strand transfer activities of MLV IN in the presence of 3 to 24 μM WT or mutant BRD2 ET domain. The donor DNA was omitted in lane 1. (F) Quantification of relative band intensities corresponding to the concerted integration products as shown in panel E. Error bars represent standard deviations from at least two independent experiments. o.c., open circular (nicked) DNA always present in DNA preparations.
As all of the tested ET domains showed similar effects on MLV integration, we continued our further studies only with the BRD2 ET domain. We next investigated if BRD2 ET domain residues that we had shown to be involved in the interaction with IN (Fig. 2 and 4) are essential for the stimulation of MLV integration activity. Compared with the WT BRD2 ET domain, the IN-binding-defective mutants L662E and D687A/E689A did not stimulate concerted integration activity (Fig. 6C, compare lanes 3, 5, and 7), whereas F688Y and S651E, which had a reduced ability to bind to MLV IN (Fig. 2C), showed a reduced stimulation of concerted integration (Fig. 6C, lanes 9 and 11). Quantification of these results by densitometry indicated that the WT BRD2 ET domain stimulated concerted integration activity of IN ∼4.5-fold, whereas L662E and D687A/E689A did not enhance integration activity (Fig. 6D). In contrast, an ∼2.3-fold stimulation of MLV IN activity was observed in the presence of the F688Y and S651E mutants (Fig. 6D). The stimulation of concerted integration with the WT BRD2 ET domain increased with higher protein concentrations, whereas the BRD2 ET L662E mutant failed to enhance integration even at the highest tested concentration (Fig. 6E, compare lanes 2 to 6 and 7 to 10, and F). Taken together, these results demonstrate that an interaction of the ET domain with IN stimulates MLV IN activity in vitro.
Overexpression of a C-terminal fragment of BRD2 containing ET domain increases MLV integration in HEK293T cells.
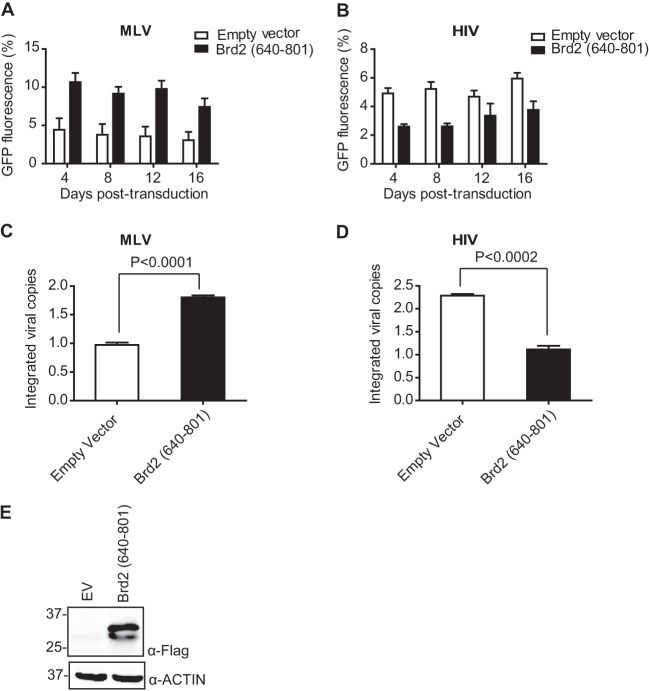
As described above, the BRD2 ET domain stimulates strand transfer activity of recombinant MLV IN in vitro. We next tested the effects of the BRD2 ET domain on MLV integration in HEK293T cells. To this end, we selected stable HEK293T cell lines overexpressing the C-terminal fragment of BRD2 containing the ET domain [BRD2(640–801)] together with a control cell line transfected with empty vector (Fig. 7E). Stable cell lines were then transduced with GFP-expressing MLV and HIV-1 vectors. We observed increased GFP expression in MLV-infected cells overexpressing the BRD2(640–801) fragment compared to cells transfected with an empty vector (Fig. 7A). In contrast, HIV-1-directed GFP expression was reduced in cells expressing the BRD2(640–801) fragment in comparison to the control cell line (Fig. 7B). In addition, when we quantified the number of integrated proviral copies by qPCR at 14 days postinfection, we observed a significant increase in the number of integrated proviral genomes in the case of BRD2(640–801)-expressing cells following infection with MLV (Fig. 7C), while the number of integrated proviral genomes was reduced in HIV-1-infected cells expressing BRD2(640–801) compared to the control cell line (Fig. 7D). The striking negative effect on HIV-1 replication is likely explained by the involvement of BET proteins in P-TEFb functions (79).
