Abstract
Lower eukaryotes like the kinetoplastid parasites are good models to study evolution of cellular pathways during steps to eukaryogenesis. In this study, a kinetoplastid parasite, Leishmania donovani, was used to understand the process of mitochondrial translocation of a nucleus-encoded mitochondrial protein, the mitochondrial tryparedoxin peroxidase (mTXNPx). We report the presence of an N-terminal cleavable mitochondrial targeting signal (MTS) validated through deletion and grafting experiments. We also establish a novel finding of calmodulin (CaM) binding to the MTS of mTXNPx through specific residues. Mutation of CaM binding residues, keeping intact the residues involved in mitochondrial targeting and biochemical inhibition of CaM activity both in vitro and in vivo, prevented mitochondrial translocation. Through reconstituted import assays, we demonstrate obstruction of mitochondrial translocation either in the absence of CaM or Ca2+ or in the presence of CaM inhibitors. We also demonstrate the prevention of temperature-driven mTXNPx aggregation in the presence of CaM. These findings establish the idea that CaM is required for the transport of the protein to mitochondria through maintenance of translocation competence posttranslation.
INTRODUCTION
Subcellular mislocalization of proteins results in pathogenesis of many diseases, and selective errors in targeting might be an attractive alternative to therapeutic intervention (1). Therefore, the understanding of regulatory mechanisms of protein targeting is crucial for comprehending disease pathogenesis. It is interesting to study early-branching organisms in eukaryotic evolution because they can illuminate how more complex regulatory systems in higher eukaryotes developed. In addition to being model systems for evolutionary studies, lower eukaryotes can also serve as systems that can be manipulated more easily than higher eukaryotes if similarities in their structural-functional organizations are established (2, 3). Parasites of the order Kinetoplastida, trypanosomatids are the most ancient eukaryotic lineage to have a mitochondrion (4) and show mitochondrial biochemistry representing an intermediary evolutionary stage reminiscent of that of the last eukaryotic common ancestor (5). Formation of mitochondria in eukaryotic cells was the result of engulfment of an ancestor of present-day alphaproteobacteria, providing the host with new metabolic pathways, such as aerobic respiration (6, 7). The events in establishment of a symbiotic relationship between the host and the bacterium catalyzed evolution of the eukaryotic way of life (8, 9). Most mitochondrial proteins retained from the original eubacterial endosymbiont came to be encoded by the nuclear genome during evolution and needed to be transported to the mitochondria after translation on the ribosomes (10). Many mitochondrial proteins, especially the inner membrane and matrix proteins, normally contain a transient cleavable N-terminal extension of about 20 to 50 amino acids functioning as a mitochondrial targeting signal (MTS) (11). Although significant data on the nature of these targeting signals and their import into the mitochondria exist for higher eukaryotes (12, 13), studies in lower eukaryotes are necessary for understanding their evolution. MTSs from different taxa do not show much sequence identity but are rich in positively charged, hydroxylated, and hydrophobic residues with a potential to form an amphiphilic α-helix (14, 15).
Mitochondrial import of proteins is a multistep process involving transport of the protein from the ribosomes through the cytosol to the mitochondria and subsequently localization to different destinations within the mitochondria. Leishmania donovani, a parasite causing the potentially fatal disease visceral leishmaniasis, belongs to the trypanosomatid group of parasites. Detailed studies on RNA import into the mitochondria are available for Leishmania (16, 17); however, the mechanisms of protein import remain poorly understood. Studies on the different candidate proteins of the monophyletic group Euglenozoa, to which the trypanosomatids belong, show that mitochondrial precursors contain 5- to 110-amino-acid-long cleaved targeting sequences (5). Further, other studies indicate that trypanosomatids contain both canonical (18, 19) and noncanonical (20) presequences. Recent reports indicate a functional conservation of mitochondrial protein import in the course of evolution by experiments showing Leishmania tarentolae marker proteins being efficiently imported into yeast mitochondria and protist marker proteins being successfully imported into isolated L. tarentolae mitochondria (21). Although the transport of proteins from the cytosol to the mitochondria has not been adequately explored in lower eukaryotes, the general paradigm is that the HSP70 and HSP90 family of proteins bind as chaperones to maintain translocation competence in higher eukaryotes (22). However, experimental studies actually demonstrating the role of MTS in transport and possible mechanisms involved in the transport in lower eukaryotes are scarce.
This study provides evidence of a 30-amino-acid stretch at the N terminus of mitochondrial tryparedoxin peroxidase (mTXNPx) serving as a signal peptide responsible for targeting the protein to the Leishmania mitochondria. Interestingly, the designated MTS contains a calmodulin (CaM) binding site, CaM being primarily a Ca2+-sensing signaling moiety in the cell (23, 24). We report a novel observation with the help of mutagenesis and other studies, an interaction between MTS of mTXNPx and CaM. We show that interference with CaM binding prevents the translocation of mTXNPx to the mitochondria both in vitro and in vivo, suggesting a crucial role of CaM in translocation of a mitochondrial protein. Based on the aggregation and unfolding assays, it can be hypothesized that CaM is responsible for maintaining the translocation competence of mTXNPx in Leishmania.
MATERIALS AND METHODS
Ethics statement.
All procedures in this study were carried out per the guidelines laid down by the Institutional Biosafety Committee via permit no. IBSC#12508.
Strains and plasmids.
Promastigotes of Leishmania donovani were used for the study. For transformation of ligation products, Escherichia coli DH5α cells were used, and for protein expression, E. coli BL21(DE3) was used. The vectors for expression of the recombinant protein in bacteria and for Leishmania spp. were pET-28a(+) and pXG-GFP+, respectively. The genes intended for bacterial expression with N-terminal His tag were cloned in BamHI and NotI sites, while those with a C-terminal His tag were cloned in NcoI and NotI sites. For overexpression in Leishmania, BamHI and EcoRV sites were used. The primer sequences were designed based on the sequence data in GenBank with the following accession numbers: mTXNPx, EF507797.1; mTXN, FJ824132.1; CaM, EU490524.1. The appropriate enzyme sites were added at the 5′ end of the primers. The required mutations were introduced through PCR mutagenesis to alanine, using the overlap extension method, replacing the respective codons with GCC or GCG sequences during the primer design (25). Detailed primer sequences can be provided upon request. The recombinant proteins were purified using Ni2+-nitrilotriacetic acid (NTA) affinity chromatography. Transfection of promastigotes with the pXG-GFP+ vector containing mTXNPx and its mutants was carried out using the high-voltage protocol, with a few modifications (26). Briefly, promastigotes (5 × 107) in mid-log phase were transfected in Cytomix buffer (120 mM KCl, 0.15 mM CaCl2, 10 mM KH2PO4, 25 mM HEPES, 2 mM EDTA, 2 mM MgCl2, pH 7.6) with 25 μg of the plasmid DNA and electroporated at 1,500 V and 25 mF with a second pulse given after a gap of 10 s. After 10 min of incubation on ice, cells were incubated at 23°C for 24 h in M199 (Sigma) with 10% fetal calf serum (FCS) (South American origin; Gibco), following which G418 was added and clones were selected with progressively increasing G418 levels to a final concentration of 100 μg ml−1. Metacyclic promastigotes were separated using peanut agglutinin (PNA)-dependent aggregation of procyclic promastigotes. Briefly, cells were washed with phosphate-buffered saline (PBS) and incubated with 100 μg PNA per 107 cells per ml at room temperature for 1 h. The cells were spun at 100 × g for 3 min to pellet out procyclic promastigotes. Metacyclic promastigotes from the supernatant were then pelleted at 2,300 rpm, washed with PBS, and used for further experimentation.
Reagents and cell treatments.
All chemicals were from Sigma, unless otherwise indicated. Reverse transcription was carried out using the SuperScript II system (Invitrogen) per the protocol mentioned in the material sheet. Taq polymerase (NEB) was used for all the PCRs. Cells treated for the required duration with trifluoroperazine dihydrochloride (TFP), N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide hydrochloride (W7), and calmidazolium chloride (CZ), well-characterized inhibitors of CaM activity (27), were washed posttreatment and analyzed by different methods such as fluorometry, flow cytometry, or Western blotting.
Cellular staining and confocal microscopy.
For estimation of cell viability on the basis of membrane permeabilization, propidium iodide was used at a concentration of 2 mg ml−1 and nuclear staining was measured by fluorescence-activated cell sorting (FACS). Mitotracker Deep Red and Hoechst 33342 were used to stain mitochondria and nucleus, respectively. All of the above-mentioned probes were obtained from Molecular Probes. Cells were stained with respective dyes in M199 with 10% FCS for 10 min at 23°C, followed by two washes of plain medium. For confocal microscopy, cells were mounted on poly-l-lysine-coated slides and imaged using a Leica SPII 5 microscopy system. Images were acquired using a 63× PlanApo oil objective, in point averaging mode at zoom value 7, with system-optimized gain and offset values (nearly equal for all sets of images). Colocalization scatter plots were generated using the LAF software, which indicated almost similar dynamic ranges of gray levels along the two channels. Bleed-through was avoided due to acousto-optical beam splitter (AOBS)-assisted point-based sequential scanning. Colocalization was calculated only after removal of basal noise, and only incidents with Pearson coefficients of above 0.7 were considered true.
CaM-Sepharose affinity precipitation and IP.
