Abstract
Sirtuins are a family of protein deacetylases, deacylases, and ADP-ribosyltransferases that regulate life span, control the onset of numerous age-associated diseases, and mediate metabolic homeostasis. We have uncovered a novel role for the mitochondrial sirtuin SIRT4 in the regulation of hepatic lipid metabolism during changes in nutrient availability. We show that SIRT4 levels decrease in the liver during fasting and that SIRT4 null mice display increased expression of hepatic peroxisome proliferator-activated receptor α (PPARα) target genes associated with fatty acid catabolism. Accordingly, primary hepatocytes from SIRT4 knockout (KO) mice exhibit higher rates of fatty acid oxidation than wild-type hepatocytes, and SIRT4 overexpression decreases fatty acid oxidation rates. The enhanced fatty acid oxidation observed in SIRT4 KO hepatocytes requires functional SIRT1, demonstrating a clear cross talk between mitochondrial and nuclear sirtuins. Thus, SIRT4 is a new component of mitochondrial signaling in the liver and functions as an important regulator of lipid metabolism.
INTRODUCTION
The liver contributes to energy homeostasis, in part, by adapting lipid metabolism to match the energetic demands of the organism. Under nutrient-rich conditions, hepatic lipogenesis and lipoprotein secretion are stimulated, while during fasting hepatic fatty acid oxidation is enhanced to maintain bioenergetics and to provide the organism with ketone bodies as an alternative fuel for glucose in the brain. One of the key mediators of the hepatic response to fasting is peroxisome proliferator-activated receptor α (PPARα) (1), a ligand-activated transcription factor that promotes the transcription of genes involved in fatty acid catabolism (2, 3). Mice lacking PPARα fail to maintain metabolic homeostasis during fasting and display reduced levels of fatty acid oxidation (2, 3).
Sirtuins are a family of proteins intimately connected to the metabolic state of an organism because their enzymatic activities, deacetylation, deacylation, and ADP-ribosylation, require NAD+ (4–8). Mammals contain seven sirtuins (SIRT1 to SIRT7) (9) that have pleiotropic roles in biology. SIRT1 is the most studied sirtuin and has been shown to deacetylate dozens of protein substrates to promote metabolic homeostasis and cell survival (10). In the liver, SIRT1 promotes lipid catabolism through binding and activating PGC-1α and PPARα (11). Three of the mammalian sirtuins (SIRT3, SIRT4, and SIRT5) are located in the mitochondria (12–14), and SIRT4 is one of the least characterized sirtuins. Knockdown of SIRT4 expression enhances fatty acid oxidation in hepatocytes, which is accompanied by a change in gene expression of mitochondrial and fatty acid metabolism enzymes (15). We recently reported that SIRT4 is an important regulator of lipid metabolism in muscle and white adipose tissue (WAT) by repressing malonyl coenzyme A (malonyl-CoA) decarboxylase, thereby controlling the level of malonyl-CoA, an inhibitor of fat oxidation (16). Notably, we did not observe changes in malonyl-CoA levels in liver, suggesting that another mechanism was involved in this organ. In this study, we analyzed the mechanism by which SIRT4 represses hepatic fatty acid oxidation in the liver. We show that SIRT4 decreases PPARα activity and consequently expression of PPARα target genes in a cell-autonomous manner. We further demonstrate that SIRT4 deletion activates PPARα activity through activation of SIRT1 by NAD+. Thus, our findings highlight a novel connection between SIRT4 and signaling to PPARα.
MATERIALS AND METHODS
Cell lines.
Primary hepatocytes were isolated using a two-step perfusion protocol, based on a previous method (17). Fatty acid oxidation assays were performed 1 day after isolation. For adenoviral infection, hepatocytes were incubated with adenovirus expressing either a scrambled short hairpin RNA (shRNA) or a shRNA targeting SIRT1 (18) for 3 h. For pharmacological inhibition of SIRT1, hepatocytes were incubated overnight with 10 μM Ex527 (Tocris Biosciences). Mouse embryonic fibroblasts (MEFs) were isolated on embryonic day 12.5 to 14.5 from heterozygous females that were mated with heterozygous males. MEF lines were genotyped before use. Primary MEFs were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and 0.1 mM beta-mercaptoethanol (BME) and were used between passages 2 and 5. HepG2 cells were infected by retrovirus expressing a hemagglutinin (HA)-tagged SIRT4 and selected by puromycin.
Luciferase assay.
Transactivation assays were carried out in mouse H2.35 hepatoma cells and human HEK293T cells transfected with PPARα and retinoid X receptor alpha (RXRα). Cells were cultured in 60-mm dishes and transfected with pCMV (where CMV is cytomegalovirus), SIRT4, and SIRT4 mutant constructs using Lipofectamine 2000 reagent according to the protocol of the manufacturer. After 24 h, cells were cotransfected with 3×PPRE-luciferase reporter vector (a luciferase reporter under the control of three tandem repeats of a consensus PPAR response element [PPRE]; 2 μg) or pGL3 firefly (FF)-Luc reporter control and Renilla (Ren)-Luc (200 ng). The following day, cells were transferred to a 96-well plate. Following 12 h of treatment with WY14643 (1 μM) or dimethyl sulfoxide (DMSO), cell lysates were assayed for luciferase activity (luciferase assay system; Promega) and normalized using coexpressed Renilla luciferase activity.
Gene expression.
RNA was isolated from frozen liver tissue of overnight fasted mice or from cells using TRIzol (Invitrogen) according to the manufacturer's instructions and further purified using RNeasy columns (Qiagen). Amplification and detection of target and reference cDNA samples were performed on a LightCycler 480 (Roche) using LightCycler 480 SYBR green I Mastermix (Roche). A standard curve was generated for all genes using serial dilutions of a pool prepared from all cDNA samples. mRNA levels of target genes were normalized using the genes for beta-2-microglobulin (B2m), peptidyl-prolyl isomerase (Ppia), and ribosomal protein 16 (Rps16) as reference genes. Primer sequences are listed in Table S2 in the supplemental material.
Microarray analysis.
SIRT4 knockout (KO) and wild-type (WT) littermates (male; n = 6 per genotype; 7- to 8-month-old littermates) were sacrificed after a 16-h overnight fast. Samples were individually hybridized on Affymetrix Mouse Genome 430 2.0 GeneChips by the Biopolymers Facility (Harvard Medical School). Data analysis was performed using dChIP software (19). Differentially expressed genes between WT and SIRT4 KO mice were ranked according to the dChIP calculated P value that takes into account measurement errors (19). ErmineJ (20) was used to calculate overrepresentation of gene ontology (GO) terms in the data set using dChIP P values as gene scores. Transcriptome similarity analysis was performed based on previously published methods (21, 22). In brief, all analyzed data sets were downloaded from Gene Expression Omnibus (GEO) or ArrayExpress, and significantly differential expressed genes were compared to SIRT4 KO differentially expressed genes (P < 0.1). To exclude confounding factors associated with between-platform and between-tissue comparisons, we selected adult mouse liver data sets that used Affymetrix GeneChip Mouse Genome 430 2.0 or Affymetrix GeneChip Murine Genome U74 platforms from Gene Expression Omnibus and ArrayExpress. Similarity was scored using two criteria: a gene should be significantly different in both gene sets and the direction of regulation should be the same. Statistical significance was calculated by permutation among annotated unique genes using 10,000 permutations. All comparisons were performed in Microsoft Excel using Visual Basic to automate calculations. Studies were performed according to protocols approved by the Institutional Animal Care and Use Committee, the Standing Committee on Animals at Harvard.
