Abstract
The ubiquitin ligase CHIP (carboxyl terminus of Hsp70-interacting protein) regulates protein quality control, and CHIP deletion accelerates aging and reduces the life span in mice. Here, we reveal a mechanism for CHIP's influence on longevity by demonstrating that CHIP stabilizes the sirtuin family member SirT6, a lysine deacetylase/ADP ribosylase involved in DNA repair, metabolism, and longevity. In CHIP-deficient cells, SirT6 protein half-life is substantially reduced due to increased proteasome-mediated degradation, but CHIP overexpression in these cells increases SirT6 protein expression without affecting SirT6 transcription. CHIP noncanonically ubiquitinates SirT6 at K170, which stabilizes SirT6 and prevents SirT6 canonical ubiquitination by other ubiquitin ligases. In CHIP-depleted cells, SirT6 K170 mutation increases SirT6 half-life and prevents proteasome-mediated degradation. The global decrease in SirT6 expression in the absence of CHIP is associated with decreased SirT6 promoter occupancy, which increases histone acetylation and promotes downstream gene transcription in CHIP-depleted cells. Cells lacking CHIP are hypersensitive to DNA-damaging agents, but DNA repair and cell viability are rescued by enforced expression of SirT6. The discovery of this CHIP-SirT6 interaction represents a novel protein-stabilizing mechanism and defines an intersection between protein quality control and epigenetic regulation to influence pathways that regulate the biology of aging.
INTRODUCTION
Although aging was once perceived as a consequence of “wear and tear” on tissues and cells, the process of aging is now understood to be a coordinated decline in cellular energy metabolism, DNA repair, protein quality control (PQC), and antioxidant capacity (1–3). Understanding the cell biology behind aging has led to the identification of proteins actively involved in processes that prevent or promote an overall aging phenotype. CHIP (carboxyl terminus of Hsp70-interacting protein) is a protein that exhibits both cochaperone and ubiquitin ligase activities and is an integral component of PQC (4). We have previously reported that CHIP−/− mice exhibit reduced life span and many aging hallmarks, including muscle wasting, osteoporosis, early cellular senescence, and accumulation of misfolded proteins and oxidized lipids (5). Although this phenotype clearly implies a protective role for CHIP in the process of aging, the mechanism behind CHIP's support of cellular longevity remains unknown.
Many cellular stressors have deleterious effects on protein stability, causing either misfolding or aggregation (6), and PQC mediates the removal of damaged or aggregated proteins. CHIP is a key player in PQC and prevents proteotoxicity through dual mechanisms. At the onset of acute stress, CHIP activates heat shock factor 1 (HSF1), the transcription factor responsible for upregulating expression of chaperone proteins (7). CHIP nuclear localization increases dramatically during acute stress, but CHIP's nuclear functions beyond HSF1 activation are currently unclear. CHIP then partners with newly transcribed heat shock proteins via CHIP's tetracopeptide repeat (TPR) domain, which promotes substrate refolding into proper conformation. CHIP also contains a C-terminal U-box domain that confers ubiquitin ligase activity and promotes ubiquitination of chaperone-associated proteins (8). After misfolded protein substrates have been depleted via proteasomal degradation, CHIP is responsible for the ubiquitination and degradation of excess heat shock proteins (9).
CHIP catalyzes the formation of canonical ubiquitin chains (linked through ubiquitin lysine 48 [K48] residues) to promote degradation of a broad array of targets, including cystic fibrosis transmembrane receptor (10), HER2/neu receptor (11), SMAD1/4 (12), and aggregate-prone proteins such as α-synuclein (13) and tau (14). However, CHIP can also form noncanonical ubiquitin chains that are linked through other lysine residues (15) that have diverse, nonproteasomal signaling functions (16). Recently, we described CHIP's noncanonical ubiquitination of the transcriptional corepressor Daxx, which promotes Daxx's translocation to an insoluble cellular compartment and prevents Daxx sumoylation, required for Daxx activity (17). However, it remains to be seen whether CHIP noncanonically ubiquitinates other targets and if there is a functional consequence of this activity.