Fig 7.
Overexpression of IN binding domain of BRD2 (residues 640 to 801) increases MLV integration. (A and B) HEK293T-based cell lines stably expressing BRD2(640–801) were challenged with MLV-based (A) and HIV-1-based (B) retroviral vectors expressing EGFP (MOI, 1). Cells were harvested at indicated time points for flow cytometry analysis. Overall GFP fluorescence over time is calculated as mean fluorescence intensity × % gated cells. Error bars reflect duplicate measurements in each experiment. (C and D) Integrated proviral vector copies were quantified by qPCR at 16 days postinfection in cells transduced with MLV-based (C) and HIV-based (D) vectors. Values represent the means ± standard errors of the means; n is >3 throughout. Results were analyzed by unpaired t tests. (E) Expression of the BRD2(640–801) fragment and control cell lines used in this experiment was analyzed by Western blotting using anti-FLAG antibody.
BRD2 binding site on IN.
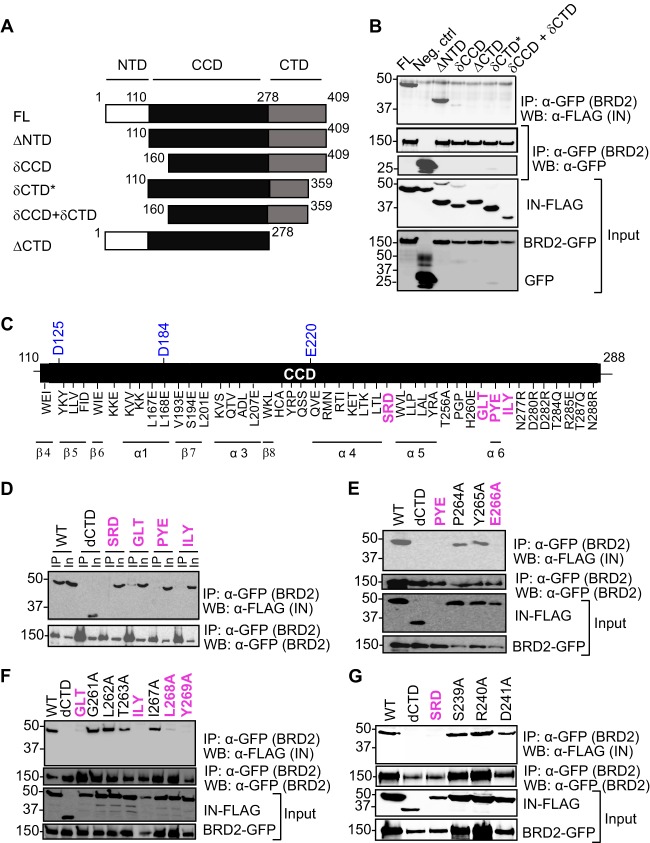
In order to identify the region(s) of MLV IN involved in the interaction with BRD2, we first prepared a series of IN deletion mutants (Fig. 8A). The mutants were FLAG tagged and tested for their ability to coimmunoprecipitate EGFP-tagged BRD2. As can be seen in Fig. 8B, both the full-length IN (residues 1 to 409) and the mutant lacking the N-terminal domain (residues 110 to 409, ΔNTD) readily bound BRD2. In contrast, a more severe deletion extending into the catalytic core domain disrupted the interaction with BRD2 (residues 160 to 409, δCCD construct, Fig. 8B). A complete deletion of the C-terminal domain of MLV IN (residues 1 to 278, ΔCTD) also resulted in a failure to bind BRD2 (Fig. 8B). Similarly, a deletion of the very C-terminal 50 residues, expected to disrupt the β-barrel fold of the CTD, was sufficient to abrogate the interaction with MLV IN (compare ΔNTD and δCTD constructs, Fig. 8B). These observations suggested that the CCD and the CTD of MLV IN are involved in the interaction with BRD2.
Fig 8.
