Abstract
Variation of core amino acid (aa) 70 of hepatitis C virus (HCV) has been shown recently to be closely correlated with liver disease progression, suggesting that the core region might be present as a quasispecies during persistent infection and that this quasispecies nature might have an influence on the progression of disease. In our investigation, the subjects were 79 patients infected with HCV genotype 1b (25 with chronic hepatitis [CH], 29 with liver cirrhosis [LC], and 25 with hepatocellular carcinoma [HCC]). Deep sequencing of the HCV core region was carried out on their sera by using a Roche 454 GS Junior pyrosequencer. Based on a plasmid containing a cloned HCV sequence (pCV-J4L6S), the background error rate associated with pyrosequencing, including the PCR procedure, was calculated as 0.092 ± 0.005/base. Deep sequencing of the core region in the clinical samples showed a mixture of “mutant-type” Q/H and “wild-type” R at the core aa 70 position in most cases (71/79 [89.9%]), and the ratio of mutant residues to R in the mixture increased as liver disease advanced to LC and HCC. Meanwhile, phylogenetic analysis of the almost-complete core region revealed that the HCV isolates differed genetically depending on the mutation status at core aa 70. We conclude that the core aa 70 mixture ratio, determined by deep sequencing, reflected the status of liver disease, demonstrating a significant association between core aa 70 and disease progression in CH patients infected with HCV genotype 1b.
INTRODUCTION
Hepatitis C virus (HCV)-related liver disease gradually advances from chronic hepatitis (CH) to liver cirrhosis (LC) and to hepatocellular carcinoma (HCC) over 20 to 30 years (1). However, the rates of disease progression differ: some patients develop HCC over several years, while others show persistently normal alanine aminotransferase (PNALT) levels for decades, and the cause of the difference remains poorly understood.
The involvement of viral and host factors in the progression of liver disease and hepatocarcinogenesis is complex (2). With regard to viral factors, the relationship between the viral core region and disease progression in HCV genotype 1b infection has attracted clinical attention. Specifically, it has been reported that the core amino acid (aa) 70 residue, identified as a variable related to the outcome of interferon (IFN) therapy (3), is closely associated with the progression of hepatitis and hepatocarcinogenesis in Japan and North America (4–7). Those previous studies and our own have reported that core aa 70 variation is strongly linked to carcinogenesis and that substitutions in the core aa 70 region aggravate hepatitis and heighten the risk of hepatocarcinogenesis during the clinical course, corroborating the relationship between the status of the core region and disease progression (8, 9).
With regard to host factors, a genomewide association study (GWAS) has recently shown that single nucleotide polymorphisms (SNPs) around the interleukin 28B (IL28B) gene (rs12979860 and rs8099917), encoding the type III IFN IFN-λ3, are strongly correlated with the outcomes of therapy with pegylated IFN-α plus ribavirin for chronic hepatitis C (CH-C) (10–13). Interestingly, in contrast to the core region, consensus has not been reached as to the relationship between disease progression and the IL28B SNP, which is associated with IFN resistance (14–17). Previously, we reported that there was no correlation between the onset of HCC and the IL28B SNP (9). However, it was reported that the IL28B rs8099917 TG/GG allele was markedly correlated with the presence of Q (glutamine) or H (histidine) instead of the R (arginine) residue at core aa 70, while the IL28B rs8099917 TT allele was correlated with core aa 70R (8). Moreover, the core amino acid R70Q/H change occurs more often in patients with IL28B rs8099917 TG/GG than in those with IL28B rs8099917 TT (9). In this manner, the contributions of the core aa 70 residue and the IL28B SNP, which were found to be IFN sensitivity factors, to the progression of liver disease have gradually been elucidated.
HCV exists in a host as a swarm of variants, known as a “quasispecies,” and this quasispecies nature has been considered to play a critical role in pathogenesis (18). However, detailed analysis has been technically difficult, and the clinical significance has not been clarified in detail thus far. Considering the observation of core aa 70 gene changes over time, it is assumed that a variety of core aa 70 isolates may exist as a quasispecies, and this could be related to pathogenesis as described above.
Recently, deep-sequencing technology has advanced rapidly and has enabled us to analyze viral quasispecies in association with the status of the disease (19–22). In this study, we investigated how the quasispecies of the HCV core gene, either at the hot spot of the core aa 70 residue or in the almost-entire core gene, is created and is involved in disease progression in patients with CH-C.
PATIENTS AND METHODS
Patients.
The subjects were 79 patients persistently infected with HCV genotype 1b who were followed up at Yamanashi University Hospital. The patients all fulfilled the following criteria: (i) they were negative for hepatitis B surface antigen; (ii) they had no other forms of hepatitis, such as primary biliary cirrhosis, autoimmune liver disease, or alcoholic liver disease; (iii) they were free of coinfection with human immunodeficiency virus; and (iv) signed consent was obtained for the study protocol. The study protocol had been approved by the Human Ethics Review Committee of Yamanashi University Hospital and conformed to the ethical guidelines of the Declaration of Helsinki.
The breakdown was as follows: 25 patients with CH, 29 with LC, and 25 with HCC. The patients' clinical backgrounds, including histories of interferon-based antiviral therapy, are shown in Table 1. Deep-sequencing analysis was performed using serum samples taken at the most recent visit from patients with chronic hepatitis or liver cirrhosis and at the first diagnosis of HCC from patients with HCC. A direct-sequencing method, which determines the dominant viral sequence, was performed as described previously (9) to determine the dominant viral sequences of the core region, the interferon sensitivity-determining region (ISDR), and the interferon-ribavirin resistance-determining region (IRRDR) from the serum of each patient.
Table 1.
