Abstract
DNA viruses often target cellular proteins to modulate host cell cycles and facilitate viral genome replication. However, whether proliferation of white spot syndrome virus (WSSV) requires regulation of the host cell cycle remains unclear. In the present study, we show that two WSSV paralogs, IE1 and WSV056, can interact with Litopenaeus vannamei retinoblastoma (Rb)-like protein (lv-RBL) through the conserved LxCxE motif. Further investigation revealed that IE1 and WSV056 could also bind to Drosophila retinoblastoma family protein 1 (RBF1) in a manner similar to how they bind to lv-RBL. Using the Drosophila RBF-E2F pathway as a model system, we demonstrated that both IE1 and WSV056 could sequester RBF1 from Drosophila E2F transcription factor 1 (E2F1) and subsequently activate E2F1 to stimulate the G1/S transition. Our findings provide the first evidence that WSSV may regulate cell cycle progression by targeting the Rb-E2F pathway.
INTRODUCTION
White spot syndrome virus (WSSV), the only species of the genus Whispovirus, family Nimaviridae, is a major pathogen of shrimp. It is a large double-stranded DNA virus with a broad host range among crustaceans (1). Although WSSV was first discovered 20 years ago, the mechanism by which this virus modulates cellular pathways during infection remains to be explored.
The immediate early (IE) genes of DNA viruses encode regulatory proteins critical for the initiation of primary infection and the switch from latent to lytic infection (2). Twenty-one IE genes have been identified from WSSV so far (3–5). IE1 is the most studied WSSV IE protein. It has been found to exhibit transactivation and DNA-binding activities (6), suggesting that it has a role as a transcription factor. Based on detailed mapping of IE1 promoters, progress has been made in understanding the cellular regulation of IE1 expression. Several transcription factors have been found to affect IE transcription, including shrimp homologs of STAT (7, 8), NF-κB (9, 10), and a TATA box binding protein (11). JAK-STAT and NF-κB pathways are believed to be critical for antiviral defense (12, 13); therefore, the findings for WSSV indicate that this virus takes advantage of the host immune system to assist its own proliferation. The biological significance of IE1 during WSSV infection was demonstrated by RNA interference experiments which revealed that depletion of IE1 strongly inhibits the replication of WSSV (11). A recent report showed that the shrimp thioredoxin PmTrx can bind to IE1 and restore its DNA-binding activity under oxidizing conditions, indicating a role for IE1 in WSSV pathogenicity (14). However, since little is known about the targets (proteins or genes) of IE1, how this viral protein functions during infection remains unknown.
Interestingly, a new member of the WSSV IE proteins, WSV056 (5), is highly homologous to IE1 in its amino acid sequence, implying a potential similarity in function. Further analysis showed that both of the proteins contain a conserved LxCxE motif at the C terminus, which is characteristic of proteins that bind to proteins in the retinoblastoma (Rb) family (15). Rb proteins are central regulators of the cell cycle and are best known for controlling the G1/S transition, through dynamic interaction with E2F transcription factors (16). Because of their pivotal roles, the Rb proteins have become popular targets for both DNA and RNA viruses which need to generate an intracellular environment suitable for the proliferation and maturation of progeny virions. Adenovirus E1A, human papillomavirus E7, and polyomavirus large T antigen are three well-studied DNA oncoproteins that repress the function of Rb proteins either by impairing the interaction between Rb and E2F or by inducing Rb degradation (17–19). In addition, proteins encoded by RNA viruses, such as hepatitis C virus NS5B (20, 21) and coronavirus Nsp15 (22), were also found to downregulate pRB and to enhance cell cycle progression. Compared with the work done on vertebrate viruses, the interaction between invertebrate viruses and the Rb family proteins is less known.
In the present study, we demonstrate that the WSSV proteins IE1 and WSV056 are both linked to the Rb-E2F pathway and can affect the cell cycle by interacting with a Drosophila Rb family protein.
MATERIALS AND METHODS
Cloning of the retinoblastoma-like protein from Litopenaeus vannamei.
Total RNA was extracted from the hepatopancreas of Litopenaeus vannamei and reverse transcribed into cDNA by use of an oligo(dT)18 primer. To clone the Rb homolog from Litopenaeus vannamei, three pairs of primers (orf-1 forward plus orf-1 reverse, orf-2 forward plus orf-2 reverse, and middle forward plus middle reverse) (Table 1) were designed according to the sequences of two shrimp cDNA fragments similar to those for an Rb family protein, provided by Lingwei Ruan. Full-length cDNA of the retinoblastoma-like protein from Litopenaeus vannamei (lv-RBL) was obtained using a SMARTer RACE cDNA amplification kit (Clontech) with the 5′-race and 3′-race primers (Table 1). The amino acid sequence of lv-RBL was analyzed by NCBI BLAST software, and the potential functional domains were predicted.