Mapping the BRD2 binding site on MLV IN. (A) Domains of MLV IN and its deletion mutants. The N-terminal HHCC zinc binding domain, the catalytic core containing the DDE motif, and the positively charged C-terminal domain are indicated. (B) BRD2 interacts with the catalytic core and the C-terminal domain of MLV IN. HEK293T cells transfected with FLAG-tagged IN constructs and GFP-tagged BRD2 were lysed. BRD2 precipitated with anti-GFP affinity beads and detected with mouse anti-GFP antibody after extensive washing with lysis buffer and MLV IN WT or deletion mutants were detected with mouse anti-FLAG antibody. Two percent of each sample was used to confirm the expression of GFP-BRD2 or IN-FLAG (input, lower panels). A result representative of several experiments is presented. (C) A schematic of the catalytic core domain showing the positions of IN mutants constructed for this study. IN mutants that are defective for binding to BRD2 are highlighted in pink. The three catalytically important residues, D125, D184, and E220, are highlighted in blue. All the residues are mutated to alanine, except where indicated otherwise. (D to G) Results of coimmunoprecipitation of FLAG-tagged MLV IN WT and its point mutants with GFP-tagged BRD2.
To narrow down the regions of MLV IN involved in the interaction with the ET domain, we carried out an extensive scanning mutagenesis, in which we tested the contribution of 103 CCD residues (Fig. 8C). The residues targeted for mutagenesis were selected to be surface exposed, based on a structural model of MLV IN constructed using the homologous PFV structure (16). The IN mutants were screened for their ability to interact with BRD2 in a coimmunoprecipitation assay. Four triple alanine mutants, S239A/R240A/D241A (designated SRD and located between the predicted α4 and α6 helices of MLV IN), G261A/L262A/T263A (GLT), P264A/Y265A/E266A (PYE), and I267A/L268A/Y269A (ILY) (all predicted to be located near the α6 helix in MLV IN CCD), failed to interact with BRD2 (Fig. 8D). By mutating individual amino acids in these four triplets, we could identify three residues of MLV IN that were required for the interaction with BRD2. While the P264A and Y265A mutants efficiently bound BRD2, the E266A mutant failed to interact with BRD2 (Fig. 8E). Similarly, L268A and Y269A failed to coimmunoprecipitate BRD2 (Fig. 8F). However, although the triple mutants GLT (Fig. 8F) and SRD (Fig. 8G) failed to interact with BRD2, individual mutation of the 6 amino acids was compatible with binding to BRD2 at normal levels (Fig. 8F and G). Collectively, our results indicate that the α6 helix of the CCD along with the CTD of MLV IN is involved in the interaction with the ET domain.
BET bromodomain inhibitors reduce and redirect MLV integration.
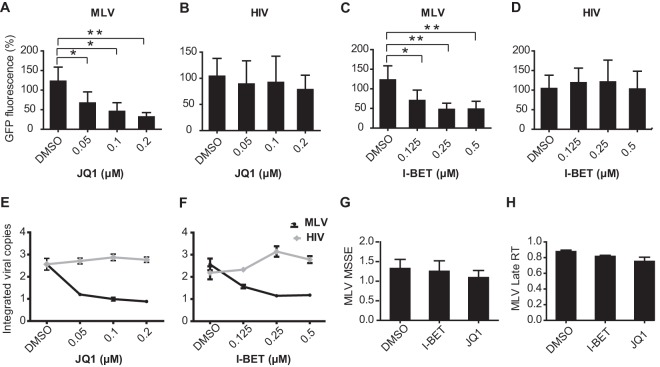
I-BET and JQ1 are small molecules that selectively disrupt the binding of the BET bromodomains to acetylated histones (48, 80). When we infected HEK293T cells with a GFP-expressing MLV in the presence of increasing JQ1 concentrations, we observed a dose-dependent reduction in MLV-driven GFP expression 3 days after infection (Fig. 9A). In contrast, no decrease in GFP expression was observed in HIV-1 vector-transduced cells (Fig. 9B), indicating that JQ1 does not affect the persistence of HIV-1. We also quantified the number of integrated proviral genomes after 3 weeks in this experiment and found them to be reduced in JQ1-treated, MLV-transduced cells but not in HIV-1-transduced cells (Fig. 9E). Reduction of MLV integration by JQ1 was observed to be dose dependent, with strong inhibition at submicromolar concentrations. Similar results were obtained with a second BET inhibitor, I-BET (Fig. 9C, D, and F). To exclude the possibility that these inhibitors affected early stages of viral entry, including reverse transcription, we quantified minus-strand strong-stop extension products (MSSEs) and late reverse transcripts (RTs). Inhibitor treatment did not alter MSSEs (Fig. 9G) or late RT products (Fig. 9H), indicating that MLV reverse transcription was not affected. Together, these data support the conclusion that BET inhibition specifically affects MLV integration in HEK293T cells. Finally, we isolated and sequenced a small number of integration sites from cells infected with an MLV vector in the absence (96 unique integration sites) or presence (52 unique sites) of 0.2 μM JQ1. As expected, under the control condition, the virus displayed a pronounced preference to integrate in the vicinity of transcription start sites, and approximately 46% of proviruses were found within 1.5 kbp of a RefSeq gene start site. Strikingly, this frequency dropped to 5% in the presence of the compound. Although the sample size was very modest, the observed effect was highly statistically significant (P < 0.00001, Fisher's exact test).