Patient characteristics classified by disease progression
| Characteristica | Value for patients with: |
P | ||
|---|---|---|---|---|
| CH (n = 25) | LC (n = 29) | HCC (n = 25) | ||
| No. male/female | 14/11 | 10/19 | 12/13 | 0.278 |
| Age (yr) (mean ± SD) | 63.4 ± 14.6 | 66.5 ± 9.2 | 68.4 ± 8.2 | 0.539 |
| Platelets (10−4/mm3) (mean ± SD) | 16.1 ± 4.7 | 9.8 ± 3.9 | 11.1 ± 5.1 | <0.001 |
| Albumin (g/dl) (mean ± SD) | 4.4 ± 0.3 | 3.9 ± 0.6 | 3.5 ± 0.5 | <0.001 |
| γ-GTP (IU/liter) (median [range]) | 53.5 (12–230) | 38.0 (12–108) | 40.8 (15–110) | 0.845 |
| T. chol. (mg/dl) (mean ± SD) | 161 ± 28 | 148 ± 30 | 140 ± 28 | 0.053 |
| HCV RNA (kIU/ml) (median [range]) | 7,047 (501–19,953) | 5,369 (126–25,119) | 8,421 (110–25,119) | 0.288 |
| Alpha-fetoprotein (ng/ml) (median [range]) | 5.0 (1.1–16.5) | 32.2 (1.0–252.6) | 614.8 (1.9–13,418) | <0.001 |
| AST (IU/liter) (mean ± SD) | 42.4 ± 17.8 | 51.1 ± 23.5 | 59.1 ± 28.2 | 0.046 |
| ALT (IU/liter) (mean ± SD) | 46.6 ± 23.1 | 43.9 ± 29.3 | 60.7 ± 52.7 | 0.263 |
| No. with R/(Q/H) at core aa 70b | 19/6 | 12/17 | 8/17 | 0.005 |
| No. with L/(M/C) at core aa 91b | 17/8 | 19/10 | 15/10 | 0.834 |
| No. of ISDR mutations (median [range])b | 0.8 (0–6) | 1.2 (0–7) | 0.9 (0–8) | 0.799 |
| No. of IRRDR mutations (median [range])b | 4.9 (1–10) | 4.7 (2–9) | 5.2 (1–12) | 0.962 |
| No. with TT/non-TT at IL28B SNP (rs8099917) | 18/7 | 17/12 | 14/11 | 0.458 |
| No. without/with a history of interferon therapy | 14/11 | 16/13 | 15/10 | 0.933 |
T. chol., total cholesterol; AST, aspartate transaminase; ALT, alanine aminotransferase.
Core aa 70, core aa 91, the interferon sensitivity-determining region (ISDR), and the interferon-ribavirin resistance-determining region (IRRDR) were dominant viral sequences determined by direct sequencing.
Deep sequencing.
Deep sequencing of the viral core region was performed for each of 79 patients. Briefly, RNA was extracted from the stored sera of these patients and was reverse transcribed to cDNA. Then two-step nested PCR was carried out with primers specific for the core region of the HCV genome (23). The primers for the second-round PCR had barcodes attached, were 10 nucleotides (nt) long, and differed for each sample, so that PCR products from each sample were identifiable (see Table S1 in the supplemental material). After the band densities of the PCR products were quantified using a Bioanalyzer (Agilent Technologies, Palo Alto, CA), the concentrations of the samples were adjusted to a common value, and pooled samples were prepared. Libraries were then subjected to emulsion PCR, the enriched DNA beads loaded onto a picotiter plate, and pyrosequencing carried out with a Roche GS Junior/454 sequencing system using titanium chemistry (Roche, Branford, CT). In order to determine the error rate of the procedure, deep sequencing was carried out under similar conditions with a plasmid containing a cloned HCV sequence (pCV-J4L6S) (24). Amplicon Variant Analyzer software, version 2.5p1 (Roche), was used for analysis.
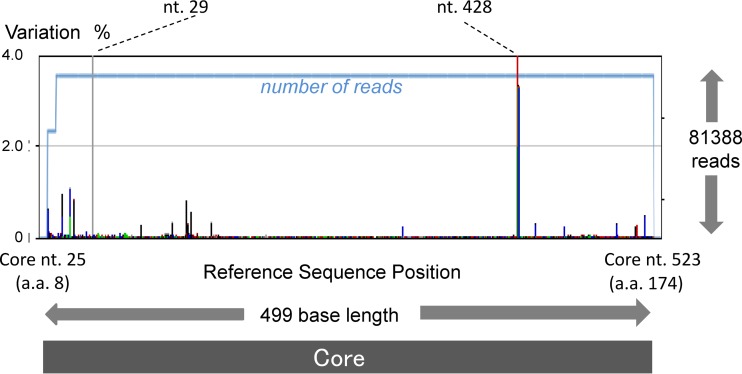
A dominant sequence of the core region for each patient was deposited in GenBank. Although the study amplified 499 nucleotides, from the 25th to the 523rd nucleotide of the core region, by PCR (Fig. 1), information for only 459 of the 499 nucleotides was uploaded for each patient, since some minor PCR amplicons obtained by deep sequencing did not include the full 499 nucleotides.
Fig 1.
The core region of pCV-J4L6S (genotype 1b) from the 25th to the 523rd nucleotide, and from the 9th to the 174th codon, was subjected to deep sequencing, and the background error rate of pyrosequencing was calculated. In order to show rare background errors, those errors with low percentages are magnified.
Phylogenetic tree analysis.
Phylogenetic trees were constructed from the sequences by using the neighbor-joining method with BioEdit and MEGA5.05, and bootstrapping was performed with 1,000 replicates (25). In constructing phylogenetic trees, the three bases of the core codon 70 were removed in the analysis of all trees, since the mutation rate of other parts of the core region is known to be rather low, and it was possible that the influence of the core aa 70 mutations might be overestimated in the phylogenetic trees. In addition, using genetic distance data obtained from the phylogenetic analysis, the genetic distances between every two HCVs with core aa 70R, between every two HCVs with a residue other than R at core aa 70 (core aa 70non-R), and between every two HCVs with different residues at core aa 70 (one HCV with R and one with a non-R residue) were also compared statistically in order to reveal the genetic associations among those HCV core subgroups.
Statistical analysis.
Statistical differences in the parameters, including all available patients' demographic, biochemical, hematological, virological, and SNP data in the three groups (CH, LC, and HCC), were determined using the Kruskal-Wallis test. The Mann-Whitney U test was used for statistical differences in numerical variables between two groups. Trends for categorical data were evaluated using the Cochran-Armitage trend test. All P values of <0.05 by the two-tailed test were considered significant in comparisons of genetic distances.
Nucleotide sequence accession numbers.
The dominant sequences of the core regions from the patient samples have been deposited in GenBank under accession numbers AB822372.1 to AB822459.1.
RESULTS
Calculation of background errors in deep sequencing.
First, the background error rate of pyrosequencing was calculated with a plasmid containing a cloned HCV sequence (pCV-J4L6S). Figure 1 shows the results of deep sequencing from the 25th to the 523rd nucleotide of the core region (499 nt) in pCV-J4L6S. Among 81,388 reads, each 499 nt long, the maximum error rate was 99.14% at the 428th base, and the next highest error rate was 64.17% at the 29th base. A six-C homopolymer region ending at the 428th base was read as a five-C homopolymer and a five-A homopolymer region ending at the 29th base was read as a four-A homopolymer in most of the obtained sequences; these homopolymer sequences are a weak point of pyrosequencing (26, 27).
A base appearing six times consecutively was the longest sequence of identical repeated nucleotides and was found only at the 428th nt position in the core region of pCV-J4L6S. Five consecutive bases were found at two sites (the 336th and 436th nt) in addition to the 29th nt position, but this error (i.e., the miscounting of homopolymer length) occurred only at the 29th base closest to the end of the sequence. Excluding the 428th and 29th nt positions, the error rate was ∼1% or lower, as shown in Fig. 1. There was no single nucleotide error in the codons for aa 70 and aa 91in the repeated control experiments. From repeated deep sequencing of the plasmid, the overall nucleotide error rate was calculated as 0.092 ± 0.005 (mean ± standard deviation [SD])/base. Based on this analysis, a mixture of bases detectable above the background error of 0.102% (mean background error rate + 2 SDs) was defined as a real mixture.
