Abstract
Prohibitins (PHBs) are ubiquitously expressed conserved proteins in eukaryotes that are associated with apoptosis, cancer formation, aging, stress responses, cell proliferation, and immune regulation. However, the function of PHBs in crustacean immunity remains largely unknown. In the present study, we identified a PHB in Procambarus clarkii red swamp crayfish, which was designated PcPHB1. PcPHB1 was widely distributed in several tissues, and its expression was significantly upregulated by white spot syndrome virus (WSSV) challenge at the mRNA level and the protein level. These observations prompted us to investigate the role of PcPHB1 in the crayfish antiviral response. Recombinant PcPHB1 (rPcPHB1) significantly reduced the amount of WSSV in crayfish and the mortality of WSSV-infected crayfish. The quantity of WSSV in PcPHB1 knockdown crayfish was increased compared with that in the controls. The effects of RNA silencing were rescued by rPcPHB1 reinjection. We further confirmed the interaction of PcPHB1 with the WSSV envelope proteins VP28, VP26, and VP24 using pulldown and far-Western overlay assays. Finally, we observed that the colloidal gold-labeled PcPHB1 was located on the outer surface of the WSSV, which suggests that PcPHB1 specifically binds to the envelope proteins of WSSV. VP28, VP26, and VP24 are structural envelope proteins and are essential for attachment and entry into crayfish cells. Therefore, PcPHB1 exerts its anti-WSSV effect by binding to VP28, VP26, and VP24, preventing viral infection. This study is the first report on the antiviral function of PHB in the innate immune system of crustaceans.
INTRODUCTION
Prohibitin-1 (PHB1), a member of the stomatin, prohibitin, flotillin, and high frequency of lysogenization C/K (SPFH) superfamily, is highly conserved, widely expressed, and present in different cellular compartments (1). The PHB domains (also called SPFH or Band-7 domains) of the SPFH superfamily all share a high degree of similarity (2). PHB was originally considered a negative regulator of cell proliferation and a tumor suppressor (3). However, this antiproliferative activity was later found to be associated with the 3′-untranslated region (UTR) of the gene, not with the actual protein (4). The 3′-UTR of the PHB mRNA has been recently reported to carry the direct binding site of the oncogene miR-27a, which is related to its antiproliferative mechanism (5). PHBs are present in divergent species, including both prokaryotes and eukaryotes (6). Two members of the PHB family, namely, PHB1 and PHB2, are expressed in eukaryotic cells and are localized in the inner mitochondrial membrane of various organisms (7). However, the subcellular locations of PHB homologues have also been reported in the nuclear and plasma membranes in different cell types (7). Although PHB1 has no signal peptide, it can be detected in the circulation (8). The PHBs present in the circulation can be internalized by cells when added to the culture (6).
Early studies on PHBs have focused on their functions in the mitochondria. PHBs are reportedly associated with mitochondrial biogenesis, cell cycle control, differentiation, senescence, apoptosis, aging, stress response, and immune regulation (2, 9, 10). Some studies have shown that PHB1 has a crucial role in signaling pathways, such as the insulin signal pathway and the signal transducers and activators of transcription signaling (STAT) pathway (9–14). PHB1 was recently reported to be a receptor protein of dengue virus 2 (DENV-2), assisting the virus in entering the cells (15). Although PHBs are involved in many biological processes, the molecular basis of their functions remains largely unknown.
PHBs are reportedly involved in plant immunity. The overexpression of a pepper hypersensitive induced reaction (HIR [a PHB domain-containing protein]) gene in Arabidopsis enhances its resistance to Pseudomonas syringae pv. tomato DC3000 (16, 17). PHBs in Arabidopsis thaliana (AtHIR) are significantly induced by microbe-associated molecular patterns and are involved in ribosomal protein S2 (RPS2)-mediated effector-triggered immunity (18). However, the functions of PHBs in animal immunity, especially in invertebrates, remain unclear. In the present study, we obtained the PHB1 cDNA from the red swamp crayfish Procambarus clarkii, designated PcPHB1, and found that its expression was significantly upregulated in crayfish after challenge with white spot syndrome virus (WSSV). Thus, we studied the functions of PcPHB1 in crayfish immunity by injecting recombinant PcPHB1 (rPcPHB1) and by conducting RNA interference (RNAi) and a rescue experiment. We also performed pulldown, coimmunoprecipitation (co-IP), far-Western overlay assays, and colloidal gold immunoelectron microscopy to analyze the interactions of PcPHB1 with the envelope proteins of WSSV. Our results showed that PcPHB1 had an important role in the crayfish antiviral response.
MATERIALS AND METHODS
Crayfish challenge and tissue collection.
Red swamp crayfish (P. clarkii) (8 g to 12 g each), were bought from a fishery market in Jinan, Shandong Province, China. The crayfish were kept for 3 days in an aerated water tank at about 24°C. Afterward, each healthy crayfish was injected in the abdomen with 50 μl of WSSV (1 × 105 virions) obtained from infected shrimp tissues. The WSSV inoculum was prepared as previously reported (19). The control groups were injected with phosphate-buffered saline (PBS) (140 mM NaCl, 10 mM sodium phosphate [pH 7.4]) or homogenates of the corresponding normal tissues. The samples of hemolymph, hemocytes, heart, hepatopancreas, gills, stomach, and intestines were collected by a previously described method (20).
cDNA cloning and sequence analysis.
Expressed sequence tags (ESTs) were obtained through random sequencing of a cDNA library constructed using different tissues from WSSV-challenged crayfish. After a BLAST analysis using the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi/), we found an EST sequence highly similar to the complete PHB sequence of the Pacific white shrimp, Litopenaeus vannamei (around 90% amino acid sequence identity). SMART was used to analyze the domain architecture of PcPHB1 (http://smart.embl-heidelberg.de/).
Tissue distribution and expression profiles of PcPHB1 under semiquantitative RT-PCR and qRT-PCR.
The tissue distribution of PcPHB1 was analyzed using semiquantitative reverse transcription-PCR (RT-PCR) with the PcPHB1-specific primers F4B (5′-TGT CCT TCC TTC CA TCA CAA AT-3′) and R4 (5′-GCC ACC TGC TTC AAC TCA AC-3′). Six tissues, namely, the hemocytes, heart, hepatopancreas, gills, stomach, and intestines, were used in this assay. The 18S RNA was used as the reference gene and was amplified with the primers 18SF2 (5′-TCT TCT TAG AGG GAT TAG CGG-3′) and 18SR2 (5′-AAG GGG ATT GAA CGG GTT A-3′). The amplification procedure consisted of an initial step at 94°C for 3 min, followed by 21 or 28 cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 20 s, and 72°C for 10 min.
Quantitative real-time PCR (qRT-PCR) was used to detect the PcPHB1 expression patterns after WSSV infection following previously described methods (21). Two pairs of primers (F4B and R4 for PcPHB1 and 18SF2 and 18SR2 for 18S RNA) were used in this assay. The qRT-PCR was programmed at 95°C for 5 min, followed by 40 cycles at 95°C for 10 s, 60°C for 30 s, and 72°C for 20 s. The plate was read at 78°C for each cycle. The final product was analyzed via a DNA melting analysis from 65°C to 95°C. The expression profiles of PcPHB1 in the hemocytes and gills of the WSSV-challenged crayfish were detected. A mock crayfish challenge was performed with PBS as the control. All experiments were repeated at least three times using individual templates. The data obtained were statistically analyzed and calculated using the threshold cycle (2−ΔΔCT) method as previously described (22). Differences in the unpaired sample t test were considered significant at a P value of <0.05.
Recombinant expression, purification, and antiserum preparation of PcPHB1.
His-tagged PcPHB1 was recombinantly expressed in Escherichia coli. PcPHB1 was amplified from hemocytes using the ExEcoRF and ExXhoR primers with EcoRI and XhoI restriction sites (ExEcoRF, 5′-TAC TCA GAA TTC ATG GGG CAG ATT GGC TTC GGT-3′; ExXhoR, 5′-TAC TCA CTC GAG TCA CTG AGG AAG AGA TAA G-3′). The PCR procedure was as follows: 1 cycle at 95°C for 3 min, 35 cycles at 94°C for 30 s, 55°C for 45 s, and 72°C for 45 s, and 1 cycle at 72°C for 10 min. The PCR products from the ExEcoRF and ExXhoR primers were then cloned into the pET30a vector (Novagen). The pET30a-PcPHB1 expression vector was transformed into competent E. coli BL21(DE3) cells. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM to induce protein expression at 28°C for 10 h. The His-tagged recombinant PcPHB1 protein was purified according to a previously described method (23). The rabbit antiserum preparation of PcPHB1 with His-PHB1 was performed as previously described (24).
Tissue distribution and expression profiles of PcPHB1 via Western blot analysis.