Fig 9.
BET bromodomain inhibitors, JQ1 and I-BET, reduce integration of MLV, but not of HIV-1, in HEK293T cells. (A to D) HEK293T cells were transduced with MLV-based (A and C) and HIV-1-based (B and D) vectors carrying EGFP as a reporter gene at an MOI of 0.1 in the presence of bromodomain inhibitors, I-BET and JQ1, at the concentrations indicated. Cells were treated with inhibitor or DMSO (control) for 36 h. Reporter gene activity was determined by FACS at 3 days following vector transduction. Overall GFP fluorescence is plotted. (E and F) qPCR analysis for integrated viral copies at 20 days postinfection in inhibitor- or DMSO-treated cells transduced with MLV- and HIV-1-based vectors. The graph indicates the integrated vector relative to DMSO sample. (G and H) qPCR analysis of JQ1- and I-BET-treated or DMSO-treated HEK293T cells transduced with MLV vector. Graphs indicate the amounts of PCR products relative to DMSO-treated sample (control) at 3 h postinfection for MSSEs (G) and 8 h postinfection for late RT (H) products. All bars represent the means ± standard errors of the means; n = 3; *, P < 0.05; **, P < 0.01. Results were analyzed by unpaired t tests and are representative of at least three independent experiments.
DISCUSSION
Mammalian BET proteins are defined by the presence of two bromodomains followed by the highly conserved extraterminal (ET) domain. Previous studies have shown that the ET domain is involved in interactions with a number of viral proteins. A recent study from Studamire and Goff (36) that identified BRD2 as a potential interactor with MLV IN prompted us to investigate a potential role of BET proteins in retroviral integration. While the manuscript was in progress, Sharma et al. (67) reported a confirmation that BET proteins do indeed interact with MLV IN but not with HIV-1 IN. They further demonstrated that BRD4 stimulates MLV IN activity in vitro and that BET proteins contribute to the efficiency of MLV integration and the propensity of the virus to target transcription start sites during viral infection (67). By and large, our findings are in excellent agreement with and significantly extend their observations. Here, we show that the INs of gammaretroviruses (MLV and FeLV), but not those of other retroviral genera (alpha-, beta-, delta-, spuma-, and lentiviruses), interact with the ET domain of the cellular BET proteins. The ET domains are ∼90% identical among mammalian BET family members (37, 81), and we therefore focused our studies on BRD2. Using coimmunoprecipitation and GST pulldown assays, we moreover narrowed down the interaction interface between MLV IN and the BRD2 ET domain and showed that BRD2 Leu662 and a pair of acidic residues, Asp687/Glu689, are critical for binding to MLV IN (Fig. 2 and 3C). We showed that MLV IN relocalizes from cytoplasm to the nuclei of live cells when co-overexpressed with BRD2. In contrast to MLV IN, HIV-1 IN displays a nuclear localization when expressed ectopically (82, 83). This property, conserved among lentiviral INs, fully depends on endogenous LEDGF (26, 83), a protein with a strong classical nuclear localization signal (84). Predictably, an HIV-1 IN mutant with a reduced affinity for LEDGF required overexpression of the host factor to display nuclear accumulation, while inactivation of the nuclear localization signal in LEDGF redirected IN to the cytoplasm (26, 84). Thus, the variations in cellular distributions of ectopically expressed retroviral INs may well be due to different affinities for their respective nuclear host factors. Intracellular levels of the specific host factors and competition with their endogenous binding partners will likely affect intracellular distribution of INs. Indeed, the affinity of the MLV IN-BRD2 interaction appears to be lower than that of HIV-1 IN-LEDGF (P. Cherepanov, unpublished observations). Furthermore, the combined intracellular level of BRD2 and BRD4 proteins is approximately 20-fold lower than that of LEDGF (85). By extension, the property of a retroviral IN to localize in the nucleus may have little to do with nuclear import of the preintegration complex.