In an effort to better understand the anti-aging function of CHIP, we sought to identify novel CHIP-interacting proteins that may provide an understanding of CHIP's effects on longevity and senescence. To this end, we have identified an interaction between CHIP and the sirtuin family isoform Sirtuin 6 (SirT6). In this report, we demonstrate that SirT6 interacts with CHIP and serves as a noncanonical ubiquitination substrate for CHIP, which prevents SirT6 protein degradation and allows SirT6 to participate in its previously described roles of DNA repair and histone deacetylation. This newly uncovered mechanism expands the current understanding of the biology of both CHIP and SirT6 and offers an exciting interpretation of CHIP's ability to influence longevity.
MATERIALS AND METHODS
Reagents and antibodies.
All chemicals and reagents were obtained from Sigma unless otherwise noted. Western blots were performed using NuPage Bis-Tris gels and an electrophoresis system (Invitrogen), transferred to 0.45-μm-pore-size polyvinylidene difluoride (PVDF) membranes (Millipore), and visualized using ECL Advance (GE Healthcare) on an EpiChemi 3 Darkroom (UVP), and band densitometry was analyzed using ImageJ. A table of antibodies used in these experiments can be found in the supplemental material.
Cell culture, plasmid transfection, and immunoprecipitation.
CHIP+/+ and CHIP−/− mouse embryonic fibroblasts (MEFs) were cultured as previously described (7). HEK 293 cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS). Transfections in 293 cells were performed using FuGENE 6 (Roche) or X-TremeGene 9 (Roche). Plasmids were introduced into MEFs through electroporation with the Amaxa MEF Nucleofector 2 kit (Lonza) using program A23. Empty vector (pcDNA3.1), FLAG-tagged plasmids expressing sirtuin family isoforms (Addgene plasmids 13812 to 13818 [18]), Myc-tagged wild-type (WT) CHIP, ΔTPR CHIP, H260Q CHIP, ΔU-box CHIP, hemagglutinin (HA)-WT ubiquitin, and HA-K48R ubiquitin constructs were described previously (8, 17). His-tagged WT SirT6 cloned into pQE-80 (Addgene plasmid 13739 [19]) and FLAG-tagged SirT6 cloned into pCMV (Addgene plasmid 13817 [18]) were used as templates with the QuikChange II site-directed mutagenesis kit (Agilent) to produce the K170R SirT6 plasmids that were expressed in bacterial and mammalian cells, respectively. Cell lysates were harvested on ice with lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, protease inhibitor [Complete; Roche]), and insoluble material was pelleted by centrifugation at 8,000 × g. Phosphatase inhibitor was added to lysis buffer for phosphorylated replication protein A (RPA) measurements (PhosStop; Roche). FLAG and Myc immunoprecipitations were performed for 16 h in lysis buffer using EZView anti-FLAG affinity gel and EZView anti-c-Myc affinity gel (Sigma). For measurement of HA-tagged ubiquitination, cells were transfected as described above, and all cells were cultured in 5 μM the proteasome inhibitor MG132 for 16 h prior to harvest and then harvested in lysis buffer containing 10 mM N-ethylmaleimide. Nuclear fractions were prepared using the NE-PER kit (Pierce).
Immunofluorescence.
Cell staining was performed as previously described (20) with modifications described in the supplemental material.
Stable cell lines.
Stable HEK 293 cell lines were created via transduction with Mission lentiviral short hairpin RNA (shRNA) (Sigma) targeting CHIP or a scrambled control. Cells were selected with puromycin (1 μg/ml) for 14 days, and CHIP knockdown was verified via quantitative PCR (qPCR) and Western blotting. The cells were then transfected with pcDNA3.1 or FLAG-SirT6 using FuGene 6, and stable expression was selected using neomycin (Gibco; 700 μg/ml) for 14 days.
in vitro ubiquitination.