At least three samples of hemocytes, heart, hepatopancreas, gills, stomach, and intestines of healthy crayfish were homogenized, respectively, in buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 3 mM EDTA, 1 μl/ml cocktail of protein inhibitors) and then centrifuged at 12,000 × g for 15 min at 4°C to collect the supernatant. The protein concentration was assayed following the Bradford method (25). Each sample (50 μg protein) was analyzed using 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli (26). The proteins were then transferred onto a nitrocellulose membrane. The membrane was blocked with 2% nonfat milk in Tris-buffered saline (TBS) (10 mM Tris-HCl [pH 7.5], 150 mM NaCl) for an hour, incubated overnight with antiserum against PcPHB1 diluted 300×, and then visualized via the colorimetric reaction catalyzed using peroxidase-conjugated goat anti-rabbit IgG (1/10,000 diluted in TBS) (ZSGB-Bio, Beijing, China). The signals were detected using 4-chloro-1-naphthol (4-CIN) and H2O2.
The hemocytes and gills of the WSSV-challenged crayfish were collected at 6, 12, 24, 48, and 72 h postinjection. The time course expression of the PcPHB1 protein in crayfish was analyzed via Western blot analysis, as described above.
WSSV inhibition assay.
To analyze the function of PcPHB1, we performed a WSSV inhibition assay with four groups of crayfish (20 individuals for each group). In group 1, the crayfish were first injected with 50 μg of recombinant PcPHB1 (rPcPHB1) and then injected with WSSV (1 × 105 virions) within 1 h after the first injection. In group 2, the same amount of empty vector pET30a-expressed protein PET30A (empty pET30a vector carrying an N-terminal His tag-thrombin-S tag-enterokinase configuration and a C-terminal His tag) and WSSV was injected into the crayfish, similar to group 1. In group 3, equal amounts of bovine serum albumin (BSA) and WSSV were injected into the crayfish, as described above. In the fourth group, the crayfish were only injected with WSSV. At 60 h after the WSSV challenge, genomic DNA was extracted from the gills of the four groups of crayfish, using a genomic DNA extraction kit (Toyobo, Japan). Proteins were also extracted from the gills. qRT-PCR was used to quantify the amount of WSSV in the crayfish with the VP28 primers VP28F/R (VP28F, 5′-CTC CGC AAT GGA AAG TCT GA-3′; VP28R, 5′-GGG TGA AGG AGG AGG TGT T-3′). Western blot analysis was similarly performed using rabbit VP28 polyclonal antiserum prepared in our laboratory.
Crayfish mortality test.
The crayfish were divided into six groups (35 crayfish per group, 8 to 12 g per crayfish) to determine whether PcPHB1 has anti-WSSV activity in vivo. All crayfish were kept in tanks at approximately 24°C. The crayfish in the experimental group were injected with 50 μl of rPcPHB1 (1 μg/μl) and then injected with WSSV (1 × 106 virions) 1 h after the first injection. Recombinant CDK10 (cyclin-dependent kinase 10 from Helicoverpa armigera), expressed in E. coli with the pET-30a(+) vector in our laboratory, and BSA were used as control proteins. The crayfish in control groups 1 and 2 were injected with 50 μl of CDK10 (1 μg/μl) or 50 μl of BSA (1 μg/μl) and the same amount of WSSV, respectively, similar to the experimental group. We also used three other controls: untreated crayfish, crayfish injected with same amount of WSSV, and crayfish injected with rPcPHB1 or PBS (50 μl). The number of dead crayfish was monitored every day, and the cumulative survival rates of the six groups of crayfish were calculated. The experiments were repeated three times.
RNAi assay. (i) dsRNA preparation.
Gene-specific primers for PcPHB1 and green fluorescent protein (GFP) were incorporated with the T7 promoter at their 5′ ends (RNAi-PHB-F, 5′-GCG TAA TAC GAC TCA CTA TAG GCT TCC TTC CAT CAC AAA T-3′; RNAi-PHB-R, 5′-GCG TAA TAC GAC TCA CTA TAG GCC CTG GTT GCC TGG TAG A-3′; RNAi-GFP-F, 5′-GCG TAA TAC GAC TCA CTAT AGG TGG TCC CAA TTC TCG TGG AAC-3′; RNAi-GFP-R, 5′-GCG TAA TAC GAC TCA CTA TAG GCT TGA AGT TGA CCT TGA TGC C-3′). These primer sets were used to amplify the corresponding PCR products as the templates for double-stranded RNA (dsRNA) synthesis. The dsRNA was synthesized using T7 polymerase (Fermentas) based on the method by Wang et al. (27).
(ii) RNAi in crayfish.
Crayfish (9 g to 11 g fresh weight) were randomly divided into three groups. All crayfish in this assay were kept in tanks at about 24°C. The first group was injected with 100 μg of dsPcPHB1 RNA and then injected with another 100 μg of dsPcPHB1 RNA after 24 h. The second group was injected with 100 μg of dsGFP RNA and then injected with another 100 μg of dsGFP RNA after 24 h. The untreated third group was used as another control. Total RNA and proteins were extracted from hemocytes and gills at 2 days and 5 days after the first dsRNA injection. RT-PCR and Western blotting (anti-PcPHB1 antibodies) were used to detect the efficiency of the RNAi. The primers RNAi-PHB-RT-Fi and RNAi-PHB-RT-Ri were used in the RT-PCR (RNAi-PHB-RT-Fi, 5′-CCG AGT GCT GTT CCA TCC-3′; RNAi-PHB-RT-Ri, 5′-GCT GCC TTT TTC TCT TGC-3′). The experiments were repeated three times.
(iii) Rescue assay with PcPHB1 after RNAi.
After setting up the RNAi assay, the function of PcPHB1 in anti-WSSV was analyzed. The crayfish were divided into six groups (10 crayfish per group) to determine the amount of WSSV after PcPHB1 knockdown. The first group was left untreated and used as the control. The second group was injected with dsGFP RNA, as mentioned previously. The third, fourth, fifth, and sixth groups were injected with dsPcPHB1 RNA, as previously mentioned. After PcPHB1 knockdown (48 h after the first injection), the fourth group was injected with 50 μg of rPcPHB1 to rescue the PcPHB1 function, the fifth group was injected with 50 μg of rPET30A, and the sixth group was injected with 50 μg of BSA. Finally, all six groups were injected with WSSV (1 × 105 virions) 1 h after groups 3 to 6 were injected with protein. The genomic DNA and proteins were extracted from the gills of the crayfish 60 h after WSSV injection. qRT-PCR and Western blot analysis were used to detect WSSV at the DNA and protein levels.
Colloidal gold labeling for PcPHB1 and TEM.
PcPHB1 was labeled with 10-nm-diameter gold nanoparticles (Sigma) following a previously reported method (28). Briefly, the pH of the colloidal gold was adjusted to at least 0.5 higher than the pI of PcPHB1 by using 0.1 N HCl. Then, the saturation isotherm was used to determine the protein/gold ratio for the protein and colloidal gold. (The minimal amount of protein necessary to stabilize the gold was determined by adding 1 ml of the colloidal gold to 0.1 ml of serial aqueous dilutions of the protein.) Approximately 0.1 ml of colloidal gold was added to 0.1 mg of PcPHB1 dissolved in 0.2 ml of water for 10 nM of gold. The solution was left to stand for 10 min, and then 1% polyethylene glycol (PEG) was added to a final concentration of 0.04%. The solution was left to stand for 30 min and centrifuged for 45 min at 50,000 × g. The supernatant was then removed, and the soft pellet was resuspended in 1.5 ml of PBS containing 0.04% PEG and stored at 4°C. The colloidal gold-labeled PcPHB1 was diluted 1:10 in PBS containing 0.02% PEG. The PET30A expressed in E. coli with empty pET30a vector was used as the control protein.
WSSV was isolated by a previously reported method (29). The purified WSSV virion suspension was adsorbed onto Formvar-supported and carbon-coated nickel grids, which were incubated for 5 min at room temperature, and then the virus-coated grids were washed three times in TBS (20 mM Tris, 150 mM NaCl [pH 7.2]). Afterward, the grids were incubated on a drop of TBS-diluted PcPHB1-gold solution. The grids were finally washed with TBS, rinsed three times with distilled water, counterstained with 2% sodium phosphotungstate at pH 7.0, and then observed by TEM (JEM-100CXII). The specificity of the labeling was demonstrated using controls, including colloidal gold-labeled PET30A and incubation with colloidal gold alone.
Far-Western overlay assay.
A far-Western overlay assay was performed to determine the interaction of PcPHB1 with WSSV proteins. A 20-μl aliquot of purified WSSV (about 15 μg) and 20 μg of BSA were subjected to SDS-PAGE. The proteins in the SDS-PAGE gel were transferred onto a nitrocellulose membrane. The membrane was then blocked with 2% BSA (dissolved in TBS buffer) for 2 h at room temperature. After three washes with TBS, the membrane was incubated with purified His-PcPHB1 (0.1 mg/ml) at 4°C for 24 h. After being washed three times with TBS (10 ml, with 10 min for each wash), the membrane was incubated with 1:300-diluted antiserum to PcPHB1 for 4 h. The membrane was washed again three times with 10 ml of TBS (each for 10 min). The protein complex on the membrane was visualized via a colorimetric reaction using peroxidase-conjugated goat anti-rabbit IgG (1/10,000 diluted in TBS). The signals were detected using 4-CIN and H2O2.