Western blotting and immunoprecipitation.
Western blotting was performed using antibodies directed against SIRT1 (07-131; Upstate), HA (sc-7392; Santa Cruz), and actin (A2066; Sigma). Antibodies raised against SIRT4 were described previously (12). For immunoprecipitations, 293T cells were transfected with either a pCMV empty vector and PPARα-HA vector or SIRT1-FLAG vector and PPARα-HA vector. Cells were harvested 48 h after transfection, and lysates were immunoprecipitated using anti-FLAG resin (F2426; Sigma). Immunoprecipitates were analyzed by Western blotting and probed with HA and FLAG antibodies.
Fatty acid oxidation.
Cells were incubated overnight in culture medium containing 100 μM palmitate (C16:0) and 1 mM carnitine. In the final 2 h of incubation, cells were pulsed with 1.7 μCi of [9,10(n)-3H]palmitic acid (GE Healthcare), and the medium was collected to analyze the released 3H2O formed during cellular oxidation of [3H]palmitate (23, 24). In brief, medium was trichloroacetic acid (TCA) precipitated, and supernatants were neutralized with NaOH and loaded onto ion exchange columns packed with Dowex 1X2-400 resin (Sigma). The radioactive product was eluted with water and quantitated by liquid scintillation counting. Oxidation of [3H]palmitate was normalized to protein content using a Bio-Rad detergent-compatible (DC) protein assay. Etomoxir (Sigma), a specific inhibitor of CPT1a, was used to inhibit mitochondrial fatty acid oxidation.
NAD+/NADH.
NAD+ and NADH were measured in cells using an NAD/NADH quantitation kit (K337-100; Biovision). In tissue, NAD+ levels were measured by high-performance liquid chromatography (HPLC) (25). NADH was determined as previously described (26).
Electron microscopy.
Tissues were fixed in 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4, for at least 1 h at room temperature. Next, tissues were washed in 0.1 M cacodylate buffer and fixed secondarily in 1% osmium tetroxide for 1 h at 4°C. Tissues were then washed in deionized (DI) water before being immersed in 2% aqueous uranyl acetate to be contrast fixed overnight at 4°C. The following day, tissues were washed and dehydrated. Infiltration proceeded with 1:1 propylene oxide and LX112 Epon resin. Tissues were then embedded and cured at 60°C over 48 h. Cured blocks were sectioned at 80-nm thickness and put on coated copper slot grids that had been carbon coated and glow discharged. Grids were contrast stained with 2% uranyl acetate for 10 min and lead citrate for 5 min. Grids were imaged on a JEOL 1400 transmission electron microscope (TEM) equipped with a side mount Gatan Orius SC1000 digital camera. Quantification was performed using Velocity software.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) analysis was performed as described by Cell Signaling (9002S; SimpleChIP enzymatic chromatin IP kit) with antibodies against SIRT1 (Sigma), PPARα (Pierce), H3 (Cell Signaling), or normal rabbit IgG. DNA fragments were subjected to quantitative PCR (qPCR) using primers flanking PPRE on various PPARα targets.
Statistical analysis.
Analysis was performed using an unpaired Student's t test.
RESULTS
SIRT4 loss enhances hepatic lipid catabolic gene expression.
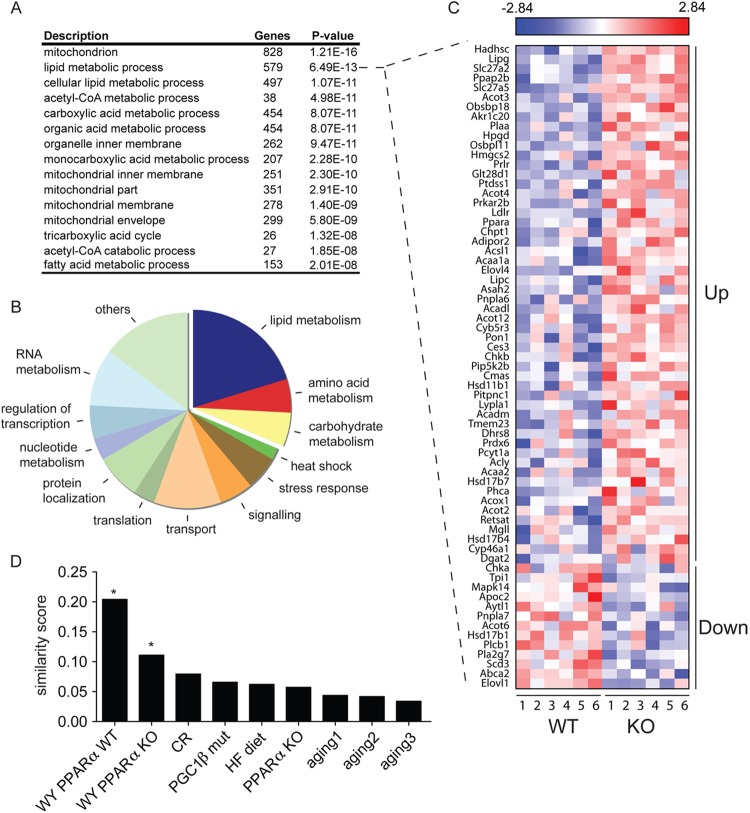
It has previously been shown that SIRT4 knockdown modifies the expression of some mitochondrial and fatty acid metabolism enzymes in mouse hepatocytes (15). To gain insight into global gene expression changes regulated by SIRT4, we analyzed genome-wide gene expression profiles in SIRT4 knockout (KO) and SIRT4 wild-type (WT) mouse livers (n = 6). Of the 22,094 unique genes on the microarray, only 654 genes were significantly different between SIRT4 KO and WT mice (P < 0.05) (see Table S1 in the supplemental material). The majority of differentially expressed genes in the SIRT4 KO livers encoded mitochondrial localized proteins (Fig. 1A) functioning in lipid, acetyl-CoA, and tricarboxylic acid metabolism. These data support the hypothesis that SIRT4 loss regulates the expression of genes involved in fatty acid metabolic pathways. Functional classification of differentially expressed genes with a P value smaller than 0.01 demonstrated that 20% of the most significantly changed genes were involved in metabolism of lipids (Fig. 1B), the majority of which were upregulated by loss of SIRT4 (Fig. 1C). For example, the expression of fatty acid oxidation genes (Acadm, Acadl, Hadhsc, Acaa1a, Acaa2, and Acox1), lipase genes (Lipg and Lipc), and thioesterase genes (Acot2, Acot3, and Acot4) was enhanced by loss of SIRT4 (Fig. 1C). Consistent with a profile of increased fatty acid catabolism, genes encoding proteins involved in fatty acid synthesis were suppressed (Elovl, Scd3, and Abca2). In sum, the microarray data indicate a focused, coordinated metabolic shift in SIRT4 KO liver toward fatty acid catabolism.