Each in vitro ubiquitination reaction was performed using 2.5 μM recombinant CHIP, 1 μM recombinant WT SirT6, or recombinant K170R SirT6 (both purified using a HisTalon gravity purification kit [Clontech]), and the following reagents where indicated: 50 nM Ube1, 2.5 μM UbcH5c, 250 μM WT or mutant lysine ubiquitin, 20 nM purified 26S proteasome, and 2.5 mM Mg-ATP (all from Boston Biochem) in reaction buffer (50 mM Tris-HCl, pH 7.6, 50 mM KCl, 0.2 mM dithiothreitol [DTT]). Reactions were performed for 1 h at 37°C. Samples were separated and visualized via Western blot analysis.
mRNA analysis.
mRNA was isolated using the RNeasy Mini kit (Qiagen) and reverse transcribed using an Iscript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using Roche LightCycler 480 Probes Master Mix, Roche Universal Probe Library probes, and primers targeting the indicated genes (Integrated DNA Technologies) (see Table S4 in the supplemental material). Amplification was measured on a Roche LightCycler 480 instrument.
SirT6 half-life measurement.
Half-life was measured via pulse-chase labeling with 35S-labeled amino acids as previously described (21) with modifications shown in the supplemental material.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed using the Magna ChIP A/G kit (Millipore) (see the supplemental material for details).
Viability assays.
Stable HEK 293 cell lines were cultured in the presence of 100 to 300 μM methyl methanesulfonate (MMS) or 200 to 600 μM hydrogen peroxide (H2O2) for 6 days. The cells were then stained with trypan blue and counted with a hemocytometer. Immortalized CHIP+/+ and CHIP−/− MEFs were grown in 96-well plates and incubated in 3-aminobenzamide (10 mM), camptothecin (1 μM), or paraquat (0.5 mM) for 16 h. Viability was assessed using an MTS assay (Promega).
Statistics.
Unless otherwise indicated, Student's t test was used to assess significance (Excel). The Fisher exact test was performed for analysis of γH2AX focus distribution (R-Stat). Excluding ChIP-sequencing (ChIP-seq) data, all experiments were performed on at least three independent occasions, with representative Western blots shown. Graphed results represent means and standard errors of the mean (SEM).
RESULTS
SirT6 protein stability is decreased in the absence of CHIP.
We sought to understand the molecular mechanisms responsible for the severe aging phenotype of the CHIP−/− mouse (5, 7). Because CHIP activates HSF1, we hypothesized that if the aging phenotype was mediated entirely through decreased HSF1 activity, the HSF1−/− mouse should show pathologies similar to those of the CHIP−/− mouse. Although there are some similarities reported in HSF1−/− mice, such as growth retardation, decreased Hsp70 expression, and neural protein aggregation (22–24), the range of unique pathologies observed in the CHIP−/− mice indicated that CHIP might regulate aging through multiple mechanisms. Several aging-related phenotypes observed in SirT6−/− mice, including small size, decreased bone mineral density, lordokyphosis, loss of subcutaneous fat, senescence-associated β-galactosidase staining, spontaneous cardiac hypertrophy, and reduced life span (25–27), are shared by CHIP−/− mice. Therefore, we hypothesized that SirT6 protein activity might be affected by the loss of CHIP. We measured SirT6 mRNA and protein levels in several tissues from 12-month-old CHIP+/+ and CHIP−/− mice. Although SirT6 mRNA levels were unchanged in tissues from CHIP−/− mice (unpublished observations), SirT6 protein levels were dramatically reduced in most CHIP−/− tissues (Fig. 1A). The exception to this was skeletal muscle, in which SirT6 levels were unaffected by the absence of CHIP, suggesting that the proteins involved in SirT6 regulation may not be active in this tissue. To further study the connection between CHIP and SirT6, HEK 293 cells were developed to stably express a scrambled shRNA (shControl) or an shRNA pool targeting CHIP (shCHIP). We examined SirT6 mRNA in 293 cells and in immortalized CHIP+/+ and CHIP−/− MEFs, and in accordance with the decreased SirT6 protein expression observed in CHIP−/− tissues, we observed decreased SirT6 protein levels in both the CHIP-depleted 293 cells and MEFs (Fig. 1B). However, CHIP depletion had no effect on SirT6 mRNA in either cell type (Fig. 1B), indicating a completely posttranslational effect. We also transiently reduced expression of CHIP in 293 cells using two individual CHIP-targeting small interfering RNAs (siRNAs) and found that a reduction in CHIP correlated with a reduction in SirT6 protein levels; however, CHIP siRNA had no effect on SirT6 mRNA levels (Fig. 1C). Together, these data suggest that SirT6 stability is positively related to the presence of CHIP. To better characterize the mechanism by which SirT6 protein is stabilized by CHIP, SirT6 protein levels were measured in shControl and shCHIP cells stably expressing FLAG-SirT6 following 16 h of treatment with chemical inhibitors of transcription, translation, proteasome activity, and autophagy (Fig. 1D). The inhibitors showed no substantial effects on SirT6 protein levels in the cells expressing CHIP. However, in shCHIP cells, the proteasome inhibitor MG132 increased expression of FLAG-SirT6, indicating that CHIP prevents proteasome-dependent degradation of SirT6. We also found that endogenous SirT6 accumulated with MG132 treatment in shCHIP cells (Fig. 1E). Together, these data indicate that CHIP depletion increases proteasome-mediated SirT6 degradation and suggest that CHIP protects SirT6 protein from proteasomal degradation.