Pulldown assays and co-IP analysis.
Pulldown assays for His-PcPHB1 with overexpressed VP28 in crayfish were also performed. The vector pIevp28, constructed in our laboratory (30), was first injected into crayfish for in vivo VP28 overexpression. The His-PcPHB1 pulldown assay of crayfish overexpressing VP28 was performed as follows. Purified His-PcPHB1 (200 μg) was incubated for 10 min with His · Bind resin (Amersham Biosciences, England) and then washed with 15 ml of binding buffer, 10 ml of wash buffer, and 5 ml of PBS. About 500 μl (4 μg/μl) of hemocyte homogenate from the crayfish injected with the pIevp28 vector was then added to the resin and incubated at 4°C for 2 h. After being washed thoroughly with 20 ml of PBS, the proteins were eluted with elution buffer. The elution solution was analyzed via Western blot analysis using antiserum against His-VP28, which was also prepared in our laboratory (31). The His-VP28 antiserum recognizes the His tag; therefore, it would recognize the His-PcPHB1 recombinant protein.
To analyze further the in vivo interaction of PcPHB1 with VP28, co-IP assays were performed using either rPcPHB1 antiserum or rVP28 antiserum. Protein A resin (20 μl; GenScript) was added to two 1.5-ml Eppendorf tubes and washed twice with 1 ml of PBS. Afterward, 20 μl of rPcPHB1 antiserum or rVP28 antiserum was added to each tube. The tubes were shaken for 30 min at 4°C. After being washed thoroughly with 1 ml of PBS, 800 μl of hemocyte homogenate from the crayfish injected with pIevp28 (30) was added to each tube and shaken at 4°C for 2 h. After being washed thoroughly three times with 1 ml of PBS by centrifugation (3,000 × g, 1 min), 20 μl of water and 10 μl of SDS-PAGE loading mix were added to each tube, and the mixture was boiled for 5 min in a water bath. About 10 μl of the samples from each tube was analyzed via Western blot assay using rVP28 antiserum and rPcPHB1 antiserum. A tube with antiserum-free protein A resin was used as the control.
Pulldown assays were performed to confirm the interaction of PcPHB1 with VP28, VP26, and VP24. The VP24 and VP26 of WSSV and PcPHB1 were recombinantly expressed in E. coli BL21 using the pGEX4T-1 vector. The primers ExVP24F and ExVP24R were used to amplify VP24 (ExVP24F, 5′-TAC TCA GAA TTC AAC ATA GAA CTT AAC AAG AAA T-3′; ExVP24R, 5′-TAC TCA CTC GAG GCC AGG AGA AAA ACG CAT-3′), whereas ExVP26F and ExVP26R were used to amplify VP26 (ExVP26F, 5′-TAC TCA GAA TTC ACA CGT GTT GGA AGA AGC GT-3′; ExVP26R, 5′-TAC TCA CTC GAG CTT CTT CTT GAT TTC GTC CTT G-3′). Glutathione S-transferase (GST)-VP28 was previously expressed in our laboratory (31). The PHB domain and the C terminal of PcPHB1 (PHB-CT) were also recombinantly expressed in E. coli BL21(DE3), using the pET-32A vector. The primers ExPHB-domain-F and ExPHB-domain-R were used to amplify the PHB domain of PcPHB1 (ExPHB-domain-F, 5′-GCG GAA TTC CTA TAC AAT GTT GAT GCA-3′; ExPHB-domain-R, 5′-G GCG CTC GAG TTA CTG CTT CAA CTC AAC AGC-3′). ExPHB-CT-F and Ex PHB-CT-R were used to amplify the C terminal of PcPHB1 (ExPHB-CT-F, 5′-GCG GAA TTC GTG GCT CAG CAG GAG GCA-3′; Ex PHB-CT-R, 5′-GCG CTC GAG TTA TCA CTG AGG AAG AGA TAA-3′). The VP19 gene of WSSV was amplified with the primer pair VP19-F and VP19R (VP19-F, 5′-CGC GGA TCC ATG GCC ACC ACG ACT AAC AC-3′; VP19R, 5′-CCG CTC GAG TTA ATC CCT GGT CCT GTT CTT AT-3′), and the recombinant VP19 protein was expressed in E. coli with the pET-32a vector used as the control.
Approximately 200 μg of purified GST-VP24 was incubated with GST · Bind resin (GenScript, Nanjing, China) and then washed with 15 ml of PBS. About 400 μg of purified His-PcPHB1, PHB domain, or PHB-CT was added to the resin and incubated at 4°C for 2 h. After being washed thoroughly with 10 ml of PBS, the proteins were eluted with elution buffer (10 mM reduced glutathione, 50 mM Tris-HCl) and then analyzed by 12.5% SDS-PAGE. The GST-VP26 and GST-VP28 pulldown assays using His-PcPHB1, the PHB domain, and PHB-CT, as well as the GST-PHB1 and GST-PHB domain pulldown assays using His-VP19, were performed as described above, and a GST-PHB1 pulldown assay using His-VP24 was performed as the positive control.
Immunocytochemistry analysis to detect rPcPHB1 in hemocytes.
Immunocytochemistry was performed to determine whether rPcPHB1 could enter hemocytes. The crayfish were separated into two groups: the first group was injected with 20 μl (1 μg/μl) of recombinant His-PcPHB1, whereas the second group was injected with 20 μl (1 μg/μl) of BSA. After 3 h, hemolymph was extracted and centrifuged to collect hemocytes. Immunocytochemistry was conducted by a previously described method using anti-His tag antibodies as the primary antibody (32).
Three-dimensional model analysis.
A three-dimensional model of VP28-PcPHB1 heterodimer was predicted using the GRAMM-X (33) and ZDOCK (34) software tools, which are rigid body protein-protein docking programs, to present the molecular interactions between VP28 and PcPHB1. Each of the programs returned the 10 most probable models out of thousands of candidates based on their geometry, hydrophobicity, and electrostatic complementarity. The first-ranked model of GRAMM-X and the second-ranked model of ZDOCK exhibited highly similar interacting modes; thus, they were selected as the final model for further analysis. The structure of VP28 used for docking was obtained from the Protein Data Bank (PDB) (35) under PDB code 2DE6. Considering the structure of PcPHB1 remains unknown, we predicted its structural model with the I-TASSER server (36). The molecular interface analysis was performed using PDBePISA (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html).
RESULTS
PcPHB1 is highly similar to PHB1 from other organisms.
The full-length PcPHB1 cDNA was 973 bp, including a 21-bp 5′-UTR and a 124-bp 3′-noncoding region with a poly(A) tail. Its 828-bp open reading frame encodes a 275-amino-acid (aa) protein with a predicted molecular mass of 30.1 kDa and a theoretical isoelectric point of 6.15 (GenBank accession no. KC195862). A PHB domain was identified in this protein. PcPHB1 is highly similar (around 90% amino acid sequence identity) to the PHBs from the Pacific white shrimp, L. vannamei (GenBank accession no. DQ907946.1), and the Chinese mitten crab, Eriocheir sinensis (GenBank accession no. HM347446.1). PcPHB1 is also highly similar to human and other animal PHB1 proteins and was clustered with other PHB1 sequences in a phylogenetic tree analysis (data not shown).
PcPHB1 is ubiquitously distributed in crayfish tissues.
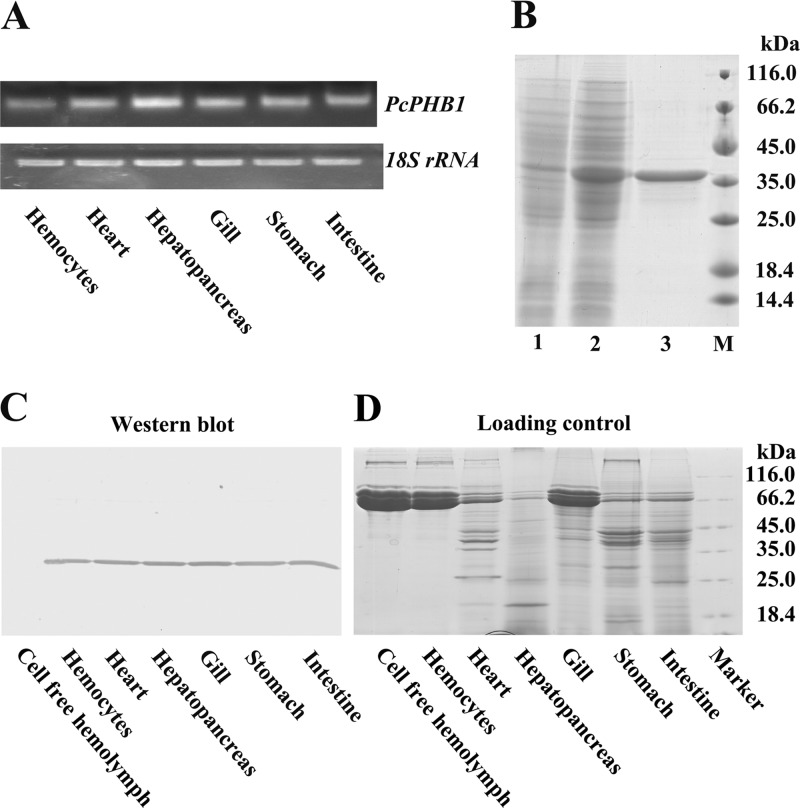
RT-PCR was used to analyze the distribution of PcPHB1 transcripts. The results revealed that PcPHB1 was expressed in all six tested tissues, namely, the hemocytes, heart, hepatopancreas, gills, stomach, and intestine (Fig. 1A). PcPHB1 antiserum was prepared after its recombinant expression and purification (Fig. 1B). The PcPHB1 protein distribution was also determined via Western blot analysis, and the same result was obtained with the RT-PCR (Fig. 1C). PcPHB1 was not detected in the cell-free hemolymph of healthy crayfish (Fig. 1C).