Recombinant protein samples (10 μg) were loaded into 50 μl CaM-Sepharose solution preequilibrated with 50 M Tris-HCl (pH 7.5) containing 0.5 M NaCl, 2 mM CaCl2, and 0.05% Triton X-100. After 12 h of incubation at 4°C, using an end-to-end rotor, the Sepharose beads were pelleted by centrifugation and the pellet was washed three times with the same buffer prior to resuspension in 50 μl Laemmli buffer. The samples were prepared by boiling at 99°C for 10 min and analyzed by being run on a 12% SDS-PAGE gel. CaM was immunodepleted using 2 μg anti-CaM antibody (Novus) with the help of the protein G immunoprecipitation (IP) kit (Sigma) according to the protocol provided in the technical bulletin of the manufacturer (Sigma). Immunodepletions of HSP70 and HSP83 were carried out using rabbit sera raised against respective Leishmania proteins at a 1:50 ratio in cytosolic fractions. Antibody complexes were separated using protein A magnetic beads (Cell Signaling Tech) using the prescribed protocol. Coimmunoprecipitations (co-IPs) of green fluorescent protein (GFP)-tagged proteins overexpressed in Leishmania were carried out using camel GFP-trap agarose beads (Chromotek) according to the protocol suggested in the technical sheet (Chromotek). Protein pulldowns were carried out using the Sigma protein G immunoprecipitation kit protocol with minor modifications. The recombinant proteins were partially heat denatured and then incubated with the cytosolic fraction in IP buffer (Sigma) for 4 h at 4°C following addition of the anti-His tag antibody (Becton, Dickinson) and protein G beads. After 1 h of incubation of the complete mixture, beads were washed and analyzed by SDS-PAGE and Western blotting. Silver staining of SDS-PAGE gels was carried out by a modified Vorums method (28).
Flow cytometry and spectrophotometry.
For detecting GFP expression in cells, measuring cellular viability, or determining Δψm, a BD-Calibur flow cytometer (Becton, Dickinson) equipped with a 15-mV, 488-nm air-cooled argon ion laser was used. Analyses were performed on 10,000 gated events, and numeric data were processed using FlowJo software. Steady-state fluorescence spectroscopy was carried out in a Cary Eclipse fluorescence spectrophotometer (Varian Inc.). Binding of 4,4′-Bis (1-anilinonaphthalene 8-sulfonate) (bis-ANS) was studied by excitation of the dye at 385 nm, and the relative emission spectra were recorded from 400 to 550 nm with a slit width set at 5 nm. An average of 3 scans was calculated for each temperature and reading. Kinetic absorbance readings at 600 nm for aggregation assays were performed using a UV-2450 spectrophotometer (Shimadzu) equipped with a temperature-controlled cuvette holder. The scattering readouts were carried out at 1-min intervals with an averaging of 10 scans per readout in 4-mm path-length cuvettes in 50 mM phosphate buffer (pH 8.0) with an ionic strength of 300 mM.
Preparation of cell lysates, SDS-PAGE, and Western and far-Western blotting.
Cells were lysed in Laemmli buffer (20 mM Tris-HCl at pH 7.4, 5 mM EDTA, 150 mM NaCl, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 1 mM aprotinin, and 1 M leupeptin), and protein content was determined using the CBX protein assay kit (G-Biosciences, St. Louis, MO). SDS-PAGE and Western blot assays were carried out as described previously (29, 30). Transblotted proteins were probed with the mouse anti-GFP antibody (Santa Cruz) at 1:15,000, rabbit anti-β-tubulin (Neomarkers) at 1:10,000, rabbit anti-mTXNPx at 1:500, rabbit anti-MTS at 1:1,000, rabbit anti-cTXNPx at 1:10,000, rabbit anti-HSP70/83 at 1:5,000, and mouse-anti CaM (Calbiochem) at 1:5,000 dilutions in phosphate-buffered saline containing 0.1% Tween 20. Goat anti-rabbit and anti-mouse horseradish peroxidase-conjugated immunoglobulin G secondary antibody (1:10,000) was used to detect reactivity of the blots to the primary antibody with enhanced chemiluminescence (ECL) using the FemtoLucent ECL kit (G-Biosciences). Recombinant proteins (0.5 μg) were loaded onto the SDS-PAGE gels and transblotted onto a nitrocellulose membrane for far-Western analysis. Biotinylated CaM (1:600) was used as the primary antibody, while streptavidin-horseradish peroxidase (HRP) (1:10,000) was used as secondary antibody for detection of the CaM bound to the recombinant proteins on the blot. Densitometric measurements for quantitation of signals on immunoblots were performed using GeneTools software (Syngene). β-Tubulin was used as a loading control wherever required.
Isolation of mitochondria and cytosol from Leishmania donovani and in vitro import assay.
Mitochondrial and cytosolic fractions were isolated from L. donovani promastigotes by using the Qproteome mitochondrial isolation kit (Qiagen) according to the protocol provided in the kit handbook. Fresh isolation of mitochondria was preferred for each experiment; however, the isolated mitochondria were stored in mitochondrial storage buffer (Qiagen) at −70°C for other analyses such as SDS-PAGE and Western blotting. C-terminally His-tagged recombinant mTXNPx precursor was used for the assays of in vitro import. A typical import assay reaction mixture consisted of precursor recombinant protein incubated along with mitochondria isolated from L. donovani in the presence of import buffer (10 mM HEPES-KOH, pH 7.4, 80 mM KCl, 5 mM MgCl2, 2 mM KH2PO4, 5 mM l-methionine, 10 mM creatine phosphate, 0.1 mg/ml creatine kinase, 2 mM NADH, 5 mM ATP, 250 mM sorbitol, 3% [wt/vol] bovine serum albumin [BSA]) at 25°C for 60 min with intermittent mixing by tapping. The reactions were stopped by adding 3 μg valinomycin. Half the reaction mixture was treated with proteinase K (50 μg/ml) for 30 min on ice to remove nonimported precursor protein, while the other half was left untreated. In order to demonstrate that the processed protein was in principle protease sensitive, 0.5% Triton X-100 was added during the digestion by proteinase K. For denaturing analysis, Laemmli buffer was added to the reaction mixtures and the proteins were separated by 15% SDS-PAGE. C-terminally His-tagged proteins were detected by anti-His mouse monoclonal antibody (1:7,500) and secondary anti-mouse IgG antibody with HRP conjugate using the ECL method (FemtoLucent kit; G-Biosciences).
Statistical analysis.
Unless mentioned in the figure legends, at least three independent repeats were performed for all the experiments. P values were calculated (by t test and one-way analysis of variance [ANOVA] as required), and graphs were plotted using SigmaPlot (Systat Software Inc.) and Prism (GraphPad Software, Inc.).
RESULTS
An N-terminal signal peptide is responsible for the mitochondrial localization of mTXNPx.
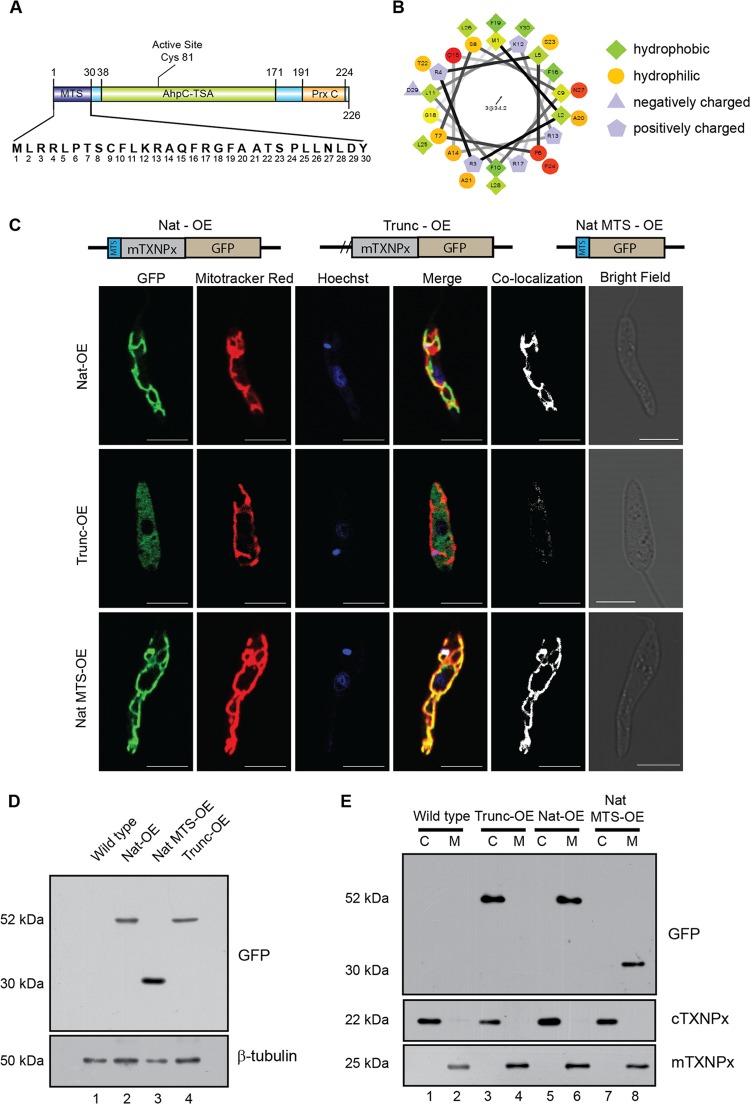
Our earlier studies and others have shown that cytosolic tryparedoxin peroxidase, a trypanothione-dependent redox enzyme, is critical for the survival of Leishmania spp. (26, 31, 32). The mitochondrial counterpart of the cytosolic form, the mitochondrial tryparedoxin peroxidase (mTXNPx; accession no. ABP68406), showed an extra stretch of a 30-amino-acid-long peptide at the N terminus that was identified as the putative mitochondrial targeting signal (MTS) (Fig. 1A) by bioinformatic analysis. Helical wheel plot analysis demonstrated the prevalence of hydrophobic amino acids of about 53% in the MTS, and predictions showed that the region of 4 to 22 amino acids had high α-helix-forming propensity (Fig. 1B) that was comparatively much greater than that of other trypanosomatids. The MTS contained an MLRR sequence found in other trypanosomatids, like Leishmania major, Trypanosoma brucei, and Trypanosoma cruzi, which is speculated to be responsible for recognition by the mitochondrial outer membrane import apparatus (5).
Fig 1.
