Abstract
Comprehensive virome analysis of RNA sequence (RNA-seq) data sets from 118 non-Hodgkin's B-cell lymphomas revealed a small subset that is positive for Epstein-Barr virus (EBV) or human herpesvirus 6B (HHV-6B), with one coinfection. EBV transcriptome analysis revealed expression of the latency genes RPMS1, LMP1, and LMP2, with one sample additionally showing a high level of early lytic expression and another sample showing a high level of EBNA2 expression. HHV-6B transcriptome analysis revealed that the majority of genes were transcribed.
TEXT
Herpesviridae is a large family of DNA viruses that can infect and cause disease in humans. Epstein-Barr Virus (EBV) and human herpesvirus 6 (HHV-6) are two members of this family that are highly ubiquitous and have been associated with mononucleosis and exanthema subitum (roseola), respectively. In addition, EBV is a well-known oncovirus that is associated with several malignancies, including nasopharyngeal carcinoma, gastric carcinoma, and lymphomas. HHV-6 is an emerging pathogen that has not been defined as an oncogenic pathogen but has been variably associated with lymphomas using traditional detection methods (e.g., PCR, Southern blotting, and immunohistochemistry [IHC]) (1).
For many years, associations between cancers and infectious agents have been made through epidemiological approaches and methods such as IHC and PCR. Although IHC and PCR approaches have been important for the detection of infectious agents in cancers, they have also led to false discovery and/or controversy. Several groups, including ours, have utilized RNA sequencing (RNA-seq) for the discovery and investigation of infectious agents; for example, Merkel cell virus was linked to Merkel cell carcinoma (2), Fusobacterium was associated with colorectal carcinoma (3, 4), EBV was studied with gastric carcinoma samples (5), murine leukemia virus (MuLV) was detected in human B-cell lines (6), and large sequencing databases were screened for oncoviruses (7). Next-generation sequencing (NGS) approaches have several advantages over previous detection methods for this type of study. In addition to being highly sensitive, NGS is highly specific, since the sequence for each read represents a fingerprint for a particular organism. Another key advantage is that a broad, relatively unbiased assessment of all known organisms can be performed in a single assay. This technology not only helps better identify etiological agents, but it can also better define cancers and/or specimens that are truly not associated with any known viruses.
Previous associations between EBV and non-Hogkin's lymphomas (8–11) prompted us to explore the links between diffuse large B-cell lymphomas (DLBCLs) and human viruses using next-generation sequencing. Using this approach, we comprehensively assessed the virome of a large non-AIDS non-Hodgkin's lymphoma (NHL) RNA-seq cohort from the Cancer Genome Characterization Initiative (CGCI).
EBV and HHV-6B are detected in a small percentage of diffuse large B-cell lymphomas.
RNA-seq data sets from 118 NHLs (105 DLBCLs and 13 follicular lymphomas [FL]) (12) were downloaded from the NIH database of genotypes and phenotypes (dbGap; http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap) using accession code phs000235.v2.p1 (additional details pertaining to the samples can be obtained through controlled access). Virome analysis of these polyA-selected RNA-seq data sets was performed by running roughly 27 million reads from each sample through our automated RNA-seq exogenous-organism analysis software, RNA CoMPASS (G. Xu, M. J. Strong, M. R. Lacey, C. Baribault, E. K. Flemington, and C. M. Taylor, submitted for publication). Within RNA CoMPASS, reads were aligned to the human reference genome, hg19 (UCSC), plus a splice junction database (which was generated using the Make Transcriptome application from Useq [13]; the splice junction radius was set to the read length minus 4) using Novoalign version 3.00.05 (www.novocraft.com) (-o SAM, default options). Nonmapped reads were isolated and subjected to consecutive BLAST version 2.2.27 searches against the human reference sequence (RefSeq) RNA database (an additional “preclearing” step) and then the NCBI nucleotide (NT) database to identify reads corresponding to known exogenous organisms (14). Results from the NT BLAST searches were filtered to eliminate matches with an E value of less than 10e−6. The results were then fed into MEGAN 4 version 4.70.4 (15) for visualization of taxonomic classifications.