Fig 1.
Tissue distribution of PcPHB1 and expression of the PcPHB1 recombinant. (A) Tissue distribution of PcPHB1 in crayfish analyzed via semiquantitative RT-PCR. 18S RNA was used as the control. (B) PcPHB1 recombinant expression and purification. Lane 1, total proteins from E. coli with pET30a without IPTG induction; lane 2, total proteins from E. coli induced with IPTG; lane 3, purified recombinant PcPHB1; lane M, protein molecular mass standard. (C) Tissue distribution of PcPHB1 in normal crayfish determined using Western blot analysis. (D) Protein loading control. The proteins from different tissues were first quantified by the Bradford method and then analyzed by SDS-PAGE.
PcPHB1 was upregulated by WSSV challenge.
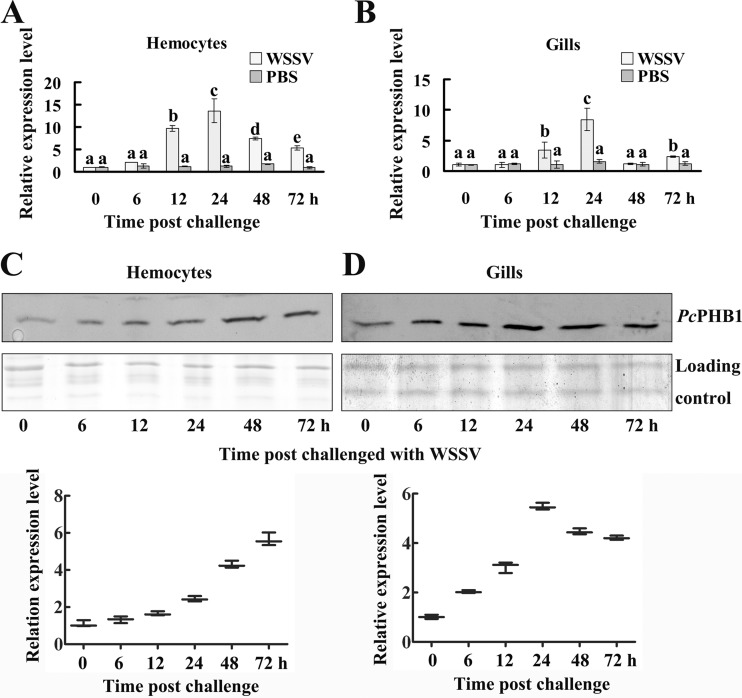
Both qRT-PCR and Western blot analysis were performed to determine the PcPHB1 expression patterns in crayfish after WSSV challenge. The qRT-PCR results showed that PcPHB1 was upregulated in hemocytes and gills after WSSV challenge (Fig. 2A and B). PcPHB1 achieved the highest expression levels in the hemocytes and gills at 24 h after WSSV injection. The results of the Western blot analysis also showed that PcPHB1 was upregulated in hemocytes and in the gills of WSSV-infected crayfish (Fig. 2C and D). Therefore, PcPHB1 is upregulated by WSSV challenge at both the RNA and protein levels.
Fig 2.
Expression patterns of PcPHB1 after WSSV challenge. (A and B) Expression patterns were detected in hemocytes (A) and gills (B) after WSSV challenge with qRT-PCR. PBS injection was used as the control. The data were calculated using the 2−ΔΔCT method. Bars represent the means of three individual measurements ± standard errors of the means (SEM). Differences between groups were analyzed using one-way analysis of variance (ANOVA) followed by a Tukey's multiple-comparison test. Different letters indicate significant differences (P < 0.05). (C) PcPHB1 expression pattern in hemocytes after WSSV challenge analyzed by Western blotting. The bottom part of panel C shows the statistical analysis for three replicates. (D) PcPHB1 expression pattern in the gills after WSSV challenge, determined using Western blot analysis. The bottom part of panel D shows the statistical analysis for three replicates.
PcPHB1 reduced the quantity of WSSV.
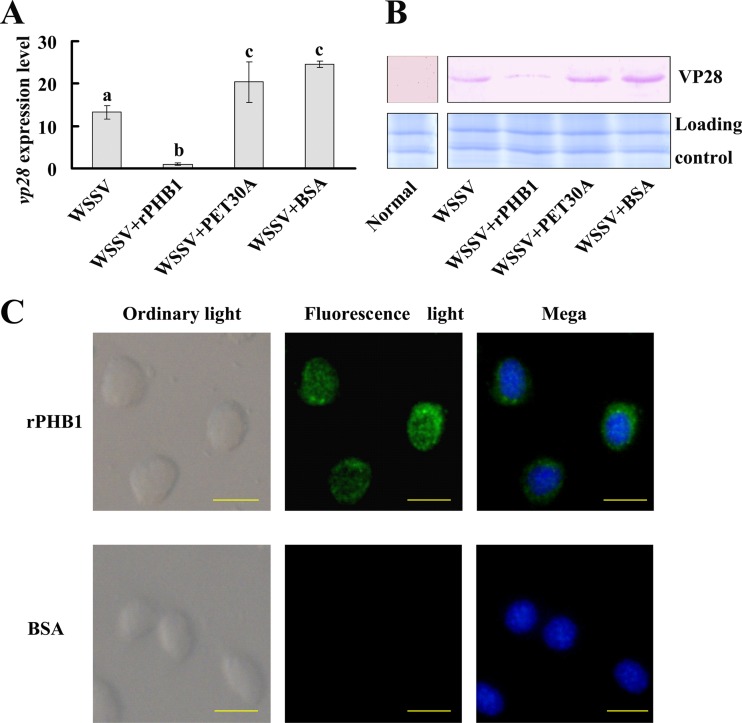
To determine whether the PcPHB1 has antiviral properties, the recombinant PcPHB1 (rPcPHB1) was injected into crayfish, and the amount of WSSV in the gills was detected via qRT-PCR using the VP28 gene as a marker at 60 h after WSSV challenge. The WSSV levels in the rPcPHB1 injection group were markedly lower than those in the controls: the PET30A-WSSV, BSA-WSSV, and WSSV-only groups (Fig. 3A). To confirm these results, the amount of WSSV was also determined via Western blot analysis using anti-VP28 serum. The WSSV levels in the rPcPHB1 injection group were markedly lower than those in the other groups (Fig. 3B). We conjectured that the injected rPcPHB1 might enter the cell to induce the viral inhibition activity, so we performed immunocytochemistry assays after rPcPHB1 injection using anti-His tag antibodies as the primary antibody. The result showed that the injected rPcPHB1 could enter the crayfish hemocytes (Fig. 3C). These data revealed that PcPHB1 reduced the quantity of WSSV in the crayfish.
Fig 3.
rPcPHB1 prevents WSSV replication. The crayfish were assigned into four groups, namely, PcPHB1 plus WSSV, PET30A plus WSSV, BSA plus WSSV, and WSSV, and the amounts of virus were analyzed via qRT-PCR using VP28 as a marker (A), as well as Western blot analysis using anti-VP28 serum (B), after 60 h of WSSV injection. (A) VP28 expression in different groups was analyzed by qRT-PCR. Bars represent the means of three individual measurements ± SEM. Differences between groups were analyzed by one-way ANOVA followed by a Tukey's multiple-comparison test. Different letters indicate significant difference (P < 0.05). (B) VP28 protein in different groups determined using Western blot analysis. (C) The recombinant protein PcPHB1 in the hemocytes was detected via immunocytochemistry using anti-His IgG (ABGNT, China) as the primary antibody. Bar, 20 μm.
