Abstract
J paramyxovirus (JPV) was first isolated from moribund mice with hemorrhagic lung lesions in Australia in the 1970s. Recent sequencing of JPV (JPV-LW) confirms that JPV is a paramyxovirus with several unique features. However, neither JPV-LW nor a recombinant JPV based on its sequence (rJPV-LW) caused obvious illness in mice. In this work, we analyzed a different JPV isolate (JPV-BH), which behaved differently from JPV-LW; JPV-BH grew more slowly in Vero cells and had less of a cytopathic effect on tissue culture cells but caused severe disease in mice. We have determined the whole genome sequence of JPV-BH. There were several nucleotide sequence differences between JPV-BH and JPV-LW, one in the leader sequence, one in the GX gene, and three in the L gene. The high sequence identity between JPV-BH and JPV-LW suggests that JPV-BH and JPV-LW are the same virus strain but were obtained at different passages from different laboratories. To understand the roles of these nucleotide sequence differences in pathogenicity in mice, we generated a recombinant JPV-BH strain (rJPV-BH) and hybrid rJPV-BH strains with sequences from the leader sequence (rJPV-BH-Le-LW), the GX gene (rJPV-BH-GX-LW), and the L gene (rJPV-BH-L-LW) of JPV-LW and compared their pathogenicities in mice. We have found that rJPV-BH-L-LW was attenuated in mice, indicating that nucleotide sequence differences in the L gene play a critical role in pathogenesis.
INTRODUCTION
Members of the family Paramyxoviridae are nonsegmented, negative-sense, single-stranded RNA viruses. The family Paramyxoviridae includes two subfamilies, Paramyxovirinae and Pneumovirinae. Most of the viruses in Paramyxovirinae are categorized into five genera, Avulavirus, Henipavirus, Morbillivirus, Respirovirus, and Rubulavirus. There are many important human and animal pathogens among the paramyxoviruses, such as measles virus, parainfluenza viruses, mumps virus, respiratory syncytial virus (RSV), and the deadly Hendra and Nipah viruses. J paramyxovirus (JPV) was isolated on four separate occasions from moribund wild mice (Mus musculus) trapped in northern Queensland, Australia, in 1972 (1, 2). The four mice had extensive hemorrhagic lung lesions. Syncytium formation was observed in kidney autoculture monolayers, and electron microscopy revealed a nucleocapsid structure typical of paramyxoviruses. The full-length genome of JPV has been sequenced, and it contains 18,954 nucleotides (nt) (3). The organization of the JPV genome is 3′-N-P/V/C-M-F-SH-TM-G-L-5′. JPV contains a small hydrophobic (SH) gene, which is not present in all paramyxoviruses. The novel transmembrane (TM) protein is predicted to be a type II glycosylated integral membrane protein. Surface biotinylation of intact cells and deglycosylation of cell lysates have confirmed that TM is a surface expression protein containing N-linked glycosylation sites (4). The G gene encodes a putative 709-amino-acid glycoprotein and distally contains a 2,115-nt second open reading frame (ORF), termed ORF X, which has not been detected in virus-infected cells (3, 4). Probes specific to the G protein coding region and ORF X both identified an mRNA species corresponding to the predicted length of the G gene (the gene is renamed GX in this work). Because of its unique features, JPV does not fit in any known paramyxovirus genus and is therefore an unclassified paramyxovirus.
Although JPV was originally isolated from moribund mice in Australia and infection with the virus reportedly caused disease in mice (1), JPV-LW (named after Lin-Fa Wang, who determined its sequence) did not cause any disease in JPV-LW-infected mice (L.-F.W., unpublished data). Likewise, in our previous study, there were no clinical signs of illness or weight loss and no pathological changes in organs such as the lungs, kidneys, heart, or liver in recombinant rJPV-LW (rJPV-LW)-infected mice (5). In this work, we obtained and analyzed a different JPV isolate, JPV-BH (named after Biao He) that behaved differently from JPV-LW; JPV-BH had less of a cytopathic effect (CPE) on tissue culture cells but caused severe disease in mice. We have determined the whole genome sequence of JPV-BH and found several nucleotide sequence differences between JPV-BH and JPV-LW. We generated recombinant hybrid rJPV-BH strains containing sequences from JPV-LW. We investigated the roles of these nucleotide sequence differences in pathogenicity in mice.
MATERIALS AND METHODS
Cells.
Monolayer cultures of HEK293T, L929, and Vero cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 μg/ml streptomycin. BSR-T7 cells were maintained in DMEM containing 10% FBS, 10% tryptose phosphate broth, and 400 μg/ml G418. All cells were incubated at 37°C in 5% CO2. Virus-infected cells were grown in DMEM containing 2% FBS. Plaque assays were performed with Vero cells.
Mice.
Female 6-week-old BALB/c mice (Charles River Labs, Frederick, MD) were used for all studies. Mouse infection with JPV was performed in enhanced biosafety level 2 facilities in HEPA-filtered isolators. All animal studies were conducted under guidelines approved by the Animal Care and Use Committee at the University of Georgia.
Construction of recombinant plasmids.
To generate a recombinant JPV-BH plasmid, JPV-BH RNA was extracted and reverse transcription (RT)-PCRs were performed. PCR products were ligated stepwise into a backbone plasmid from a parainfluenza virus 5 infectious cDNA clone (6). To differentiate wild-type and recombinant JPV-BH, a PvuI restriction site was introduced into the N gene without changing the amino acid residues. The recombinant plasmids containing the leader sequence, GX genes, and L gene of rJPV-LW within the JPV-BH backbone (designated JPV-BH-Le-LW, JPV-BH-GX-LW, and JPV-BH-L-LW, respectively) were constructed by standard molecular biology techniques. Three helper expression plasmids containing the JPV-BH N, P, and L genes in a pCAGGs vector were constructed by using appropriate oligonucleotide primer pairs. The sequences of the primers used for JPV-BH, JPV-BH-Le-LW, JPV-BH-GX-LW, and JPV-BH-L-LW and the three helper plasmids are available on request.
Virus rescue and sequencing.