Fig 1.
SirT6 protein is stabilized in the presence of CHIP. (A) SirT6 expression was analyzed via Western blot analysis in multiple tissues from 12-month-old CHIP+/+ and CHIP−/− mice. Skel., skeletal. (B) Endogenous SirT6 and CHIP protein expression was measured in shControl and shCHIP 293 cells (left) and in CHIP+/+ and CHIP−/− MEFs (right) via Western blot analysis. SirT6 protein quantification (middle) and mRNA quantification (bottom) are shown (n = 4). (C) Normal 293 cells were transiently transfected with control siRNA, two independent siRNAs targeting CHIP, or SirT6 siRNA. CHIP and SirT6 protein were measured via Western blot analysis, and SirT6 protein quantification (middle) and mRNA quantification (bottom) are shown (n = 3). (D) shControl and shCHIP 293 cells stably expressing FLAG-SirT6 were exposed to dimethyl sulfoxide (DMSO), actinomycin, chloroquine, cycloheximide, MG132, or ammonium chloride (NH4Cl) for 16 h. SirT6 protein levels were visualized in Western blots, quantified, and normalized to the DMSO control for each cell type (n = 3). (Right) Representative Western blot from cells treated with MG132. The proteasome-regulated protein p27 is shown to indicate MG132 effectiveness. (E) shCHIP cells were exposed to DMSO or MG132 for 16 h, and Endogenous SirT6 protein levels were measured via Western blotting. p27 indicates MG132 treatment. (Top) Representative Western blot. (Bottom) Quantification (n = 4). Results represent means and SEM. *, P < 0.05. Graphed results represent SirT6 protein relative to β-tubulin and normalized to control in arbitrary units.
CHIP interacts with SirT6, and CHIP ubiquitin ligase activity increases SirT6 expression.
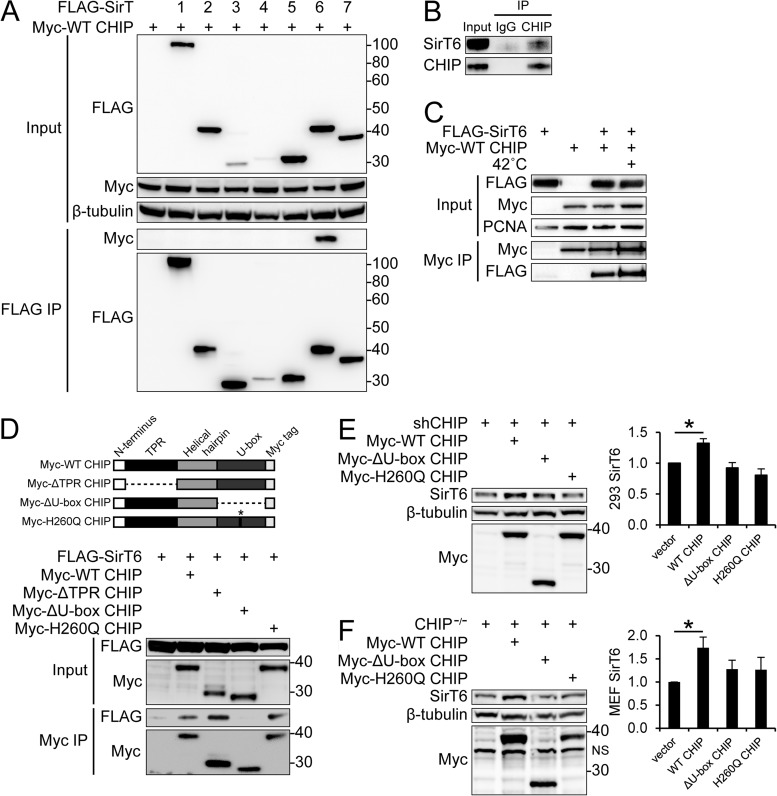
Since CHIP has many substrates, it is possible that CHIP's effect on SirT6 stability is mediated through an indirect mechanism, such as inhibition of a negative regulator of SirT6. However, it is also possible that CHIP interacts directly with SirT6 to influence protein stability. We screened CHIP for interactions with all seven sirtuin isoform members, a family of proteins that generally promote cell homeostasis and affect the rate of aging by regulating the cell cycle, DNA repair, and metabolism through NAD-dependent lysine deacetylase (KDAC) or ADP ribosylase activity (28). Using coexpression and immunoprecipitation of tagged CHIP and sirtuin proteins, we found that CHIP stably interacted solely with SirT6 (Fig. 2A). We also found that