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| RBL-HA forward | TGAAGCTTTACCCATACGATGTTCCAGATTACGCTATGAGTGACACTGATCTAGGCTC |
| RBL-HA reverse | ATGCTCGAGCTAAGGTTGCTCTGGTGGGA |
| WSV056 C217G-FLAG forward | ATGGGATCCATGGACTACAAGGATGACGATGACAAGG |
| WSV056 C217G-FLAG reverse | ATGGGATCCTTATTGTACCAAAAACTCAGAAATCTCATCTCCTGA |
| IE1 C217G-FLAG forward | ATGGGATCCATGGACTACAAGGATGACGATGACAA |
| IE1 C216G-FLAG reverse | ATGGGATCCTTATACAAAGAATCCAGAAATCTCATCTCCTGT |
| hRluc forward | ATGGGATCCGCTTCCAAGGTGTACGACCC |
| hRluc reverse | ATGCTCGAGCTGCTCGTTCTTCAGCACGC |
| EGFP forward | ATGAAGCTTGTGAGCAAGGGCGAGGA |
| EGFP reverse | ATGCTCGAGTTACTTGTACAGCTCGTCCATG |
| WSV056-EGFP forward | ATGGGATCCCGCCTCAGTCTTTGAAGACCC |
| WSV056-EGFP reverse | ATGGGTACCTTGTACCAAAAACTCAGAAATCTC |
| IE1-EGFP forward | ATGGGATCCCGCCTTTAATTTTGAAGACTCTACA |
| IE1-EGFP reverse | ATGGGTACCTACAAAGAATCCAGAAATCTCATC |
| WSV056 C217G-EGFP forward | ATGGGATCCCGCCTCAGTCTTTGAAGACCC |
| WSV056 C217G-EGFP reverse | ATGGGTACCTTGTACCAAAAACTCAGAAATCTC |
| IE1 C216G-EGFP forward | ATGGGATCCC GCCTTTAATTTTGAAGACTCTACA |
| IE1 C216G-EGFP reverse | ATGGGTACCTACAAAGAATCCAGAAATCTCATC |
| orf-1 forward | AATTACGAGAACATCCAGCTGA |
| orf-1 reverse | GAATCTTTCTGAGATCGTTGAA |
| orf-2 forward | GAATTGATGCGAGACCGTCA |
| orf-2 reverse | TGTGGCAGAGAGCCTATCCC |
| middle forward | ATTTGCCAAGTTAAGATTACAAA |
| middle reverse | GCTGGTTCCTATAGTGCTTCATA |
| 5′-race primer | ACAGCTGACAAGATCCTGAGGAATACAT |
| 3′-race primer | TCTTTCTCCATTGCCAGTAGTACGTCAC |
Plasmid construction.
The open reading frame (ORF) of lv-RBL was cloned into the pIEx-4 vector (Novagen), with a hemagglutinin (HA) tag fused to the N terminus. WSV056, the WSV056C217G mutant (cysteine in the LxCxE motif replaced by glycine), IE1, and the IE1C216G mutant (cysteine in the LxCxE motif replaced by glycine) were cloned into the pIEx-4 vector, with a FLAG tag fused to the N terminus or enhanced green fluorescent protein (EGFP) fused to the C terminus. The Renilla luciferase gene (hRluc) was cloned into pIEx-4 to generate the control plasmid pIE-hRluc for the dual-luciferase assay. Primers used for plasmid construction are listed in Table 1. Plasmids pIE-RBF1 (expressing HA-tagged Drosophila retinoblastoma family protein 1 [RBF1]) and pIE-E2F1 (expressing c-myc-tagged Drosophila E2F transcription factor 1 [E2F1]) were kindly provided by Nicholas Dyson (23), and the PCNA-luc reporter (a firefly luciferase reporter gene containing the Drosophila proliferating cell nuclear antigen [PCNA] gene promoter) was provided by Masamitsu Yamaguchi (24).
Cell culture and transfection.
Trichoplusia ni High Five cells (BTI-TN-5B1-4) were maintained in SFX medium (HyClone) at 27°C. Drosophila melanogaster S2 cells were grown in Sf-900 II SFM medium (Life Technology) supplied with 10% fetal bovine serum (Life Technology), 100 U/ml penicillin, and 100 μg/ml streptomycin at 27°C.
High Five cells were transfected with Cellfectin II reagent (Life Technology), and S2 cells were transfected with Effectene transfection reagent (Qiagen) according to the manufacturers' instructions.
Western blotting.
The protein samples were separated in SDS-PAGE gels, transferred to nitrocellulose membranes, and detected with appropriate antibodies. The primary antibodies (monoclonal anti-FLAG M2 antibody, monoclonal anti-c-myc antibody, monoclonal anti-HA antibody, and anti-α-tubulin antibody) were purchased from Sigma. Horseradish peroxidase (HRP)- or alkaline phosphatase (AP)-conjugated secondary antibodies were purchased from Pierce.
Coimmunoprecipitation.
High Five cells (cultured in 100-mm plates) were harvested at 48 h posttransfection (p.t.) and lysed with 1 ml lysis buffer (1% NP-40, 500 mM NaCl, 50 mM HEPES-KOH, pH 7.8, 5 mM EDTA, 3 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 50 mM NaF) for 10 min on ice. After centrifugation at 12,000 × g for 10 min at 4°C, the supernatant was incubated with 30 μl anti-FLAG M2 affinity gel (Sigma) overnight at 4°C with gentle rotation. The beads were washed sequentially with lysis buffer and washing buffer (20 mM HEPES, pH 7.8, 15% glycerol, 250 mM KCl, 0.2 mM EDTA, 0.1% NP-40, 1 mM DTT, 0.5 mM PMSF). The FLAG-tagged proteins were then eluted with FLAG peptide (1 mg/ml) in a total volume of 30 μl.
Dual-luciferase assay.
The dual-luciferase assay was performed with a dual-luciferase reporter assay system (Promega). At 48 h p.t., S2 cells were lysed with the passive lysis buffer supplied in the kit. The activities of firefly (Photinus pyralis) and Renilla (Renilla reniformis) luciferases were measured sequentially for each sample. The activity of the firefly luciferase reporter was correlated with the effect of specific experimental conditions, while the activity of the Renilla luciferase reporter was provided as an internal control. The relative luciferase activity was calculated with the following formula: relative luciferase activity = firefly luciferase activity/Renilla luciferase activity. The experiment was repeated three times, and the standard deviations were calculated.