To generate viable recombinant strains, the following plasmids were cotransfected into HEK293T cells at 95% confluence in 6-cm plates with Jetprime (Polyplus-Transfection, Inc., New York, NY): a plasmid expressing T7 polymerase (pT7P); three plasmids, pJPV-N, pJPV-P, and pJPV-L, encoding the N, P, and L proteins, respectively; and a plasmid encoding the full-length JPV genome, JPV-BH, JPV-BH-Le-LW, JPV-BH-GX-LW, or JPV-BH-L-LW. The amounts of plasmids used were as follows: 5 μg of full-length JPV plasmids, 1 μg of pT7P, 1 μg of pJPV-N, 0.3 μg of pJPV-P, and 1.5 μg of pJPV-L. Two days posttransfection, 1/10 of the HEK293T cells were cocultured with 1 × 106 Vero cells in a 10-cm plate. The media were harvested on days 5 to 7 after coculture, and cell debris was pelleted by low-speed centrifugation (3,000 rpm, 10 min). Plaque assays on Vero cells were used to obtain single clones of recombinant strains.
The full-length genomes of plaque-purified JPV-BH strains were sequenced. Total RNAs from JPV-BH-, rJPV-BH-, rJPV-BH-Le-LW-, rJPV-BH-GX-LW-, and rJPV-BH-L-LW-infected Vero cells were purified with the RNeasy minikit (Qiagen, Valencia, CA). cDNAs were prepared by using random hexamers and then amplified by PCR with appropriate oligonucleotide primer pairs as described previously (5). Improved rapid amplification of cDNA ends PCR was used to amplify the leader and trailer sequences (7). The sequences of all of the primers used to sequence the complete genomes of JPV-BH, rJPV-BH, rJPV-BH-Le-LW, rJPV-BH-GX-LW, and rJPV-BH-L-LW are available on request. DNA sequences were determined with an Applied Biosystems sequencer (ABI, Foster City, CA).
Growth kinetics.
Vero or L929 cells in six-well plates were infected with rJPV-LW, JPV-BH, rJPV-BH, rJPV-BH-Le-LW, rJPV-BH-GX-LW, or rJPV-BH-L-LW at a multiplicity of infection (MOI) of 5 or 0.1. The cells were then washed with phosphate-buffered saline (PBS) and maintained in DMEM–2% FBS. Medium samples were collected at 24-h intervals. Virus titers were determined by plaque assay on Vero cells.
Detection of viral protein expression.
The P expression levels in virus-infected cells were compared. Vero cells in six-well plates were mock infected or infected with rJPV-LW, JPV-BH, rJPV-BH, rJPV-BH-Le-LW, rJPV-BH-GX-LW, or rJPV-BH-L-LW at an MOI of 5. The cells were collected at 2 dpi and fixed with 0.5% formaldehyde for 1 h. The fixed cells were resuspended in FBS-DMEM (50:50) and then permeabilized in 70% ethanol overnight. The cells were washed once with PBS and then incubated with rabbit anti-P antibody in PBS–1% BSA (1:200) for 1 h at 4°C. The cells were stained with anti-rabbit antibody labeled with phycoerythrin (1:200) for 1 h at 4°C in the dark and then washed once with PBS–1% BSA. The fluorescence intensity was measured with a flow cytometer (BD LSR II; BD Biosciences, San Jose, CA).
Real-time PCR.
To compare viral mRNA and genome levels in infected cells, real-time PCR was carried out as described previously (8). Briefly, Vero cells in six-well plates were infected with rJPV-LW or JPV-BH at an MOI of 5. At 24 h postinfection (hpi), the total RNA was extracted from the infected cells with the RNeasy minikit (Qiagen). One-fifth of the total RNA from each sample was used for RT with Superscript III reverse transcriptase (Invitrogen). Oligo(dT)15 was used for RT to detect viral mRNA levels. Primer 376, which anneals to a region of the genomic RNA in the N gene, was used for RT to measure viral genomes. Two percent of the cDNA was used for a real-time PCR with a Step One Plus real-time PCR system with TaqMan universal PCR master mix and custom-made TaqMan gene expression assays for the M gene of JPV with 6-carboxyfluorescein dye and nonfluorescent quencher (Applied Biosystems). Relative viral mRNA and genome levels were determined by calculating the change in the cycle threshold and normalized to the level of the input genome, which was defined as the genome level at 2 hpi. Three replicates of each sample were used for statistical analysis.
Apoptosis assay.
L929 cells in six-well plates were mock infected or infected with rJPV-LW, JPV-BH, rJPV-BH, rJPV-BH-Le-LW, rJPV-BH-GX-LW, or rJPV-BH-L-LW at an MOI of 5. At 2 days postinfection (dpi), L929 cells were trypsinized and combined with floating cells in the media. The harvested cells were centrifuged, washed with PBS, fixed, and permeabilized. The cells were then incubated with 25 μl of terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) reaction mixture (Cell Death Detection kit; Roche Diagnostics Corp., Mannheim, Germany) for 2 h in the dark at 37°C. The cells were analyzed by flow cytometry (BD LSR II).
Infection of mice with JPV.
All animal experiments were carried out by strictly following the protocol approved by IACUC. To study the pathogenesis of JPV in mice, 6-week-old wild-type BALB/c mice were inoculated intranasally with 100 μl of PBS or 5 × 105 PFU of rJPV-LW, JPV-BH, rJPV-BH, rJPV-BH-Le-LW, rJPV-BH-GX-LW, or rJPV-BH-L-LW. Body weights were monitored daily for 15 dpi. Mice were euthanized at 1, 3, 5, and 7 dpi, and their lungs were collected. Virus titers in the lungs were determined by plaque assay on Vero cells.
Histology studies.
BALB/cJ mice from the infection study were euthanized by CO2 asphyxiation. The lungs were inflated with 4% paraformaldehyde and collected. Samples were routinely processed, embedded, and sectioned for hematoxylin and eosin (H&E) staining. Alveolar infiltrates and perivascular cuffing were scored on a scale of 1 (minimal) to 4 (severe) in a blinded fashion by a board-certified veterinary pathologist. Photomicrographs were taken with an Olympus BX41 microscope, an Olympus DP70 microscope digital camera, and DP Controller imaging software.
RESULTS
Sequence analysis of JPV-BH.