The N-terminal signal peptide is responsible for the mitochondrial localization of mTXNPx. (A) A cartoon of mTXNPx showing various domains as predicted by PROSITE and Pfam servers. Residues 1 to 30 constitute the predicted MTS. (B) Helical wheel plot of the first 30 amino acids of mTXNPx showing preponderance of positively charged amino acids. (C) Parasites were stained with Mitotracker Red, a mitochondrion-specific dye, and Hoechst 33342, a nuclear and kinetoplast stain, showing native mTXNPx-GFP fusion protein (Nat-OE) and the GFP fusion protein with MTS (Nat MTS-OE) localized to the mitochondria, whereas the MTS-deleted truncated mTXNPx (Trunc-OE) localizes to the cytosol. Mask of colocalization is shown in white. Bars, 5 μm. (D) Lysates of transfectants overexpressing Nat-OE and Trunc-OE probed with anti-GFP antibody show the expression of the recombinant protein compared to GFP only. β-Tubulin served as a loading control. (E) Fractionation profile of wild-type and Nat-OE- and Trunc-OE-expressing cells show recombinant Trunc-OE in the cytoplasm (C), while Nat-OE and MTS-GFP localize to the mitochondria (M). The same blot was probed with anti-cTXNPx and anti-mTXNPx serum as shown below, indicating the identity of the cytosolic and mitochondrial fractions, respectively.
Further analysis by ClustalW multiple sequence alignment (33) of the mTXNPx showed significant similarity with respect to peroxiredoxins of human, mouse, Drosophila, and Trypanosoma species, with all sequences showing the presence of MTS (data not shown). To validate if this region actually serves as an MTS in vivo, mTXNPx constructs expressing a C-terminal GFP with (Trunc-OE) or without (Nat-OE) a 30-amino-acid deletion from the N terminus were episomally overexpressed in Leishmania promastigotes (Fig. 1C). Transfectants showed high GFP expression confirming episomal overexpression, which was further established by detecting the presence of the fusion mRNA through cDNA amplification (data available on request). The transfectants were analyzed for mitochondrial localization of the episomally expressed GFP fusion protein through colocalization using the mitochondrion-specific dye Mitotracker Red. Transfectants overexpressing the native mTXNPx-GFP fusion protein (Nat-OE) showed clear colocalization with Mitotracker Red on the mitochondria (Fig. 1C, Nat-OE), whereas the truncated recombinant protein (Trunc-OE) demonstrated predominant localization within the cytoplasm (Fig. 1C, Trunc-OE). This observation clearly demonstrated the functional role of MTS in translocation of the mTXNPx to the mitochondria. Western blot analysis of transfected cell lysates showed both Nat-OE and Trunc-OE as higher-molecular-mass proteins than GFP only (Fig. 1D). Similar molecular masses of Nat-OE and Trunc-OE showed that cleavage of MTS of the Nat-OE protein occurred upon localization to the mitochondria, resulting in the acquisition of similar size to the Trunc-OE protein. This corroborates other observations of MTS cleavage upon mitochondrial translocation in many organisms (18, 19). Analysis of subcellular fractions of mitochondria and cytosol probed with anti-GFP antibody on Western blots showed complete cytosolic localization of the Trunc-OE (Fig. 1E, lane 3) compared to the mitochondrial localization of the Nat-OE protein (Fig. 1E, lane 6). The competency of the MTS for mitochondrial translocation was established by overexpressing a fusion protein of N-terminal MTS sequence only tagged to GFP that colocalized with Mitotracker Deep Red in the mitochondria (Fig. 1C, Nat MTS-OE). The above data clearly demonstrated that the MTS of mTXNPx was functionally competent to translocate proteins to the mitochondria.
The MTS encodes a CaM binding site, and inhibition of CaM activity prevents translocation.
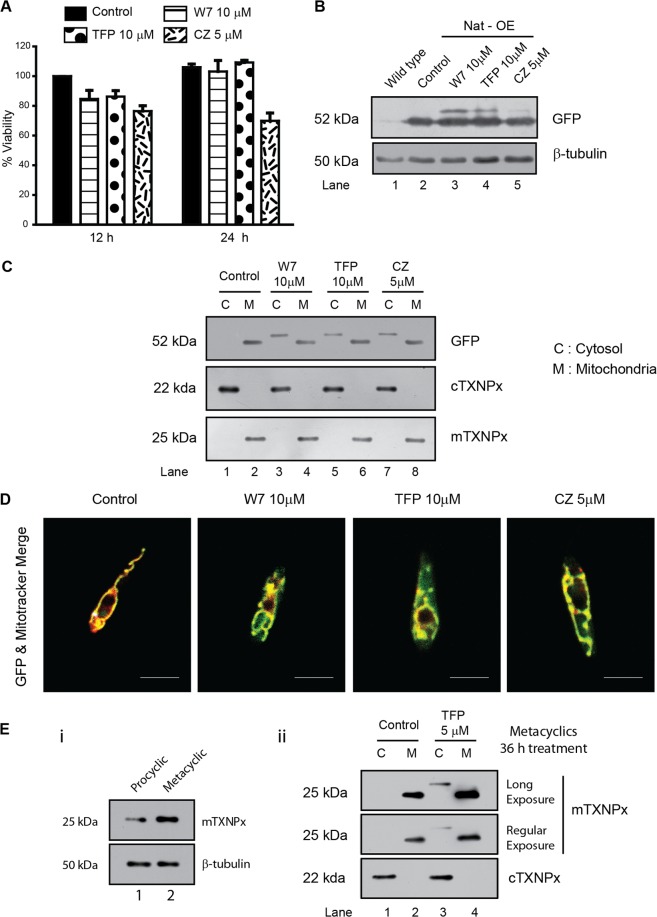
Bioinformatic analysis showed that the MTS of mTXNPx contained a putative CaM binding site (residues 8 to 22, SCFLKRAQFRGFAAT). This observation generated the hypothesis that CaM binding to the MTS could be essential for the targeting of mTXNPx to the mitochondria. If CaM did have a role in targeting of the protein, arguably, inhibition of CaM activity would prevent translocation. To prove this, functional inhibition of CaM binding to the mitochondria was achieved through the use of CaM inhibitors in cells overexpressing the native protein (Nat-OE). Several doses of CaM inhibitors (W7, TFP, and CZ) and several incubation times were tested for their toxicity by viability assay (Fig. 2A). The selected doses of 5 to 10 μM CaM inhibitors and treatment time of 12 h were used for further studies. All three inhibitors were partially able to prevent mTXNPx translocation as shown by the presence of a high-molecular-mass protein band representing the precursor protein (Fig. 2B, lanes 3 to 5). Among all inhibitors, CZ was least effective in preventing translocation, as the bulk of the protein was cleaved and the high-molecular-mass form was less (Fig. 2B, lane 5). Analysis of subcellular fractions of these cells showed a higher-molecular-mass unprocessed band in the cytosolic fraction except in untreated cells (Fig. 2C, lanes 3, 5, and 7). Microscopic colocalization showed the protein perfectly colocalized at the mitochondria of untreated cells, but GFP label was visible mostly in the cytoplasm of the treated cells (Fig. 2D), iterating that CaM inhibition prevented transport of mTXNPx to the mitochondria in vivo. The CaM inhibitors apparently do not seem to affect the levels of endogenous mTXNPx in the above-mentioned studies. This may be due to the slow turnover of mTXNPx, and hence, the precursor form may not be detectable at 12 h (Fig. 2C). Since expression of mTXNPx increases in the metacyclic promastigotes (Fig. 2E, i), we used 3- to 4-day-old culture for the treatments followed by purification of metacyclic promastigotes from the entire treated population. Treatment with 5 μM TFP, however, did not affect either viability or mitochondrial potential, and we were able to detect the presence of the untargeted form in the endogenous mTXNPx in wild-type cells treated with TFP as against control (Fig. 2E, ii). This indicated that the endogenous protein, too, is sensitive to CaM inhibition and reiterates the possible role of CaM in the transport of mTXNPx.
Fig 2.
CaM inhibition in vivo leads to block in transport of mTXNPx to mitochondria. (A) Viability of Nat-OE cells treated with CaM inhibitors such as W7, TFP, and CZ at 12 and 24 h. Note no major change in cell viability except with CZ at 24 h. (B) Western blots of lysates of the wild-type and Nat-OE cells treated with CaM inhibitors (W7, TFP, and CZ) for 12 h showing the presence of higher-molecular-mass mTXNPx isoform. (C) Cytoplasmic fractions of CaM inhibitor-treated (12 h) Nat-OE cells show the presence of a higher-molecular-mass isoform in lanes C. In controls, the protein predominantly localizes to the mitochondria (M). The same blots reprobed with anti-cTXNPx and anti-mTXNPx serum confirm cytosolic and mitochondrial fractions, respectively. (D) Confocal microscopic analysis of colocalization of Mitotracker Deep Red and GFP-mTXNPx after treatment with CaM inhibitors for 12 h showing protein in the treated cytoplasm compared to control. Bars, 5 μm. (E) (i) Metacyclic promastigotes express higher levels of endogenous mTXNPx than do procyclic promastigotes. (ii) Subcellular fractionation of wild-type metacyclic promastigotes isolated from 36-h TFP treatment shows unprocessed band in the metacyclic cytoplasm, indicating the sensitivity of endogenous mTXNPx transport to CaM inhibition. Blots with β-tubulin are loading controls.
CaM binding is disrupted when specific residues on the MTS are modified.