Further extending the observations of Sharma et al. (67), who reported that MLV IN CTD is essential for interaction with BRD3, we showed that CCD of MLV IN greatly contributes to the interaction with BET proteins. Our mutagenesis data suggest that MLV IN Glu266, Leu268, and Tyr269, predicted to reside on the α6 helix of the MLV IN catalytic core domain, are crucial for the interaction with BRD2 in vitro. Interestingly, MLV IN residues Glu266, Leu268, and Tyr269 are well conserved among gammaretroviruses but not across other retroviral genera, which supports the notion that the binding of integrase to BET proteins is a feature conserved among and specific to gammaretroviruses.
Sharma et al. observed a stimulatory effect of full-length BRD4 on half-site and concerted MLV integration in vitro (67). Here, we demonstrated that isolated ET domains of various BET proteins, BRD2/RING3, BRD3/ORFX, and BRD4/HUNK-1, are sufficient for this function. These in vitro results are further supported by our observation that a cell line overexpressing the BRD2 ET domain shows increased MLV integration (Fig. 7). This is in contrast to the effects of isolated LEDGF IBD on HIV-1 integration: not only does IBD lacks a stimulatory effect on HIV-1 IN in vitro (25), but it also inhibits HIV-1 replication when overexpressed in target cells (86, 87).
Similar to the report by Sharma et al. (67), we found that bromodomain inhibitor JQ1, which disrupts the binding of BET/BRD bromodomains to acetylated histones and thereby displaces BET proteins from chromatin (44, 48), inhibits MLV integration (Fig. 9A). We also used a second bromodomain inhibitor, I-BET, with similar results (Fig. 9C). Thus, BET proteins may exert a dual effect on gammaretroviral replication: allosteric activation of IN enzymatic activity via the ET domains and chromatin tethering via their bromodomain modules.
Integration site selection has gained increased interest in human gene therapy. Retroviral vectors are important tools and have been widely used to deliver cDNAs for therapeutic purposes. However, retroviral integration can be associated with insertional activation of oncogenes (88), and several cases of leukemia caused by the integration of gammaretroviral vectors near proto-oncogenes were reported in two gene therapy trials (89–92). The safety of gene therapy could be improved if these retroviral vectors could be targeted to specific sites in the genome. The strong bias toward integration into transcription start sites, CpG islands, and promoter regions of genes appears to be a unique feature of gammaretroviruses such as MLV (93). In view of the results presented here and by Sharma et al. (67), the interaction of BET proteins with MLV IN could be exploited to control the integration pattern of gammaretroviral gene therapy vectors.
ACKNOWLEDGMENTS
We are grateful to Sabrine Woltemeyer (Institute of Microbiology, Hannover Medical School) for performing 454 pyrosequencing and to Benno Woelk (Institute of Virology, Hannover Medical School) for providing mCherry-tagged lentiviral vector p189 and for help with fluorescence microscopy.
This work was supported by the International Research Training group IRTG 1273 and the Collaborative Research Centre 900 “Chronic infections: Mechanisms of Microbial Persistence and its Control” project C1, both funded by the Deutsche Forschungsgemeinschaft (DFG). The work in P.C.'s laboratory was supported by the EU FP7 HIVINNOV consortium grant 305137.