We obtained a sample of a potential new JPV isolate (J virus) from Robert Tesh at the University of Texas Medical Branch. The tube was labeled “J virus.” However, it was not certain that it was JPV because it might be an arbovirus, which was of interest to Robert Tesh's lab. The putative virus was incubated with Vero cells. For logistical reasons, we were unable to obtain wild-type JPV-LW, which was stored in a high-security lab in Australia. As a control, Vero cells were incubated with rJPV-LW. Large syncytia were produced in rJPV-LW-infected cells but not in mock- or J virus-infected cells. However, J virus had a CPE on Vero cells, suggesting an infectious agent in the original sample (Fig. 1). To determine whether the infectious agent was JPV, RNA was purified from J virus-inoculated medium and RT-PCR was performed with primers based on the JPV-LW sequence. The RT-PCR was positive, suggesting that the virus was closely related to JPV-LW (data not shown). We named this J virus the JPV BH strain (JPV-BH). The entire genome of JPV-BH was amplified by overlapping RT-PCR and then sequenced (Table 1). JPV-BH has the same genome structure and number of nucleotides as JPV-LW and contains one nucleotide difference in the leader sequence and three nucleotide differences in the L gene compared to the rJPV-LW and JPV-LW sequences (Table 1). rJPV-LW contains one extra nucleotide difference in ORF G and three nucleotide differences in ORF X compared to the JPV-LW sequence deposited in GenBank (accession no. NC_007454) (3).
Fig 1.
Comparison of the rJPV-LW and JPV-BH strains in vitro. Shown are the CPEs of the rJPV-LW and JPV-BH strains on Vero cells. Vero cells were mock infected or infected with rJPV-LW or JPV-BH at an MOI of 0.1. At 3 dpi, the cells were photographed.
Table 1.
Sequence differences between JPV-LW and JPV-BH strains
| Genome position (region or ORF) | rJPV-LW | JPV-LW | JPV-BH | Amino acid residue change |
|---|---|---|---|---|
| 16 (leader) | G | G | U | NAa |
| 9568 (G) | A | C | C | Lys-Thr |
| 10417 (X) | C | A | A | Ser-Tyr |
| 10726 (X) | G | A | A | Arg-Gln |
| 11753 (X) | A | U | U | Lys-Asn |
| 13932 (L) | A | A | G | Ile-Val |
| 13997 (L) | C | C | G | Asp-Glu |
| 15260 (L) | A | A | C | Glu-Asp |
NA, not applicable.
Analysis of JPV-BH in tissue culture cells.
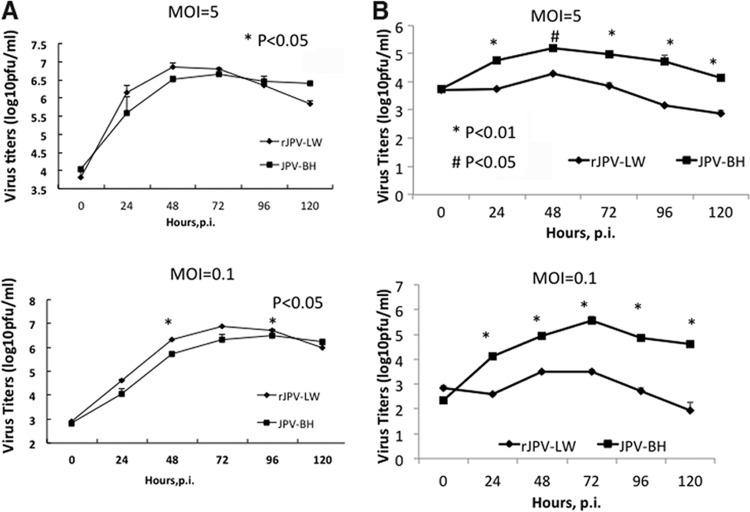
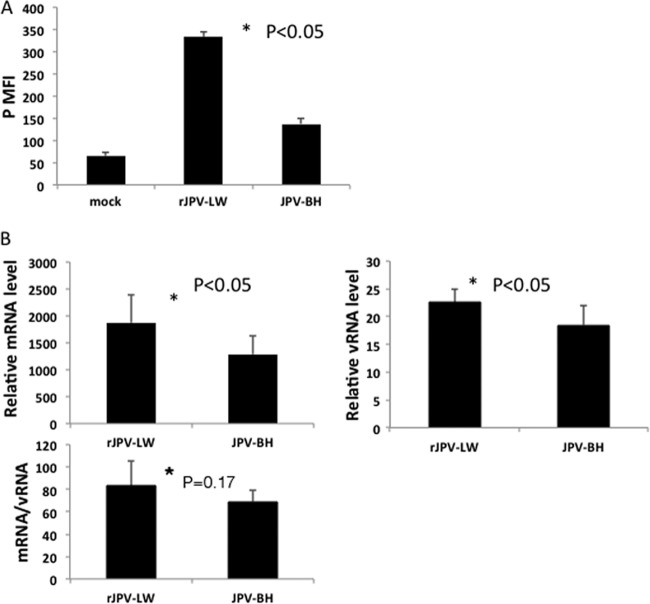
To compare the growth kinetics of rJPV-LW and JPV-BH, single- and multiple-step growth curves were obtained with Vero cells, an interferon (IFN)-deficient simian cell line, and L929 cells, a murine cell line. Interestingly, rJPV-LW replicated faster than JPV-BH in Vero cells in single- and multiple-step growth curves (Fig. 2A); however, rJPV-LW replicated 1 to 2 logs (5) lower than JPV-BH in L929 cells (Fig. 2B). To examine whether there was a difference in JPV viral protein expression levels, the mean fluorescence intensity (MFI) of the P protein in infected cells was examined. Consistent with the growth curves, P protein expression was higher in rJPV-LW-infected Vero cells than in JPV-BH-infected Vero cells (Fig. 3A). Real-time PCR was used to determine viral genome RNA and mRNA levels. rJPV-LW produced higher levels of viral genome RNA and mRNA transcripts than JPV-BH. rJPV-LW also produced more mRNA transcripts per viral genome copy, as determined by the ratio of viral mRNA to viral genome RNA, suggesting that rJPV-LW viral mRNA transcription was increased in infected cells relative to that in JPV-BH-infected cells (Fig. 3B). These results were consistent with the observation of higher viral protein expression levels in rJPV-LW-infected cells than in JPV-BH-infected cells.