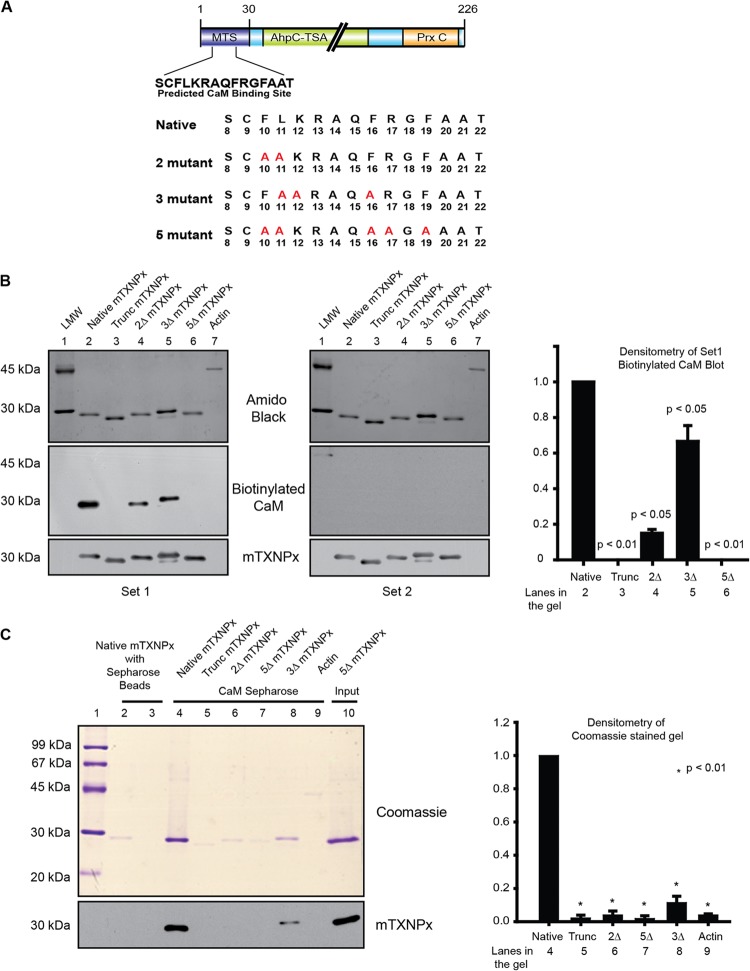
To prove the working hypothesis that CaM binding may be essential for translocation of the protein, we sought to identify the residues important for binding of CaM through mutagenesis studies. Because the predicted CaM binding site overlapped with the mitochondrial targeting site as predicted by online server iPSORT (http://hc.ims.u-tokyo.ac.jp/iPSORT/) (Fig. 3A), mutants needed to be designed in such a way that only the binding to CaM and not the targeting to mitochondria was interfered with. Accordingly, specific residues of importance to CaM binding but dispensable for mitochondrial targeting were chosen for mutagenesis by analysis through iPSORT and the CaM target database. Based on these analyses, the following mutant constructs of mTXNPx were made: 2ΔmTXNPx (replacement of residues 10 and 11), 3ΔmTXNPx (replacement of residues 11, 12, and 16), and 5ΔmTXNPx (replacement of residues 10, 11, 16, 17, and 19). The residues (Fig. 3A) were mutated to alanine by PCR mutagenesis and cloned into a bacterial expression system, and the expressed recombinant native and mutant proteins were used for enzyme assays and other studies.
Fig 3.
Calmodulin binding is disrupted when specific residues on the MTS are modified. (A) Cartoon of mTXNPx showing the location of the MTS where residues 8 to 22 denote the predicted CaM binding site. The residues mutated to alanine to generate the required mutants of CaM binding are shown in red. (B) Far-Western blots show in vitro binding of biotinylated CaM to denatured recombinant native as well as mutant proteins. The top blot is the protein stained with 0.1% amido black, indicating equal loading. The middle blot shows reactivity of biotinylated CaM to streptavidin-HRP (densitometry plot on the right). The blot on the right was processed for biotinylated CaM binding in buffers containing Ca2+ chelators such as EDTA and EGTA, showing abrogation of signal. (C) in vitro binding of CaM-Sepharose beads to native recombinant proteins in solution shown on SDS-PAGE. Native mTXNPx incubated with only Sepharose beads served as controls (lanes 2 and 3). Lanes 4 to 9 show native and mutant proteins incubated with CaM-Sepharose beads. The last lane (Input) indicates the protein loaded on the beads. The identity of the protein was confirmed with immunoblotting using polyclonal mTXNPx antibody as shown below.
Subsequently, the expressed mutant proteins and the native protein were analyzed for their ability to bind to CaM by binding of biotinylated CaM to native and mutant mTXNPxs attached to solid support on far-Western blots and to CaM-Sepharose affinity columns. Biotinylated CaM was able to bind to native mTXNPx (Fig. 3B, set 1, lane 2) but bound with lesser affinity to 2Δ, 3Δ, 5Δ, and Trunc mTXNPx (Fig. 3B, set 1, lanes 3 to 6). These data were normalized against mTXNPx intensity on Western blots, and data for set 1 are represented in the bar graph in Fig. 3B. This part of the data indicated that CaM interacts with mTXNPx using the predicted binding sites on the N terminus of the mTXNPx. Actin is known not to interact with CaM and hence served as a negative control. Absence of signal with actin was taken as specificity (Fig. 3B, set 1, lane 7). Since CaM binding to any protein is dependent on retention of CaM structure in the presence of Ca2+, abrogation of signal on processing of the blots in the presence of Ca2+ chelators like EDTA and EGTA (Fig. 3B, set 2) indicated CaM binding to mTXNPx to be Ca2+ reliant and CaM conformation dependent. To further verify the results obtained by the far-Western experiments, CaM-Sepharose affinity precipitation with the recombinant proteins was used. Recombinant proteins were incubated with the CaM-Sepharose beads where Sepharose beads served only as controls. CaM-Sepharose beads were able to pull down the native mTXNPx (Fig. 3C, lane 4), but the pulldowns with 2Δ, 3Δ, 5Δ, and Trunc mTXNPx and actin were significantly less (Fig. 3C, lanes 5 to 9). In summary, the above data established that modification of CaM binding residues on the MTS of mTXNPx interfered with binding of CaM to the MTS.
Mutations in the CaM binding site result in interference with mitochondrial targeting in vivo.
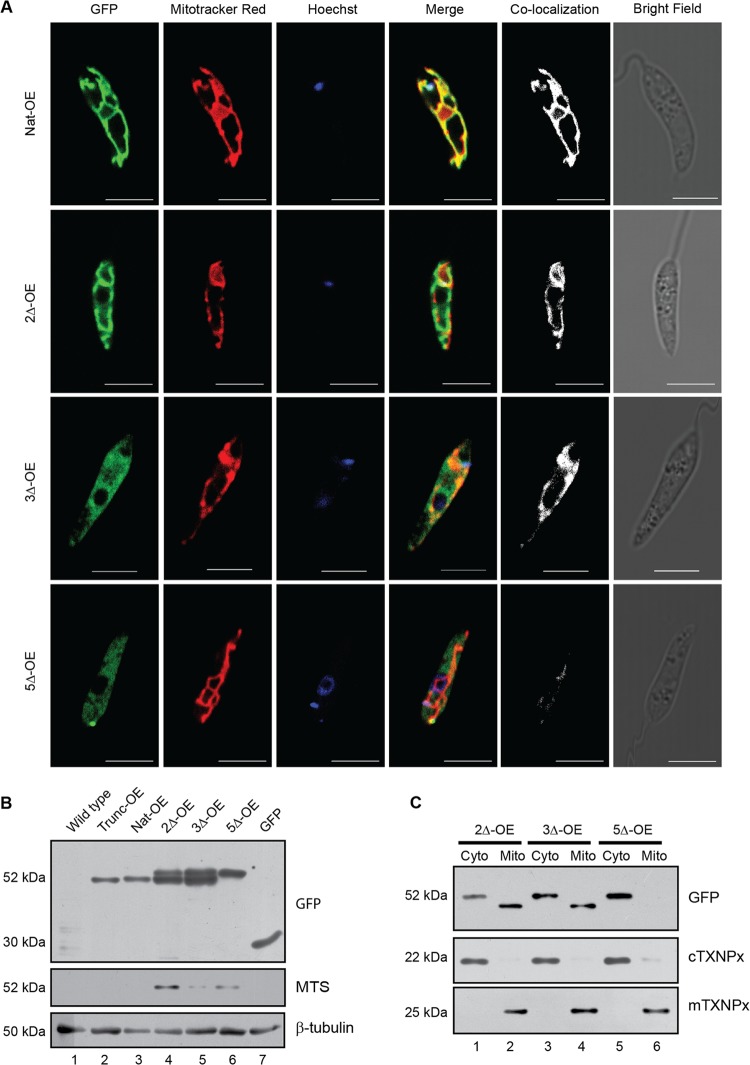
Having established in vitro that CaM interacts with mTXNPx, the biological significance of this interaction was studied in vitro. Leishmania promastigotes were transfected with native and mutant mTXNPx-pXG-GFP+ constructs so as to overexpress the recombinant proteins in fusion with C-terminal GFP. High expression of GFP confirmed protein overexpression. Further validation of overexpression was carried out by fusion mRNA detection showing that the constructs transfected were being transcribed (data available on request). Subsequent visualization of localization of the overexpressed proteins showed colocalization of the native protein (Nat-OE) expressing GFP and Mitotracker Red, a mitochondrion-specific dye (Fig. 4A), showing mitochondrial localization of the expressed protein. This colocalization pattern decreased in transfectants overexpressing 2Δ mTXNPx (2Δ-OE) and 3Δ mTXNPx (3Δ-OE) (Fig. 4A). Transfectants overexpressing 5Δ mTXNPx (5Δ-OE) showed a predominant localization of the protein to the cytosol. These observations clearly indicated that replacement of these 5 amino acids in the CaM binding site did interfere with transport to the mitochondria (Fig. 4A) while replacement of either 2 or 3 amino acids resulted in translocation with decreased efficiency.
Fig 4.