Baseline characteristics.
The baseline characteristics of the 79 patients are shown in Table 1. The values for viral factors core aa 70 and aa 91, NS5A-ISDR, and NS5A-IRRDR are the results of the direct-sequencing study. As shown in Table 1, the results for the variables platelets, albumin, alpha-fetoprotein, and core aa 70 differed significantly according to disease progression. On the other hand, no difference was observed in core aa 91 and IL28B SNP (rs8099917) according to disease progression.
Quasispecies nature of core amino acid 70 and disease progression.
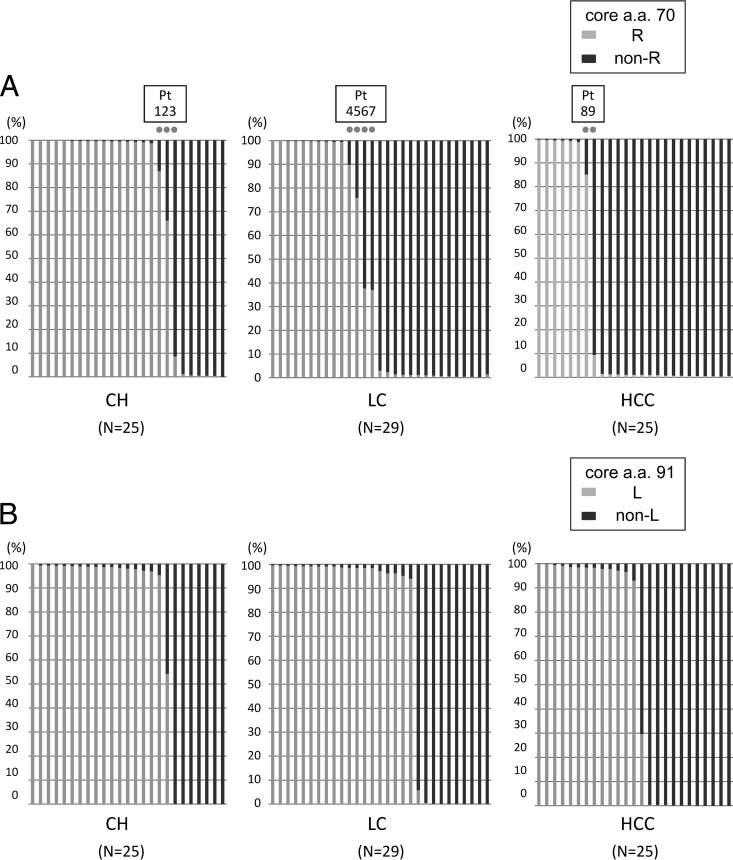
Deep sequencing of the core region was carried out with a variety of clinical samples. Simultaneous analysis was carried out using the barcoded primers, and approximately 950 reads were obtained per sample (Table 2). When the analysis was focused on core aa 70, the proportion of non-R (Q/H) sequences increased as disease severity advanced from CH to LC to HCC, as shown in Fig. 2A and Table 3 (P = 0.018). When a mixture of 0.102% or more was defined as a real mixture, deep sequencing showed the presence of a mixture at core aa 70 in 71 of the 79 patients (89.9%).
Table 2.
Amplicon read numbers obtained by deep sequencing of samples from 79 patients
| Group | No. of patients | Total no. of reads | Avg no. of reads ± SD (range)/sample |
|---|---|---|---|
| CH | 25 | 22,365 | 894.6 ± 222.8 (367–1,486) |
| LC | 29 | 28,537 | 982.5 ± 258.1 (660–1,528) |
| HCC | 25 | 24,284 | 971.4 ± 242.5 (405–1,749) |
| Total | 79 | 75,186 | 951.7 ± 240.9 (367–1,749) |
Fig 2.
The core regions of the HCV genomes from 79 patients persistently infected with HCV genotype 1b (25 patients with chronic hepatitis [CH], 29 with liver cirrhosis [LC], and 25 with hepatocellular carcinoma [HCC]) were subjected to deep sequencing. Each bar represents the result for a single patient. At the specific locations of core aa 70 and aa 91, no single nucleotide mutation was observed in the previous control plasmid experiment. (A) Disease stages and percentages of mutations at core aa 70. Nine dots indicate the nine patients with a high mixture rate (between 5% and 95%) at core aa 70 (R and non-R). (B) Disease stages and percentages of mutations at core aa 91. (C) IL28B SNP and percentages of mutations at core aa 70. (D) IL28B SNP and percentages of mutations at core aa 91.
Table 3.
Correlation between quasispecies composition and disease progression
| Patient group (n) | Median % (range) with: |
|
|---|---|---|
| R at core aa 70a | L at core aa 91b | |
| CH (25) | 70.35 (0.00–100.00) | 69.12 (0.00–99.40) |
| LC (29) | 43.22 (0.24–100.00) | 64.54 (0.00–99.50) |
| HCC (25) | 28.20 (0.00–99.80) | 52.23 (0.00–100.00) |
P, 0.018.
P, 0.630.
The relationship between disease progression and the occurrence of a quasispecies was also analyzed at the codon for core aa 91, which has also been reported to be associated with the outcomes of IFN therapy and the occurrence of HCC. As with core aa 70, a quasispecies was recognized at this site, and mixtures were observed in most patients. However, in contrast to the core aa 70 codon, there was no clear relationship with disease progression (Fig. 2B and Table 3).