Fig 2.
Growth rates of JPV-BH in tissue culture cells. Single- and multiple-step growth curves of rJPV-LW and JPV-BH in Vero (A) and L929 (B) cells are shown. Vero or L929 cells were infected with rJPV-LW or JPV-BH at an MOI of 5 or 0.1. Medium samples were collected at 24-h intervals. Virus titers were determined by plaque assay on Vero cells. All P values were calculated by using a t test.
Fig 3.
Viral gene transcription and expression. (A) Viral protein expression of rJPV-LW and JPV-BH in Vero cells. Vero cells were mock infected or infected with rJPV-LW or JPV-BH at an MOI of 5. At 2 dpi, the cells were collected and the MFI of P was determined by flow cytometry. (B) Viral RNA (vRNA) and mRNA synthesis and ratio of mRNA to viral RNA in Vero cells. Vero cells were infected with rJPV-LW or JPV-BH at an MOI of 5. At 24 hpi, a real-time PCR was performed to determine viral RNA and mRNA levels and mRNA/viral RNA ratios. rJPV-LW and JPV-BH genome levels at 2 hpi were used as baselines for normalization.
JPV-BH was pathogenic in mice.
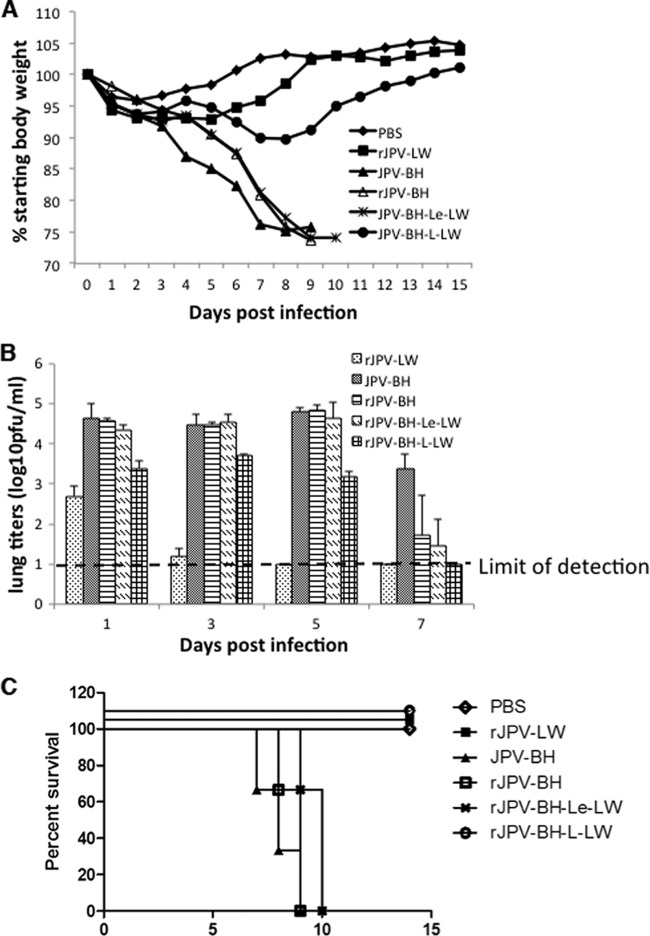
Previously, we reported that rJPV-LW was not pathogenic in mice (5). To investigate whether JPV-BH was pathogenic in mice, mice were infected intranasally with rJPV-LW and JPV-BH (Fig. 4A and B). rJPV-LW-infected mice lost little body weight (about 5%) and recovered quickly. rJPV-LW replicated poorly in mouse lungs, as there was no detectable virus in the lungs at 5 dpi. JPV-BH-infected mice lost up to 25% of their body weight. JPV-BH replicated steadily at 104 PFU/ml for 5 days in the lungs with a small drop in virus titers at 7 dpi. Further analysis of lungs infected with JPV-BH and rJPV-LW showed no histopathologic differences between rJPV-LW- and mock-infected lungs, while JPV-BH resulted in perivascular cuffing, as well as interstitial and alveolar infiltrates (Fig. 5). These results indicate that JPV-BH, unlike rJPV-LW, is pathogenic in mice.
Fig 4.
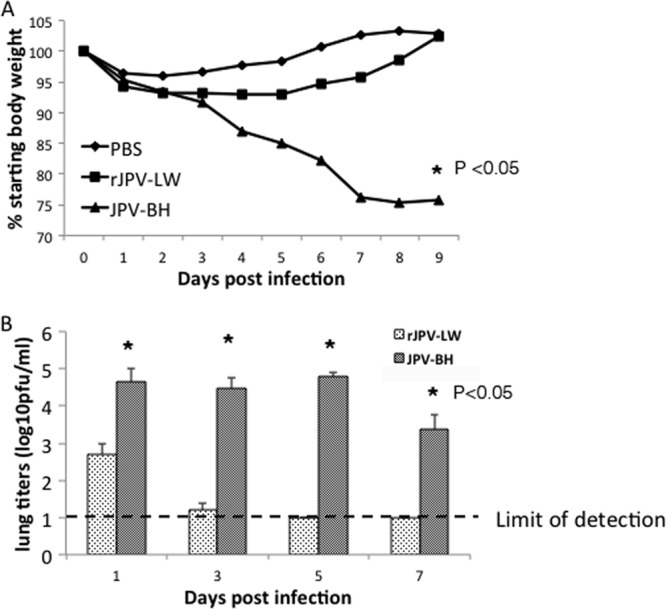
Pathogenicity of JPV-BH in mice. Mice were inoculated intranasally with PBS or 5 × 105 PFU of rJPV-LW or JPV-BH. (A) Body weight change. Mice were monitored daily, and weight loss was graphed as the average percentage of the original weight (on the day of infection). (B) Lung titers of mice. Mouse lungs were collected at 1, 3, 5, and 7 dpi. Virus titers were determined by plaque assay on Vero cells.
Fig 5.