Footnotes
Published ahead of print 18 September 2013
REFERENCES
- 1.Engelman A, Mizuuchi K, Craigie R. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211–1221 [DOI] [PubMed] [Google Scholar]
- 2.Hare S, Maertens GN, Cherepanov P. 2012. 3′-processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 31:3020–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujiwara T, Craigie R. 1989. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc. Natl. Acad. Sci. U. S. A. 86:3065–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holman AG, Coffin JM. 2005. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl. Acad. Sci. U. S. A. 102:6103–6107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodarzi G, Im GJ, Brackmann K, Grandgenett D. 1995. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J. Virol. 69:6090–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushman FD, Fujiwara T, Craigie R. 1990. Retroviral DNA integration directed by HIV integration protein in vitro. Science 249:1555–1558 [DOI] [PubMed] [Google Scholar]
- 7.Li X, Krishnan L, Cherepanov P, Engelman A. 2011. Structural biology of retroviral DNA integration. Virology 411:194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan E, Mack JP, Katz RA, Kulkosky J, Skalka AM. 1991. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 19:851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. U. S. A. 90:3428–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman A, Bushman FD, Craigie R. 1993. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 12:3269–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelman A, Craigie R. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I. 2003. Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 278:1323–1327 [DOI] [PubMed] [Google Scholar]
- 13.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. 2005. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 33:977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. 2006. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 25:1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren G, Gao K, Bushman FD, Yeager M. 2007. Single-particle image reconstruction of a tetramer of HIV integrase bound to DNA. J. Mol. Biol. 366:286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maertens GN, Hare S, Cherepanov P. 2010. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468:326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. 2005. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3:848–858 [DOI] [PubMed] [Google Scholar]
- 19.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521–529 [DOI] [PubMed] [Google Scholar]
- 20.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:E234. 10.1371/journal.pbio.0020234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Li Y, Crise B, Burgess SM. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749–1751 [DOI] [PubMed] [Google Scholar]
- 22.Trobridge GD, Miller DG, Jacobs MA, Allen JM, Kiem HP, Kaul R, Russell DW. 2006. Foamy virus vector integration sites in normal human cells. Proc. Natl. Acad. Sci. U. S. A. 103:1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman A, Cherepanov P. 2008. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 4:e1000046. 10.1371/journal.ppat.1000046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherepanov P. 2007. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 35:113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cherepanov P, Devroe E, Silver PA, Engelman A. 2004. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 279:48883–48892 [DOI] [PubMed] [Google Scholar]
- 26.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528–33539 [DOI] [PubMed] [Google Scholar]
- 27.Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, Poirier MG, Foster MP, Kvaratskhelia M. 2013. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 41:3924–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Nuland R, van Schaik FM, Simonis M, van Heesch S, Cuppen E, Boelens R, Timmers HM, van Ingen H. 2013. Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics Chromatin 6:12. 10.1186/1756-8935-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. 2009. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 41:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. 2007. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2:e1340. 10.1371/journal.pone.0001340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287–1289 [DOI] [PubMed] [Google Scholar]
- 32.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21:1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silvers RM, Smith JA, Schowalter M, Litwin S, Liang Z, Geary K, Daniel R. 2010. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum. Gene Ther. 21:337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z. 2010. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 18:552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A, Hughes SH. 2010. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc. Natl. Acad. Sci. U. S. A. 107:3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studamire B, Goff SP. 2008. Host proteins interacting with the Moloney murine leukemia virus integrase: multiple transcriptional regulators and chromatin binding factors. Retrovirology 5:48. 10.1186/1742-4690-5-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Florence B, Faller DV. 2001. You bet-cha: a novel family of transcriptional regulators. Front. Biosci. 6:D1008–D1018 [DOI] [PubMed] [Google Scholar]