Competitive binding assay.
Plasmids pIE-RBF1 and pIE-E2F1 were cotransfected with pIE-WSV056 or pIE-IE1 into High Five cells. The vector pIEx-4 was used to normalize the amount of DNA in each transfection mixture. At 48 h p.t., cells were lysed with lysis buffer, the HA-tagged protein was immunoprecipitated with anti-HA affinity gel (Sigma), and the amount of each component was analyzed by Western blotting.
Flow cytometry analysis.
S2 cells were harvested at 24 h p.t., washed once with phosphate-buffered saline (PBS), and fixed with 70% ethanol at −20°C for 1 h. After fixation, the cells were washed once with PBS, followed by RNase A (0.5 mg/ml in PBS) treatment for 20 min at 37°C, and then were stained with 40 μg/ml propidium iodide (PI) solution for 15 min. The cellular DNA content was determined with a BD FACSCalibur flow cytometer. Ten thousand events in the EGFP-positive gate were read for each sample. Mofit LT software (Verity Software House) was used to determine the distribution of cells in each phase of the cell cycle (G1/G0, S, and G2/M). The experiment was repeated three times, and the standard deviations were calculated.
Nucleotide sequence accession number.
The lv-RBL gene was deposited in GenBank under accession number KC181921.
RESULTS
IE1 and WSV056 interact with the shrimp Rb homolog through the LxCxE motif.
WSSV immediate early proteins IE1 and WSV056 share ∼56% identity in their amino acid sequences (Fig. 1). A search for motifs within these two proteins identified the LxCxE motif, which is characteristic for proteins that bind to the Rb family proteins (15) (Fig. 1, boxed area). To test if these two proteins do bind to host Rb family proteins, a shrimp Rb homolog was cloned from Litopenaeus vannamei (GenBank accession number KC181921). The shrimp Rb homolog comprises 1,080 amino acids (aa) and contains a pocket domain formed by domain A (aa 393 to 588) and domain B (aa 766 to 923). These A and B domains each represent a single cyclin fold, and the LxCxE Rb binding motif is presented in domain B. A domain of unknown function (DUF3452; aa 64 to 208) that is conserved in Rb family proteins of humans and Drosophila is also present in the N terminus of lv-RBL (Fig. 2A). Sequence analysis showed that this protein is more closely related to mammalian Rb-like proteins p107 and p130 than to pRB (data not shown). Therefore, we named it Litopenaeus vannamei Rb-like protein (lv-RBL). To investigate the interaction between IE1, WSV056, and lv-RBL, HA-tagged lv-RBL was coexpressed with FLAG-tagged IE1 or WSV056 in High Five cells. The FLAG-tagged proteins were immunoprecipitated at 48 h posttransfection, and the protein complex was probed for HA-tagged lv-RBL. The results showed that both IE1 and WSV056 were physically associated with lv-RBL (Fig. 2B, lanes 3 and 5). To examine whether this interaction depends on the conserved LxCxE motif, the cysteine of the LxCxE motifs of IE1 and WSV056 was replaced by glycine to generate the WSV056C217G and IE1C216G mutants. Immunoprecipitation analysis revealed that these two mutants no longer bound to lv-RBL (Fig. 2B, lanes 2 and 4), demonstrating that IE1 and WSV056 interact with lv-RBL through their LxCxE motif.
Fig 1.
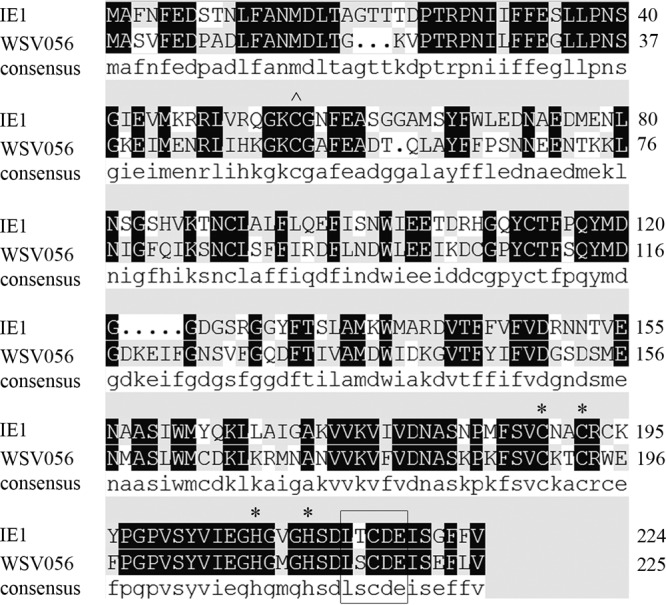
Alignment of the amino acid sequences of IE1 and WSV056. The amino acid sequences of IE1 and WSV056 were aligned using DNAMAN software (Lynnon Biosoft). The LxCxE motif is boxed. The C2H2 zinc finger motif is marked with asterisks. ∧, residue C55, which is required for DNA binding.
Fig 2.