Fig 1.
SIRT4 loss enhances hepatic lipid catabolic gene expression. (A) Gene ontology terms overrepresented in the gene expression profile of SIRT4 KO mouse livers (n = 6 per genotype). Microarray data were analyzed using dChIP, and expression profiles were analyzed for overrepresentation using ErmineJ. Shown are the top significantly overrepresented 15 gene ontology terms (out of 58; P < 0.001), taking into account multiple testing using Benjamini-Hochberg. (B) Classification of pathways and metabolic processes of all annotated differentially expressed genes with a P value of <0.01. (C) Relative expression of genes (P < 0.1) associated with the GO term lipid metabolic process. SIRT4 KO and SIRT4 WT liver expression profiles are represented in a heat map. Genes are ordered according to direction of regulation and P value. (D) Similarity between SIRT4 KO liver transcriptome and published liver transcriptomes from Gene Expression Omnibus (GEO) and ArrayExpress. WY PPARα WT, WT mice treated for 5 days with WY14643 (GSE8295); WY PPARα KO, PPARα KO mice treated for 5 days with WY14643 (GSE8295); PPARα KO, WT versus PPARα KO mice (GSE8295); CR, long-term caloric restriction mice compared with mice on a control diet (GSE2431); PGC-1β mut, PGC-1β hypomorph mutant mice versus WT mice (GSE6210); HF diet, WT mice under a high-fat diet; aging1, 22- versus 4-month-old WT Snell dwarf mice (GSE3129); aging2, 22- versus 4-month-old WT Ames dwarf mice (GSE3150); aging3, 130- versus 13-week-old WT mice (ArrayExpress experiment E-MEXP-1504). Significance was calculated using permutation. *, P < 0.0001.
Because PPARα is the major transcriptional activator of fatty acid catabolism in the liver (2, 3), we examined whether PPARα target genes were upregulated in SIRT4 null livers by comparing the gene expression profile of SIRT4 KO livers with published liver gene expression profiles from PPARα KO mice and WT mice treated with WY14643, a chemical agonist of PPARα activity (27). We observed a significant overlap between gene expression profiles of PPARα activation (mice treated for 5 days with WY14643) (GSE8295) and SIRT4 KO expression profiles (Fig. 1D). In contrast, the similarity was much lower in PPARα KO mice treated with WY14643, and the transcriptome of PPARα KO mice without activation by WY14643 did not overlap the SIRT4 KO transcriptome (Fig. 1D). Furthermore, the SIRT4 KO gene expression profile did not significantly overlap the liver gene expression profiles from PGC-1β hypomorph mutant (28), caloric restriction (CR) (29), high-fat diet, or aging transcriptomes (30–32) (Fig. 1D), demonstrating a unique and specific overlap between gene expression changes in SIRT4 KO mice and in mice where PPARα is activated. These results suggest that a subset of PPARα-driven pathways may be repressed by SIRT4.
SIRT4 represses PPARα target gene expression.
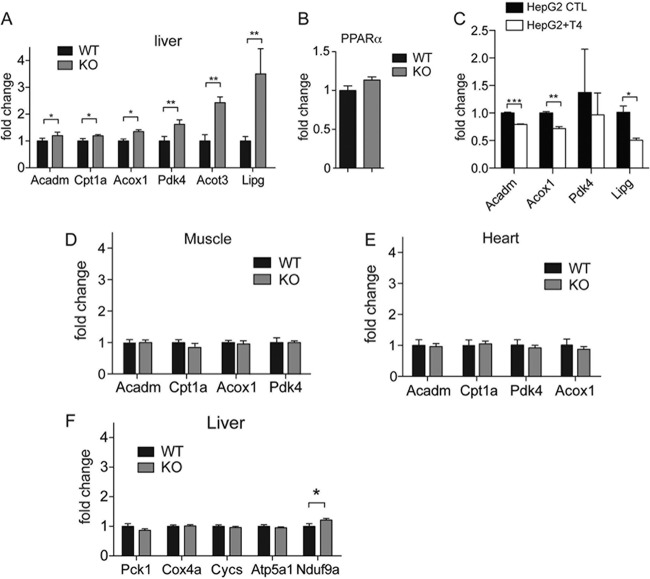
To validate whether PPARα activation is heightened in SIRT4 KO mice, we analyzed a set of canonical PPARα target genes by quantitative real-time reverse transcription-PCR (RT-PCR) (33). The expression levels of PPARα target genes Lipg, Acot3, Pdk4, Acox1, Cpt1a, and Acadm were significantly elevated (1.3- to 3.5-fold) in SIRT4 KO livers compared to the WT livers (Fig. 2A). We did not detect induction of PPARα itself (Fig. 2B), indicating that modulation of coregulators is most likely responsible for changes in PPARα signaling. To confirm these results, we analyzed the expression of PPARα target genes in a human hepatic cell line, HepG2, overexpressing SIRT4. We found that their expression was significantly decreased in HepG2 cells overexpressing SIRT4 (Fig. 2C). Together, these data confirm that SIRT4 represses PPARα target gene expression.
Fig 2.
PPARα target genes are upregulated in the liver of SIRT4 KO mice. Quantitative real-time PCR of PPARα target genes (33) in SIRT4 KO and SIRT4 WT livers. (A and B) Expression of PPARα and its target genes in SIRT4 KO and WT livers. (C) Expression of PPARα target genes in HepG2 control cells (HepG2 CTL) and HepG2 cells overexpressing SIRT4 (HepG2+T4). (D and E) Expression of PPARα target genes in muscle and heart. (F) Expression of mitochondrial genes in SIRT4 WT and KO livers. Target gene expression is represented relative to the expression of reference genes for beta-2-microglobulin (B2m) and ribosomal protein 16 (Rps16) genes. Experiments were performed in mice fasted for 16 h (n = 6 per genotype). Data are represented as mean ± standard errors of the means (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
We next analyzed expression of PPARα target genes in heart and skeletal muscle, which also utilize fatty acid oxidation. However, fatty acid oxidation genes, Cpt1a, Acadm, Acox1, and Pdk4, were not upregulated in heart or muscle tissue of SIRT4 KO mice (Fig. 2D and E). These results demonstrate that a network of PPARα targets is upregulated in the liver upon loss of SIRT4. Moreover, numerous target genes (Pck1, Cox4a, Cycs, Atp5a1, and Nduf9a) of the master regulator of mitochondrial biogenesis, PGC-1α, were not altered in SIRT4 KO liver (Fig. 2F). These results demonstrate that SIRT4 loss increases expression of fatty acid oxidation genes in the liver in the absence of general changes in mitochondrial gene expression.