- 38.Rhee K, Brunori M, Besset V, Trousdale R, Wolgemuth DJ. 1998. Expression and potential role of Fsrg1, a murine bromodomain-containing homologue of the Drosophila gene female sterile homeotic. J. Cell Sci. 111:3541–3550 [DOI] [PubMed] [Google Scholar]
- 39.Trousdale RK, Wolgemuth DJ. 2004. Bromodomain containing 2 (Brd2) is expressed in distinct patterns during ovarian folliculogenesis independent of FSH or GDF9 action. Mol. Reprod. Dev. 68:261–268 [DOI] [PubMed] [Google Scholar]
- 40.Guo N, Faller DV, Denis GV. 2000. Activation-induced nuclear translocation of RING3. J. Cell Sci. 113:3085–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LeRoy G, Rickards B, Flint SJ. 2008. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell 30:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denis GV. 2001. Bromodomain motifs and “scaffolding”? Front. Biosci. 6:D1065–D1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng J, Dong W, Chen L, Zou T, Qi Y, Liu Y. 2007. Brd2 is a TBP-associated protein and recruits TBP into E2F-1 transcriptional complex in response to serum stimulation. Mol. Cell. Biochem. 294:45–54 [DOI] [PubMed] [Google Scholar]
- 44.Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. 2004. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell 13:33–43 [DOI] [PubMed] [Google Scholar]
- 45.Nakamura Y, Umehara T, Nakano K, Jang MK, Shirouzu M, Morita S, Uda-Tochio H, Hamana H, Terada T, Adachi N, Matsumoto T, Tanaka A, Horikoshi M, Ozato K, Padmanabhan B, Yokoyama S. 2007. Crystal structure of the human BRD2 bromodomain: insights into dimerization and recognition of acetylated histone H4. J. Biol. Chem. 282:4193–4201 [DOI] [PubMed] [Google Scholar]
- 46.Huang H, Zhang J, Shen W, Wang X, Wu J, Wu J, Shi Y. 2007. Solution structure of the second bromodomain of Brd2 and its specific interaction with acetylated histone tails. BMC Struct. Biol. 7:57. 10.1186/1472-6807-7-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umehara T, Nakamura Y, Wakamori M, Ozato K, Yokoyama S, Padmanabhan B. 2010. Structural implications for K5/K12-di-acetylated histone H4 recognition by the second bromodomain of BRD2. FEBS Lett. 584:3901–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. 2010. Selective inhibition of BET bromodomains. Nature 468:1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., 3rd 2011. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. U. S. A. 108:16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muller S, Filippakopoulos P, Knapp S. 2011. Bromodomains as therapeutic targets. Expert Rev. Mol. Med. 13:e29. 10.1017/S1462399411001992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146:904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Li J, Schowalter RM, Jiao J, Buck CB, You J. 2012. Bromodomain protein Brd4 plays a key role in Merkel cell polyomavirus DNA replication. PLoS Pathog. 8:e1003021. 10.1371/journal.ppat.1003021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin YJ, Umehara T, Inoue M, Saito K, Kigawa T, Jang MK, Ozato K, Yokoyama S, Padmanabhan B, Guntert P. 2008. Solution structure of the extraterminal domain of the bromodomain-containing protein BRD4. Protein Sci. 17:2174–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Platt GM, Simpson GR, Mittnacht S, Schulz TF. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789–9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weidner-Glunde M, Ottinger M, Schulz TF. 2010. WHAT do viruses BET on? Front. Biosci. 15:537–549 [DOI] [PubMed] [Google Scholar]
- 56.McPhillips MG, Ozato K, McBride AA. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 79:8920–8932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schweiger MR, You J, Howley PM. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 80:4276–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. 2006. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 80:9530–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Senechal H, Poirier GG, Coulombe B, Laimins LA, Archambault J. 2007. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology 358:10–17 [DOI] [PubMed] [Google Scholar]
- 60.Wu SY, Lee AY, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang CM. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 20:2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith JA, White EA, Sowa ME, Powell ML, Ottinger M, Harper JW, Howley PM. 2010. Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl. Acad. Sci. U. S. A. 107:3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.You J, Srinivasan V, Denis GV, Harrington WJ, Jr, Ballestas ME, Kaye KM, Howley PM. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 80:8909–8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viejo-Borbolla A, Ottinger M, Bruning E, Burger A, Konig R, Kati E, Sheldon JA, Schulz TF. 2005. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 79:13618–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ottinger M, Christalla T, Nathan K, Brinkmann MM, Viejo-Borbolla A, Schulz TF. 2006. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J. Virol. 80:10772–10786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ottinger M, Pliquet D, Christalla T, Frank R, Stewart JP, Schulz TF. 2009. The interaction of the gammaherpesvirus 68 orf73 protein with cellular BET proteins affects the activation of cell cycle promoters. J. Virol. 83:4423–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin A, Wang S, Nguyen T, Shire K, Frappier L. 2008. The EBNA1 protein of Epstein-Barr virus functionally interacts with Brd4. J. Virol. 82:12009–12019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, Kessl JJ, Shkriabai N, Coward E, Aiyer SS, Green PL, Wu L, Roth MJ, Bushman FD, Kvaratskhelia M. 2013. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc. Natl. Acad. Sci. U. S. A. 110:12036–12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372–381 [DOI] [PubMed] [Google Scholar]