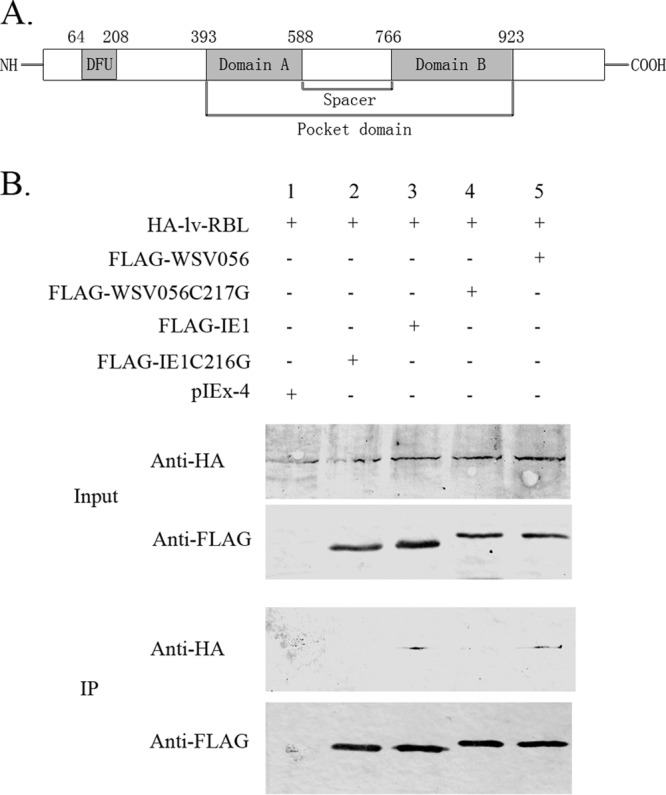
Interaction between IE1, WSV056, and lv-RBL. (A) Structure of the shrimp Rb homolog lv-RBL. The conserved domains in lv-RBL were predicted by NCBI BLAST. Domains A and B of the pocket domain and a domain of unknown function (DUF3452) conserved in Rb family proteins are shaded. (B) Coimmunoprecipitation of IE1, WSV056, and lv-RBL. HA-lv-RBL was coexpressed in High-Five cells with FLAG-IE1, FLAG-IE1C216G, FLAG-WSV056, or FLAG-WSV056C217G, separately. The pIEx-4 vector was used as a negative control. Cells were harvested, the FLAG-tagged proteins were immunoprecipitated (IP) with anti-FLAG M2 affinity gel, and then the immunoprecipitated complexes were analyzed by Western blotting with anti-HA antibody and anti-FLAG antibody, as indicated.
IE1 and WSV056 modulate the Rb-E2F pathway.
Rb binding proteins encoded by mammalian viruses are known to regulate Rb functions in different ways to support viral replication. We asked if WSSV IE1 and WSV056 could affect the host Rb-E2F pathway as well. However, it is difficult to analyze the functions of WSSV IE1 and WSV056 in the natural hosts of WSSV because of the limited knowledge of the Rb-E2F pathway in crustaceans and the lack of cell lines for in vitro analysis. Therefore, Drosophila RBF1 and E2F1 and a reporter gene containing the PCNA promoter were used instead to investigate the relationship between IE1, WSV056, and the Rb-E2F pathway.
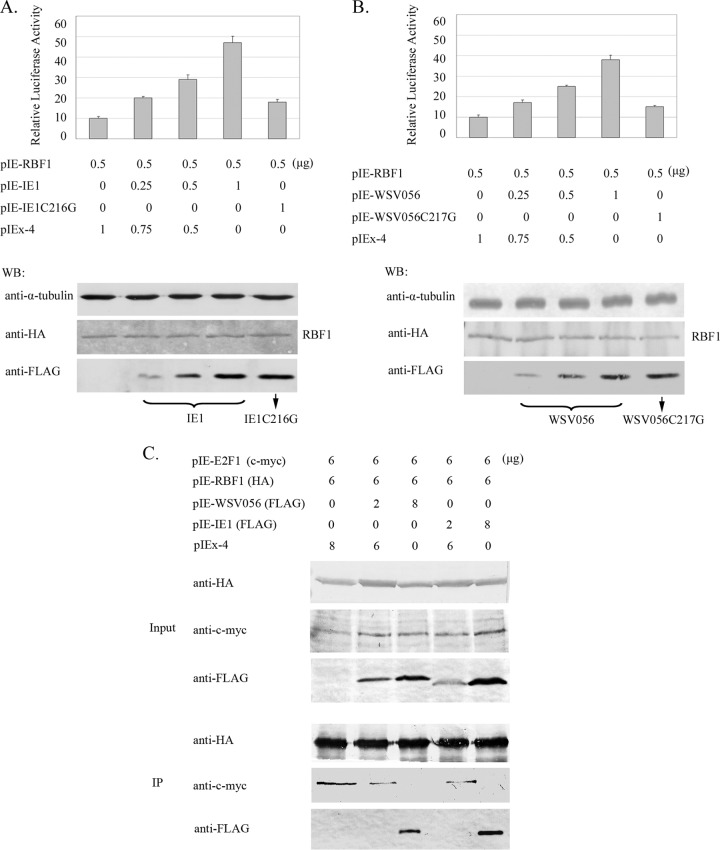
We first tested if IE1 and WSV056 could bind to RBF1. As shown in Fig. 3, RBF1 was coimmunoprecipitated with IE1 (lane 3) and WSV056 (lane 1) but not with the IE1C216G (lane 4) or WSV056C217G (lane 2) mutant, which indicated that WSSV IE1 and WSV056 could physically interact with Drosophila RBF1 in a way similar to how they bind to lv-RBL. Rb proteins are well known to regulate cell cycle progression by binding to E2F transcription factors and repressing their function. Here we showed that overexpression of IE1 and WSV056 could promote the activity of Drosophila E2F1. Different amounts of IE1 and WSV056 (0 to 1.0 μg/well of a 24-well plate) were cotransfected with RBF1 into Drosophila S2 cells, while the pIEx-4 vector was used to normalize the amount of DNA in each transfection mixture. The activity of E2F1 was measured using a firefly luciferase reporter construct containing the Drosophila PCNA promoter that can be activated by E2F1 and repressed by RBF1 (23). The relative luciferase activity was indicated by the ratio of firefly luciferase activity to control Renilla luciferase activity. As shown in Fig. 4, overexpression of both IE1 (Fig. 4A) and WSV056 (Fig. 4B) increased firefly luciferase levels in a dose-dependent manner, which means that IE1 and WSV056 stimulated E2F1 activity in the cells and strongly induced the transcription driven by the PCNA promoter. In contrast, no significant increase of the reporter activity was observed in the cells overexpressing IE1C216G (Fig. 4A) or WSV056C217G (Fig. 4B), even when the mutant constructs were transfected into the cells at the highest dose (1 μg/well of a 24-well plate). These results suggest that IE1 and WSV056 alter E2F1 activity through their interaction with RBF1.