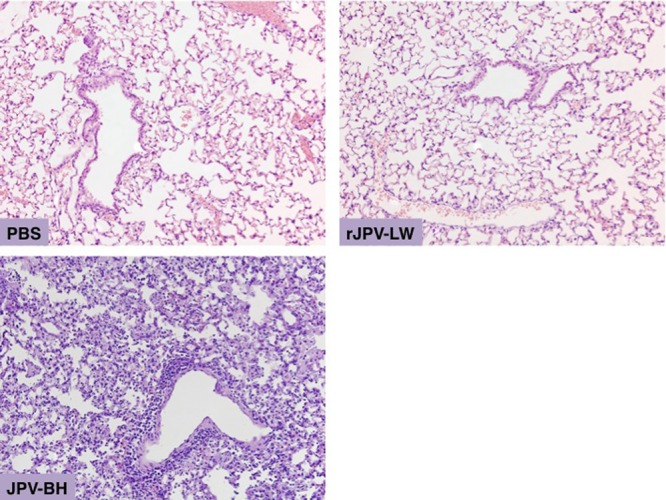
Histopathology of lungs of JPV-infected mice. Lungs were collected from infected and mock-infected animals at 7 dpi and stained with H&E. Photomicrographs were taken at a magnification of ×20.
A reverse genetics system for JPV-BH.
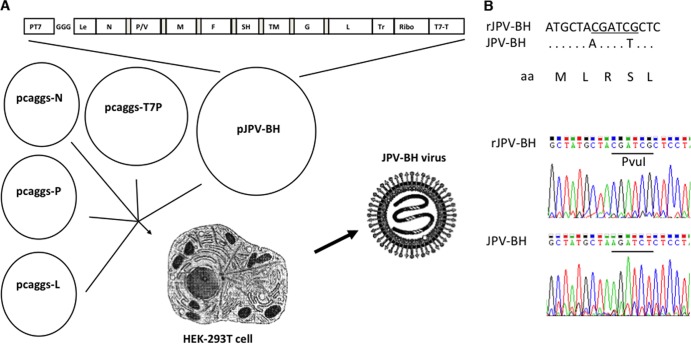
To understand the pathogenesis of JPV-BH, we generated a recombinant JPV-BH by using a reverse genetics system. The recombinant JPV-BH plasmid (JPV-BH) and three helper plasmids, JPV-BH N, P, and L, encoding the N, P, and L proteins, respectively, were cotransfected into BSR-T7 cells as described previously (5). However, the rescue efficiency was very low. HEK293T cells had a higher transfection efficiency and had been used for measles virus and vesicular stomatitis virus recovery (9, 10). We compared HEK293T cells to BSRT7 cells and found that HEK293T cells had a higher JPV infection rate than BSRT7 cells (data not shown). To use HEK293T cells in the recovery of rJPV-BH, a plasmid expressing T7 RNA polymerase was added to the mixture of plasmids (Fig. 6A). At 2 days posttransfection, HEK293T cells were mixed with Vero cells. The mixed cells were cocultured for less than 1 week for plaque purification of a single virus clone. In our previous report, BSRT7 and Vero cells were cocultured for up to 2 weeks to recover virus (5). After the rescued virus was obtained, RT-PCR was used to confirm the recovery of the recombinant virus with the correct genome sequence as described previously (5). A PvuI restriction site was introduced into the N gene of rJPV-BH without changing the amino acid residues to differentiate wild-type and recombinant JPV-BH (Fig. 6B). This novel strategy of recovery of rJPV-BH allows us to recover infectious virus without the requirement of a stable T7 RNA polymerase-expressing cell line.
Fig 6.
Generation of rJPV-BH. (A) Schematic of rescue of rJPV-BH strains. The JPV-BH plasmid, containing the full-length genome of JPV-BH, a plasmid expressing T7 polymerase, and three helper plasmids (pJPV-N, pJPV-P, and pJPV-L, encoding the N, P, and L proteins, respectively) were cotransfected into HEK293T cells. HEK293T cells were mixed with Vero cells. Plaque assays on Vero cells were used to obtain single clones of recombinant strains. (B) Introduction of a PvuI sequence into the N gene of rJPV-BH. Top, schematic diagram of the PvuI site introduced into the N gene without changing amino acid (aa) residues. Bottom, sequencing result of the PCR product. The PvuI recognition sequence, CGATCG, is underlined.
Generation of JPV-BH/LW hybrids.
To determine which sequence element in the genome was responsible for the difference in pathogenicity between JPV-BH and rJPV-LW, JPV-BH/LW hybrids were generated (Fig. 7). rJPV-BH-Le-LW containing the backbone of JPV-BH and the leader sequence of rJPV-LW, rJPV-BH-GX-LW containing the backbone of JPV-BH and the GX sequence of rJPV-LW, and rJPV-BH-L-LW containing the backbone of JPV-BH and the L sequence of rJPV-LW were recovered from HEK293T cells. Single plaques of each virus were obtained, and the genome sequences of the viruses were confirmed to match the input cDNA sequences (data not shown).
Fig 7.
Generation of rJPV-BH/LW hybrids. rJPV-BH-Le-LW, rJPV-BH-GX-LW, and rJPV-BH-L-LW containing the backbone of rJPV-BH and the leader (Le), GX, and L sequences of rJPV-LW are shown. The sequences from rJPV-LW are shaded.
Analysis of JPV-BH/LW hybrids in tissue culture cells.
To compare the growth rates of the JPV-BH/LW hybrids in tissue culture cells, single- or multiple-step growth curves were obtained with Vero cells and L929 cells. In Vero cells, JPV-BH and rJPV-BH grew similarly, as expected. rJPV-BH-Le-LW, rJPV-BH-GX-LW, and rJPV-BH-L-LW were similar to rJPV-LW and slightly faster than JPV-BH and rJPV-BH in both single- and multiple-step growth curves (Fig. 8A). In L929 cells, JPV-BH, rJPV-BH, and rJPV-BH-GX-LW grew at similar rates, which were faster than those of rJPV-LW and rJPV-BH-L-LW, as shown in single- and multiple-step growth curves. rJPV-BH-Le-LW replicated similarly to rJPV-LW and rJPV-BH-L-LW at early time points but replicated to titers similar to those of JPV-BH, rJPV-BH, and rJPV-BH-GX-LW by later time points. rJPV-BH-L-LW replicated faster than rJPV-LW in the multiple-step growth curve (Fig. 8B).