Mutations result in interference of mitochondrial targeting. (A) Parasites stained with Mitotracker Red, a mitochondrion-specific dye, and Hoechst 33342, a nuclear and kinetoplast stain, respectively, show a decrease in protein localization in transfectants overexpressing 2Δ mTXNPx (2Δ-OE) and 3Δ mTXNPx (3Δ-OE). Transfectants overexpressing 5Δ mTXNPx (5Δ-OE) show no significant colocalization between the GFP-labeled recombinant protein and Mitotracker Red. Bars, 5 μm. (B) Western blots of parasite cell lysates episomally overexpressing native as well as the CaM binding mutants of mTXNPx show high- as well as low-molecular-mass isoforms in 2Δ-OE and 3Δ-OE cells. 5Δ-OE cells show only the high-molecular-mass band while native full-length (Nat-OE) and truncated mTXNPx (Trunc-OE) show the processed band. The last lane shows the molecular mass of only GFP. β-Tubulin was the loading control. The middle blot with anti-MTS specific serum which recognizes the MTS only indicates the higher-molecular-mass isoform in 2Δ-OE and 5Δ-OE, showing that MTS is retained in these 2 groups where either some proteins or the bulk of the proteins are in the cytosol. The absence of recognition of the Trunc-OE and Nat-OE clearly indicates the absence of MTS in these groups. (C) Subcellular fractionation shows overexpressed recombinant proteins present in both the cytosolic (Cyto) and mitochondrial (Mito) fractions in the 2Δ-OE and 3Δ-OE cells, whereas in the case of 5Δ-OE the overexpressed protein is detected only in the cytosolic fraction when immunoblotted with anti-GFP antibody. The anti-cTXNPx and anti-mTXNPx sera were used to detect the cytosolic and the mitochondrial fractions, respectively. Loss of MTS upon localization to mitochondria is indicated by the presence of the low-molecular-mass band in 2Δ-OE and 3Δ-OE cells which is absent in 5Δ-OE cells, showing retention of MTS in these cells.
The microscopy data were verified by Western blotting assays of cell lysates of the respective groups. While Trunc-OE and Nat OE showed a lower-molecular-mass protein of 52 kDa as a single band (Fig. 4B, lanes 2 and 3), the 2Δ-OE and the 3Δ-OE mutants showed two bands in that region (Fig. 4B, lanes 4 and 5), confirming the presence of proteins with both cleaved and uncleaved MTS. The disparity in molecular masses is due to a high-molecular-mass protein with an uncleaved MTS in the absence of transport to the mitochondria and a low-molecular-mass protein with a cleaved MTS due to cleavage at mitochondria after transport. N-terminal sequencing confirmed the identity of the lower-molecular-mass protein band, showing the protein as a mature form of the mTXNPx, with the sequence starting at QMYRTATVR. The 5Δ-OE lysate showed the recombinant fusion protein as a single band with a higher molecular mass (Fig. 4B, lane 6), demonstrating the presence of the uncleaved form only. Therefore, it is evident that the 2Δ and 3Δ proteins were partially able to reach mitochondria, where they were cleaved, showing both the cleaved and uncleaved form of the protein, whereas the 5Δ protein did not reach the mitochondria at all and remained an intact protein. The same blot was processed with a specific anti-MTS serum that would detect proteins containing the MTS region only and not proteins that did not have the MTS. This antibody recognized 2Δ-OE, 5Δ-OE, and, with lesser reactivity, 3Δ-OE (Fig. 4B), indicating that proteins retained in the cytosol contained the MTS. Various degrees of recognition between overexpressed proteins of 2Δ-OE, 3Δ-OE, and 5Δ-OE by anti-MTS sera possibly indicate the differences in the MTS epitopes due to mutations generated. To pinpoint the subcellular protein localization, cytosolic and mitochondrial fractions were probed with anti-GFP antisera. Overexpressed proteins with the 2Δ-OE and 3Δ-OE mutations were detected in both the cytosolic and mitochondrial fractions (Fig. 4C, lanes 1 to 4), with the cytosolic fraction isoform being larger than the mitochondrial fraction isoform, which was smaller due to the cleavage of MTS upon localization. The overexpressed 5Δ-OE protein was detected only in the cytosolic fraction (Fig. 4C, lane 5), with no low-molecular-mass form in the mitochondria (Fig. 4C, lane 6). In summary, these experiments clearly showed mislocalization of mTXNPx when CaM binding sites were interfered with, indicating a role for CaM in transport of mTXNPx to the mitochondria.
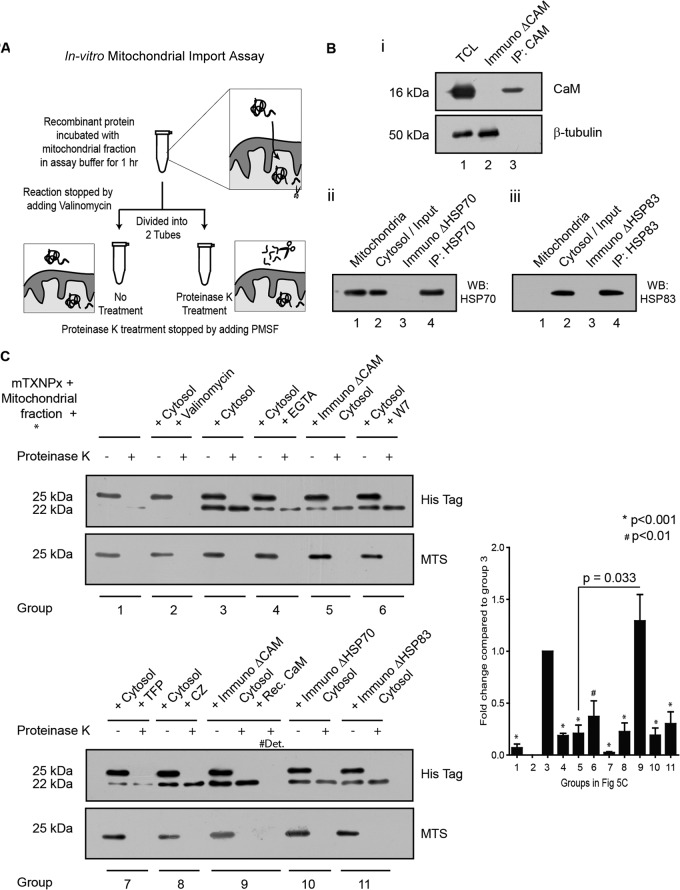
CaM insufficiency interferes with targeting of mTXNPx to isolated mitochondria.
Since we established that interference with CaM binding sites influenced the translocation of mTXNPx, the next step was to investigate whether the binding between the two proteins was directly responsible or required for the transport mechanism. An in vitro protein import assay (34) was designed where intact mitochondria isolated from the L. donovani cells were incubated with the recombinant proteins (with a C-terminal His tag) under different reaction conditions. Subsequently, the reaction mixture was divided into identical halves and one half was treated with proteinase K to digest the nontransported protein (Fig. 5A). Figure 5B shows depletion of CaM (ImmunoΔCaM), HSP70 (ImmunoΔHSP70), and HSP83 (ImmunoΔHSP83) in the cytosolic fraction using anti-CaM (i, lane 2) or anti-HSP antibodies (ii and iii, lane 3). The various reaction mixtures were analyzed in Western blot assays using anti-His tag antibody that detected both the precursor and the processed or mature form of the recombinant protein as shown in Fig. 5C. When the K+ ion channel-opening agent valinomycin, known for induction of dissipation of mitochondrial membrane potential (ψ), was used in the reaction along with the cytosolic fraction, no mitochondrial transport occurred (Fig. 5C, group 2). This indicated that the transport was a potential dependent phenomenon despite the presence of all cytosolic components present in the reaction. As shown in Fig. 5C, the reaction mixture with cytosol only showed two protein bands, one that had migrated to the mitochondria and another that had remained in the cytosol (Fig. 5C, group 3). Proteinase K treatment removed the cytosolic protein (Fig. 5C, group 3), and only the mitochondrial protein remained (group 3, lane 2). The presence of EGTA, a Ca2+ chelator, resulted in reduced targeting of mTXNPx as seen by the lower quantity of cleaved protein in the mixture (Fig. 5C, group 4), suggesting Ca2+ involvement in the process. Reduced mTXNPx translocation to the mitochondria was also recorded in CaM-immunodepleted cytosol (Fig. 5B, i) compared to the only-cytosol group (Fig. 5C, group 5). When recombinant CaM was added back to the CaM-immunodepleted cytosol (group 9), the transport was reinstated and was significantly higher than the CaM-immunodepleted cytosol (group 5). Statistically significant reduction was also obtained from experiments where CaM inhibitors were present during the experiment (Fig. 5C, groups 6 to 8). Similarly, there was a significant decrease in the amount of protein transported in reactions with cytosolic fractions (groups 10 and 11) where HSP70 and HSP83 were individually immunodepleted (Fig. 5B, ii and iii) compared to the one with cytosol only. The lower blot in Fig. 5C processed with anti-MTS sera showed the cytosolic mTXNPx present in the reaction mixture not treated with proteinase K.
Fig 5.
CaM is responsible for targeting mTXNPx to isolated mitochondria. (A) Representation of the in vitro mitochondrial protein import assay in a cartoon. (B) Immunodepletion of CaM from cytosolic fraction of L. donovani cells. (i) TCL indicates the total cell lysate, ImmunoΔCaM represents CaM-depleted cytosolic fraction, and IP:CaM denotes the immunoprecipitated fraction. (ii and iii) Similar nomenclature was followed to indicate immunodepletion of HSP70 and HSP83. (C) Both panels show the results of in vitro translocation of recombinant mTXNPx, under different conditions, to mitochondria isolated from Leishmania. All reaction conditions had the mitochondrial fraction and C-terminally His-tagged mTXNPx in a transport-competent buffer. Additional components added are indicated on top accordingly. Group numbers are indicated at the bottom, where each group was divided into two fractions and one fraction of each group was treated with proteinase K. Immunoblotting with anti-His antibody detected both the precursor (25 kDa) and the processed or mature form (22 kDa) of the recombinant protein. Anti-MTS antibody detected only the unprocessed protein left out in fractions that were not treated with proteinase K. #Det denotes the fraction of the import assay reaction mixture treated with 0.5% Triton X-100 along with proteinase K. The bar graph shows the densitometry of the processed (22-kDa) form in proteinase K-treated lanes as detected by the anti-His antibody. P values were calculated by one-way ANOVA comparing different groups to group 3. A paired t test was conducted to compare group 9 to group 5.
The above findings clearly showed the requirement of CaM and HSPs in the cytosol for targeting of the recombinant protein to isolated mitochondria in vitro.