endogenous CHIP and SirT6 stably interacted (Fig. 2B). Given that SirT6 is a nuclear protein and that CHIP's nuclear localization increases at the onset of stress (20), we hypothesized that the CHIP-SirT6 interaction would increase with stress due to CHIP nuclear accumulation. Nuclear extracts freshly prepared from cells exposed to elevated culture temperatures demonstrated increased CHIP nuclear localization, which allows more SirT6 to coimmunoprecipitate with CHIP within the nuclear compartment (Fig. 2C).
Fig 2.
CHIP and SirT6 stably interact through the CHIP U-box domain. (A) HEK 293 cells were transfected with Myc-tagged CHIP (Myc-WT CHIP) and FLAG-tagged sirtuin isoforms 1 to 7 (FLAG-SirT). Input (above) and FLAG immunoprecipitation (IP) (below) were performed on cell lysates and visualized via Western blotting with the indicated antibodies. (B) Endogenous CHIP was immunoprecipitated from HEK 293 cell lysates, and endogenous SirT6 was detected and visualized via Western blotting. (C) HEK 293 cells transfected with Myc-WT CHIP and FLAG-SirT6 were incubated at 37°C or 42°C for 30 min. Nuclear fractions were immediately prepared, and input and Myc immunoprecipitations were visualized via Western blotting with the indicated antibodies. (D) The indicated Myc-tagged CHIP constructs (diagrammed) were transfected into HEK 293 cells with FLAG-SirT6. Input and Myc immunoprecipitations were performed on cell lysates and visualized via Western blotting. (E and F) shCHIP 293 cells (E) and CHIP−/− MEFs (F) were transfected with Myc-WT CHIP, Myc-ΔU-box CHIP, or Myc-H260Q CHIP. Lysates were collected, and endogenous SirT6 expression was quantified via Western blot analysis (right; n = 4 of each). NS, nonspecific band. The results represent means and SEM. *, P < 0.05. Graphed results represent SirT6 protein relative to β-tubulin and normalized to control in arbitrary units.
We hypothesized that the CHIP domain responsible for the interaction with SirT6 might hint at the nature of CHIP's regulatory effect on SirT6. In coexpression/immunoprecipitation experiments using WT CHIP and CHIP domain mutants, we found that SirT6 associates with a CHIP construct lacking the TPR domain (ΔTPR CHIP) and with a CHIP mutant that has an inactivating point mutation in the U-box domain (H260Q CHIP) but not with a CHIP mutant lacking the entire U-box domain (ΔU-box CHIP [Fig. 1D]). This indicates that CHIP's intact U-box domain is required for interaction with SirT6.