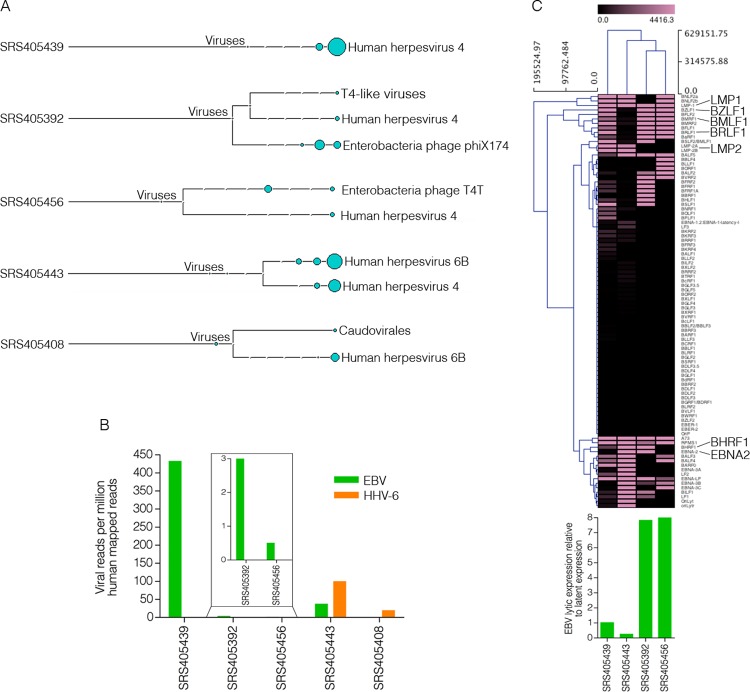
Most of the samples analyzed contained low levels of bacteriophage sequences, which likely represent either environmental contamination or quality control spike-ins (Fig. 1A). Of the 118 samples analyzed, 113 showed no evidence of eukaryotic viral polyadenylated-RNA expression, suggesting a different mechanism for tumor progression in these cases. Nevertheless, five DLBCL samples were positive for EBV (4 samples, 3.4%) or HHV-6B (2 samples, 1.7%), with one of these samples, SRS405443, being coinfected (Fig. 1A).
Fig 1.
EBV and HHV-6B detection in diffuse large B-cell lymphomas. (A) Virus branch of the taxonomy trees for the five virus-positive DLBCL samples. (B) Numbers of viral reads per million human mapped reads. (C) Cluster analysis of EBV transcripts along with a bar graph representing the ratio of EBV lytic-to-latent gene expression for each positive sample.
The findings for virus-positive samples were further analyzed by combining all sequencing runs for each EBV- and/or HHV-6B-positive tumor and aligning them directly to the human reference genome (hg19; UCSC) plus the Akata strain of the EBV genome (GenBank accession number KC207813) (16) and the HHV-6B genome (GenBank accession number NC000898). Alignments were performed using the Spliced Transcripts Alignment to a Reference (STAR) version 2.3.0 aligner (default options) (17). From this analysis, samples SRS405439 and SRS405443 were found to have the highest EBV read numbers (432 and 37 reads per million human mapped reads, respectively), while samples SRS405392 and SRS405456 had relatively low EBV read numbers (3 and 0.5 reads per million human mapped reads, respectively) (Fig. 1B). Samples SRS405408 and SRS405443 showed 19 and 99 HHV-6B reads per million human mapped reads, respectively (Fig. 1B).
Viral transcriptome analysis.