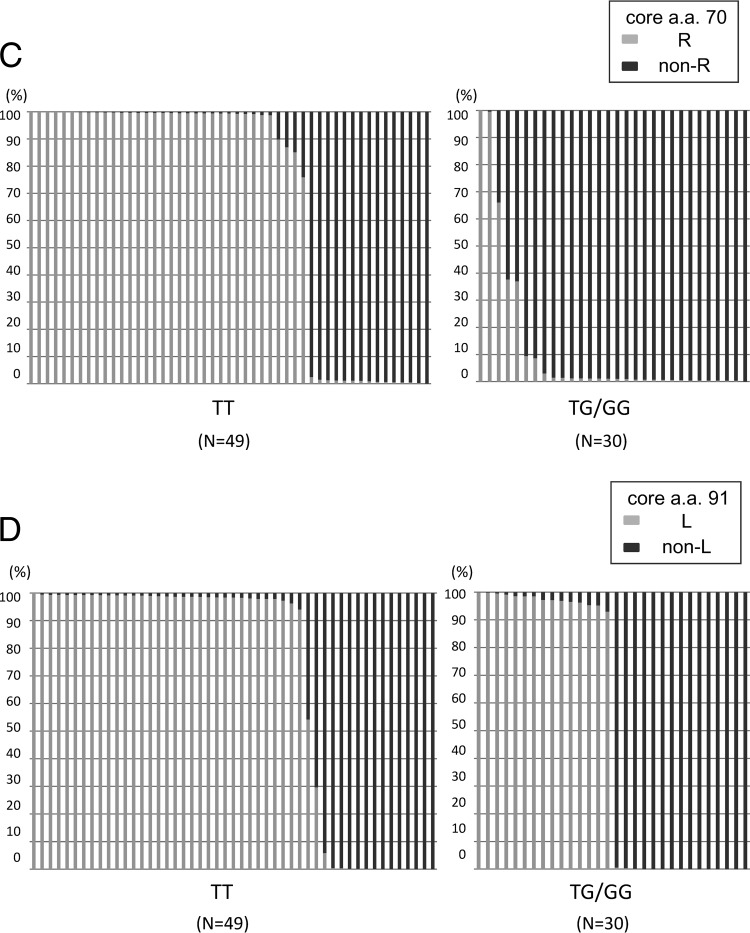
Figure 2C and D show the correlation between mixtures in the core aa 70 and 91 regions and IL28B SNPs. As shown in Fig. 2C and Table 4, the proportion of mutations in the core aa 70 codon was highly dependent on the IL28B SNP (P, <0.005 [Table 4]). Such a relationship was also found between the proportion of mutations in core aa 91 and IL28B SNPs (Fig. 2D and Table 4), although its significance was rather weaker (P, 0.010).
Table 4.
Correlation between quasispecies composition and IL28B SNP rs8099917
| Group (sequence at IL28B SNP rs8099917) | Median % (range) with: |
|
|---|---|---|
| R at core aa 70a | L at core aa 91b | |
| TT (n = 49) | 68.15 (0.00–100.00) | 68.24 (0.00–100.00) |
| TG/GG (n = 30) | 12.64 (0.00–100.00) | 48.76 (0.00–100.00) |
P, <0.005.
P, 0.010.
Since direct sequencing has also shown an association of several sites other than core aa 70 and 91 with the occurrence of HCC (6), those sites were also investigated for such an association. However, there was no clear relationship between these sites and disease progression, except for G209A (core aa R70Q) (see Fig. S1 and Table S2 in the supplemental material).
Phylogenetic tree analysis of HCV core region focusing on the core aa 70 residue.
Because it was clear that the core aa 70 quasispecies state was significantly associated with disease progression, our next interest was to determine how this single hot spot is correlated with the remainder of the (almost-entire) core region. Therefore, phylogenetic tree analysis was performed, and genetic distances among aa 70-associated core sequences were also compared statistically. In constructing all phylogenetic trees, the three bases of the core 70 codon were removed, since the mutation rate of other parts of the core region is known to be rather low, and it was possible that the influence of the core aa 70 mutations might be overestimated in the phylogenetic trees.
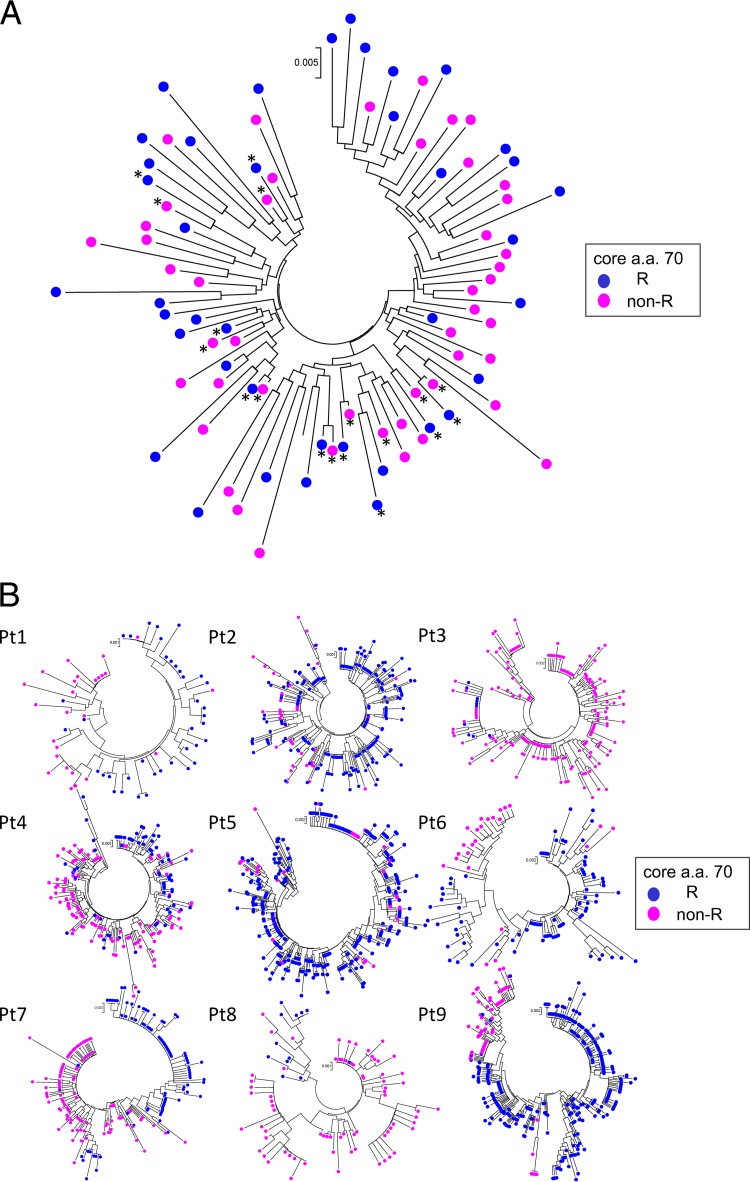
At first, to determine the associations among the remainder of the core sequences across different patients, a phylogenetic tree was constructed for all 79 patients using dominant core sequences obtained from each patient. In constructing the tree, two dominant sequences (the dominant core sequence in isolates with aa 70R and the dominant core sequence in isolates with aa 70non-R) were included in the analysis for each of the nine patients with high mixture rates (5% or more) of R and non-R at core aa 70 (Fig. 2A), while one dominant sequence each was included for other patients. As shown in Fig. 3A and Table 5, genetic distances calculated between every two core sequences with aa 70R (R-R) were significantly larger than those between two core sequences with aa 70non-R (non-R–non-R) or those between a core sequence with aa 70R and a core sequence with aa 70non-R (non-R–R), demonstrating that core sequences with aa 70R were heterogeneous, while core sequences with aa 70non-R were homogeneous.
Fig 3.