We next tested whether reintroduction of CHIP in CHIP-depleted cells could increase SirT6 protein levels. As shown in Fig. 2E and F, transient transfection of WT CHIP into shCHIP cells (Fig. 2E) or CHIP−/− MEFs (Fig. 2F) demonstrated that reintroduction of WT CHIP increased SirT6 protein expression. Furthermore, ectopic expression of WT CHIP had no effect on SirT6 mRNA levels (data not shown), indicating that the manner in which WT CHIP overexpression increases SirT6 protein expression is mediated at a posttranslational level. Expression of ΔU-box CHIP, which can interact with heat shock proteins but has no ubiquitin ligase activity and does not interact with SirT6, had no effect on SirT6 protein levels. However, expression of H260Q CHIP, which stably interacts with SirT6 but has no ubiquitin ligase activity, also had no effect on SirT6 protein levels, demonstrating that the CHIP-SirT6 interaction alone is not sufficient to increase SirT6 stability. Taken together, these data demonstrate that SirT6 protein stability is positively correlated with the presence of CHIP and requires CHIP's ubiquitin ligase activity. This suggests that CHIP could stabilize SirT6 indirectly by degrading a negative regulator of SirT6, or alternatively, CHIP could stabilize SirT6 through noncanonical ubiquitination, a mechanism previously described for CHIP's effect on Daxx (17).
CHIP noncanonically ubiquitinates SirT6 at K170.
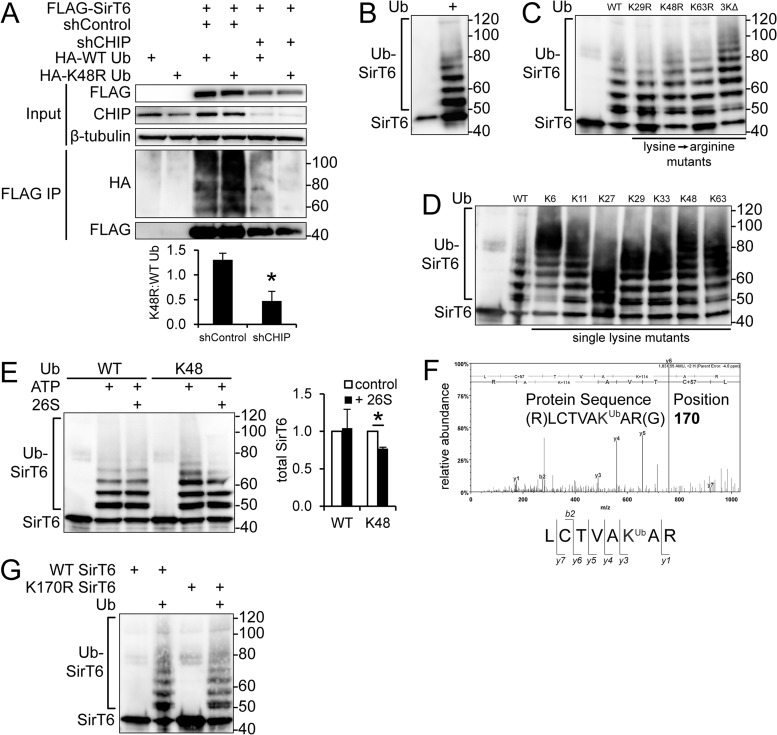
Because CHIP and SirT6 stably interact and CHIP ubiquitin ligase activity is required to enhance SirT6 stability, we hypothesized that CHIP noncanonically ubiquitinates SirT6. To determine the contribution of CHIP to SirT6 ubiquitination, shControl and shCHIP cells stably expressing FLAG-SirT6 were transiently transfected with HA-tagged WT ubiquitin or K48R mutant ubiquitin, which cannot form canonical linkages associated with proteasomal degradation. All cells were incubated in proteasome inhibitor prior to harvest and lysed in the presence of a deubiquitinase inhibitor to preserve SirT6 ubiquitination. As shown in Fig. 3A, immunoprecipitation analysis demonstrated SirT6 ubiquitination in both shControl and shCHIP cells transfected with WT ubiquitin (lanes 3 and 5). However, polyubiquitination involving K48R ubiquitin (noncanonical ubiquitination; lanes 4 and 6) occurred in shControl cells only, suggesting that CHIP is required for noncanonical ubiquitination of SirT6. Furthermore, the canonical ubiquitination of SirT6 in the absence of CHIP suggests the involvement of another, as yet unidentified ubiquitin ligase. Taken together, these results indicate that CHIP competes with other ubiquitin ligases to regulate SirT6 stability.