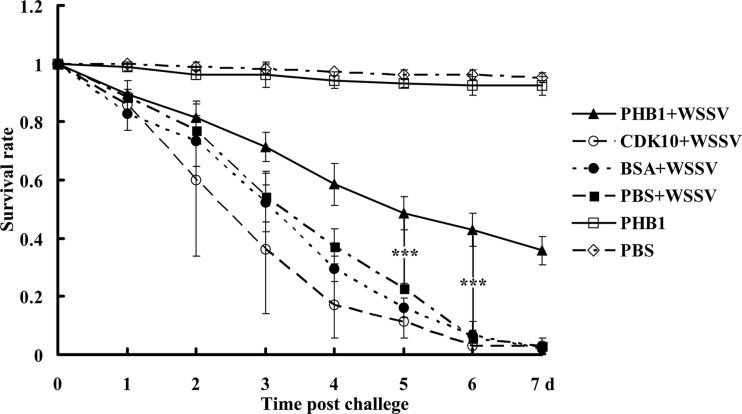
The survival rate of WSSV-infected crayfish increased after rPcPHB1 injection.
To evaluate the effect of PcPHB1 in vivo, the survival rate was investigated after the crayfish were injected with the recombinant protein and subsequently infected with WSSV. The injections of CDK10 expressed in the same E. coli system and BSA served as the controls. The WSSV injection group and the other control groups exhibited survival rates of approximately 40% to 65% by the third day of WSSV injection, whereas the group injected with rPcPHB1 plus WSSV exhibited a survival rate of about 70% on the same day. On the fifth to seventh days, the survival rate of the rPcPHB1-WSSV-injected group was significantly higher than those of the three other WSSV-injected groups (Fig. 4). These data indicate that rPcPHB1 injections significantly increase the survival rate of the WSSV-infected crayfish.
Fig 4.
PcPHB1 protein increases the survival rate of WSSV-infected crayfish. Six groups of crayfish were used in the experiment. Three groups were injected with PcPHB1, BSA, and CDK10 and then with WSSV after 1 h. The other three groups were injected with only WSSV, PcPHB1, or PBS. The crayfish were monitored daily for mortality, and the survival rate was calculated. This assay was repeated three times in 1 month under the same conditions.
PcPHB1 knockdown decreased the antiviral activity and was rescued by PcPHB1 reinjection.
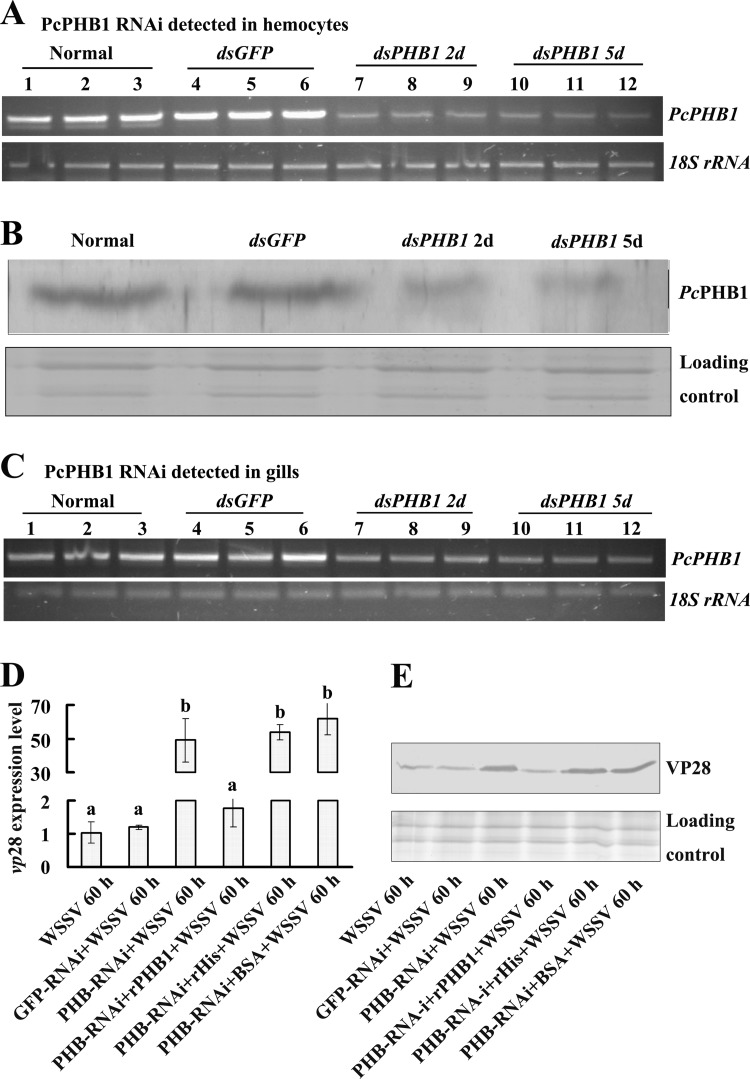
RNA interference was performed to confirm the antiviral effect of PcPHB1. PcPHB1 was knocked down in hemocytes and gills after PcPHB1 dsRNA was injected into the crayfish from 2 days to 5 days postinjection (Fig. 5A and C). The PcPHB1 protein levels in the RNA-knockdown crayfish were also determined via Western blot analysis, and the results showed that the PcPHB1 levels in hemocytes were lower than those in the controls (Fig. 5B).
Fig 5.
PcPHB1 knockdown increases WSSV in crayfish. The crayfish were subjected to RNA interference with dsGFP as the control. (A and B) Effect of PcPHB1 RNAi on hemocytes determined using RT-PCR and Western blot analysis. (C) Effect of PcPHB1 RNAi in the gills analyzed via RT-PCR. The experiment was repeated tree times, and the lane numbers 1 to 12 in panels A and C represent different crayfish groups: lanes 1 to 3 show three replicates of the untreated (normal) groups, lanes 4 to 6 show three replicates of the dsGFP groups, and lanes 7 to 9 and 10 to 12 show three replicates of the dsPHB1 groups detected on the 2nd and 5th days (2d and 5d, respectively), with at least three crayfish in each group. (D) The crayfish were infected with WSSV after PcPHB1 knockdown, and the amount of WSSV was detected via qRT-PCR using VP28 as a marker. The rescue experiment was also performed through the injection of rPcPHB1 into the PcPHB1-silenced crayfish; PET30A and BSA were injected into the PcPHB1-silenced crayfish as controls. Bars represent the means of three individual measurements ± SEM. Differences between groups were analyzed using one-way ANOVA followed by a Tukey's multiple-comparison test. Different letters indicate significant difference (P < 0.001). (E) The PcPHB1-silenced crayfish were infected with WSSV, and the VP28 of WSSV was detected via Western blot analysis. The rescue experiment and controls were performed with the same method as panel D.
After PcPHB1 knockdown, the crayfish were infected with WSSV. We further analyzed the amount of WSSV in the PcPHB1-knockdown crayfish using qRT-PCR and Western blot analysis. The results showed that the amount of WSSV increased in the gills of PcPHB1-knockdown crayfish compared with those in the normal (untreated) control and the dsGFP injection group at the DNA level (Fig. 5D) and the protein level (Fig. 5E). After injection of rPcPHB1 into the RNA-knockdown crayfish, the amount of WSSV decreased compared with that in the RNA-silenced crayfish (Fig. 5D and E). In the groups injected with PET30A and BSA after PcPHB1 knockdown, the amount of WSSV was unchanged compared with that in the RNA-silenced crayfish. These results suggest that the antiviral effects of PcPHB1 knockdown in crayfish declined after PcPHB1 silencing, and the deactivation of the antiviral effects could be rescued by injection of rPcPHB1 into the RNA-silenced crayfish.
PcPHB1 binds to WSSV by interacting with VP24, VP26, and VP28.