Fig 8.
Growth rates of rJPV-BH/LW hybrids in tissue culture cells. Single- and multiple-step growth curves of rJPV-BH/LW hybrids in Vero (A) and L929 (B) cells are shown. Vero or L929 cells were infected with JPV at an MOI of 5 or 0.1. Medium samples were collected at 24-h intervals. Virus titers were determined by plaque assay on Vero cells. P < 0.05 for JPV-BH or rJPV-BH versus rJPV-LW or rJPV-BH-L-LW in Vero cells at 24 hpi in high-MOI infection, P < 0.05 for JPV-BH versus rJPV-LW in Vero cells at 48 hpi in low-MOI infection, P < 0.05 for rJPV-BH versus rJPV-LW in Vero cells at 120 hpi in low-MOI infection, and P < 0.05 for JPV-BH or rJPV-BH versus rJPV-LW or rJPV-BH-L-LW in L929 cells in low- and high-MOI infections at 120 hpi. (C) Viral protein expression of rJPV-BH/LW hybrids. Vero cells were mock infected or infected with JPV at an MOI of 5. At 2 dpi, the MFI of P was determined.
To examine whether there was a difference in the viral protein expression levels of JPV-BH/LW hybrids, the MFI of the P protein was examined. The P expression levels in rJPV-LW- and rJPV-BH-L-LW-infected Vero cells were higher than those in Vero cells infected with other viruses (Fig. 8C).
The CPE on Vero cells infected with the rJPV-BH/LW hybrids was examined. rJPV-BH-GX-LW and rJPV-BH-L-LW produced larger syncytial cells than other viruses, and rJPV-BH-Le-LW produced more syncytial cells than JPV-BH and rJPV-BH (data not shown). To examine whether rJPV-BH/LW hybrids also had different CPEs on L929 cells, these cells were infected with the JPV. In our previous report (5), rJPV-LW had a CPE on and induced apoptosis in L929 cells. In that study, rJPV-LW had the most severe CPE, following by rJPV-BH-L-LW, which had the second-most-severe CPE (data not shown). The TUNEL assay results were consistent with the CPE results; rJPV-LW induced the highest level of apoptosis, followed by rJPV-BH-L-LW. JPV-BH, rJPV-BH, rJPV-BH-le-LW, and rJPV-BH-GX-LW induced similar levels of apoptosis (Fig. 9).
Fig 9.
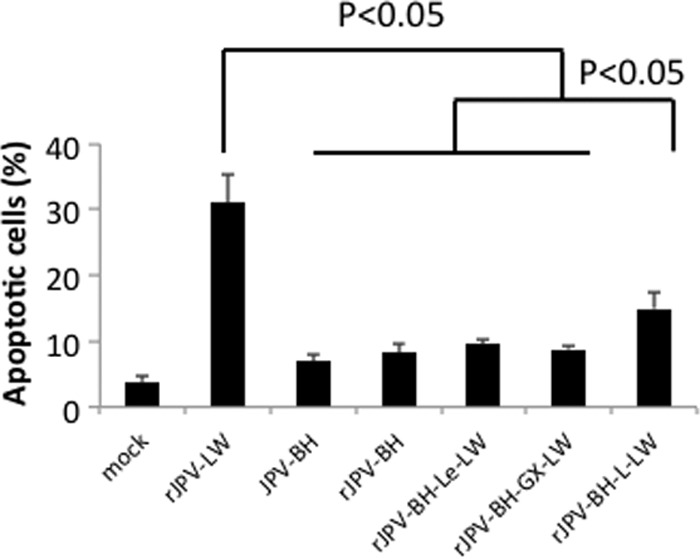
CPEs of rJPV-BH/LW hybrids on tissue culture cells. (A) CPEs of rJPV-BH/LW hybrids on Vero cells. Vero cells were mock infected or infected with JPV at an MOI of 0.1. At 3 dpi, the cells were photographed. (B) CPEs of rJPV-BH/LW hybrids. L929 cells were mock infected or infected with JPV at an MOI of 5. At 2 dpi, the cells were photographed. (C) Induction of apoptosis by rJPV-BH/LW hybrids. L929 cells were mock infected or infected with JPV at an MOI of 5. At 2 dpi, a TUNEL assay was performed.
Analysis of rJPV-BH/LW hybrids in vivo.
Mice were infected intranasally with rJPV-LW, JPV-BH, rJPV-BH, rJPV-BH-Le-LW, and rJPV-BH-L-LW (Fig. 10A to C). rJPV-LW-infected mice lost the least body weight and recovered quickly. rJPV-LW replicated poorly in mouse lungs; there was no detectable virus in the lungs at 5 dpi. rJPV-BH-L-LW-infected mice lost approximately 10% of their body weight and recovered by 7 dpi. rJPV-BH-L-LW replicated better than rJPV-LW, the virus titers in lungs were about 103 PFU/ml by 5 dpi, and there was no detectable virus in the lungs at 7 dpi. JPV-BH-, rJPV-BH-, and rJPV-BH-Le-LW-infected mice lost up to 25% of their body weight. The viral titers in their lungs stayed similar for 5 days and dropped by 7 dpi. Infection with JPV-BH, rJPV-BH, or rJPV-BH-Le-LW was lethal to mice. All infected mice died by 10 dpi. In contrast, mock-, rJPV-LW-, and rJPV-BH-L-LW-infected mice survived the study, suggesting that rJPV-BH-L-LW was attenuated in mice. This was supported by histopathology, which showed greater fibrinonecrotizing lesions and infiltrates within the alveoli of JPV-BH-, rJPV-PH-, and rJPV-BH-Le-LW-infected mice, than in those of mock-, rJPV-LW-, and rJPV-BH-L-LW-infected mice (Fig. 11).
Fig 10.
Analysis of rJPV-BH/LW hybrid growth in vivo. Mice were inoculated intranasally with PBS or 5 × 105 PFU of JPV. (A) Body weight loss. Mice were monitored daily, and weight loss was graphed as the average percentage of their original weight (on the day of infection). (B) Lung virus titers of mice. Mouse lungs were collected at 1, 3, 5, and 7 dpi. Virus titers were determined by plaque assay on Vero cells. (C) Survival rate (n = 3).