Fig 3.
CHIP ubiquitinates SirT6 with noncanonical chains at K170. (A) shControl and shCHIP 293 cells stably expressing FLAG-SirT6 were transfected with HA-tagged WT ubiquitin (Ub) or HA-tagged K48R ubiquitin and cultured in MG132 for 16 h. Protein levels of input and FLAG-immunoprecipitated samples were visualized via Western blot analysis. The K48R/WT ubiquitination ratio is shown below (n = 3). (B to E) in vitro ubiquitination reactions were performed using recombinant CHIP, SirT6, and ubiquitin. Reaction mixtures containing WT ubiquitin (B), WT ubiquitin or ubiquitin mutants with the indicated lysines mutated to arginine (C), WT ubiquitin or ubiquitin mutants in which the lysine indicated is the only lysine available for ubiquitination (D), or WT ubiquitin or K48-only ubiquitin with the addition of purified 26S proteasome (E) were used to perform ubiquitination reactions and visualized via Western analysis; representative images of at least 3 experiments are shown. (F) Monoubiquitinated SirT6 was analyzed using LC–MS-MS, and a single ubiquitination modification was discovered at K170. (G) in vitro ubiquitination reactions were performed using CHIP, WT ubiquitin, and WT SirT6 or K170R SirT6. For all ubiquitination reactions, representative images are shown (n = 3). The results represent means and SEM. *, P < 0.05. Graphed results represent SirT6 protein relative to β-tubulin and normalized to control in arbitrary units.
To test whether CHIP directly ubiquitinates SirT6, we performed in vitro ubiquitination reactions with recombinant CHIP, SirT6, and ubiquitin proteins. As shown in Fig. 3B, we found that SirT6 is a substrate for CHIP-mediated polyubiquitination. We then employed a variety of ubiquitin mutants to determine whether CHIP demonstrates a preferential ubiquitin linkage. As shown in Fig. 3C, we first used ubiquitin mutants in which the indicated lysine had been mutated to arginine and found that SirT6 could be ubiquitinated by any of these arginine mutation molecules, including K48R (indicative of a noncanonical chain), or even by a ubiquitin molecule lacking K29, K48, and K63 (3KΔ). Furthermore, in vitro reactions with ubiquitin mutants expressing a single lysine demonstrated that CHIP can polyubiquitinate SirT6 with chains linked through any individual lysine (Fig. 3D). This is entirely consistent with previous data demonstrating that CHIP and its partner E2 enzyme, UbcH5, can ubiquitinate a model substrate with a variety of linkages (15) and suggests that CHIP might directly decorate SirT6 with noncanonical ubiquitin chains. Furthermore, as shown in Fig. 3E, addition of purified 26S proteasome to an in vitro ubiquitination assay mixture containing WT ubiquitin did not lead to the disappearance of ubiquitinated SirT6; however, there was a reduction in ubiquitinated SirT6 when a canonical chain was added by including a ubiquitin mutant expressing lysine at position 48 only (K48). Together these data indicate that CHIP can ubiquitinate SirT6 and most likely does so with noncanonical chains.
To identify the target residue within SirT6 that is modified by CHIP-dependent ubiquitination, in vitro ubiquitination reactions using CHIP, SirT6, and WT ubiquitin were separated by gel electrophoresis, and the resulting gel was stained. Trypsin digestion of the band corresponding to monoubiquitinated SirT6 followed by liquid chromatography-tandem mass spectrometry (LC–MS-MS) analysis revealed a single ubiquitination site at K170 (Fig. 3F), which is found within the core sirtuin domain (29) and is conserved across human, rodent, chicken, and bovine SirT6 ortholog protein sequences. We mutated the lysine at position 170 in WT SirT6 to arginine (K170R SirT6) to determine whether this residue contributes to overall SirT6 protein stability. in vitro ubiquitination reactions demonstrated that recombinant K170R SirT6 is capable of being polyubiquitinated (Fig. 3G). This was somewhat surprising, as we expected to observe less ubiquitination on K170R SirT6. However, there are several possible interpretations of these results. One possibility is that CHIP's physical interaction with SirT6 promotes ubiquitination on one of the 18 other available lysine residues when K170 is not available, which may or may not be important in determining SirT6 stability. Because K170 is the preferred ubiquitination site, another possibility is that the remaining lysine residues in K170R SirT6 are not as readily ubiquitinated, but the reaction kinetics of in vitro ubiquitination are too rapid to detect a difference in ubiquitination in the short time frame of this experiment. In either case, the next logical step is to determine the stability of K170R SirT6 in cells with varying CHIP expression. Together, the data in Fig. 3 support the notion that CHIP ubiquitinates SirT6 at K170 with noncanonical chains and suggest that this might be a regulatory mechanism in determining SirT6 stability.