Fig 3.
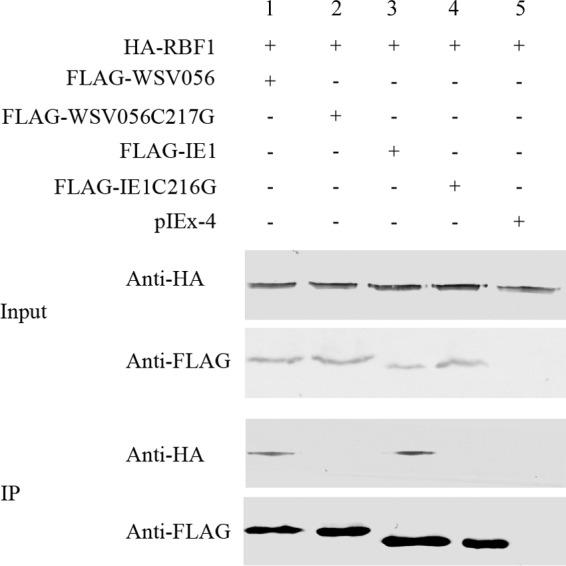
Interaction between IE1, WSV056, and Drosophila RBF1. HA-RBF1 was coexpressed in High Five cells with FLAG-IE1, FLAG-IE1C216G, FLAG-WSV056, or FLAG-WSV056C217G, separately. The pIEx-4 vector was used as a negative control. Cells were lysed, the FLAG-tagged proteins were immunoprecipitated with anti-FLAG M2 affinity gel, and then the immunoprecipitated complexes were analyzed by Western blotting with anti-HA antibody and anti-FLAG antibody.
Fig 4.
IE1 and WSV056 regulate the Rb-E2F pathway in Drosophila cells. (A and B) Dual-luciferase assays. S2 cells were cotransfected with pIE-RBF1 (500 ng/well of a 24-well plate), the reporter plasmid PCNA-luc (500 ng/well of a 24-well plate), and different amounts of pIE-IE1 (FLAG tagged) (A) or pIE-WSV056 (FLAG tagged) (B), as indicated. The mutant constructs, pIE-IE1C216G and pIE-WSV056C217G, served as controls, and the pIEx-4 vector was used to normalize the amount of DNA in each transfection mixture. The Renilla luciferase reporter plasmid pIE-hRluc (100 ng/well of a 24-well plate) was introduced into each transfection mixture to correct the transfection efficiency. At 48 h p.t., the luciferase activity in each well was measured by use of the Promega dual-luciferase reporter assay system. The relative luciferase activity was calculated with the following formula: relative luciferase activity = firefly luciferase activity/Renilla luciferase activity. The experiment was repeated three times, and the standard deviations were calculated. Western blot analysis (WB) was carried out to measure expression of RBF1, IE1 (A, lower panel), and WSV056 (B, lower panel) in each experiment. α-Tubulin was used as a loading control. (C) IE1 and WSV056 release E2F1 from the RBF1-E2F1 complex. pIE-RBF1 and pIE-E2F1 were cotransfected into High Five cells with pIE-WSV056 or pIE-IE1. The vector pIEx-4 was used to normalize the amount of DNA in each transfection mixture. At 48 h p.t., cells were harvested, the HA-tagged protein was immunoprecipitated with anti-HA affinity gel, and then the amount of each component was analyzed by Western blotting.
Some viral Rb binding proteins are known to repress Rb function by targeting it for degradation (19), but no obvious reduction of the amount of RBF1 was observed in the cells expressing IE1 or WSV056 (Fig. 3). Hence, we proposed that instead of degrading RBF1, IE1 and WSV056 might repress RBF1 by disrupting its interaction with E2F1. An in vivo competition assay was carried out to answer this question. Briefly, RBF1 and E2F1 were cotransfected with IE1 or WSV056 into High Five cells. RBF1 was immunoprecipitated, and the amount of each component incorporated into the protein complex was evaluated by Western blotting. We found that with the increase of the levels of IE1 and WSV056 in the cells, more IE1 and WSV056 but less E2F1 was coimmunoprecipitated with similar amounts of RBF1 (Fig. 4C), suggesting that E2F1 was readily displaced from the RBF1-E2F complex by the introduction of IE1 and WSV056, in a dose-dependent manner. These findings indicate that binding of IE1 and WSV056 to RBF1 may activate E2F1 by releasing it from the Rb-E2F complex.
IE1 and WSV056 modulate cell cycle progression via binding to Rb protein.