Phylogenetic trees were constructed using core sequences covering almost the entire core region. In the construction of those trees, the three bases of the core 70 codon were removed in the analysis of all 79 patients. Branches with core aa 70R are indicated by blue circles, while those with core aa 70non-R are indicated by pink circles. (A) Phylogenetic trees were constructed for all 79 patients by using dominant core sequences obtained from each patient. In the construction of the tree, two dominant sequences (a dominant core sequence in isolates with aa 70R and a dominant core sequence in isolates with aa 70non-R) were included in the analysis for each of the nine patients with high mixture rates (5% or more) of R and non-R at core aa 70 (Fig. 2A), while one dominant sequence each was included for other patients. (B) A phylogenetic tree of the core region was constructed for each patient with a high mixture rate (5% or more) of core aa 70R and core aa 70non-R. A total of nine patients were included in this analysis (patients 1 to 3 had CH; patients 4 to 7 had LC; and patients 9 and 10 had HCC). Pt, patient.
Table 5.
Comparison of genetic distances among core subgroups related to aa 70 residues
| Patient(s)a | Genetic distance (mean ± SD) between two core sequencesb |
Comparison of genetic distance measurements |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-R–non-R | Non-R–R | R-R | Non-R–R vs non-R–non-R |
Non-R–R vs R-R |
Non-R–non-R vs R-R |
||||
| Larger genetic distance | P | Larger genetic distance | P | Larger genetic distance | P | ||||
| All (n = 79) | 0.0349 ± 0.0101 | 0.0379 ± 0.0109 | 0.0401 ± 0.0113 | Non-R–R | <0.001 | R-R | <0.001 | R-R | <0.001 |
| CH | |||||||||
| Pt 1 | 0.0086 ± 0.0042 | 0.0098 ± 0.0037 | 0.0064 ± 0.0042 | Non-R–R | <0.001 | Non-R–R | <0.001 | Non-R–non-R | <0.001 |
| Pt 2 | 0.0097 ± 0.0048 | 0.0104 ± 0.0041 | 0.0087 ± 0.0038 | Non-R–R | <0.001 | Non-R–R | <0.001 | Non-R–non-R | 0.009 |
| Pt 3 | 0.0107 ± 0.0058 | 0.0137 ± 0.0050 | 0.0034 ± 0.0022 | Non-R–R | <0.001 | Non-R–R | <0.001 | Non-R–non-R | <0.001 |
| LC | |||||||||
| Pt 4 | 0.0078 ± 0.0036 | 0.0103 ± 0.0038 | 0.0053 ± 0.0029 | Non-R–R | <0.001 | Non-R–R | <0.001 | Non-R–non-R | <0.001 |
| Pt 5 | 0.0118 ± 0.0090 | 0.0232 ± 0.0085 | 0.0159 ± 0.0170 | Non-R–R | <0.001 | Non-R–R | <0.001 | No difference | 0.991 |
| Pt 6 | 0.0115 ± 0.0057 | 0.0121 ± 0.0055 | 0.0108 ± 0.0056 | Non-R–R | <0.001 | Non-R–R | <0.001 | Non-R–non-R | <0.001 |
| Pt 7 | 0.0141 ± 0.0085 | 0.0146 ± 0.0070 | 0.0136 ± 0.0067 | Non-R–R | 0.002 | Non-R–R | <0.001 | Non-R–non-R | <0.001 |
| HCC | |||||||||
| Pt 8 | 0.0124 ± 0.0063 | 0.0225 ± 0.0060 | 0.0181 ± 0.0094 | Non-R–R | <0.001 | Non-R–R | <0.001 | R-R | <0.001 |
| Pt 9 | 0.0082 ± 0.0042 | 0.0162 ± 0.0047 | 0.0078 ± 0.0050 | Non-R–R | <0.001 | Non-R–R | <0.001 | Non-R–non-R | <0.001 |
Pt, patient.
Non-R–non-R, comparison of two core sequences with residues other than R at aa 70; Non-R–R, comparison of a core sequence with a residue other than R at aa 70 and a core sequence with aa 70R; R-R, comparison of two core sequences with aa 70R. Genetic distances were calculated for all patients by using dominant sequences and for a single patient by using quasispecies sequences.
Next, to determine the association of the remainder of the core sequences in a single patient, phylogenetic trees were also constructed for each of the nine patients with high mixture rates (5% or more) of R and non-R residues at core aa 70 (Fig. 3B). As shown in Fig. 3B, HCV isolates with core aa 70R and those with core aa 70non-R formed distinctly clustered subgroups on the phylogenetic tree, according to the mutation status at core aa 70. Comparison of genetic distances also proved the finding that HCV isolates with core aa 70R and those with core aa 70non-R form distinctly clustered subgroups on the phylogenetic tree in a single patient, since genetic distances calculated between every two core sequences with aa 70R (R-R) or between every two core sequences with aa 70non-R (non-R–non-R) were significantly smaller than those between a core sequence with aa 70R and a core sequence with aa 70non-R (non-R–R). On the other hand, no significant difference was found when the genetic distance between two core sequences with aa 70non-R (non-R–non-R) and that between two core sequences with aa 70R (R-R) were compared in a single patient (Table 5).
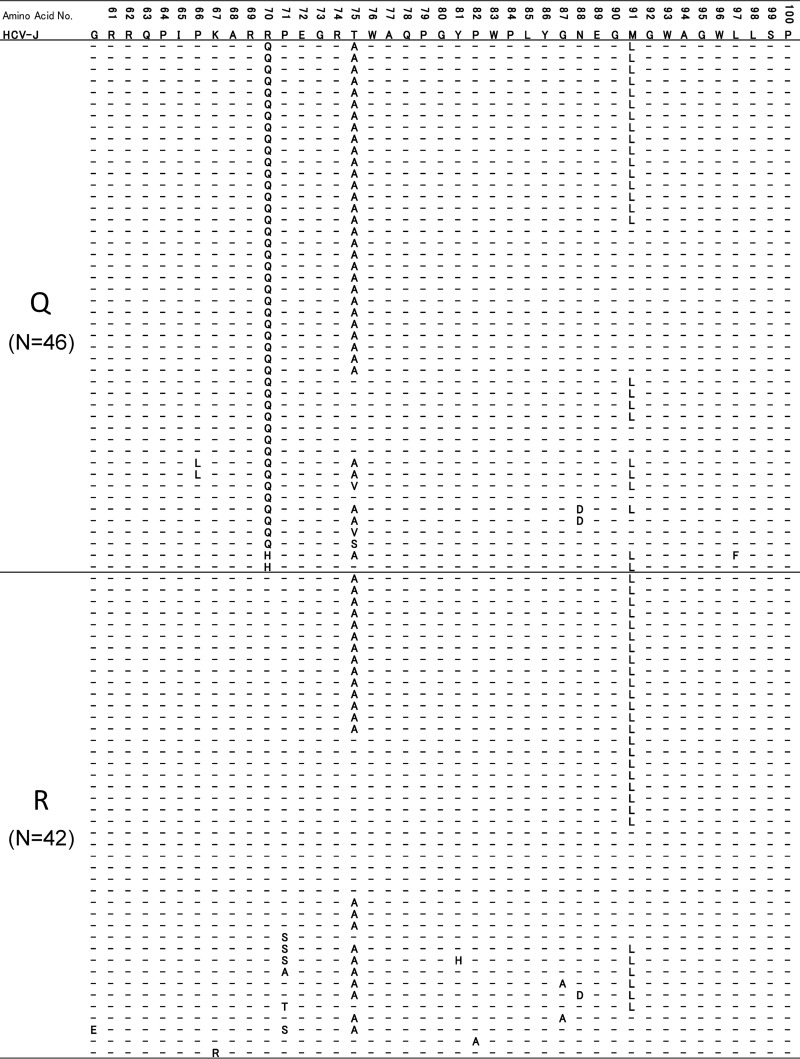
Since the genetic relationships of the remainder of the core sequences were found to differ significantly according to the core aa 70 residue, we then investigated whether there are any common haplotypic sequences specific to each residue. In the comparison of dominant sequences in all 79 patients, most amino acid substitutions clustered in three amino acids (aa 70, aa 75, and aa 91) both in core sequences with aa 70R and in those with aa 70nonR, but no other substitutions specific to each core aa 70 residue were found (Fig. 4).