In a recent study, we showed that gastric carcinomas with high EBV read numbers exhibited signaling effects on cellular and microenvironmental pathways that were not observed for samples with either low or no EBV gene expression (5). Cluster analysis of these EBV-positive samples based on EBV gene expression alone showed unique clustering of the samples with high-versus-low EBV read counts. The distinct EBV gene expression patterns in these two groups suggested distinct infection types, which may partly explain differences in signaling effects. To similarly assess global differences in EBV gene expression patterns in the EBV-positive DLBCL samples, we performed cluster analysis. Transcript quantification of EBV genes was performed using SAMMate (18). Transcript counts and reads per kilobase per million mapped reads (RPKMs) were imported into Multiple Experiment Viewer (MeV) (19) for hierarchical clustering analysis. The Manhattan distance matrix was computed for the samples and used as input for hierarchical clustering using the complete linkage-clustering algorithm. The samples with low EBV read counts, SRS405392 and SRS405456, were found to cluster together (Fig. 1C). Visualization of reads across the EBV genome using the Integrative Genomics Viewer (IGV) (20) showed latency-gene peaks in the two samples with high EBV read counts (Fig. 2A). In contrast, only scattered reads were observed across the entire genome in the two samples with low EBV read counts (data not shown). This observation is illustrated by the finding of high lytic-to-latent read ratios in the samples with low EBV read counts versus those with high EBV read counts (Fig. 1C, bottom). The lack of distinct latency-gene expression along with the observed overall low EBV transcript levels for the samples with low EBV read numbers raises the possibility that the finding of EBV in these samples is less consequential than it is in samples SRS405439 and SRS405443, possibly reflecting low-level reactivation in infiltrating latent B-cells.
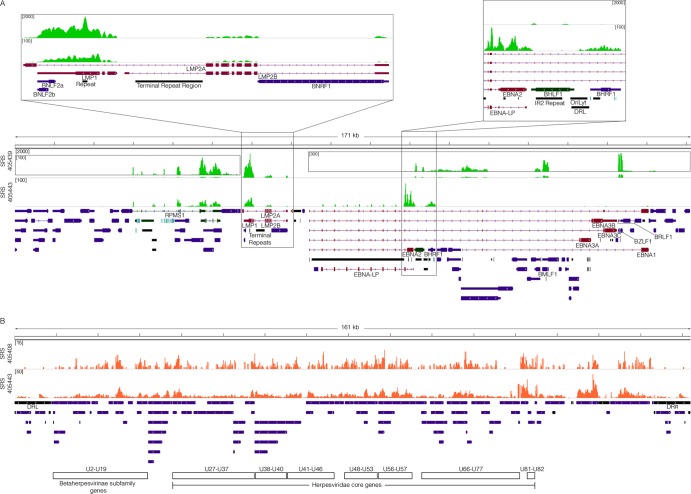
Fig 2.
Viral transcriptome. (A) EBV genome coverage data for EBV-positive DLBCLs. The y axis represents the number of reads at each nucleotide position. The modified EBV Akata genome was split between the BBLF2/3 and the BGLF3.5 lytic genes rather than at the terminal repeats to accommodate coverage of splice junctions for the latency membrane protein LMP2. The scale for sample SRS405439 is set to a maximum read level of 2,000 reads, with scales for the inset displays set to maximum read levels of 100 and 300 reads. The scale for sample SRS405443 is set to a maximum read level of 100 reads. Blue features represent lytic genes, red features represent latent genes, green features represent potential noncoding genes, aquamarine features represent microRNAs, and black features represent nongene features (e.g., repeat regions). (B) HHV-6B genome coverage data for HHV-6B-positive DLBCLs. The scale for sample SRS405408 is set to a maximum read level of 15 reads, while the scale for SRS405443 is set to 50 reads.
Detailed analysis of gene expression in the two EBV-positive samples with higher read counts showed expression of the EBV latency genes RPMS1, LMP1, and LMP2 in both cases (Fig. 2A). In contrast to the similarities in expression of these genes, expression of EBNA2 differed; sample SRS405443 expressed EBNA2 and sample SRS405439 did not (Fig. 2A). On the other hand, sample SRS405439 was unique in the detection of lytic transcripts with a disproportionately high level of the immediate early/early genes BZLF1 and BMLF1 relative to the levels of the bulk of other lytic genes (Fig. 2A). This predominant expression of early genes without other lytic genes is suggestive of an abortive lytic cycle, which has previously been linked to tumor progression (5, 21, 22). In contrast to the hallmark distinctive expression of EBV latency genes in samples SRS405439 and SRS405443, HHV-6B gene expression showed a broader expression profile across the entire genome, consistent with lytic transcription (Fig. 2B).