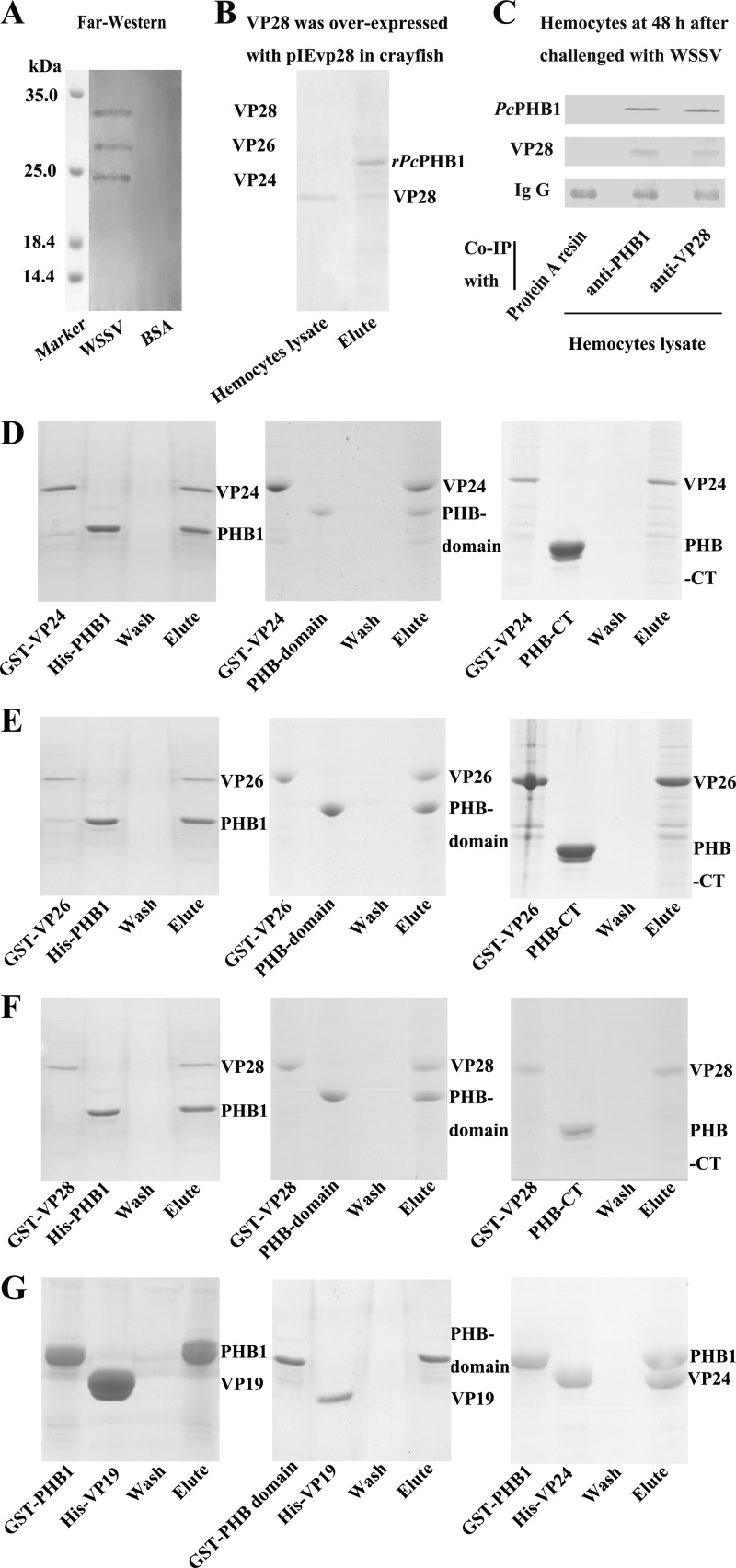
The results suggest that PcPHB1 has antiviral activity, but its mechanism remains unclear. The interaction of PcPHB1 with WSSV was detected. We first purified the WSSV to determine whether the recombinant protein binds to the native WSSV by using a far-Western overlay assay. The results show that PcPHB1 interacts with the WSSV envelope proteins VP24, VP26, and VP28 (Fig. 6A). We then performed an assay using rPcPHB1 to pull down the VP28 expressed in crayfish injected with pIevp28 overexpression vector, which expresses VP28 in vivo (30). We found that rPcPHB1 could bind to the VP28 expressed in the hemocytes (Fig. 6B). A co-IP assay was also performed using hemocytes from WSSV-challenged crayfish to detect the interaction of PcPHB1 and VP28. The results show that native PcPHB1 and native VP28 in the hemocytes interact with each other (Fig. 6C). Given that we do not have antibodies against VP26 and VP24, we performed an in vitro pulldown assay to confirm the interactions of VP26 and VP24 with PcPHB1. VP24, VP26, and VP28 with GST tag were expressed in E. coli using the pGEX-4T-1 vector, and PcPHB1, the PHB domain, and the His-tagged C terminal of PcPHB1 were expressed in E. coli using the pET-32A vector. The results of the pulldown assay show that the PcPHB1 and PHB domains interact with the recombinant VP24, VP26, and VP28, but the PcPHB1 C-terminal did not (Fig. 6D, E, and F). These results suggest that PcPHB1 physically interacts with VP24, VP26, and VP28 and that the PHB domain is responsible for the interaction. VP19, another major WSSV envelope protein, was also used for the in vitro pulldown analysis. The results show that both PcPHB1 and the PHB domain do not interact with VP19 (Fig. 6G).
Fig 6.
PcPHB1 interacts with VP24, VP26, and VP28 of WSSV. (A) A far-Western overlay assay was used to analyze the interaction between the PcPHB1 and WSSV proteins. The PcPHB1 antiserum revealed 24-, 26-, and 28-kDa bands. (B) The pIevp28 overexpression vector was injected into crayfish to overexpress VP28. Hemocytes were then collected. Hemocyte lysate was subjected to His · Bind resin binding with His-PcPHB1. (C) The co-IP assay was performed using PcPHB1 antiserum and VP28 antiserum with hemocytes from WSSV-challenged crayfish. A tube with antiserum-free protein A resin was used as the control. (D to F) In vitro pulldown assays were performed to confirm the interactions of rVP24, rVP26, and rVP28 with PcPHB1. (D) Interactions of His-PcPHB1, PHB domain, and the C terminal of PcPHB1 (PHB1-CT) with GST-VP24. (E) Interactions of His-PcPHB1, His-PHB domain, and PHB1-CT with GST-VP26. (F) Interactions of His-PcPHB1, His-PHB domain, and PHB1-CT with GST-VP28 (G) Interactions of His-VP19 with GST-PHB and GST-PcPHB1 and the interaction of His-VP24 with GST-PHB.
A three-dimensional model of the VP28-PcPHB1 heterodimer was constructed to present the molecular interactions between VP28 and PcPHB1 (Fig. 7). As illustrated, the C-terminal side of the β-barrel domain of VP28 interacts tightly with the N-terminal domain of PcPHB1, producing a broad interface of 694.1 Å2. This interface is composed of 33 residues from VP28 and 27 residues from PcPHB1 with six hydrogen bonds (data not shown).
Fig 7.
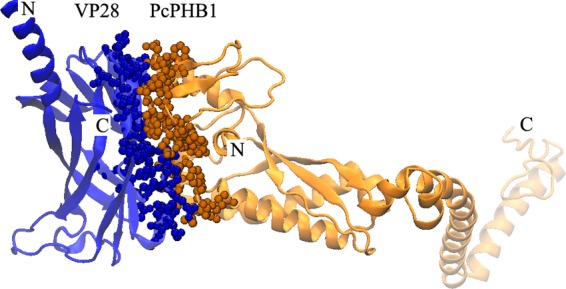
Structural model of the VP28-PcPHB1 heterodimer. The blue spheres represent the interface residues from VP28, and the orange spheres represent those from PcPHB1.
The above data of interactions among PcPHB1 and envelope proteins were all obtained by protein chemistry methods. We further performed colloidal gold electron microscopy to confirm the interactions of PcPHB1 with envelope proteins. The result shows that all of the colloidal gold-labeled PcPHB1 is located on the outer surface of WSSV (Fig. 8), suggesting that PcPHB1 could specifically bind to the envelope proteins on the WSSV envelope. Thus, these findings indicate that PcPHB1 may bind to WSSV through interaction with its envelope proteins VP28, VP26, and VP24.
Fig 8.
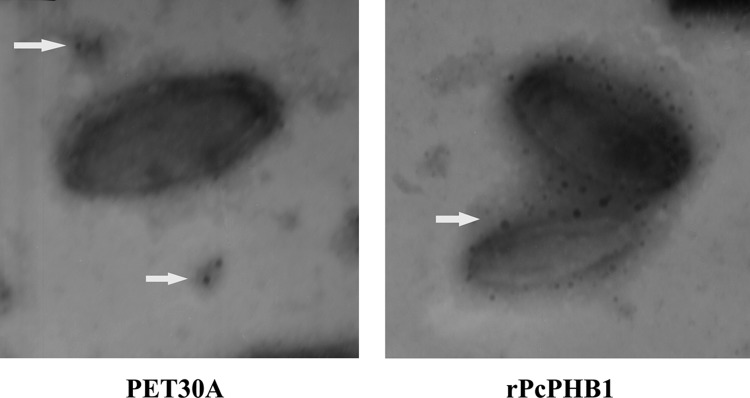
rPcPHB1 binds to the WSSV envelope. PcPHB1 was first labeled with colloidal gold and then incubated with purified WSSV. After being stained with phosphotungstic acid, the viral suspension was adsorbed onto carbon-coated nickel grids and observed under transmission electron microscopy (TEM). PET30A was used as the control. Arrows show PET30A (A) or PcPHB1 (B) labeled with colloidal gold.
DISCUSSION
In the present study, PcPHB1 has been identified in red swamp crayfish. PcPHB1 is significantly upregulated by WSSV challenge, and the recombinant PcPHB1 protein reduced the quantity of WSSV and increased the survival rate of the crayfish infected with WSSV. After PcPHB1 knockdown, the viral inhibitory effect of PcPHB1 was reduced, but it could be rescued with PcPHB1 reinjection. To analyze the mechanism of PcPHB1 in reducing the quantity of WSSV, the interaction of PcPHB1 with WSSV was analyzed by far-Western, pulldown, and co-IP assays, as well as colloidal gold electron microscopy. The results showed that PcPHB1 could bind to WSSV through interaction with envelope proteins VP24, VP26, and VP28. Given that the envelope proteins interact with the host cell membrane for effective transfer of the viral infection, PcPHB1 may block the viral infection by interacting with these proteins.