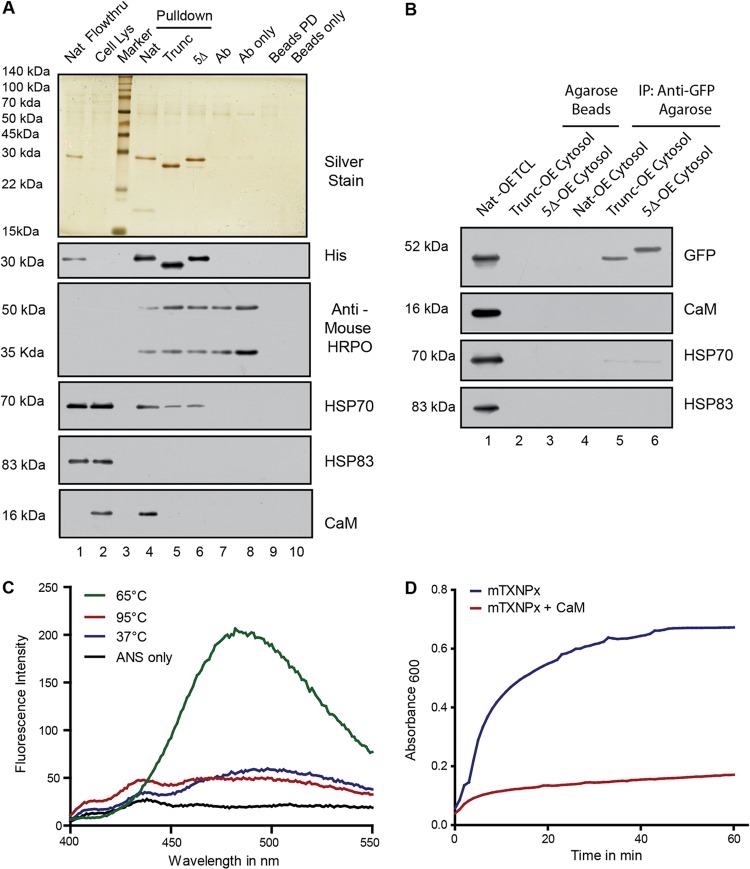
CaM from Leishmania cells interacts with the native protein at the predicted residues.
Finally, to establish that endogenous CaM from Leishmania reacts with endogenous mTXNPx, immunoprecipitation (IP) of mTXNPx and CaM was attempted; however, endogenous mTXNPx inside a cell would already have localized to mitochondria and lost the targeting signal, thereby reducing the chances of cleaved mTXNPx to interact with CaM. Therefore, experiments were carried out with cellular cytosolic fractions incubated with recombinant native, 5Δ-OE protein and truncated mTXNPx proteins followed by immunoprecipitation with anti-His tag antibody. Endogenous CaM was precipitated down only in case of native protein and not with the 5Δ-OE protein and the truncated protein (Fig. 6A, anti-CaM, lane 4). Interestingly, Western blot assays with anti-HSP70 and anti-HSP83 indicated that only HSP70 interacted with mTXNPx. The interaction was strongest in the case of the native protein and decreased significantly in the case of truncated and 5Δ protein (Fig. 6A, HSP70, lanes 4 to 6). Anti-HSP83 blot assays did not show any interaction (Fig. 6A, HSP83). The efficiency of the immunoprecipitation was checked by analyzing the amounts of recombinant proteins that interacted with the IP antibody (anti-His), which were found to be roughly equal (Fig. 6A, anti-His, lanes 4 to 6). Both the Western blot assays were carried out with a secondary antibody that recognized only the native form of the primary antibody so as to avoid signals arising out of the denatured IP antibody present in the lysates (as detected by a direct immunoblot assay with anti-mouse antibody–HRP) (Fig. 6A). Thus, endogenous CaM in the cytosol was able to bind to the mTXNPx, and this observation lends credence to the observations that CaM influences mTXNPx transport to the mitochondria. These observations were further investigated in Trunc-OE and 5Δ-OE cells to check for the ability of these mutant proteins to bind CaM or HSPs. Cytosolic fractions from these cells were subjected to coimmunoprecipitation using camel anti-GFP–agarose beads (camel antibodies ensure the noninterference of heavy-chain bands during subsequent Western blot assays). CaM, as expected, did not bind to these cytosolic (GFP-fused) recombinant proteins (Fig. 6B) because they lacked an effective MTS. HSP83 also did not bind, and HSP70 weakly interacted with only 5Δ protein and not the truncated one (Fig. 6B, HSP70, lanes 5 and 6). Nat-OE cytosol has been shown not to contain any GFP-fused protein because of complete transport to the mitochondria and served as a control for anti-GFP interactions and incubations of the Trunc-OE and 5Δ-OE cytosols with agarose beads and as a control for nonspecific binding to the beads. These results strongly indicate that interaction of HSP70 with mTXNPx is dependent on the interaction of CaM with the protein.
Fig 6.
CaM from Leishmania cells interacts with the native protein with the predicted residues. (A) Silver-stained gel shows immunoprecipitates from cytosolic fractions with different recombinant proteins such as native, Trunc, and 5Δ mTXNPx as baits (lanes 4, 5, and 6, respectively). Immunoblotting with anti-CaM, anti-HSP70, and anti-HSP83 antibodies shows the degree of interaction of the respective proteins with mTXNPx. Immunoblotting with anti-His antibody shows the amount of bait protein pulled down by the IP antibody. Both the blots were processed with Trueblot anti-mouse secondary antibody which does not recognize the denatured form of the primary antibody. Blot with anti-mouse antibody–HRP indicates the heavy chain (50 kDa) and light chain (30 kDa) of the mouse anti-His IP antibody. Pulldown with just the IP antibody or protein G beads (lanes 7 and 9) served as control for determination of nonspecific proteins from the cytosol. IP antibody or protein G beads (lanes 8 and 10) were loaded to check if they themselves contributed to any nonspecific bands on the gel. The presence of bait protein in the flowthrough fraction supports the use of bait protein in saturating amounts with respect to the IP antibody. (B) Coimmunoprecipitations using camel anti-GFP–agarose beads from the cytosols of Trunc-OE and 5Δ-OE cells to probe for cytosolic factors interacting with the overexpressed proteins. Immunoblotting with anti-CaM, anti-HSP70, and anti-HSP83 shows the degree of interaction of the respective proteins. Pulldowns with Nat-OE cytosol and with agarose beads serve as a control for anti-GFP interactions and nonspecific binding to the beads. (C) Fluorescence emission spectra of 120 μM bis-ANS in the presence of 4 μM mTXNPx at different temperatures show a marked increase in fluorescence emission when bound to partially unfolded protein at 65°C compared with the emission of the native one at 37°C, the completely unfolded one at 95°C, or free bis-ANS. (D) Aggregation of mTXNPx monitored as absorbance at 600 nm in a 4-mm-path-length cuvette at 69°C (blue) compared to that in the presence of equimolar concentrations of CaM (red).
CaM prevents the native protein from aggregation and thus maintains translocation competence.
The fact that the binding between MTS and CaM was essential for targeting of mTXNPx to mitochondria and that CaM was able to bind to the denatured protein (far-Western experiments) led us to test the possibility that CaM was involved in maintenance and translocation competence of mTXNPx. CaM is known to assist the transport of small secretory proteins that are not cotranslationally targeted to the endoplasmic reticulum of mammalian cells (35). Denatured proteins expose their hydrophobic regions and are prone to aggregation through beta sheet interactions, which cause such events to be irreversible (36). To determine the interaction of denatured mTXNPx with CaM, a time-based aggregation assay was designed where recombinant native mTXNPx to be used for such assays was first checked for its native conformation at room temperature using circular dichroism (CD). Spectra obtained for native mTXNPx had a characteristic shape and magnitude of CD spectrum for alpha-helical proteins (data available on request), indicating its suitability for further experimentation. The thermodynamics of protein unfolding was studied through ellipticity values collected as a function of temperature to determine the melting temperature of mTXNPx (data available on request). Results obtained were further validated through a conformation-sensitive fluorescent probe, bis-ANS. Bis-ANS bound to exposed hydrophobic surfaces in partially folded intermediates has much higher affinity and fluorescence than those to native or completely unfolded proteins (37). Binding of bis-ANS was monitored at different temperatures to determine the transition from partial to complete unfolding (Fig. 6C). Using the approximate temperature ascribed by both these experiments, the aggregation was observed over time. It was observed that denatured mTXNPx is prone to aggregation which is significantly reduced in the presence of recombinant CaM (Fig. 6D). This indicated a possible role for CaM to maintain the translocation competence of nascent mTXNPx polypeptides by preventing aggregation.
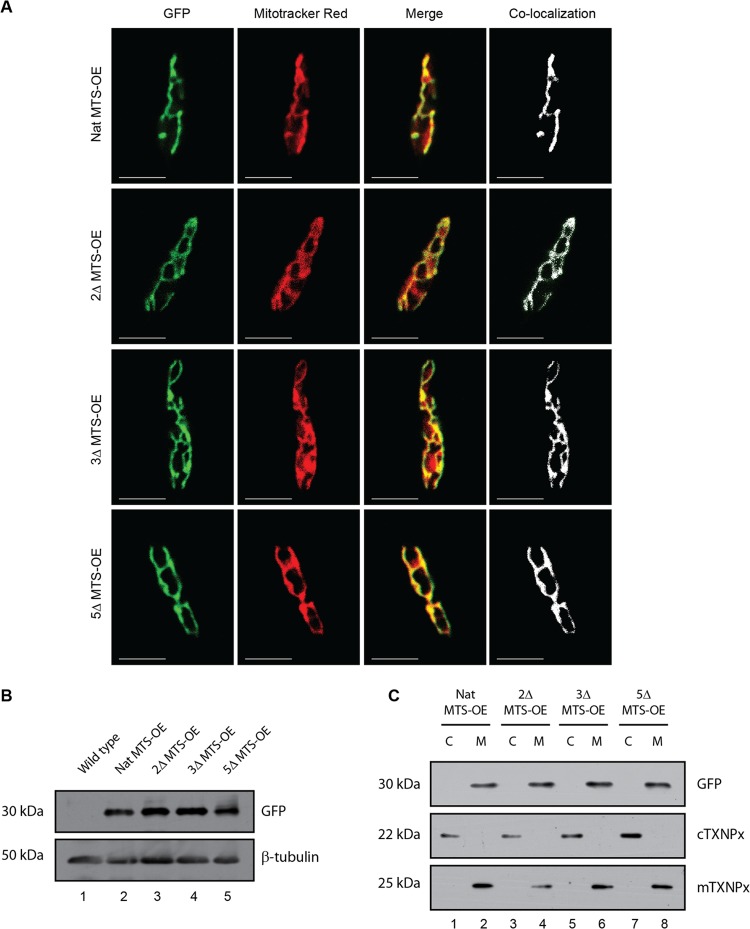
Mutations in the MTS do not compromise the competence for mitochondrial targeting.