In conclusion, based on the samples tested here, most non-AIDS NHLs are free of known eukaryotic viruses expressing polyadenylated RNAs. In the two EBV-positive DLBCL samples, SRS405439 and SRS405443, the high read numbers in conjunction with the finding of clear expression of oncogenic latency genes (23–26) are consistent with an etiological role for EBV in these cases. In contrast, it is much less clear whether EBV contributes to the tumor phenotype in the two samples with lower read numbers where there is a lack of pronounced oncogenic latent-gene expression. Similarly, the general observation of broad lytic HHV-6B gene expression in the two HHV-6B-positive samples rather than expression of any particular potentially oncogenic latency gene suggests that at a minimum any contribution of HHV-6B to tumor progression likely occurs through a different mechanism (e.g., through a mechanism involving persistent smoldering stimulation of an inflammatory response to HHV-6B lytic antigens).
It is possible that moderate disease-related immunosuppression could lead to HHV-6B reactivation in HHV-6B-positive tumors, which may or may not contribute to the tumor phenotype. The finding of EBV and HHV-6 coinfection in one case raises the possibility that this patient may in fact have some level of immunosuppression. The expression of the highly immunogenic EBNA2 gene in this case further supports the suspicion of immunosuppression. Despite this possibility, it seems likely that EBV latency genes contribute to tumor progression (23–26) in this patient. Whether HHV-6B plays a role in tumor progression or whether expression is just a bystander effect of possible immunosuppression is unclear and will require further investigation. Regardless, HHV-6B is a component of the tumor microenvironment and it is appropriate to consider its presence in potential future tailored therapeutic design.
ACKNOWLEDGMENTS
We thank the Cancer Genome Characterization Initiative, all tissue donors, and the investigators for acquiring and sequencing the samples analyzed in this study.
This work was supported by National Institutes of Health grants R01CA124311 and R01CA138268 to E.K.F., F30CA177267 to M.J.S., and P20GM103518 to Prescott Deininger.
Footnotes
Published ahead of print 18 September 2013
REFERENCES
- 1.Ogata M. 2009. Human herpesvirus 6 in hematological malignancies. J. Clin. Exp. Hematopathol. 49:57–67 [DOI] [PubMed] [Google Scholar]
- 2.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castellarin M, Warren R, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. 2012. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostic AD, Ojesina AI, Pedamallu CS, Jung J, Verhaak RGW, Getz G, Meyerson M. 2011. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat. Biotechnol. 29:393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strong MJ, Xu G, Coco J, Baribault C, Vinay DS, Lacey MR, Strong AL, Lehman TA, Seddon MB, Lin Z, Concha M, Baddoo M, Ferris M, Swan KF, Sullivan DE, Burow ME, Taylor CM, Flemington EK. 2013. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PLoS Pathog. 9:e1003341. 10.1371/journal.ppat.1003341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Z, Puetter A, Coco J, Xu G, Strong MJ, Wang X, Fewell C, Baddoo M, Taylor C, Flemington EK. 2012. Detection of murine leukemia virus in the Epstein-Barr virus-positive human B-cell line JY, using a computational RNA-seq-based exogenous agent detection pipeline, PARSES. J. Virol. 86:2970–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khoury JD, Tannir NM, Williams MD, Chen Y, Yao H, Zhang J, Thompson EJ, Network T, Meric-Bernstam F, Medeiros LJ, Weinstein JN, Su X. 2013. Landscape of DNA virus associations across human malignant cancers: analysis of 3,775 cases using RNA-seq. J. Virol. 87:8916–8926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Sanjosé S, Bosch R, Schouten T, Verkuijlen S, Nieters A, Foretova L, Maynadié M, Cocco PL, Staines A, Becker N, Brennan P, Benavente Y, Boffetta P, Meijer CJ, Middeldorp JM. 2007. Epstein-Barr virus infection and risk of lymphoma: immunoblot analysis of antibody responses against EBV-related proteins in a large series of lymphoma subjects and matched controls. Int. J. Cancer 121:1806–1812 [DOI] [PubMed] [Google Scholar]