SIRT4 loss enhances palmitate oxidation in liver cells, and SIRT4 expression is regulated by nutritional state.
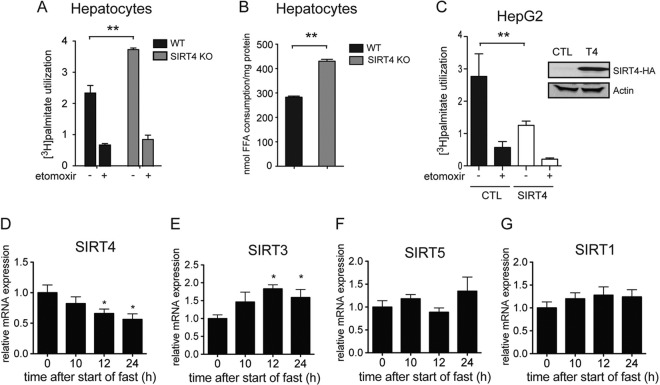
To probe the function of altered fatty acid oxidation gene expression, we analyzed the rate of oxidation of palmitate, a saturated long-chain fatty acid, using primary hepatocytes isolated from SIRT4 WT and KO animals. The CPT1a inhibitor etomoxir, which specifically blocks mitochondrial import of fatty acids, was used as a control. Oxidation rates were higher (59%) in primary hepatocytes isolated from SIRT4 KO mice than in hepatocytes from WT mice (Fig. 3A). We also examined the effect of SIRT4 on fatty acid consumption by analyzing the levels of fatty acids in culture medium before and after exposure to palmitate. Consumption of palmitate from the culture medium was significantly higher in SIRT4 KO hepatocytes than in WT hepatocytes, confirming that utilization of palmitate was indeed higher in SIRT4 KO hepatocytes (Fig. 3B). These data clearly show that loss of SIRT4 leads to enhanced fatty acid consumption due to increased rates of fatty acid oxidation. To test whether SIRT4 could actively repress fatty acid oxidation, we analyzed fatty acid oxidation in SIRT4-overexpressing HepG2 cells. As expected, SIRT4 overexpression repressed fatty acid oxidation (Fig. 3C). Taken together, these data demonstrate that SIRT4 represses fatty acid oxidation in hepatocytes.
Fig 3.
SIRT4 expression is regulated by nutritional state repressing fatty acid oxidation in the fed state. (A) Palmitate oxidation rates in SIRT4 KO primary hepatocytes are higher than in SIRT4 WT primary cells. Treatment with etomoxir, a fatty acid oxidation inhibitor, was used as a control. (B) Consumption of palmitate from culture medium in SIRT4 KO and WT primary hepatocytes. (C) Oxidation of [3H]palmitate (nmol of [3H]palmitate/h/mg protein) from hepatocyte cell line HepG2 stably overexpressing SIRT4 and a control cell line. Data represent means ± standard errors of the means (n = 6 per genotype; **, P < 0.01). (D to G) Relative hepatic mRNA expression levels of Sirt4, Sirt3, Sirt5, and Sirt1 at 0, 10, 12, and 24 h after start of the fast. Expression of target genes was normalized to beta-2-microglobulin (B2m), peptidyl-prolyl isomerase (Ppia), and ribosomal protein 16 (Rps16) genes. Experiments were performed in 4-month-old 129/Sv males. Data are presented as means ± standard errors of the means (n = 4; *, P < 0.05).
To gain insight into the physiological regulation of hepatic fat oxidation by SIRT4, we investigated whether modulation of SIRT4 activity participates in the response to nutrient deprivation in the liver. To do so, we analyzed Sirt4 expression in livers of WT animals during fasting. Sirt4 expression decreased following 10 h of fasting and was reduced to one-half of the initial fed levels after 24 h (Fig. 3D). We next examined whether the other mitochondrial sirtuins, SIRT3 and SIRT5, were similarly regulated. In contrast to the downregulation of Sirt4 upon fasting, nutrient deprivation induced Sirt3 by 1.8-fold (Fig. 3E) and did not alter Sirt5 levels (Fig. 3F). In addition, we observed a trend toward increased Sirt1 mRNA expression during fasting, as was reported previously (18) (Fig. 3G). As a control, we monitored the expression of metabolic genes known to change with nutrient status. The 24-h fasting period suppressed glucokinase gene (Gk) expression (see Fig. S1A in the supplemental material) and induced the expression of gluconeogenic genes (for glucose 6-phosphatase [G6pc] and phosphoenolpyruvate carboxykinase 1 [Pck1]) (see Fig. S1B and C) and lipid catabolism genes (for carnitine palmitoyltransferase 1a [Cpt1a] and acyl-CoA thioesterase 3 [Acot3]) (see Fig. S1D and E). Taken together, our data demonstrate that Sirt4 levels are downregulated in liver during fasting, which raises the possibility that SIRT4 participates in the metabolic shift in liver during nutrient deprivation.
SIRT4 loss induces mitochondrion enlargement and decreases the number of mitochondria in the periportal zone.
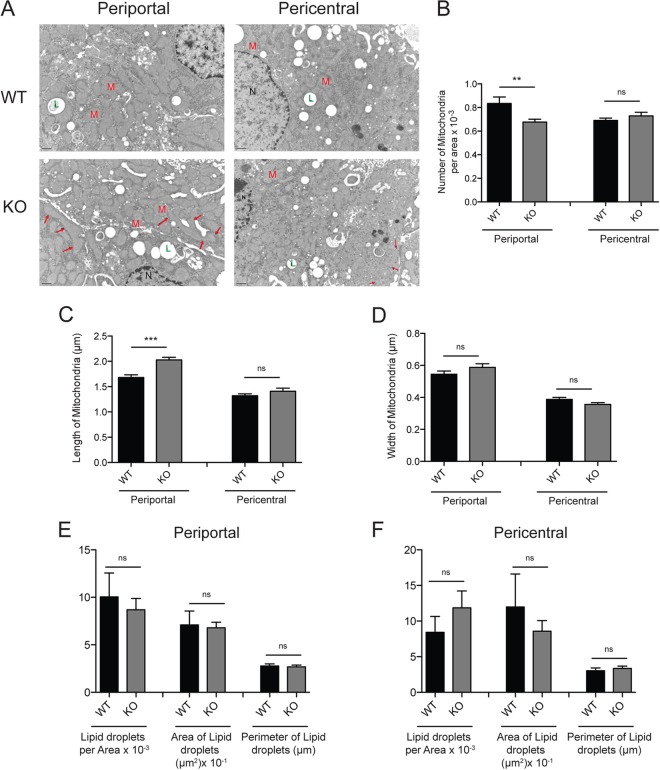
To begin to examine the consequence of SIRT4 deletion on mitochondria in liver, we evaluated the changes in hepatocyte and mitochondrial ultrastructure by electron microscopy. Two histological zones define the liver. The periportal zone is close to the entering vascular supply (periportal vein) and receives the most oxygenated blood and contains higher concentrations of substrates and hormones. Conversely, the pericentral zone has the poorest oxygenation and lower substrate concentrations. Functionally, the periportal zone is specialized for oxidative functions such as gluconeogenesis, β-oxidation of fatty acids, and cholesterol synthesis, whereas the pericentral zone is more important for glycolysis and lipogenesis (34). We observed a decrease in the number of mitochondria and an increase in their length in the periportal zone (Fig. 4A to C), whereas no changes were observed in the pericentral area (Fig. 4A to C). Changes in the mitochondrial width were not observed in either zone (Fig. 4D). Interestingly, fasting triggers similar features, i.e., mitochondrion enlargement and a decrease in mitochondrion numbers (35), suggesting that SIRT4 deletion shares similarity with the fasting-like phenotype in mitochondrial morphology in the liver. In quantifying lipid droplet number and size in our electron microscopy images, we did not observe significant differences between WT and SIRT4 KO hepatic lipid accumulation (Fig. 4E and F). Thus, our data show that SIRT4 deletion induces hepatic catabolic gene expression but has little effect on expression of mitochondrial biogenesis-associated genes and only a mild effect on mitochondrial ultrastructure.