Fig 4.
Dominant core amino acid haplotypic sequences dependent on the core aa 70 residues are given for all 79 patients. Two dominant sequences (a dominant core sequence in isolates with aa 70R and a dominant core sequence in isolates with aa 70non-R) were included for each of nine patients with high mixture rates (5% or more) of R and non-R at core aa 70, while one dominant sequence each was included for other patients. Therefore, a total of 88 haplotypic sequences are shown here. Since most amino acid mutations were observed in the limited core region, only that part of the core sequence is shown. The HCV-J isolate is used as the reference sequence for the core region.
Quasispecies at core aa 70 and clinical characteristics.
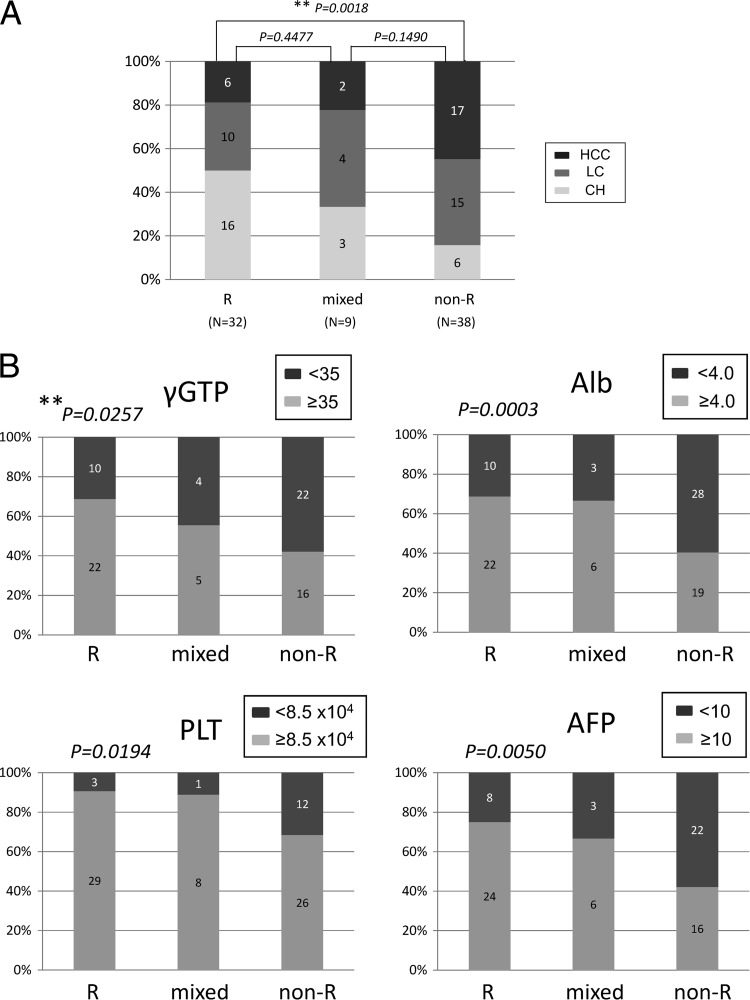
To clarify the association of the core aa 70 quasispecies with the clinical picture, levels of gamma-glutamyl transpeptidase (γ-GTP), albumin, platelets, and alpha-fetoprotein, as well as disease progression in the liver, were investigated for correlation with the core aa 70R/non-R mixture ratio. As shown in Fig. 5A and B, the values for these clinical parameters became significantly more abnormal as the proportion of non-R residues increased, showing that a high proportion of non-R residues at core aa 70 was significantly associated with disease severity and hepatocarcinogenesis.
Fig 5.
The advance of liver disease (A) and the levels of γ-GTP, albumin (Alb), platelets (PLT), and alpha-fetoprotein (AFP) (B) were investigated for correlation with the ratio of R to non-R at core aa 70. Results for R/R + non-R ratios of ≥95% (R), ≥5% and <95% (mixed), and <5% (non-R) are shown. **, Cochran-Armitage analysis.
DISCUSSION
This study examined, for the first time, the relationship between the progression of liver disease and the quasispecies nature of the HCV core region (already known to be associated with liver disease progression) by deep sequencing, with the focus on the core aa 70 residue. The analysis revealed that core aa 70 existed as a mixture of “mutant” Q/H (non-R) and “wild-type” R residues in most of the patients and that the proportion of mutant residues increased as liver disease advanced to LC and HCC. Meanwhile, phylogenetic analysis showed that the viral sequences of the almost-entire core region differed genetically depending on the status of core aa 70.
Before starting the analysis, we verified the rate of background error associated with the process of pyrosequencing by analyzing the control plasmid pCV-J4L6S (Fig. 1). Homopolymers of repeated bases, a weak point of pyrosequencing, were generated at two sites, with the same base appearing five and six times. The overall mutation rate at other sites was 0.092% ± 0.005%, and a mutation rate of 0.102% (mean + 2 SDs) or higher was defined as significant in the analysis, in order to avoid detecting background errors.
We focused our analysis on the quasispecies state of core aa 70, because the presence of a quasispecies was expected at this position, considering reports by previous studies of its association with liver disease progression and the frequent observation of time-dependent changes (8, 9). R and non-R residues were mixed in 89.9% of the 79 patients examined in this study, indicating that the absence of a mixture was rare. Furthermore, the percentage of total isolates encoding non-R residues at this position showed a relationship with the advance of chronic liver disease, as shown in Fig. 2A and Table 3. Therefore, with regard to the relationship between the advance of liver disease and core aa 70, it may be accurate to say that a change in the ratio of amino acids at core aa 70, rather than mutation of core aa 70, was related to the advance of liver disease.