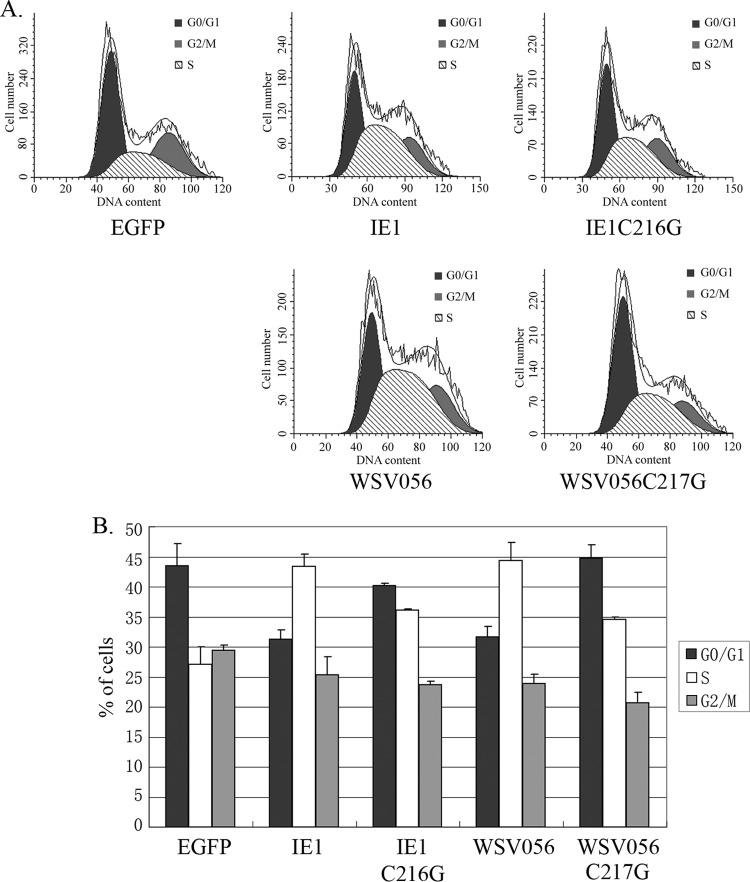
To investigate whether the interaction between the WSV056, IE1, and Rb proteins can affect the cell cycle, EGFP-IE1 and EGFP-WSV056 were overexpressed in S2 cells, and the DNA content in the cells with green fluorescence was examined by flow cytometry. As shown in Fig. 5, both IE1 and WSV056 expression caused a significant increase in the percentage of cells in S phase at 24 h p.t. Cells expressing IE1 exhibited an ∼1.5-fold increase in the percentage of S-phase cells compared to the control cells expressing EGFP alone (43% versus 27%, respectively). Correlated with the increase in S-phase cells, there was an obvious decrease in the percentage of G0/G1 cells following IE1 expression (33% versus 43%). Similar phenomena were observed in WSV056-expressing cells, which contained 31% G0/G1, 44% S-phase, and 24% G2/M cells, in comparison with the control group, which contained 43% G0/G1 cells, 27% S-phase cells, and 29% G2/M cells. Notably, mutation in the LxCxE binding motif hampered (but did not fully eliminate) the ability of IE1 and WSV056 to induce accumulation of S-phase cells, indicating that IE1 and WSV056 modulated cell cycle progression by binding to a cellular Rb protein(s).
Fig 5.
IE1 and WSV056 modulate cell cycle progression by binding to RBF1. S2 cells were harvested 24 h after transfection with EGFP, IE1-EGFP, IE1C216G-EGFP, WSV056-EGFP, or WSV056C217G-EGFP. After fixation with 70% ethanol, cells were treated with RNase A and then stained with 40 μg/ml PI solution. (A) The cellular DNA content was determined with a BD FACSCalibur flow cytometer. Ten thousand events in the EGFP-positive gate were read for each sample. Mofit LT software was used to determine the distribution of cells in each phase of the cell cycle (G1/G0, S, and G2/M). (B) Percentages (means and standard deviations) of cells in G0/G1, S, and G2/M phases in three independent experiments.
DISCUSSION
Viruses are tiny intracellular parasites that rely on the host machinery to complete their replication cycle. Many of them can interact with the host cell cycle in order to achieve high replication efficiencies and virus yields. The Rb-E2F pathway plays a central role in cell cycle progression out of G0, through G1, and into S phase (16), which makes it a popular target for viruses (25, 26). Research on mammalian DNA viruses has revealed that small DNA viruses such as adenovirus, papillomavirus, and polyomavirus encode no or very few proteins required for DNA replication. As an alternative, they code for oncoproteins that can stimulate G1/S transition of the host cells by repressing Rb family proteins and can activate the cellular DNA replication machinery (17–19, 25). In contrast, large DNA viruses belonging to the alpha- or gammaherpesvirus family encode many more enzymes required for DNA replication, nucleotide biosynthesis, and metabolism than those of the small viruses. Thus, they depend more on viral machinery for DNA replication. Instead of inducing early S-phase entry, some of the herpesviruses tend to retain the activated state of Rb proteins and arrest cells in G0, G1 phase, or at the G1/S boundary to prevent host DNA replication during viral lytic infection. Examples include (but are not limited to) herpes simplex virus, Epstein-Barr virus, and Kaposi's sarcoma-associated herpesvirus (27–29).
WSSV is a large double-stranded DNA virus with a genome of about 300 kb which encodes several proteins related to DNA replication (30, 31), including DNA polymerase (32), thymidine kinase (33), thymidylate synthase (34), ribonucleotide reductase large and small subunits (35), dUTPase (36), helicase (30), nuclease (37), and a homologous region binding protein that interacts with the potential WSSV replication origin (38). Hence, one may expect WSSV to depend less on the host's cellular apparatus for viral genome replication, like the large mammalian DNA viruses mentioned above. However, in this study, we found that two IE proteins of WSSV, IE1 and WSV056, could interact with a shrimp Rb homolog, lv-RBL, through the conserved LxCxE Rb binding motif (Fig. 2), which is similar to the case for oncoproteins encoded by small DNA tumor viruses (25). Therefore, it would be interesting to investigate how WSSV IE1 and WSV056 affect the function of host Rb family proteins.