In plants, PHBs have an active role in stress tolerance and are involved in triggering retrograde signals in response to stress (2). PHB transcript levels increase during different abiotic and biotic stress conditions in some plants, such as Arabidopsis, tobacco, and rice (37, 38). The mRNA levels of AtHIRs, PHB domain-containing proteins, are significantly induced by microbe-associated molecular patterns, such as the bacterial flagellin fragment flg22. The overexpression of AtHIR1 and AtHIR2 reduces the growth of the bacterium P. syringae pv. tomato DC3000 (39). In animals, Ross et al. reported that PHBs were upregulated upon the activation of primary human T cells (40). PHB1 also increases when challenged with viruses in human cell lines (41, 42). In our study, PcPHB1 was universally expressed in several tissues (Fig. 1A and C). PcPHB1 expression was significantly upregulated (more than 10-fold in hemocytes) in the WSSV-infected crayfish (Fig. 2A). This result indicates that PcPHB1 may be involved in the antiviral response of crayfish.
To analyze the function of PcPHB1, rPcPHB1 was injected into crayfish, which were then infected with WSSV, and the WSSV replication in vivo was monitored by analyzing the VP28 expression. We found that PcPHB1 could inhibit WSSV replication in crayfish (Fig. 3A and B), indicating that the injected rPcPHB1 entered the cells to induce an antiviral response. A previous study has shown that the PHBs present in the circulation can be internalized when added to cultured cells (6). Other reports have also shown that the recombinant protein could enter the hemocytes. For example, a recombinant immulectin-3 from the insect Manduca sexta translocates from hemolymph into hemocytes (43), and the recombinant shrimp ubiquitin-conjugating enzyme E2 enters the hemocytes by endocytosis (44). In the present study, we also found that rPcPHB1 could enter the hemocytes of crayfish (Fig. 3C).
PHBs have versatile functions through interaction with different proteins. PHB1 binds and activates complement protein 3 in humans, which has an essential role in antibacterial response (45). PHB1 and PHB2 are both required for human T cell survival (40). In Arabidopsis, AtHIR1/2, which contains a PHB domain, is physically associated with RPS2 (a typical nucleotide-binding leucine-rich repeat resistance protein) and quantitatively contributes to RPS2-mediated effector-triggered immunity (39). PHB1 is a DENV receptor that helps in DENV-2 replication (15). PHB1, together with PHB2, also binds the cytoplasmic C-terminal domains of human immunodeficiency virus glycoprotein, and PHB1 and -2 have a role in the viral replicative spread in nonpermissive cells (41). In the present study, PcPHB1 was significantly upregulated after WSSV infection in crayfish and showed antiviral function after rPcPHB1 injection, as well as in RNA interference and rescue assays.
To explore the possible mechanism of PcPHB1 against virus, we first performed far-Western blotting to detect the interaction of the protein with WSSV. The result showed that PcPHB1 interacted with VP28, VP26, and VP24 of the WSSV. We then recombinantly expressed the three envelope proteins and performed the pulldown and co-IP assays to verify the interactions. All results suggest that PcPHB1 interacts with the three envelope proteins (Fig. 6). Given that all of above results were obtained by proteochemistry methods to show the interactions, we further performed a colloidal gold electron microscopy assay. The result showed that colloidal gold-labeled PcPHB1 was located on the outer surface of WSSV virions, which suggests that PcPHB1 specifically binds to the envelope proteins on the WSSV surface.
The entrance of viruses into the host cells requires their viral envelope proteins to attach to the host cell surface (46). VP28, VP26, VP24, and VP19 are the four major envelope proteins of WSSV (47). VP28 is located on the outer surface of WSSV and is involved in viral attachment to and penetration of shrimp cells (48). VP26 and VP24 were first identified as nucleocapsid proteins (49); however, subsequent studies have demonstrated that both proteins are regarded as envelope proteins or tegument proteins (50, 51). VP28 and VP26 have 41% sequence similarity. VP24 has 46% sequence similarity to VP28 and 41% sequence similarity to VP26. VP24, VP26, and VP28 have a predicted 30-amino acid (aa) transmembrane region at the N terminus (50). The SMART analysis showed that all three envelope proteins have a 210-aa WSS_VP domain. VP19, the other major envelope protein, does not share sequence homology with VP28, VP26, and VP24. Our results showed that PcPHB1 could interact with VP28, VP26, and VP24 but not with VP19. In addition, we determined the interaction of PcPHB1 with VP28 by using structural model analysis. The structural model analysis also demonstrated almost the same results for both PcPHB1-VP26 and PcPHB1-VP24 interactions (data not shown).
Viral structural proteins have vital roles in cell targeting, viral entry, assembly, and budding (52), as well as triggering host antiviral defenses (53). Some WSSV envelope proteins involved in shrimp infection have been identified (54). For example, VP26 and VP28 anchored on the viral envelope membrane interact with the host cell membrane for effective transfer of the viral infection (50). VP24 interacts with VP28 to form a protein complex, which participates in viral infection (55). Therefore, the direct interactions of PcPHB1 with VP28, VP26, and VP24 might interfere with viral invasion, assembly, or the pervasion of WSSV in vivo. The precise mechanism needs to be clarified for further studies.
ACKNOWLEDGMENT
This work was financially supported by the National Natural Science Foundation of China (no. 31130056), National Basic Research Program of China (973 Program, no. 2012CB114405), the Ph.D. Programs Foundation of the Ministry of Education of China (no. 20110131130003), and Provincial Natural Science Foundation of Shandong, China (no. ZR2011CM014).
We thank Kenneth Söderhäll of Uppsala University for suggestions for this work.
Footnotes
Published ahead of print 18 September 2013
REFERENCES
- 1.Tavernarakis N, Driscoll M, Kyrpides NC. 1999. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 24:425–427 [DOI] [PubMed] [Google Scholar]
- 2.Van Aken O, Whelan J, Van Breusegem F. 2010. Prohibitins: mitochondrial partners in development and stress response. Trends Plant Sci. 15:275–282 [DOI] [PubMed] [Google Scholar]
- 3.McClung JK, Danner DB, Stewart DA, Smith JR, Schneider EL, Lumpkin CK, Dell'Orco RT, Nuell MJ. 1989. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem. Biophys. Res. Commun. 164:1316–1322 [DOI] [PubMed] [Google Scholar]
- 4.Jupe ER, Liu XT, Kiehlbauch JL, McClung JK, Dell'Orco RT. 1996. Prohibitin in breast cancer cell lines: loss of antiproliferative activity is linked to 3′ untranslated region mutations. Cell Growth Differ. 7:871–878 [PubMed] [Google Scholar]
- 5.Liu T, Tang H, Lang Y, Liu M, Li X. 2009. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett. 273:233–242 [DOI] [PubMed] [Google Scholar]
- 6.Mishra S, Murphy LC, Murphy LJ. 2006. The prohibitins: emerging roles in diverse functions. J. Cell. Mol. Med. 10:353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merkwirth C, Langer T. 2009. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta 1793:27–32 [DOI] [PubMed] [Google Scholar]
- 8.Mengwasser J, Piau A, Schlag P, Sleeman JP. 2004. Differential immunization identifies PHB1/PHB2 as blood-borne tumor antigens. Oncogene 23:7430–7435 [DOI] [PubMed] [Google Scholar]
- 9.Mishra S, Ande SR, Nyomba BL. 2010. The role of prohibitin in cell signaling. FEBS J. 277:3937–3946 [DOI] [PubMed] [Google Scholar]
- 10.Artal-Sanz M, Tavernarakis N. 2009. Prohibitin and mitochondrial biology. Trends Endocrinol. Metab. 20:394–401 [DOI] [PubMed] [Google Scholar]
- 11.Ande SR, Gu Y, Nyomba BL, Mishra S. 2009. Insulin induced phosphorylation of prohibitin at tyrosine 114 recruits Shp1. Biochim. Biophys. Acta 1793:1372–1378 [DOI] [PubMed] [Google Scholar]
- 12.Ande SR, Mishra S. 2009. Prohibitin interacts with phosphatidylinositol 3,4,5-triphosphate (PIP3) and modulates insulin signaling. Biochem. Biophys. Res. Commun. 390:1023–1028 [DOI] [PubMed] [Google Scholar]