Because information for almost the entire core region was obtained from each patient, our next interest was to determine whether core aa 70 is associated with other viral regions. In other words, we sought to determine whether HCVs with core aa 70R and HCVs with core aa 70non-R are phylogenetically distinct variants. To clarify the issue, phylogenetic tree analysis using dominant sequences for the (almost-entire) core regions from all 79 patients was performed at first. This analysis disclosed, after the calculation of genetic distances, that core sequences with aa 70non-R were significantly more homogeneous than those with aa 70R, demonstrating that the hot spot core aa 70 residue is significantly associated with the remainder of the core sequence. Although the underlying mechanism is unclear, we speculated that the close correlation between core aa 70 residues and IL28B SNPs might have contributed to the result. That is, since endogenous IFN levels are known to be upregulated in patients with IL28B minor types (TG/GG) relative to those in patients with the IL28B major type (TT) in its natural state (28), it is possible that HCVs with core aa 70non-R, which are closely linked to IL28B TG/GG, are under strong antiviral pressure induced by IFN, resulting in the selection of more-homogeneous HCVs, which can survive in such an environment.
Considering this possibility, we proceeded to perform phylogenetic analyses of core sequences in single patients with high-percentage mixtures (5% or more) of R and non-R residues at core aa 70 by using deep-sequencing data, since the influence of endogenous IFNs was considered equal for all HCV isolates in a single patient. The deep-sequencing data showed that the genetic heterogeneity of core sequences in a single patient did not differ according to the core aa 70 residue but that core sequences formed distinct subgroups on the phylogenetic tree according to the core aa 70 residue (Fig. 3B), and this result was also proved by the calculation of genetic distances (Table 5). However, since no common haplotypic sequences specific to each residue at core aa 70 were found across the patients (Fig. 4), we cannot determine whether core aa 70R and aa 70non-R HCVs are phylogenetically distinct variants. It is possible that the result simply reflects a major evolutional event of core aa 70 mutations followed by derivative variants; however, extension of the investigation and analysis to viral regions beyond the core region might reveal such associations. However, due to the technical limitations of second-generation sequencers, deep-sequencing analysis of the long amplicon is difficult, and new technology is needed.
With regard to the mechanism underlying the relationship between the core protein and disease progression and hepatocarcinogenesis, a study using transgenic mice showed that the core protein induces HCC (29). Fat metabolism was accelerated in the liver, leading to inflammation, iron metabolism, oxidative stress, and insulin resistance, which were considered to be the carcinogenic factors (30–32). Clinically, mutation of the core and the concentration of γ-GTP in serum, a marker of steatosis, are related, and the relationship between IL28B SNP and liver steatosis or γ-GTP has been elucidated (33). In this study, moreover, we have confirmed the correlation between the core aa 70 mixture ratio, determined by deep-sequencing analysis, and clinical parameters reflecting disease progression, illustrating the significant association of core aa 70 with disease progression (Fig. 5A and B).
In conclusion, the quasispecies state of the core region was analyzed by deep sequencing. It was found that the status of the quasispecies was closely related to the advance of HCV-associated liver disease. In order to understand the mechanism of hepatocarcinogenesis, it is desirable to elucidate pathogenesis further by detailed examination of the quasispecies of the HCV core gene.
Supplementary Material
ACKNOWLEDGMENTS
Nobuyuki Enomoto received research funding from MSD (Tokyo, Japan) and Roche (Tokyo, Japan).
This study was supported in part by grants-in-aid from the Ministry of Education, Science, Sports and Culture of Japan (grants 23390195, 23791404, 24590964, and 24590965) and in part by grants-in-aid from the Ministry of Health, Labour, and Welfare of Japan (H23-kanen-001, H23-kanen-004, H23-kanen-006, H24-kanen-002, H24-kanen-004, and H25-kanen-006).
Footnotes
Published ahead of print 14 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00826-13.
REFERENCES
- 1.Niederau C, Lange S, Heintges T, Erhardt A, Buschkamp M, Hurter D, Nawrocki M, Kruska L, Hensel F, Petry W, Haussinger D. 1998. Prognosis of chronic hepatitis C: results of a large, prospective cohort study. Hepatology 28:1687–1695 [DOI] [PubMed] [Google Scholar]
- 2.Koike K. 2005. Molecular basis of hepatitis C virus-associated hepatocarcinogenesis: lessons from animal model studies. Clin. Gastroenterol. Hepatol. 3:S132–S135 [DOI] [PubMed] [Google Scholar]
- 3.Akuta N, Suzuki F, Sezaki H, Suzuki Y, Hosaka T, Someya T, Kobayashi M, Saitoh S, Watahiki S, Sato J, Matsuda M, Arase Y, Ikeda K, Kumada H. 2005. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and non-virological response to interferon-ribavirin combination therapy. Intervirology 48:372–380 [DOI] [PubMed] [Google Scholar]
- 4.Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Kumada H. 2011. Amino acid substitutions in hepatitis C virus core region predict hepatocarcinogenesis following eradication of HCV RNA by antiviral therapy. J. Med. Virol. 83:1016–1022 [DOI] [PubMed] [Google Scholar]
- 5.Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Arase Y, Ikeda K, Kumada H. 2007. Amino acid substitutions in the hepatitis C virus core region are the important predictor of hepatocarcinogenesis. Hepatology 46:1357–1364 [DOI] [PubMed] [Google Scholar]
- 6.Fishman SL, Factor SH, Balestrieri C, Fan X, Dibisceglie AM, Desai SM, Benson G, Branch AD. 2009. Mutations in the hepatitis C virus core gene are associated with advanced liver disease and hepatocellular carcinoma. Clin. Cancer Res. 15:3205–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamoto S, Imazeki F, Fukai K, Fujiwara K, Arai M, Kanda T, Yonemitsu Y, Yokosuka O. 2010. Association between mutations in the core region of hepatitis C virus genotype 1 and hepatocellular carcinoma development. J. Hepatol. 52:72–78 [DOI] [PubMed] [Google Scholar]