Finally, to demonstrate competence of mutant signals for mitochondrial targeting, constructs expressing the native MTS protein (Nat MTS-OE) and mutant MTSs—2Δ MTS-OE, 3Δ MTS-OE, and 5Δ MTS-OE without any deletion, all with c-terminal GFPs—were episomally overexpressed (Fig. 7A). Transfectants showed high GFP expression, confirming episomal overexpression (data available on request). Colocalization of the GFP fusion protein using mitochondrion-specific dye Mitotracker Red showed clear colocalization at the mitochondria (Fig. 7A). Western blot analysis of transfectant lysates showed equal-molecular-mass proteins, suggesting similar fates inside the cell (Fig. 7B). Subcellular fractions of mitochondria and cytosol probed with anti-GFP antibody on Western blot assays showed primary localization in the mitochondrial fraction (Fig. 7C). Thus, the hypothesis that the entire length of the protein requires CaM for the transport due to its aggregation-prone nature is strengthened. However, a short length of the same (just the MTS) would not face such issues and could be transported without the assistance of CaM.
Fig 7.
Mutations in the MTS do not compromise the competence for mitochondrial targeting. (A) Parasites were stained with Mitotracker Red, a mitochondrion-specific dye showing native MTS-GFP fusion protein (Nat MTS-OE) and the GFP fusion protein with mutant MTSs (mutations indicated in Fig. 3A): 2Δ MTS-OE, 3Δ MTS-OE, and 5Δ MTS-OE completely localized to the mitochondria. Mask of colocalization is shown in white. Bars, 5 μm. (B) Western blots of whole-cell lysates of transfectants overexpressing native as well as the mutant MTS probed with GFP show the expression of the recombinant proteins of equal molecular masses. β-Tubulin was used as loading control. (C) Fractionation profile of Nat MTS-OE, 2Δ MTS-OE, 3Δ MTS-OE, and 5Δ MTS-OE shows that all the recombinant proteins, whether native or mutant, localize to the mitochondria (M). The same blot was probed with anti-cTXNPx and anti-mTXNPx serum (shown below) to indicate the identity of the cytosolic and mitochondrial fractions, respectively.
DISCUSSION
Mitochondrial protein import is a process with multiple steps that include synthesis of proteins in the cytosol, transport of the protein to the mitochondria, receptor recognition of the protein, translocation across the mitochondrial membranes, and the assembly after cleavage of the MTS. This study illuminates details of a part of the transport process from cytosol to mitochondria in a trypanosomatid parasite, L. donovani, a lower eukaryote. Most mitochondrial proteins are synthesized on the ribosomes as preproteins, and typically they contain a 20- to 50-amino-acid-long presequence, the MTS. Bioinformatic analysis shows the presence of MTS in Kinetoplastida mTXNPx, the model protein that we selected for mitochondrial targeting studies. Although presequence is shown in these parasites, no literature exists on their actual function. Our studies by truncation of the MTS resulting in abrogation of transport and attachment of the same to GFP, facilitating GFP transport to the mitochondria, provided in vivo evidence that the 30-amino-acid MTS identified by bioinformatic analysis was responsible for targeting the protein to the mitochondria. This observation confirms the in vivo functionality for an active MTS in a lower eukaryote.
There are regulatory mechanisms in place that ensure the proper translocation of the native protein from the ribosomes to the mitochondria. The identification of a putative CaM binding site on the MTS of mTXNPx generated a hypothesis that CaM could be involved in the translocation of the mTXNPx to the mitochondria. CaM, a calcium sensor protein, with conformations depending upon Ca2+ binding, is one of the most evolutionarily ancient proteins in eukaryotes with a very low evolution rate (38). CaM binds to the target proteins in an open or extended conformation, induced by Ca2+ binding to the EF motifs, and then wraps around the target, resulting in a closed conformation (39). Ca2+, on the other hand, is a universal second messenger in eukaryotes, mediating both biotic and abiotic signals. The requirement of Ca2+ in CaM binding to mTXNPx as demonstrated in this study suggests a similar conformational change in CaM for binding to mTXNPx. The ability of CaM inhibitors in vivo and CaM depletion in vitro to prevent mTXNPx translocation clearly indicated the involvement of CaM binding to the mTXNPx as an integral part of the translocation process. The binding disruption due to inhibitors also supports conformation-based interaction of CaM with mTXNPx. The identification of the MTS-located CaM binding site suggested that the effects of inhibition of CaM could be primarily mediated through the MTS region. This was confirmed by site-directed mutagenesis studies where residues on the CaM binding site on MTS were mutated, taking care that there was no interference with the mitochondrial transport. Evidence that mutagenesis did not interfere with mitochondrial transport came from translocation competence of mutated MTS residues. For the whole mTXNPx, mutagenesis of 5 residues was required to restrict translocation to mitochondria; however, mutations of 2 or 3 residues were not sufficient to completely prevent mitochondrial translocation. The ability of the 5Δ mutant MTS tagged with GFP but not the 5Δ whole mTXNPx to translocate to mitochondria suggests that the MTS may directly interact with CaM, but the region beyond the MTS on mTXNPx is also required for a successful translocation process (even if not a direct one). The involvement of Ca2+/CaM in translocation of a mitochondrial protein, alternative oxidase of Pisum sativum, has been shown through Ca2+ depletion studies (using A23187 and ionomycin) and use of CaM inhibitor ophiobolin A. However, the study did not project either CaM binding sites or any CaM interaction with the protein (40), leaving open the possibilities of involvement of other components influenced by ionophores and inhibitors.
From the data presented, only two possible functional models seemed feasible: either CaM was a part of a complex meant for posttranslational transport of polypeptides or CaM binds to the unfolded nascent polypeptide to maintain the translocation competence of the protein. The former model seemed highly improbable because of both the absence of such a mechanism across the eukaryotic kingdom and the high evolutionary costs of evolving such a mechanism only in kinetoplastids. The aggregation experiments with and without CaM in vitro clearly indicated that the nature of mTXNPx was such that unfolded forms were susceptible to aggregation, as CaM was able to reduce the aggregation in our experimental system. Hence, by the principle of Occam's razor, the latter hypothesis was the most suitable model to explain the functional significance of mTXNPx-CaM binding.
The role of CaM in transport of proteins to mitochondria of higher eukaryotes is uncertain because similar studies are lacking. However, it seems to be a key player in nucleocytoplasmic exchange through studies which have shown a regulatory role for CaM binding to the N-terminal nuclear localization signal (NLS) of the HMG box of SOX family members for their transport into the nucleus (41). The prevailing dogma for the eukaryotic translocation of mitochondrial proteins is the involvement of HSP70 proteins as chaperones (42). In the case of Leishmania, HSP70 and HSP83 (HSP90) are known to be constitutively expressed, with HSP70 found in both the cytosol and mitochondria and HSP83 found in the cytosol only (43–46). It was clear from the in vitro import assay and immunopulldowns that HSP70 interacts with mTXNPx and is required for the transport. Similarly, HSP83 too seems to be required for the transport, but its interaction could not be demonstrated; nevertheless, its role cannot be discounted. The inability of HSP70 to interact with 5Δ protein expressed in either E. coli or Leishmania indicates that CaM binding is required for the HSPs to interact with mTXNPx, suggesting the role of CaM to be a primary one. These results open up new avenues of investigation into the nature and extent of involvement of HSPs in mitochondrial translocation in lower eukaryotes. CaM itself acting as a chaperone has been described in the higher eukaryotes, where it was shown to maintain the posttranslational competence of small secretory proteins destined for the endoplasmic reticulum (35), showing an unexpected chaperone activity. The involvement of CaM in mitochondrial transport shown in this study could perhaps indicate an ancient mechanism existing in lower eukaryotes, lost in selection to a different CaM-independent translocation system in the higher eukaryotes, rather than convergent appearance of CaM-dependent translocation competence maintenance mechanisms. However, possibilities of CaM interaction with MTS in higher eukaryotes as yet unreported cannot be ruled out.
Thus, this investigation describes a novel finding of CaM binding to MTS and being responsible for translocation of a protein from the cytosol to the mitochondria in a lower eukaryote, L. donovani. Another important conclusion obtained from this study suggests that alterations in CaM or Ca2+ possibly affect the mitochondrial transport of mTXNPx, a defensive enzyme, thus compromising the survival possibilities of the parasite in the face of oxidative stress, which is a part of its environment during infection.
ACKNOWLEDGMENTS
This work was supported by grants from the Department of Biotechnology, New Delhi, India (http://dbtindia.nic.in/index.asp), to the National Institute of Immunology (grant no. BT/03/033/88); Centre for Molecular Medicine, New Delhi (grant no. BT/PR/14549/MED/14/1291); and Life Science Research Board of Defense Research Development Organization, New Delhi, India (grant no. DLS/81/48222/LSRB-169/ID2008). A.A. was supported by a fellowship from the Council of Scientific and Industrial Research, India (award no. 09/485[0174]/2008-EMR-I).