- 13.Rajalingam K, Wunder C, Brinkmann V, Churin Y, Hekman M, Sievers C, Rapp UR, Rudel T. 2005. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat. Cell Biol. 7:837–843 [DOI] [PubMed] [Google Scholar]
- 14.Zhu B, Fukada K, Zhu H, Kyprianou N. 2006. Prohibitin and cofilin are intracellular effectors of transforming growth factor beta signaling in human prostate cancer cells. Cancer Res. 66:8640–8647 [DOI] [PubMed] [Google Scholar]
- 15.Kuadkitkan A, Wikan N, Fongsaran C, Smith DR. 2010. Identification and characterization of prohibitin as a receptor protein mediating DENV-2 entry into insect cells. Virology 406:149–161 [DOI] [PubMed] [Google Scholar]
- 16.Jung HW, Hwang BK. 2007. The leucine-rich repeat (LRR) protein, CaLRR1, interacts with the hypersensitive induced reaction (HIR) protein, CaHIR1, and suppresses cell death induced by the CaHIR1 protein. Mol. Plant Pathol. 8:503–514 [DOI] [PubMed] [Google Scholar]
- 17.Jung HW, Lim CW, Lee SC, Choi HW, Hwang CH, Hwang BK. 2008. Distinct roles of the pepper hypersensitive induced reaction protein gene CaHIR1 in disease and osmotic stress, as determined by comparative transcriptome and proteome analyses. Planta 227:409–425 [DOI] [PubMed] [Google Scholar]
- 18.Jones JD, Dangl JL. 2006. The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Zhao XF, Wang JX. 2009. Molecular cloning and characterization of the translationally controlled tumor protein from Fenneropenaeus chinensis. Mol. Biol. Rep. 36:1683–1693 [DOI] [PubMed] [Google Scholar]
- 20.Li XC, Zhang RR, Sun RR, Lan JF, Zhao XF, Wang JX. 2010. Three Kazal-type serine proteinase inhibitors from the red swamp crayfish Procambarus clarkii and the characterization, function analysis of hcPcSPI2. Fish Shellfish Immunol. 28:942–951 [DOI] [PubMed] [Google Scholar]
- 21.Shi XZ, Zhao XF, Wang JX. 2008. Molecular cloning and expression analysis of chymotrypsin-like serine protease from the Chinese shrimp, Fenneropenaeus chinensis. Fish Shellfish Immunol. 25:589–597 [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 23.Du XJ, Wang JX, Liu N, Zhao XF, Li FH, Xiang JH. 2006. Identification and molecular characterization of a peritrophin-like protein from fleshy prawn (Fenneropenaeus chinensis). Mol. Immunol. 43:1633–1644 [DOI] [PubMed] [Google Scholar]
- 24.Du XJ, Zhao XF, Wang JX. 2007. Molecular cloning and characterization of a lipopolysaccharide and beta-1,3-glucan binding protein from fleshy prawn (Fenneropenaeus chinensis). Mol. Immunol. 44:1085–1094 [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 26.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Liu N, Chen AJ, Zhao XF, Wang JX. 2009. TRBP homolog interacts with eukaryotic initiation factor 6 (eIF6) in Fenneropenaeus chinensis. J. Immunol. 182:5250–5258 [DOI] [PubMed] [Google Scholar]
- 28.Oliver C. 2010. Conjugation of colloidal gold to proteins. Methods Mol. Biol. 588:369–373 [DOI] [PubMed] [Google Scholar]
- 29.Xie X, Li H, Xu L, Yang F. 2005. A simple and efficient method for purification of intact white spot syndrome virus (WSSV) viral particles. Virus Res. 108:63–67 [DOI] [PubMed] [Google Scholar]
- 30.Mu Y, Lan JF, Zhang XW, Wang XW, Zhao XF, Wang JX. 2012. A vector that expresses VP28 of WSSV can protect red swamp crayfish from white spot disease. Dev. Comp. Immunol. 36:442–449 [DOI] [PubMed] [Google Scholar]
- 31.Wang XW, Xu WT, Zhang XW, Zhao XF, Yu XQ, Wang JX. 2009. A C-type lectin is involved in the innate immune response of Chinese white shrimp. Fish Shellfish Immunol. 27:556–562 [DOI] [PubMed] [Google Scholar]
- 32.Chen AJ, Gao L, Wang XW, Zhao XF, Wang JX. 2013. SUMO-conjugating enzyme E2 UBC9 mediates viral immediate-early protein SUMOylation in crayfish to facilitate reproduction of white spot syndrome virus. J. Virol. 87:636–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tovchigrechko A, Vakser IA. 2006. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34:W310–W314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R, Li L, Weng Z. 2003. ZDOCK: an initial-stage protein-docking algorithm. Proteins 52:80–87 [DOI] [PubMed] [Google Scholar]
- 35.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujiwara M, Umemura K, Kawasaki T, Shimamoto K. 2006. Proteomics of Rac GTPase signaling reveals its predominant role in elicitor-induced defense response of cultured rice cells. Plant Physiol. 140:734–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandenabeele S, Van Der Kelen K, Dat J, Gadjev I, Boonefaes T, Morsa S, Rottiers P, Slooten L, Van Montagu M, Zabeau M, Inze D, Van Breusegem F. 2003. A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc. Natl. Acad. Sci. U. S. A. 100:16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi Y, Tsuda K, Nguyen LV, Wang VX, Lin J, Murphy AS, Glazebrook J, Thordal-Christensen H, Katagiri F. 2011. Physical association of Arabidopsis hypersensitive induced reaction proteins (HIRs) with the immune receptor RPS2. J. Biol. Chem. 286:31297–31307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross JA, Nagy ZS, Kirken RA. 2008. The PHB1/2 phosphocomplex is required for mitochondrial homeostasis and survival of human T cells. J. Biol. Chem. 283:4699–4713 [DOI] [PubMed] [Google Scholar]
- 41.Emerson V, Holtkotte D, Pfeiffer T, Wang IH, Schnolzer M, Kempf T, Bosch V. 2010. Identification of the cellular prohibitin 1/prohibitin 2 heterodimer as an interaction partner of the C-terminal cytoplasmic domain of the HIV-1 glycoprotein. J. Virol. 84:1355–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He F, Zeng Y, Wu X, Ji Y, He X, Andrus T, Zhu T, Wang T. 2009. Endogenous HIV-1 Vpr-mediated apoptosis and proteome alteration of human T-cell leukemia virus-1 transformed C8166 cells. Apoptosis 14:1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu XQ, Tracy ME, Ling E, Scholz FR, Trenczek T. 2005. A novel C-type immulectin-3 from Manduca sexta is translocated from hemolymph into the cytoplasm of hemocytes. Insect Biochem. Mol. Biol. 35:285–295 [DOI] [PubMed] [Google Scholar]
- 44.Chen AJ, Wang S, Zhao XF, Yu XQ, Wang JX. 2011. Enzyme E2 from Chinese white shrimp inhibits replication of white spot syndrome virus and ubiquitinates its RING domain proteins. J. Virol. 85:8069–8079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishra S, Moulik S, Murphy LJ. 2007. Prohibitin binds to C3 and enhances complement activation. Mol. Immunol. 44:1897–1902 [DOI] [PubMed] [Google Scholar]
- 46.van Hulten MC, Witteveldt J, Snippe M, Vlak JM. 2001. White spot syndrome virus envelope protein VP28 is involved in the systemic infection of shrimp. Virology 285:228–233 [DOI] [PubMed] [Google Scholar]
- 47.Zhou Q, Xu L, Li H, Qi YP, Yang F. 2009. Four major envelope proteins of white spot syndrome virus bind to form a complex. J. Virol. 83:4709–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yi G, Wang Z, Qi Y, Yao L, Qian J, Hu L. 2004. Vp28 of shrimp white spot syndrome virus is involved in the attachment and penetration into shrimp cells. J. Biochem. Mol. Biol. 37:726–734 [DOI] [PubMed] [Google Scholar]
- 49.van Hulten MC, Goldbach RW, Vlak JM. 2000. Three functionally diverged major structural proteins of white spot syndrome virus evolved by gene duplication. J. Gen. Virol. 81:2525–2529 [DOI] [PubMed] [Google Scholar]
- 50.Tang X, Wu J, Sivaraman J, Hew CL. 2007. Crystal structures of major envelope proteins VP26 and VP28 from white spot syndrome virus shed light on their evolutionary relationship. J. Virol. 81:6709–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie X, Xu L, Yang F. 2006. Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus. J. Virol. 80:10615–10623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chazal N, Gerlier D. 2003. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67:226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reske A, Pollara G, Krummenacher C, Chain BM, Katz DR. 2007. Understanding HSV-1 entry glycoproteins. Rev. Med. Virol. 17:205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang YS, Liu WJ, Lee CC, Chou TL, Lee YT, Wu TS, Huang JY, Huang WT, Lee TL, Kou GH, Wang AH, Lo CF. 2010. A 3D model of the membrane protein complex formed by the white spot syndrome virus structural proteins. PLoS One 5:e10718. 10.1371/journal.pone.0010718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie X, Yang F. 2006. White spot syndrome virus VP24 interacts with VP28 and is involved in virus infection. J. Gen. Virol. 87:1903–1908 [DOI] [PubMed] [Google Scholar]