Fig 4.
SIRT4 loss induces mitochondrion enlargement and decreases the number of mitochondria in the periportal zone of the liver. (A) Representative images of periportal and pericentral zone in SIRT4 WT and KO liver. M, mitochondria; L, lipid droplet; N, nucleus. The arrows point to elongated mitochondria. (B to D) Quantification of mitochondrial number (B), length (C), and width (D) in periportal and pericentral zones of the liver in SIRT4 WT and KO mice. (E and F) Lipid droplet quantification in periportal and pericentral zones of SIRT4 WT and SIRT4 KO mice. Data are generated from 10 images from 3 mice per genotype. **, P < 0.01; ***, P < 0.001; ns, not significant.
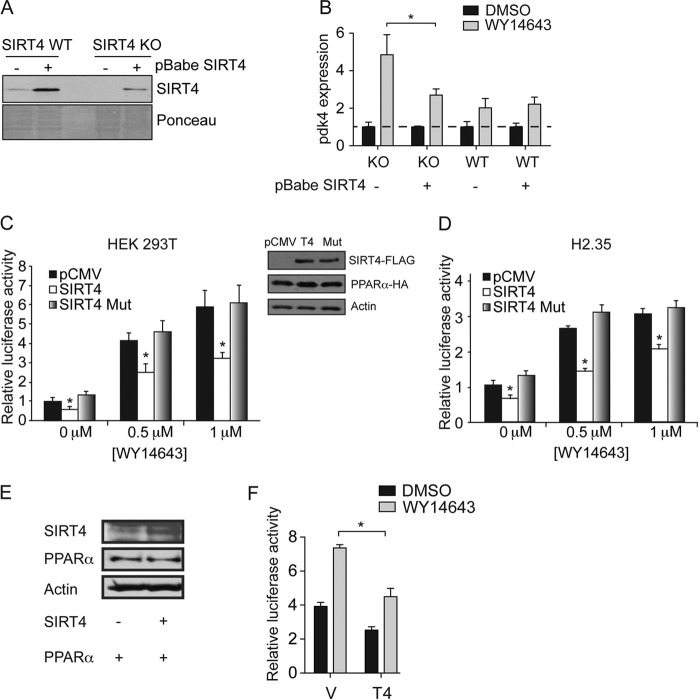
SIRT4 suppresses PPARα transcriptional activation.
Our data are consistent with the model that SIRT4 dampens fatty acid oxidation in the liver through a mechanism of repressing PPARα activity. To determine whether SIRT4 is a bona fide repressor of PPARα in a cell-autonomous manner, we examined the induction of the PPARα target gene Pdk4 in Sirt4 KO and WT mouse embryonic fibroblast (MEF) cell lines exposed to the PPARα synthetic ligand WY14643. In WT cells, activation of PPARα by WY14643 leads to a 2-fold increase in Pdk4 expression (Fig. 5A and B). In contrast, the stimulation of Pdk4 expression was more than doubled in MEFs lacking SIRT4 (Fig. 5A and B), indicating that SIRT4 negatively regulates the ability of PPARα to activate transcription of Pdk4. To test this hypothesis further, we used a retroviral vector to express SIRT4 in wild-type and SIRT4 KO MEFs (Fig. 5A and B). Reintroduction of SIRT4 into KO MEFs suppressed the induction of Pdk4 by WY14643 (Fig. 5A and B). We then examined the expression of PPARγ target genes in SIRT4 KO and WT WAT, its main organ of expression, but did not observe differences (see Fig. S2 in the supplemental material). To test further the ability of SIRT4 to regulate PPARα transcriptional activity, we monitored PPARα activity with luciferase reporter assays in cells with or without increased levels of SIRT4. Human embryonic kidney (HEK293T) cells expressing either the control vector, SIRT4, or an SIRT4 catalytic mutant variant were transfected with a luciferase reporter driven by three tandem repeats of a consensus PPAR response element (3×PPRE) together with constructs expressing PPARα, PPARα coactivator RXRα, and a control Renilla luciferase reporter (Fig. 5C). Under basal conditions, we observed a significant decrease in PPARα activity (Fig. 5C). Increased levels of wild-type SIRT4 significantly reduced ligand-induced transactivation of PPARα (Fig. 5C). In contrast, HEK293T cells that expressed the SIRT4 inactive mutant had PPARα reporter activity similar to that of cells transfected with the control plasmid (Fig. 5C). In H2.35 mouse hepatoma cells that endogenously express PPARα and RXRα, we again found repression of PPARα activity by SIRT4 under basal conditions and after treatment with WY14643 but no evidence of repression by SIRT4 mutant protein (Fig. 5D).
Fig 5.
SIRT4 suppresses PPARα activity cell autonomously. (A) Immunoblots showing SIRT4 expression in primary mouse embryonic fibroblasts (MEFs) from SIRT4 KO and SIRT4 WT mice. MEFs were stably infected with virus that contained SIRT4 (+) or a pBabe vector control (−). (B) Induction of Pdk4 expression by WY14643 is modulated by SIRT4. Primary SIRT4 KO and SIRT4 WT MEFs infected with control or SIRT4 expression virus were treated with WY14643 or with 0.5% DMSO for 24 h. Gene expression was normalized to B2m and Rps16 and normalized to the DMSO-treated conditions for each sample (dotted line). (C) SIRT4 expression decreased PPARα transcriptional activity in human embryonic kidney 293T (HEK293T) cells cotransfected with PPARα and RXRα plasmids, whereas the SIRT4 mutant did not have an effect on PPARα activity. Expression of pCMV empty vector control also showed no effect. Cells were incubated for 12 h with 0, 0.5, or 1 μM WY14643. Activity was assessed using PPARα luciferase constructs. Expression of SIRT4 and PPARα is shown in the right panel. Results are representative of two independent experiments. (D) PPARα transcriptional activity in H2.35 hepatoma cells endogenously expressing PPARα and RXRα and transfected with pCMV control (pCMV), SIRT4-FLAG (SIRT4), or mutant H125A SIRT4-FLAG (SIRT4 Mut). Cells were treated as described for panel C. (E and F) SIRT4 represses PPARα transcriptional activity. The Western blot in panel E shows expression of SIRT4-FLAG, HA-PPARα, and actin in transiently transfected MEFs. For the experiment shown in panel F, Cells were cotransfected with PPARα and SIRT4 (T4) or pCMV empty vector (V) as a control. Cells were incubated for 12 h with 1 μM WY14643. In all experiments, data are represented as means ± standard errors of the means (n = 3; *, P < 0.05).