Fig 11.
Lung histopathology of JPV-infected mice. Mice were infected as described in the legend to Fig. 10. Lungs were collected from infected and mock-infected animals at 7 dpi and stained with H&E. Photomicrographs were taken at a magnification of ×40.
DISCUSSION
Many new paramyxoviruses have been identified recently (11–15). Isolating infectious viruses and studying them in suitable animal models will enable us to understand their pathogeneses. Obtaining an infectious virus that truly reflects the original virus is a major challenge. While original reports indicated that JPV was pathogenic in mice, JPV-LW was not pathogenic in mice, as expected. Initially, we doubted the original report, since it was possible that the animals used in the original experiment over 40 years ago were not pathogen free, and they might have been coinfected with other unknown pathogens. On the basis of the fact that the genome sequences of JPV-BH and JPV-LW are almost identical, it can be concluded that JPV-BH and JPV-LW are the same virus. Although JPV was originally isolated in Australia, JPV-LW was obtained from a laboratory in the United States. We speculate that JPV-LW was from a later laboratory passage of JPV and JPV-BH was from an earlier passage. Vero cells have often been used as the cells of choice for virus isolation because they lack IFN genes. rJPV-LW grew better than JPV-BH in Vero cells, suggesting that JPV-LW was likely adapted for growth in cell culture. This work reestablishes that JPV is indeed pathogenic in mice.
Besides JPV, BeiPV and TlmPV contain the same genome organization in the order 3′-N-P/V/C-M-F-SH-TM-G(GX)-L-5′. BeiPV was isolated from rat and human mesangial cell lines (rat mesangial cells [RMC] and human mesangial cells [HMC]). The detection of BeiPV in the RMC line present in the laboratory before the HMC line was introduced strongly suggested a potential rodent origin of BeiPV (16). RT-PCR confirmed that variants of BeiPV were present in 40 kidney and 9 spleen samples from 43 rats (40/105 brown rats [Rattus norvegicus] and 3/15 black rats [Rattus rattus]). Sequence analysis showed 1 to 9 nucleotide differences in the N gene and 6 to 13 nucleotide differences in the L gene, suggesting that BeiPV originated in rodents (15). The genome replication machineries of JPV and BeiPV can be interchanged (17), suggesting that JPV and BeiPV are closely related. TlmPV was isolated from Sikkim rats (Rattus andamanensis) in a molecular epidemiology study (14). Phylogenetic analysis and protein alignment showed that these three viruses were closely related, supporting the grouping of JPV, BeiPV, and Tailam virus into a new genus of Paramyxovirinae. The existence of a pathogenic JPV-BH in a laboratory with a mouse model of infection and a reverse genetics system will enable studies of this unique virus and provide new information on this novel class of paramyxoviruses.
The GX gene of JPV is unique in that ORF X in the 3′ nontranslated region (NTR) of G is in frame with ORF G. If the stop codon for G were deleted, a GX fusion protein could be made. The existence of a large ORF (>2,000 nt) in the 3′ NTR of G is puzzling. It is hard to imagine that the virus contains such a long sequence without a function. Replacement of the GX sequence of JPV-BH with the GX sequence of JPV-LW resulted in a virus (rJPV-BH-GX-LW) similar in pathogenicity to its parental virus, JPV-BH. However, rJPV-BH-GX-LW formed larger syncytia, much like rJPV-LW, than its parental virus, rJPV-BH (data not shown), suggesting that the differences within the GX sequence are important for syncytium formation. Further analysis will lead to a better understanding of the roles of G and X in virus replication.
Viral RNA-dependent RNA polymerases of paramyxoviruses consist of two major viral components, the phosphoprotein (P) and the large (L) protein. The L protein, with >2,000 amino acid residues in a single polypeptide chain, is thought to be responsible for the enzymatic activities involved in viral RNA replication and transcription. The L genes of paramyxoviruses have been implicated in viral pathogenesis. cpts530/1009 is a live, attenuated, temperature-sensitive RSV vaccine candidate. Compared to the parental cold-passaged RSV, it contains two mutation sites at residues 521 and 1169 in the L protein (18). In Newcastle disease virus (NDV), replacement of the L gene of a mesogenic strain, Beaudette C, with that of a lentogenic strain, LaSota, significantly increased the pathogenicity of the L-chimeric virus, which had increased replication levels (19). Furthermore, attenuation of measles virus and an NDV pigeon variant (pigeon paramyxovirus 1) in chickens with enhanced virus replication and virulence has also been associated with the P and L genes (20, 21). In our study, rJPV-BH-L-LW replicated well in tissue culture cells but poorly in mouse lungs compared to rJPV-BH, indicating that L plays a critical role in viral pathogenesis. Because rJPV-BH-L-LW replicated well in tissue culture cells (Fig. 8C), we speculate that the attenuation of rJPV-BH-L-LW in vivo (Fig. 10C) is due to a function of the L gene not associated with its polymerase function. The L protein has six domains that are highly conserved among all members of the order Mononegavirales (22, 23). It has been reported that domains I and II of the parainfluenza virus 5 L protein activate NF-κB through an AKT-dependent pathway and that the mRNA of domain II of L can induce IFN-β expression through an RNase L-MDA5-dependent pathway (24, 25). Interestingly, two differences between the L genes of L-LW and L-BH are in domain II. Further analysis is needed to dissect the roles of these individual residues in pathogenesis.
While replacement of the L gene of JPV-BH with that of JPV-LW resulted in attenuation (Fig. 10), this virus (rJPV-BH-L-LW) was not as nonpathogenic as rJPV-LW, suggesting that other sequence elements are likely to play roles in the attenuation of JPV-LW. Further studies, especially with rJPV-LW containing elements from JPV-BH, will elucidate the contribution of each sequence element to the pathogenesis of JPV.
ACKNOWLEDGMENTS
We appreciate the helpful discussions with and technical assistance of all of the members of the He laboratory. We thank Shannon Phan for critical readings of the manuscript. We thank Robert Tesh for providing JPV-BH.