Using the Drosophila RB-E2F pathway as a model system, we found that both IE1 and WSV056 bound to Drosophila RBF1 (Fig. 3) and enhanced the transcription of the E2F1 target promoter (Fig. 4A and B). Knowledge from mammalian viruses indicates that viral oncoproteins can repress Rb proteins by either targeting them for degradation or directly abolishing the Rb-E2F interaction (25). In our experiments, no obvious reduction of the amount of RBF1 was observed in IE1/WSV056-expressing cells (Fig. 3). Instead, E2F1 was found to be released from the RBF1-E2F1 complex by either IE1 or WSV056, in a dose-dependent manner (Fig. 4C). Thus, we deduce that IE1 and WSV056 may activate E2F1 by sequestering RbF1 from E2F1. Furthermore, by binding to RBF1, IE1 or WSV056 substantially increased the portion of S-phase cells among the S2 cells, which correlated with a significant decrease in the number of cells in G0/G1 phase (Fig. 5). Together with the fact that no obvious reduction of cell numbers was observed in cells transfected with IE1 or WSV056, we assumed that the increase of S-phase cells was due to promotion of the G1/S transition. Taking these data together, we reason that WSSV encodes some but not all proteins required for viral DNA replication. Therefore, it expresses IE1 and WSV056 to target the host Rb protein(s), which will help to create an S-phase or pseudo-S-phase environment in the host cell to support successful replication of the viral genome. At present, we have no direct evidence that binding of IE1 and WSV056 to the Rb protein(s) can lead to cell cycle modulation in the natural host of WSSV, as they do in S2 cells. However, considering the conserved role of the Rb-E2F pathway from yeasts to mammals, it is likely that IE1 and WSV056 act in a similar way during WSSV infection.
Interestingly, IE1 and WSV056 are highly homologous both in their amino acid sequences (∼56% identity) (Fig. 1) and in their nucleic acid sequences (∼68.05% identity) (data not shown), suggesting that they may be paralogs that were generated via WSSV genome duplication. Both IE1 and WSV056 are IE proteins expressed at the very beginning of viral infection (4, 5), and they behave similarly in repressing Rb function and modulating cell cycle progression. Thus, the question of why WSSV needs to express two proteins with similar functions at the same time can be raised. One possible explanation is that manipulation of the Rb-E2F pathway is essential to WSSV replication and that the virus adopts a redundancy strategy to guarantee the repression of the Rb protein(s). Moreover, despite their similarity in modulating the Rb-E2F pathway, IE1 and WSV056 may have additional (perhaps different) functions in the replication cycle of WSSV. Indeed, WSSV IE1 has already been found to exhibit DNA-binding and transactivation activities, indicating that it has a role as a transcription factor (6). Although the C55 residue and the C2H2 zinc finger motif important for DNA binding (14) are conserved between these two proteins (Fig. 1), whether WSV056 has a similar transcriptional regulation function remains to be explored.
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (973 Program; grant 2012CB114401), the National Natural Science Foundation of China (grant 40976100), and the China Agriculture Research System (grant CARS-47).
We thank Nicholas Dyson for kindly providing the constructs for Drosophila E2F1 and RBF1, Masamitsu Yamaguchi for kindly providing the PCNA-Luc reporter, Lingwei Ruan for sharing the results of shrimp transcriptome analysis, and John van der Meer and Rei Zhang for polishing the manuscript.
Footnotes
Published ahead of print 11 September 2013
REFERENCES
- 1.Leu JH, Yang F, Zhang X, Xu X, Kou GH, Lo CF. 2009. Whispovirus. Curr. Top. Microbiol. Immunol. 328:197–227 [DOI] [PubMed] [Google Scholar]
- 2.Yuan Y. 2004. Identification and characterization of herpesviral immediate-early genes, p 231–244 In Lieberman PM. (ed), DNA viruses: methods and protocols. Humana Press Inc, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 3.Li F, Li M, Ke W, Ji Y, Bian X, Yan X. 2009. Identification of the immediate-early genes of white spot syndrome virus. Virology 385:267–274 [DOI] [PubMed] [Google Scholar]
- 4.Liu WJ, Chang YS, Wang CH, Kou GH, Lo CF. 2005. Microarray and RT-PCR screening for white spot syndrome virus immediate-early genes in cycloheximide-treated shrimp. Virology 334:327–341 [DOI] [PubMed] [Google Scholar]
- 5.Lin F, Huang H, Xu L, Li F, Yang F. 2011. Identification of three immediate-early genes of white spot syndrome virus. Arch. Virol. 156:1611–1614 [DOI] [PubMed] [Google Scholar]
- 6.Liu WJ, Chang YS, Wang HC, Leu JH, Kou GH, Lo CF. 2008. Transactivation, dimerization, and DNA-binding activity of white spot syndrome virus immediate early protein IE1. J. Virol. 82:11362–11373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu WJ, Chang YS, Wang AH, Kou GH, Lo CF. 2007. White spot syndrome virus annexes a shrimp STAT to enhance expression of the immediate-early gene ie1. J. Virol. 81:1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen WY, Ho KC, Leu JH, Liu KF, Wang HC, Kou GH, Lo CF. 2008. WSSV infection activates STAT in shrimp. Dev. Comp. Immunol. 32:1142–1150 [DOI] [PubMed] [Google Scholar]
- 9.Wang PH, Gu ZH, Wan DH, Zhang MY, Weng SP, Yu XQ, He JG. 2011. The shrimp NF-kappaB pathway is activated by white spot syndrome virus (WSSV) 449 to facilitate the expression of WSSV069 (ie1), WSSV303 and WSSV371. PLoS One 6:e24773. 10.1371/journal.pone.0024773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang XD, Zhao L, Zhang HQ, Xu XP, Jia XT, Chen YH, Wang PH, Weng SP, Yu XQ, Yin ZX, He JG. 2010. Shrimp NF-kappaB binds to the immediate-early gene ie1 promoter of white spot syndrome virus and upregulates its activity. Virology 406:176–180 [DOI] [PubMed] [Google Scholar]