- 69.Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. 2010. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J. 29:2553–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voelkel C, Galla M, Maetzig T, Warlich E, Kuehle J, Zychlinski D, Bode J, Cantz T, Schambach A, Baum C. 2010. Protein transduction from retroviral Gag precursors. Proc. Natl. Acad. Sci. U. S. A. 107:7805–7810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valkov E, Gupta SS, Hare S, Helander A, Roversi P, McClure M, Cherepanov P. 2009. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 37:243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schambach A, Bohne J, Chandra S, Will E, Margison GP, Williams DA, Baum C. 2006. Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells. Mol. Ther. 13:391–400 [DOI] [PubMed] [Google Scholar]
- 73.Maetzig T, Brugman MH, Bartels S, Heinz N, Kustikova OS, Modlich U, Li Z, Galla M, Schiedlmeier B, Schambach A, Baum C. 2011. Polyclonal fluctuation of lentiviral vector-transduced and expanded murine hematopoietic stem cells. Blood 117:3053–3064 [DOI] [PubMed] [Google Scholar]
- 74.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suerth JD, Maetzig T, Brugman MH, Heinz N, Appelt JU, Kaufmann KB, Schmidt M, Grez M, Modlich U, Baum C, Schambach A. 2012. Alpharetroviral self-inactivating vectors: long-term transgene expression in murine hematopoietic cells and low genotoxicity. Mol. Ther. 20:1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hellert J, Weidner-Glunde M, Krausze J, Richter U, Adler H, Pietrek M, Ruckert J, Ritter C, Schulz TF, Luhrs T. A structural basis for BRD2/4-mediated host chromatin interaction and oligomer assembly of Kaposi sarcoma-associated herpesvirus and murine gammaherpesvirus LANA proteins. PLoS Pathog., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sinha S, Grandgenett DP. 2005. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J. Virol. 79:8208–8216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li M, Craigie R. 2005. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 280:29334–29339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. U. S. A. 104:13690–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A. 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468:1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu SY, Chiang CM. 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 282:13141–13145 [DOI] [PubMed] [Google Scholar]
- 82.Bouyac-Bertoia M, Dvorin JD, Fouchier RA, Jenkins Y, Meyer BE, Wu LI, Emerman M, Malim MH. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025–1035 [DOI] [PubMed] [Google Scholar]
- 83.Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. 2004. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 78:9524–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maertens G, Cherepanov P, Debyser Z, Engelborghs Y, Engelman A. 2004. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J. Biol. Chem. 279:33421–33429 [DOI] [PubMed] [Google Scholar]
- 85.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. 2011. The quantitative proteome of a human cell line. Mol. Syst. Biol. 7:549. 10.1038/msb.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314:461–464 [DOI] [PubMed] [Google Scholar]
- 87.De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z. 2006. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J. Virol. 80:11498–11509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coffin JM, Hughes SH, Varmus HE. 1997. The interactions of retroviruses and their hosts, p 335–341 In Coffin JM, Hughes SH, Varmus HE. (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 89.Check E. 2002. A tragic setback. Nature 420:116–118 [DOI] [PubMed] [Google Scholar]
- 90.Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, Cavazzana-Calvo M, Fischer A. 2003. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 348:255–256 [DOI] [PubMed] [Google Scholar]
- 91.Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, Brugman MH, Pike-Overzet K, Chatters SJ, de Ridder D, Gilmour KC, Adams S, Thornhill SI, Parsley KL, Staal FJ, Gale RE, Linch DC, Bayford J, Brown L, Quaye M, Kinnon C, Ancliff P, Webb DK, Schmidt M, von Kalle C, Gaspar HB, Thrasher AJ. 2008. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 118:3143–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, Schmidt M, Kramer A, Schwable J, Glimm H, Koehl U, Preiss C, Ball C, Martin H, Gohring G, Schwarzwaelder K, Hofmann WK, Karakaya K, Tchatchou S, Yang R, Reinecke P, Kuhlcke K, Schlegelberger B, Thrasher AJ, Hoelzer D, Seger R, von Kalle C, Grez M. 2010. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 16:198–204 [DOI] [PubMed] [Google Scholar]
- 93.Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, Berry CC, Ecker JR, Bushman FD. 2006. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2:e60. 10.1371/journal.ppat.0020060 [DOI] [PMC free article] [PubMed] [Google Scholar]