Footnotes
Published ahead of print 25 September 2013
REFERENCES
- 1.Jun MH, Karabatsos N, Johnson RH. 1977. A new mouse paramyxovirus (J virus). Aust. J. Exp. Biol. Med. Sci. 55:645–647 [DOI] [PubMed] [Google Scholar]
- 2.Mesina JE, Campbell RS, Glazebrook JS, Copeman DB, Johnson RH. 1974. The pathology of feral rodents in North Queensland. Tropenmed. Parasitol. 25:116–127 [PubMed] [Google Scholar]
- 3.Jack PJ, Boyle DB, Eaton BT, Wang LF. 2005. The complete genome sequence of J virus reveals a unique genome structure in the family Paramyxoviridae. J. Virol. 79:10690–10700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jack PJ, Anderson DE, Bossart KN, Marsh GA, Yu M, Wang LF. 2008. Expression of novel genes encoded by the paramyxovirus J virus. J. Gen. Virol. 89:1434–1441 [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Xu J, Patel J, Fuentes S, Lin Y, Anderson D, Sakamoto K, Wang LF, He B. 2011. Function of the small hydrophobic protein of J paramyxovirus. J. Virol. 85:32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He B, Paterson RG, Ward CD, Lamb RA. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249–260 [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Yu M, Zhang H, Wang HY, Wang LF. 2005. Improved rapid amplification of cDNA ends (RACE) for mapping both the 5′ and 3′ terminal sequences of paramyxovirus genomes. J. Virol. Methods 130:154–156 [DOI] [PubMed] [Google Scholar]
- 8.Sun D, Luthra P, Xu P, Yoon H, He B. 2011. Identification of a phosphorylation site within the P protein important for mRNA transcription and growth of parainfluenza virus 5. J. Virol. 85:8376–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parks CL, Lerch RA, Walpita P, Sidhu MS, Udem SA. 1999. Enhanced measles virus cDNA rescue and gene expression after heat shock. J. Virol. 73:3560–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witko SE, Kotash CS, Nowak RM, Johnson JE, Boutilier LA, Melville KJ, Heron SG, Clarke DK, Abramovitz AS, Hendry RM, Sidhu MS, Udem SA, Parks CL. 2006. An efficient helper-virus-free method for rescue of recombinant paramyxoviruses and rhadoviruses from a cell line suitable for vaccine development. J. Virol. Methods 135:91–101 [DOI] [PubMed] [Google Scholar]
- 11.Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Rasche A, Yordanov S, Seebens A, Oppong S, Adu Sarkodie Y, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stocker A, Carneiro AJ, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EK, Kruppa T, Franke CR, Kallies R, Yandoko ER, Herrler G, Reusken C, Hassanin A, Kruger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C. 2012. Bats host major mammalian paramyxoviruses. Nat. Commun. 3:796. 10.1038/ncomms1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurth A, Kohl C, Brinkmann A, Ebinger A, Harper JA, Wang LF, Muhldorfer K, Wibbelt G. 2012. Novel paramyxoviruses in free-ranging European bats. PLoS One 7:e38688. 10.1371/journal.pone.0038688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YN, Lee C. 2013. Complete genome sequence of a novel porcine parainfluenza virus 5 isolate in Korea. Arch. Virol. 158:1765–1772 [DOI] [PubMed] [Google Scholar]
- 14.Woo PC, Lau SK, Wong BH, Wong AY, Poon RW, Yuen KY. 2011. Complete genome sequence of a novel paramyxovirus, Tailam virus, discovered in Sikkim rats. J. Virol. 85:13473–13474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo PC, Lau SK, Wong BH, Wu Y, Lam CS, Yuen KY. 2012. Novel variant of Beilong paramyxovirus in rats, China. Emerg. Infect. Dis. 18:1022–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Yu M, Zhang H, Magoffin DE, Jack PJ, Hyatt A, Wang HY, Wang LF. 2006. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology 346:219–228 [DOI] [PubMed] [Google Scholar]
- 17.Magoffin DE, Mackenzie JS, Wang LF. 2007. Genetic analysis of J-virus and Beilong virus using minireplicons. Virology 364:103–111 [DOI] [PubMed] [Google Scholar]
- 18.Juhasz K, Whitehead SS, Boulanger CA, Firestone CY, Collins PL, Murphy BR. 1999. The two amino acid substitutions in the L protein of cpts530/1009, a live-attenuated respiratory syncytial virus candidate vaccine, are independent temperature-sensitive and attenuation mutations. Vaccine 17:1416–1424 [DOI] [PubMed] [Google Scholar]
- 19.Rout SN, Samal SK. 2008. The large polymerase protein is associated with the virulence of Newcastle disease virus. J. Virol. 82:7828–7836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dortmans JC, Rottier PJ, Koch G, Peeters BP. 2011. Passaging of a Newcastle disease virus pigeon variant in chickens results in selection of viruses with mutations in the polymerase complex enhancing virus replication and virulence. J. Gen. Virol. 92:336–345 [DOI] [PubMed] [Google Scholar]
- 21.Takeda M, Kato A, Kobune F, Sakata H, Li Y, Shioda T, Sakai Y, Asakawa M, Nagai Y. 1998. Measles virus attenuation associated with transcriptional impediment and a few amino acid changes in the polymerase and accessory proteins. J. Virol. 72:8690–8696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poch O, Blumberg BM, Bougueleret L, Tordo N. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 71(Pt 5):1153–1162 [DOI] [PubMed] [Google Scholar]
- 23.Sidhu MS, Menonna JP, Cook SD, Dowling PC, Udem SA. 1993. Canine distemper virus L gene: sequence and comparison with related viruses. Virology 193:50–65 [DOI] [PubMed] [Google Scholar]
- 24.Luthra P, Sun D, Silverman RH, He B. 2011. Activation of IFN-beta expression by a viral mRNA through RNase L and MDA5. Proc. Natl. Acad. Sci. U. S. A. 108:2118–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luthra P, Sun D, Wolfgang M, He B. 2008. AKT1-dependent activation of NF-kappaB by the L protein of parainfluenza virus 5. J. Virol. 82:10887–10895 [DOI] [PMC free article] [PubMed] [Google Scholar]