- 11.Liu WJ, Chang YS, Huang WT, Chen IT, Wang KC, Kou GH, Lo CF. 2011. Penaeus monodon TATA box-binding protein interacts with the white spot syndrome virus transactivator IE1 and promotes its transcriptional activity. J. Virol. 85:6535–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kemp C, Imler JL. 2009. Antiviral immunity in Drosophila. Curr. Opin. Immunol. 21:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, Liu S, Chen ZJ. 2010. SnapShot: pathways of antiviral innate immunity. Cell 140:436–436.e2. 10.1016/j.cell.2010.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang JY, Liu WJ, Wang HC, Lee DY, Leu JH, Wang HC, Tsai MH, Kang ST, Chen IT, Kou GH, Chang GD, Lo CF. 2012. Penaeus monodon thioredoxin restores the DNA binding activity of oxidized white spot syndrome virus IE1. Antioxid. Redox Signal. 17:914–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahiya A, Gavin MR, Luo RX, Dean DC. 2000. Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 20:6799–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Heuvel S, Dyson NJ. 2008. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9:713–724 [DOI] [PubMed] [Google Scholar]
- 17.Berk AJ. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673–7685 [DOI] [PubMed] [Google Scholar]
- 18.White MK, Khalili K. 2006. Interaction of retinoblastoma protein family members with large T-antigen of primate polyomaviruses. Oncogene 25:5286–5293 [DOI] [PubMed] [Google Scholar]
- 19.Wise-Draper TM, Wells SI. 2008. Papillomavirus E6 and E7 proteins and their cellular targets. Front. Biosci. 13:1003–1017 [DOI] [PubMed] [Google Scholar]
- 20.Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, Nomoto A, Lemon SM. 2007. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 3:1335–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. 2005. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 102:18159–18164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhardwaj K, Liu P, Leibowitz JL, Kao CC. 2012. The coronavirus endoribonuclease Nsp15 interacts with retinoblastoma tumor suppressor protein. J. Virol. 86:4294–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevaux O, Dimova D, Frolov MV, Taylor-Harding B, Morris E, Dyson N. 2002. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 21:4927–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi M, Hayashi Y, Matsukage A. 1995. Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J. Biol. Chem. 270:25159–25165 [DOI] [PubMed] [Google Scholar]
- 25.Felsani A, Mileo AM, Paggi MG. 2006. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene 25:5277–5285 [DOI] [PubMed] [Google Scholar]
- 26.Hume AJ, Kalejta RF. 2009. Regulation of the retinoblastoma proteins by the human herpesviruses. Cell Div. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu FY, Tang QQ, Chen H, ApRhys C, Farrell C, Chen J, Fujimuro M, Lane MD, Hayward GS. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. U. S. A. 99:10683–10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flemington EK. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 75:4475–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudoh A, Fujita M, Kiyono T, Kuzushima K, Sugaya Y, Izuta S, Nishiyama Y, Tsurumi T. 2003. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high S-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. J. Virol. 77:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F, He J, Lin X, Li Q, Pan D, Zhang X, Xu X. 2001. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 75:11811–11820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Hulten MC, Witteveldt J, Peters S, Kloosterboer N, Tarchini R, Fiers M, Sandbrink H, Lankhorst RK, Vlak JM. 2001. The white spot syndrome virus DNA genome sequence. Virology 286:7–22 [DOI] [PubMed] [Google Scholar]
- 32.Chen LL, Wang HC, Huang CJ, Peng SE, Chen YG, Lin SJ, Chen WY, Dai CF, Yu HT, Wang CH, Lo CF, Kou GH. 2002. Transcriptional analysis of the DNA polymerase gene of shrimp white spot syndrome virus. Virology 301:136–147 [DOI] [PubMed] [Google Scholar]
- 33.Tsai MF, Yu HT, Tzeng HF, Leu JH, Chou CM, Huang CJ, Wang CH, Lin JY, Kou GH, Lo CF. 2000. Identification and characterization of a shrimp white spot syndrome virus (WSSV) gene that encodes a novel chimeric polypeptide of cellular-type thymidine kinase and thymidylate kinase. Virology 277:100–110 [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Pan D, Zhang JH, Yang F. 2004. Identification of the thymidylate synthase within the genome of white spot syndrome virus. J. Gen. Virol. 85:2035–2044 [DOI] [PubMed] [Google Scholar]
- 35.van Hulten MC, Tsai MF, Schipper CA, Lo CF, Kou GH, Vlak JM. 2000. Analysis of a genomic segment of white spot syndrome virus of shrimp containing ribonucleotide reductase genes and repeat regions. J. Gen. Virol. 81:307–316 [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Yang F. 2005. Identification and function of a shrimp white spot syndrome virus (WSSV) gene that encodes a dUTPase. Virus Res. 110:21–30 [DOI] [PubMed] [Google Scholar]
- 37.Li L, Lin S, Yanga F. 2005. Functional identification of the non-specific nuclease from white spot syndrome virus. Virology 337:399–406 [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Ding Q, Yang F. 2007. Characterization of a homologous-region-binding protein from white spot syndrome virus by phage display. Virus Res. 125:145–152 [DOI] [PubMed] [Google Scholar]