The pXG-GFP+ vector was a kind gift from Stephen M. Beverley, Department of Molecular Microbiology, Washington University School of Medicine. The kind gift of HSP70 and HSP83 antibodies from Jose M. Requena, Centro de Biología Molecular, Universidad Autónoma de Madrid, Spain, is acknowledged. The assistance of Manoj Mishra for mitochondrial import assays is greatly appreciated. Technical assistance from G. S. Neelaram is acknowledged. We acknowledge the help of Ayub Qadri for his suggestions and reviews.
C.S. and A.A. conceived and designed the experiments. A.A. performed the experiments. C.S. and A.A. analyzed the data. C.S. contributed reagents/materials/analysis tools. C.S. and A.A. wrote the paper.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 16 September 2013
REFERENCES
- 1.Hung MC, Link W. 2011. Protein localization in disease and therapy. J. Cell Sci. 124:3381–3392 [DOI] [PubMed] [Google Scholar]
- 2.Shpakov AO. 2006. Structural and functional organisation of G protein signal systems of amoeba Dictyostelium discoideum. Zh. Evol. Biokhim. Fiziol. 42:426–444 [PubMed] [Google Scholar]
- 3.Shpakov AO. 2007. Structural-functional organization of the adenylyl cyclases in unicellular eukaryotes and molecular mechanisms of its regulation. Tsitologiia 49:91–106 [PubMed] [Google Scholar]
- 4.Hauser R, Pypaert M, Hausler T, Horn EK, Schneider A. 1996. In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J. Cell Sci. 109:517–523 [DOI] [PubMed] [Google Scholar]
- 5.Krnacova K, Vesteg M, Hampl V, Vlcek C, Horvath A. 2012. Euglena gracilis and trypanosomatids possess common patterns in predicted mitochondrial targeting presequences. J. Mol. Evol. 75:119–129 [DOI] [PubMed] [Google Scholar]
- 6.Margulis L. 1970. Origin of eukaryotic cells: evidence and research implications for a theory of the origin and evolution of microbial, plant, and animal cells on the precambrian Earth. Yale University Press, New Haven, CT [Google Scholar]
- 7.Cavalier-Smith T. 2009. Predation and eukaryote cell origins: a coevolutionary perspective. Int. J. Biochem. Cell Biol. 41:307–322 [DOI] [PubMed] [Google Scholar]
- 8.Lane N. 2011. Energetics and genetics across the prokaryote-eukaryote divide. Biol. Direct 6:35. 10.1186/1745-6150-6-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyall SD, Brown MT, Johnson PJ. 2004. Ancient invasions: from endosymbionts to organelles. Science 304:253–257 [DOI] [PubMed] [Google Scholar]
- 10.Schatz G, Dobberstein B. 1996. Common principles of protein translocation across membranes. Science 271:1519–1526 [DOI] [PubMed] [Google Scholar]
- 11.Roise D, Schatz G. 1988. Mitochondrial presequences. J. Biol. Chem. 263:4509–4511 [PubMed] [Google Scholar]
- 12.Milenkovic D, Muller J, Stojanovski D, Pfanner N, Chacinska A. 2007. Diverse mechanisms and machineries for import of mitochondrial proteins. Biol. Chem. 388:891–897 [DOI] [PubMed] [Google Scholar]
- 13.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mossmann D, Meisinger C, Vogtle FN. 2012. Processing of mitochondrial presequences. Biochim. Biophys. Acta 1819:1098–1106 [DOI] [PubMed] [Google Scholar]
- 15.Pfanner N. 2000. Protein sorting: recognizing mitochondrial presequences. Curr. Biol. 10:R412–R415 [DOI] [PubMed] [Google Scholar]
- 16.Adhya S. 2008. Leishmania mitochondrial tRNA importers. Int. J. Biochem. Cell Biol. 40:2681–2685 [DOI] [PubMed] [Google Scholar]
- 17.Lithgow T, Schneider A. 2010. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:799–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filser M, Comini MA, Molina-Navarro MM, Dirdjaja N, Herrero E, Krauth-Siegel RL. 2008. Cloning, functional analysis, and mitochondrial localization of Trypanosoma brucei monothiol glutaredoxin-1. Biol. Chem. 389:21–32 [DOI] [PubMed] [Google Scholar]
- 19.Tasker M, Timms M, Hendriks E, Matthews K. 2001. Cytochrome oxidase subunit VI of Trypanosoma brucei is imported without a cleaved presequence and is developmentally regulated at both RNA and protein levels. Mol. Microbiol. 39:272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priest JW, Hajduk SL. 2003. Trypanosoma brucei cytochrome c1 is imported into mitochondria along an unusual pathway. J. Biol. Chem. 278:15084–15094 [DOI] [PubMed] [Google Scholar]
- 21.Eckers E, Cyrklaff M, Simpson L, Deponte M. 2012. Mitochondrial protein import pathways are functionally conserved among eukaryotes despite compositional diversity of the import machineries. Biol. Chem. 393:513–524 [DOI] [PubMed] [Google Scholar]
- 22.Young JC, Hoogenraad NJ, Hartl FU. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112:41–50 [DOI] [PubMed] [Google Scholar]
- 23.Benaim G, Cervino V, Hermoso T, Felibert P, Laurentin A. 1993. Intracellular calcium homeostasis in Leishmania mexicana. Identification and characterization of a plasma membrane calmodulin-dependent Ca(2+)-ATPase. Biol. Res. 26:141–150 [PubMed] [Google Scholar]
- 24.Chin D, Means AR. 2000. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 10:322–328 [DOI] [PubMed] [Google Scholar]
- 25.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 26.Iyer JP, Kaprakkaden A, Choudhary ML, Shaha C. 2008. Crucial role of cytosolic tryparedoxin peroxidase in Leishmania donovani survival, drug response and virulence. Mol. Microbiol. 68:372–391 [DOI] [PubMed] [Google Scholar]
- 27.Kaplan B, Davydov O, Knight H, Galon Y, Knight MR, Fluhr R, Fromm H. 2006. Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18:2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mortz E, Krogh TN, Vorum H, Gorg A. 2001. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 1:1359–1363 [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee SB, Das M, Sudhandiran G, Shaha C. 2002. Increase in cytosolic Ca2+ levels through the activation of non-selective cation channels induced by oxidative stress causes mitochondrial depolarization leading to apoptosis-like death in Leishmania donovani promastigotes. J. Biol. Chem. 277:24717–24727 [DOI] [PubMed] [Google Scholar]
- 30.Sudhandiran G, Shaha C. 2003. Antimonial-induced increase in intracellular Ca2+ through non-selective cation channels in the host and the parasite is responsible for apoptosis of intracellular Leishmania donovani amastigotes. J. Biol. Chem. 278:25120–25132 [DOI] [PubMed] [Google Scholar]
- 31.Krauth-Siegel RL, Meiering SK, Schmidt H. 2003. The parasite-specific trypanothione metabolism of trypanosoma and leishmania. Biol. Chem. 384:539–549 [DOI] [PubMed] [Google Scholar]
- 32.Castro H, Tomas AM. 2008. Peroxidases of trypanosomatids. Antioxid. Redox Signal. 10:1593–1606 [DOI] [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews GD, Gur N, Koopman WJ, Pines O, Vardimon L. 2010. Weak mitochondrial targeting sequence determines tissue-specific subcellular localization of glutamine synthetase in liver and brain cells. J. Cell Sci. 123:351–359 [DOI] [PubMed] [Google Scholar]
- 35.Shao S, Hegde RS. 2011. A calmodulin-dependent translocation pathway for small secretory proteins. Cell 147:1576–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beerten J, Schymkowitz J, Rousseau F. 2012. Aggregation prone regions and gatekeeping residues in protein sequences. Curr. Top. Med. Chem. 12:2470–2478 [DOI] [PubMed] [Google Scholar]
- 37.Bothra A, Bhattacharyya A, Mukhopadhyay C, Bhattacharyya K, Roy S. 1998. A fluorescence spectroscopic and molecular dynamics study of bis-ANS/protein interaction. J. Biomol. Struct. Dyn. 15:959–966 [DOI] [PubMed] [Google Scholar]
- 38.Friedberg F, Rhoads AR. 2001. Evolutionary aspects of calmodulin. IUBMB Life 51:215–221 [DOI] [PubMed] [Google Scholar]
- 39.Nelson MR, Chazin WJ. 1998. An interaction-based analysis of calcium-induced conformational changes in Ca2+ sensor proteins. Protein Sci. 7:270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhn S, Bussemer J, Chigri F, Vothknecht UC. 2009. Calcium depletion and calmodulin inhibition affect the import of nuclear-encoded proteins into plant mitochondria. Plant J. 58:694–705 [DOI] [PubMed] [Google Scholar]
- 41.Hanover JA, Love DC, Prinz WA. 2009. Calmodulin-driven nuclear entry: trigger for sex determination and terminal differentiation. J. Biol. Chem. 284:12593–12597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt O, Pfanner N, Meisinger C. 2010. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11:655–667 [DOI] [PubMed] [Google Scholar]
- 43.Klein KG, Olson CL, Donelson JE, Engman DM. 1995. Molecular comparison of the mitochondrial and cytoplasmic hsp70 of Trypanosoma cruzi, Trypanosoma brucei and Leishmania major. J. Eukaryot. Microbiol. 42:473–476 [DOI] [PubMed] [Google Scholar]
- 44.Argaman M, Aly R, Shapira M. 1994. Expression of heat shock protein 83 in Leishmania is regulated posttranscriptionally. Mol. Biochem. Parasitol. 64:95–110 [DOI] [PubMed] [Google Scholar]
- 45.Brandau S, Dresel A, Clos J. 1995. High constitutive levels of heat-shock proteins in human-pathogenic parasites of the genus Leishmania. Biochem. J. 310:225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quijada L, Soto M, Alonso C, Requena JM. 1997. Analysis of post-transcriptional regulation operating on transcription products of the tandemly linked Leishmania infantum hsp70 genes. J. Biol. Chem. 272:4493–4499 [DOI] [PubMed] [Google Scholar]