- 8.Akuta N, Suzuki F, Seko Y, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Hara T, Saitoh S, Arase Y, Ikeda K, Kumada H. 2012. Complicated relationships of amino acid substitution in hepatitis C virus core region and IL28B genotype influencing hepatocarcinogenesis. Hepatology 56:2134–2141 [DOI] [PubMed] [Google Scholar]
- 9.Miura M, Maekawa S, Kadokura M, Sueki R, Komase K, Shindo H, Ohmori T, Kanayama A, Shindo K, Amemiya F, Nakayama Y, Kitamura T, Uetake T, Inoue T, Sakamoto M, Okada S, Enomoto N. 2012. Analysis of viral amino acids sequences and the IL28B SNP influencing the development of hepatocellular carcinoma in chronic hepatitis C. Hepatol. Int. 6:386–396 [DOI] [PubMed] [Google Scholar]
- 10.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399–401 [DOI] [PubMed] [Google Scholar]
- 11.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, Colombo S, Cerny A, Dufour JF, Furrer H, Gunthard HF, Heim M, Hirschel B, Malinverni R, Moradpour D, Mullhaupt B, Witteck A, Beckmann JS, Berg T, Bergmann S, Negro F, Telenti A, Bochud PY. 2010. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138:1338–1345.e7 [DOI] [PubMed] [Google Scholar]
- 12.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J. 2009. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 41:1100–1104 [DOI] [PubMed] [Google Scholar]
- 13.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. 2009. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 41:1105–1109 [DOI] [PubMed] [Google Scholar]
- 14.Bochud PY, Bibert S, Kutalik Z, Patin E, Guergnon J, Nalpas B, Goossens N, Kuske L, Mullhaupt B, Gerlach T, Heim MH, Moradpour D, Cerny A, Malinverni R, Regenass S, Dollenmaier G, Hirsch H, Martinetti G, Gorgiewski M, Bourliere M, Poynard T, Theodorou I, Abel L, Pol S, Dufour JF, Negro F. 2012. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology 55:384–394 [DOI] [PubMed] [Google Scholar]
- 15.Fabris C, Falleti E, Cussigh A, Bitetto D, Fontanini E, Bignulin S, Cmet S, Fornasiere E, Fumolo E, Fangazio S, Cerutti A, Minisini R, Pirisi M, Toniutto P. 2011. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. J. Hepatol. 54:716–722 [DOI] [PubMed] [Google Scholar]
- 16.Joshita S, Umemura T, Katsuyama Y, Ichikawa Y, Kimura T, Morita S, Kamijo A, Komatsu M, Ichijo T, Matsumoto A, Yoshizawa K, Kamijo N, Ota M, Tanaka E. 2012. Association of IL28B gene polymorphism with development of hepatocellular carcinoma in Japanese patients with chronic hepatitis C virus infection. Hum. Immunol. 73:298–300 [DOI] [PubMed] [Google Scholar]
- 17.Marabita F, Aghemo A, De Nicola S, Rumi MG, Cheroni C, Scavelli R, Crimi M, Soffredini R, Abrignani S, De Francesco R, Colombo M. 2011. Genetic variation in the interleukin-28B gene is not associated with fibrosis progression in patients with chronic hepatitis C and known date of infection. Hepatology 54:1127–1134 [DOI] [PubMed] [Google Scholar]
- 18.Pawlotsky JM. 2006. Hepatitis C virus population dynamics during infection. Curr. Top. Microbiol. Immunol. 299:261–284 [DOI] [PubMed] [Google Scholar]
- 19.Hiraga N, Imamura M, Abe H, Hayes CN, Kono T, Onishi M, Tsuge M, Takahashi S, Ochi H, Iwao E, Kamiya N, Yamada I, Tateno C, Yoshizato K, Matsui H, Kanai A, Inaba T, Tanaka S, Chayama K. 2011. Rapid emergence of telaprevir resistant hepatitis C virus strain from wildtype clone in vivo. Hepatology 54:781–788 [DOI] [PubMed] [Google Scholar]
- 20.Nasu A, Marusawa H, Ueda Y, Nishijima N, Takahashi K, Osaki Y, Yamashita Y, Inokuma T, Tamada T, Fujiwara T, Sato F, Shimizu K, Chiba T. 2011. Genetic heterogeneity of hepatitis C virus in association with antiviral therapy determined by ultra-deep sequencing. PLoS One 6:e24907. 10.1371/journal.pone.0024907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verbinnen T, Van Marck H, Vandenbroucke I, Vijgen L, Claes M, Lin TI, Simmen K, Neyts J, Fanning G, Lenz O. 2010. Tracking the evolution of multiple in vitro hepatitis C virus replicon variants under protease inhibitor selection pressure by 454 deep sequencing. J. Virol. 84:11124–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang GP, Sherrill-Mix SA, Chang KM, Quince C, Bushman FD. 2010. Hepatitis C virus transmission bottlenecks analyzed by deep sequencing. J. Virol. 84:6218–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagayama K, Kurosaki M, Enomoto N, Maekawa SY, Miyasaka Y, Tazawa J, Izumi N, Marumo F, Sato C. 1999. Time-related changes in full-length hepatitis C virus sequences and hepatitis activity. Virology 263:244–253 [DOI] [PubMed] [Google Scholar]
- 24.Gates AT, Sarisky RT, Gu B. 2004. Sequence requirements for the development of a chimeric HCV replicon system. Virus Res. 100:213–222 [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becker EA, Burns CM, Leon EJ, Rajabojan S, Friedman R, Friedrich TC, O'Connor SL, Hughes AL. 2012. Experimental analysis of sources of error in evolutionary studies based on Roche/454 pyrosequencing of viral genomes. Genome Biol. Evol. 4:457–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6:639–641 [DOI] [PubMed] [Google Scholar]
- 28.Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, Nakamura M, Shirasaki T, Horimoto K, Tanaka Y, Tokunaga K, Mizokami M, Kaneko S. 2010. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology 139:499–509 [DOI] [PubMed] [Google Scholar]
- 29.Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4:1065–1067 [DOI] [PubMed] [Google Scholar]
- 30.Leandro G, Mangia A, Hui J, Fabris P, Rubbia-Brandt L, Colloredo G, Adinolfi LE, Asselah T, Jonsson JR, Smedile A, Terrault N, Pazienza V, Giordani MT, Giostra E, Sonzogni A, Ruggiero G, Marcellin P, Powell EE, George J, Negro F. 2006. Relationship between steatosis, inflammation, and fibrosis in chronic hepatitis C: a meta-analysis of individual patient data. Gastroenterology 130:1636–1642 [DOI] [PubMed] [Google Scholar]
- 31.Nishina S, Hino K, Korenaga M, Vecchi C, Pietrangelo A, Mizukami Y, Furutani T, Sakai A, Okuda M, Hidaka I, Okita K, Sakaida I. 2008. Hepatitis C virus-induced reactive oxygen species raise hepatic iron level in mice by reducing hepcidin transcription. Gastroenterology 134:226–238 [DOI] [PubMed] [Google Scholar]
- 32.Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. 2002. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology 122:366–375 [DOI] [PubMed] [Google Scholar]
- 33.Abe H, Ochi H, Maekawa T, Hayes CN, Tsuge M, Miki D, Mitsui F, Hiraga N, Imamura M, Takahashi S, Ohishi W, Arihiro K, Kubo M, Nakamura Y, Chayama K. 2010. Common variation of IL28 affects γ-GTP levels and inflammation of the liver in chronically infected hepatitis C virus patients. J. Hepatol. 53:439–443 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.