To confirm these findings with a natural PPARα binding sequence, we analyzed a reporter construct based on the promoter of the fibroblast growth factor 21 (FGF21) gene, a PPARα target gene (11). SIRT4 reduced both FGF21 reporter and synthetic reporter activity to similar degrees (Fig. 5E and F). Taken together, our results support the hypotheses that SIRT4 acts cell-autonomously to repress PPARα activity in multiple cell types.
SIRT4 acts on PPARα by signaling via NAD+ and SIRT1.
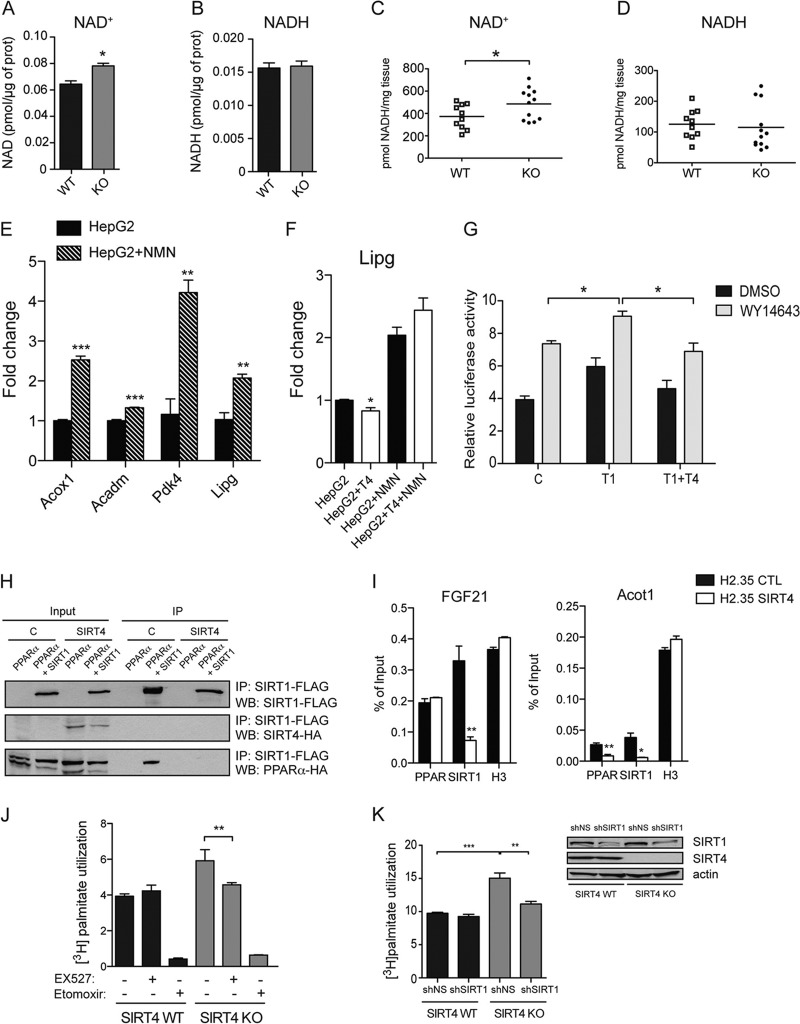
We next assessed the mechanism for repression of PPARα activity by SIRT4. Mitochondria sense environmental changes, such as redox status or energy balance, and communicate with the nucleus through retrograde signaling pathways that are poorly characterized in mammals (36). To determine whether the altered catabolism of fatty acids in SIRT4 KO cells affected the redox balance, we assessed levels of NAD+ and NADH. Consistent with a hypercatabolic phenotype, NAD+ levels were increased in SIRT4 KO fibroblasts compared to WT levels, while NADH levels were not significantly different (Fig. 6A and B). We confirmed this in livers where higher levels of NAD+ were observed in SIRT4 KO tissue than in WT tissue (Fig. 6C and D). We next assessed whether an increase in NAD+ levels could change PPARα activity by treating HepG2 cells with nicotinamide mononucleotide (NMN), a source of NAD+. After NMN treatment, PPARα target genes were increased from 1.4- to 4-fold (Fig. 6E). Importantly, treatment with NMN of HepG2 cells overexpressing SIRT4 restored the expression of PPARα target genes to their control levels (Fig. 6F). These data are consistent with the idea that SIRT4 may repress PPARα activity through a change in NAD+ levels.
Fig 6.