- 9.Hardell K, Carlberg M, Hardell L, Björnfoth H, Ericson Jogsten I, Eriksson M, Van Bavel B, Lindström G. 2009. Concentrations of organohalogen compounds and titres of antibodies to Epstein-Barr virus antigens and the risk for non-Hodgkin lymphoma. Oncol. Rep. 21:1567–1576 [DOI] [PubMed] [Google Scholar]
- 10.Mueller NE, Mohar A, Evans A. 1992. Viruses other than HIV and non-Hodgkin's lymphoma. Cancer Res. 52:5479s–5481s [PubMed] [Google Scholar]
- 11.Pozdnyakova O, Spieler PJ, Abraham J, Freedman AS, Kutok JL. 2010. Epstein-Barr virus-associated diffuse large B-cell lymphoma in an immunocompetent woman. J. Clin. Oncol. 28:e75–e78 [DOI] [PubMed] [Google Scholar]
- 12.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, Jackman S, Krzywinski M, Scott DW, Trinh DL, Tamura-Wells J, Li S, Firme MR, Rogic S, Griffith M, Chan S. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nix D, Courdy S, Boucher K. 2008. Empirical methods for controlling false positives and estimating confidence in ChIP-seq peaks. BMC Bioinformatics 9:523. 10.1186/1471-2105-9-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt KD, Tatusova T, Brown GR, Maglott DR. 2012. NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40:D130–D135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21:1552–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Z, Wang X, Strong MJ, Concha M, Baddoo M, Xu G, Baribault C, Fewell C, Hulme W, Hedges D, Taylor CM, Flemington EK. 2013. Whole-genome sequencing of the Akata and Mutu Epstein-Barr virus strains. J. Virol. 87:1172–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu G, Deng N, Zhao Z, Judeh T, Flemington E, Zhu D. 2011. SAMMate: a GUI tool for processing short read alignments in SAM/BAM format. Source Code Biol. Med. 6:2. 10.1186/1751-0473-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saeed A, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378 [DOI] [PubMed] [Google Scholar]
- 20.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat. Biotech. 29:24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong GK, Gulley ML, Feng W-H, Delecluse H-J, Holley-Guthrie E, Kenney SC. 2005. Epstein-Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J. Virol. 79:13993–14003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma S, Yu X, Mertz JE, Gumperz JE, Reinheim E, Zhou Y, Tang W, Burlingham WJ, Gulley ML, Kenney SC. 2012. An Epstein-Barr virus (EBV) mutant with enhanced BZLF1 expression causes lymphomas with abortive lytic EBV infection in a humanized mouse model. J. Virol. 86:7976–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawson CW, Port RJ, Young LS. 2012. The role of the EBV-encoded latent membrane proteins LMP1 and LMP2 in the pathogenesis of nasopharyngeal carcinoma (NPC). Semin. Cancer Biol. 22:144–153 [DOI] [PubMed] [Google Scholar]
- 24.Frappier L. 2012. Role of EBNA1 in NPC tumourigenesis. Semin. Cancer Biol. 22:154–161 [DOI] [PubMed] [Google Scholar]
- 25.Portis T, Cooper L, Dennis P, Longnecker R. 2002. The LMP2A signalosome—a therapeutic target for Epstein-Barr virus latency and associated disease. Front. Biosci. 1:d414–d426 [DOI] [PubMed] [Google Scholar]
- 26.Saha A, Robertson ES. 2013. Impact of EBV essential nuclear protein EBNA-3C on B-cell proliferation and apoptosis. Future Microbiol. 8:323–352 [DOI] [PMC free article] [PubMed] [Google Scholar]