SIRT4 suppresses PPARα by modulating SIRT1 activity. NAD+ (A) and NADH (B) levels were measured from SIRT4 WT and KO cells. (C) NAD+ was measured in SIRT4 WT and KO mice (n = 10 to 12). Each data point represents the NAD+ concentration (pmol of NAD+/mg of tissue) of one animal. The line represents the mean NAD+ concentration. (D) NADH was measured from livers of SIRT4 KO and SIRT4 WT mice (n = 10 to 12; same mice as used for the experiment shown in panel C). Each data point represents the NADH concentration (pmol of NADH/mg of tissue) in one animal. The line represents the mean NADH concentration. (E) NAD+ increases PPARα target gene expression. HepG2 cells were treated with 1 mM NMN overnight, and PPARα target gene expression was analyzed by qPCR (n = 3). (F) Lipg expression was measured in HepG2 control and SIRT4-overexpressing cells after treatment with NMN (n = 3). (G) PPARα activation by SIRT1 is repressed by SIRT4. Cells were cotransfected with PPARα and SIRT1, SIRT4 or pCMV empty vector as a control. Cells were incubated for 12 h with 1 μM WY14643. See also Fig. S1B in the supplemental material. (H) SIRT1-PPARα complex was assessed by anti-FLAG immunoprecipitations (IP) in 293T cells with or without stable expression of SIRT4-HA transiently transfected with a PPARα-HA construct together with a FLAG control or a SIRT1-FLAG construct. Immunoprecipitates and 1/100 of starting lysate (input) were analyzed by Western blotting (WB) and probed with HA or FLAG antibody. (I) SIRT4 represses the binding of SIRT1 to the PPRE of target genes. H2.35 cells were treated with 1 μM WY14643 overnight and subjected to ChIP with PPARα and SIRT1 antibodies and analyzed by qPCR with primers flanking PPRE in the promoter of FGF21 and Acot1. ChIP with H3 antibody was used as a positive control. (J) Oxidation of [3H]palmitate (nmol of [3H]palmitate/h/mg protein) was analyzed using primary hepatocytes isolated from SIRT4 WT and KO mice. Cells were cultured in the presence or absence of 10 μM Ex527, a SIRT1 inhibitor, for 16 h in the presence or absence of 200 μM etomoxir. (K) Oxidation of [3H]palmitate (nmol of [3H]palmitate/h/mg protein) was analyzed using primary hepatocytes isolated from SIRT4 WT and KO mice treated with an shRNA control or targeting SIRT1 (n = 3). At right, immunoblots show SIRT1 and SIRT4 expression in primary hepatocytes from SIRT4 KO and SIRT4 WT mice infected by adenovirus expressing a scrambled shRNA or an shRNA targeting SIRT1. In all panels, data represent means ± standard errors of the means (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Increased NAD+ levels have been proposed to contribute to elevated enzymatic activity of SIRT1, a nuclear protein, and changing NAD+ concentrations have been shown to affect SIRT1 function (37). Additionally, in the liver, SIRT1 is recruited to PPARα response elements (PPRE) where it binds to and deacetylates the PGC-1α coactivator, resulting in increased fatty acid oxidation (11). Thus, we investigated whether SIRT1 activity could play a role in the increased fatty acid oxidation observed in SIRT4 null cells. First, we examined the impact of SIRT4 and SIRT1 together on PPARα transcriptional activity. In line with previous reports, we observed that SIRT1 activates PPARα but found that this activation was repressed by SIRT4 overexpression (Fig. 6G; see also Fig. S3 in the supplemental material), suggesting a model whereby SIRT4 represses SIRT1 to regulate PPARα activity. Next, we examined the impact of SIRT4 on the stability of the SIRT1/PPARα complex by measuring the interaction between PPARα-HA and SIRT1-FLAG in HEK293T cells constitutively overexpressing SIRT4-HA. PPARα and SIRT1 formed a stable complex in control cells (Fig. 6H), which was blocked by SIRT4 overexpression (Fig. 6H), suggesting that SIRT4 interferes with the regulation of PPARα by SIRT1.
To test this idea, we analyzed the association of SIRT1 with PPAR response elements (PPREs) using a chromatin immunoprecipitation (ChIP) assay. We observed a modest decrease in PPARα binding to PPRE of Acot1 (Fig. 6I). However, in both cases, we observed a strong decrease in SIRT1 binding to PPRE (Fig. 6I), suggesting that SIRT4 disrupts the interaction between SIRT1 and PPARα at PPREs. These results suggest that SIRT4 activity inhibits the function of PPARα through SIRT1, demonstrating a novel cross talk between mitochondrial and nuclear sirtuins.
To assess directly whether SIRT1 is required for the induction of fatty acid oxidation observed in SIRT4-deficient cells, we analyzed fatty acid oxidation in primary hepatocytes isolated from SIRT4 KO and SIRT4 WT mice following inhibition of SIRT1 activity with Ex527, a small-molecule inhibitor specific for SIRT1 (38). As expected, fatty acid oxidation rates were higher in the SIRT4 KO hepatocytes than in WT cells. However, SIRT4 KO cells treated with Ex527 exhibited fatty acid oxidation rates comparable to those observed in WT hepatocytes (Fig. 6J). Thus, SIRT1 activity is critical for the upregulation of fatty acid oxidation in SIRT4 KO cells. Finally, we used shRNA to reduce SIRT1 expression in SIRT4 KO and WT primary hepatocytes (Fig. 6K). Consistent with the pharmacological results, lowering SIRT1 expression did not influence fatty acid oxidation in WT cells, whereas SIRT1 knockdown in SIRT4 KO cells reduced the elevated fatty acid oxidation rates observed by SIRT4 loss (Fig. 6K). Taken together, our data suggest that SIRT4 activity represses hepatic fatty acid oxidation by dampening SIRT1 and PPARα activity.
DISCUSSION
In this study, we demonstrate that the mitochondrial protein SIRT4 represses PPARα transcriptional activity, suppresses the expression of PPARα transcripts, and inhibits fatty acid oxidation. In addition, we have uncovered an unexpected nuclear-mitochondrial signaling pathway involving SIRT4 and SIRT1; SIRT4 null livers exhibit increased NAD+ levels, and the increase in fatty acid oxidation requires intact activity of SIRT1, a nuclear protein. Our findings highlight a unique function for SIRT4 in the coordination of lipid metabolism regulation between mitochondria and the nucleus in the liver and demonstrate an interconnection between sirtuin proteins in the regulation of nutrient sensing signaling.
The liver is a pivotal organ in the adaptation to nutrient deprivation. Transcriptional regulation in this tissue provides a key mechanism to cope with the dynamic energy needs during fasting by generating and preserving glucose via utilization of lipids and amino acids (1). PPARα is a central regulator of the hepatic fasting response (2, 3). Activation of PPARα promotes fatty acid oxidation during fasting through increased expression of fatty acid oxidation genes such as Ctp1a, Pdk4, and Acox1 (33, 39–41). Here, we demonstrate that loss of SIRT4 increases fatty acid oxidation by increasing PPARα-dependent transcription of fatty acid catabolic genes. Although the changes observed in gene expression are modest, the complete PPARα-driven regulatory program was affected in the SIRT4 KO mouse liver. Furthermore, we showed that SIRT4 overexpression blunts PPARα activity. As SIRT4 levels are modulated in vivo during the fasting response, the regulation of SIRT4 expression or activity might provide an additional layer of regulation for PPARα activation.
Mitochondrial retrograde signaling is an important mode of communication with the nucleus and is an important means by which nutrient stresses are relayed to the nucleus, enabling cells to adjust their metabolism via appropriate transcriptional responses (36). In this study, we show that the deletion of SIRT4 leads to an increase in NAD+ levels. Notably, NAD+ is an essential cofactor for sirtuin activity, and modulation of NAD+ levels has been shown to modulate SIRT1 activity (42). Since activation of PPARα by SIRT1 is necessary for the transcriptional control of fatty acid oxidation (11), we hypothesized that increased NAD+ observed in SIRT4 KO mice promotes SIRT1-mediated activation of PPARα. Indeed, we find that SIRT1 activity is definitively required for the increase in fatty acid oxidation seen in SIRT4 null cells as pharmacological and genetic inhibition of SIRT1 blocks the increase in fatty acid oxidation observed in SIRT4 null cells. We also show that SIRT4 disrupts the interaction between SIRT1 and PPARα at PPREs. Based on our findings, we propose a model whereby increased levels of NAD+ in SIRT4 KO cells activates SIRT1, which interacts with and activates PPARα. In the long-term, it will be interesting to investigate whether SIRT4 also influences the acetylation of other SIRT1 targets in this node, such as PGC-1α (43). In sum, our work has uncovered a novel link between SIRT1 and SIRT4 and establishes SIRT4 as a part of the mitochondrial retrograde signaling pathway.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jennifer Huang and Ditte Lee for technical assistance and all other members of the Haigis lab for helpful discussions. We thank Andrea Calhoun from the Morphology Core Facility at Beth Israel Deaconess Medical Center, the Biopolymers Facility at Harvard Medical School for analyzing microarray gene expression, and Joseph Rodgers for help with primary hepatocyte isolations and the generous gift of adenovirus.
G.L. was supported by a fellowship from the Human Frontier Science Program. V.C.J.D.B. was supported by a Rubicon fellowship from the Netherlands Organization for Scientific Research. M.C.H. was supported by a Brookdale Leadership in Aging Fellowship, Ellison Medical Foundation New Scholar Award, funding from NIA R01/AG032375, and the Glenn Foundation for Medical Research.
Footnotes
Published ahead of print 16 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00087-13.
REFERENCES
- 1.Desvergne B, Michalik L, Wahli W. 2006. Transcriptional regulation of metabolism. Physiol. Rev. 86:465–514 [DOI] [PubMed] [Google Scholar]
- 2.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. 1999. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Invest. 103:1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leone TC, Weinheimer CJ, Kelly DP. 1999. A critical role for the peroxisome proliferator-activated receptor alpha (PPARα) in the cellular fasting response: the PPARα-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. U. S. A. 96:7473–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai S, Armstrong CM, Kaeberlein M, Guarente L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800 [DOI] [PubMed] [Google Scholar]
- 5.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. U. S. A. 97:5807–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. U. S. A. 97:6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanner KG, Landry J, Sternglanz R, Denu JM. 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. U. S. A. 97:14178–14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanny JC, Moazed D. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. U. S. A. 98:415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frye RA. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273:793–798 [DOI] [PubMed] [Google Scholar]
- 10.Guarente L. 2006. Sirtuins as potential targets for metabolic syndrome. Nature 444:868–874 [DOI] [PubMed] [Google Scholar]
- 11.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. 2009. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 9:327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente LP. 2006. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126:941–954 [DOI] [PubMed] [Google Scholar]
- 13.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. 2005. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16:4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwer B, North BJ, Frye RA, Ott M, Verdin E. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158:647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L. 2010. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J. Biol. Chem. 285:31995–32002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurent G, German NJ, Saha AK, de Boer VCJ, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, Lovitch SB, Sharpe AH, Kurland IJ, Steegborn C, Gygi SP, Muoio DM, Ruderman NB, Haigis C. 2013. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl-CoA decarboxylase. Mol. Cell 50:686–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. 2004. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell 119:121–135 [DOI] [PubMed] [Google Scholar]
- 18.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. 2005. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434:113–118 [DOI] [PubMed] [Google Scholar]
- 19.Li C, Hung Wong W. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:RESEARCH0032. 10.1186/gb-2001-2-8-research0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HK, Braynen W, Keshav K, Pavlidis P. 2005. ErmineJ: tool for functional analysis of gene expression data sets. BMC Bioinformatics 6:269. 10.1186/1471-2105-6-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swindell WR, Huebner M, Weber AP. 2007. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8:125. 10.1186/1471-2164-8-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swindell WR, Huebner M, Weber AP. 2007. Plastic and adaptive gene expression patterns associated with temperature stress in Arabidopsis thaliana. Heredity 99:143–150 [DOI] [PubMed] [Google Scholar]
- 23.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. 2005. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 3:e101. 10.1371/journal.pbio.0030101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. 2007. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 26:1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. 2009. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324:651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang T, Sauve AA. 2006. NAD metabolism and sirtuins: metabolic regulation of protein deacetylation in stress and toxicity. AAPS J 8:E632–E643. 10.1208/aapsj080472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakhshandehroo M, Sanderson LM, Matilainen M, Stienstra R, Carlberg C, de Groot PJ, Muller M, Kersten S. 2007. Comprehensive analysis of PPARα-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007:26839. 10.1155/2007/26839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vianna CR, Huntgeburth M, Coppari R, Choi CS, Lin J, Krauss S, Barbatelli G, Tzameli I, Kim YB, Cinti S, Shulman GI, Spiegelman BM, Lowell BB. 2006. Hypomorphic mutation of PGC-1βcauses mitochondrial dysfunction and liver insulin resistance. Cell Metab. 4:453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhahbi JM, Mote PL, Fahy GM, Spindler SR. 2005. Identification of potential caloric restriction mimetics by microarray profiling. Physiol. Genomics 23:343–350 [DOI] [PubMed] [Google Scholar]
- 30.Boylston WH, Gerstner A, DeFord JH, Madsen M, Flurkey K, Harrison DE, Papaconstantinou J. 2004. Altered cholesterologenic and lipogenic transcriptional profile in livers of aging Snell dwarf (Pit1dw/dwJ) mice. Aging Cell 3:283–296 [DOI] [PubMed] [Google Scholar]
- 31.Boylston WH, DeFord JH, Papaconstantinou J. 2006. Identification of longevity-associated genes in long-lived Snell and Ames dwarf mice. Age 28:125–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schumacher B, van der Pluijm I, Moorhouse MJ, Kosteas T, Robinson AR, Suh Y, Breit TM, van Steeg H, Niedernhofer LJ, van Ijcken W, Bartke A, Spindler SR, Hoeijmakers JH, van der Horst GT, Garinis GA. 2008. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet 4:e1000161. 10.1371/journal.pgen.1000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandard S, Müller M, Kersten S. 2004. Peroxisome proliferator-activated receptor alpha target genes. Cell. Mol. Life Sci. 61:393–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bacon BR, O'Grady JG, Di Bisceglie AM, Lake JR. 2006. Comprehensive clinical hepatology. Elsevier Health Sciences, Philadelphia, PA [Google Scholar]
- 35.Salas M, Tuchweber B, Kourounakis P. 1980. Liver ultrastructure during acute stress. Pathol. Res. Pract. 167:217–233 [DOI] [PubMed] [Google Scholar]
- 36.Finley LW, Haigis MC. 2009. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res. Rev. 8:173–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imai S. 2010. “Clocks” in the NAD world: NAD as a metabolic oscillator for the regulation of metabolism and aging. Biochim. Biophys. Acta 1804:1584–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. 2006. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell. Biol. 26:28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brandt JM, Djouadi F, Kelly DP. 1998. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 273:23786–23792 [DOI] [PubMed] [Google Scholar]
- 40.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. 1992. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell 68:879–887 [DOI] [PubMed] [Google Scholar]
- 41.Tugwood JD, Issemann I, Anderson RG, Bundell KR, McPheat WL, Green S. 1992. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 11:433–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. 2010. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 31:194–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lefebvre P, Chinetti G, Fruchart JC, Staels B. 2006. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J. Clin. Invest. 116:571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.