SirT6 K170 mediates susceptibility to protein degradation in the absence of CHIP.
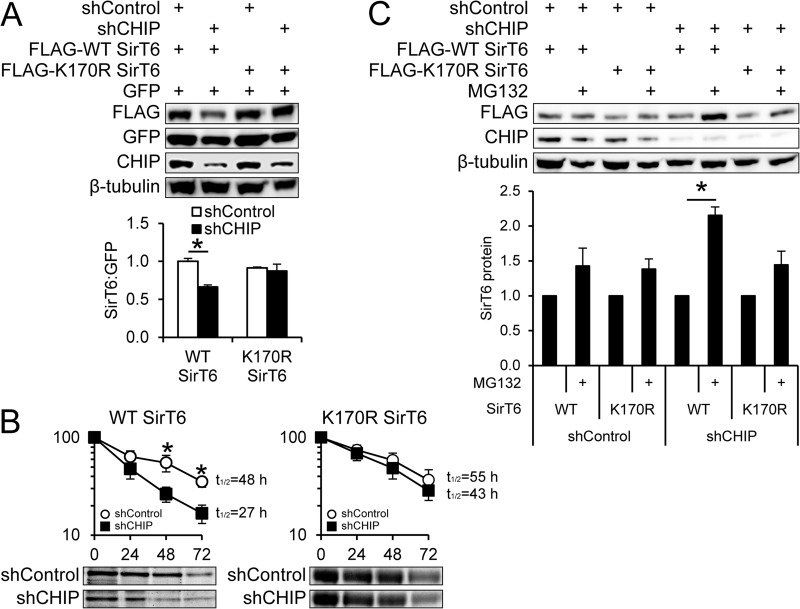
To determine whether SirT6 K170 has any effect on overall protein stability, we introduced WT SirT6 and K170R SirT6 in shControl and shCHIP cells and compared expression levels. We found that although WT SirT6 is less stable in the absence of CHIP, K170R SirT6 expression is unaffected by the presence or absence of CHIP (Fig. 4A). To directly quantify CHIP's effect on SirT6 protein stability, WT SirT6 and K170R SirT6 protein half-lives were measured via pulse-chase labeling experiments using 35S-labeled amino acids. Loss of CHIP reduced the WT SirT6 protein half-life by 43% (from 48 h in shControl cells to 27 h in shCHIP cells) (Fig. 4B). However, the K170R mutation completely rescued the SirT6 half-life in CHIP-depleted cells, so that the calculated half-life of K170R SirT6 in the presence or absence of CHIP was similar to the half-life of WT SirT6 in shControl cells (Fig. 4B).
Fig 4.
Mutant SirT6 has improved stability in CHIP-depleted cells. (A) shControl and shCHIP cells were cotransfected with green fluorescent protein (GFP) and WT SirT6 or K170R SirT6. Forty-eight hours later, the levels of transfected proteins were visualized via Western blotting, and SirT6 levels were calculated relative to GFP as an internal transfection control (below; n = 3). (B) Pulse-chase assays using 35S-labeled methionine/cysteine were used to measure protein half-lives of WT SirT6 and K170R SirT6 in shControl and shCHIP cells expressing FLAG-tagged SirT6 constructs. Following FLAG immunoprecipitation and autoradiography, protein levels were calculated at the indicated time points. Representative autoradiography images are shown below the corresponding graphs (n = 3). (C) WT SirT6 and K170R SirT6 were transiently expressed to achieve similar protein expression in shControl and shCHIP 293 cells and then split and treated for 16 h with DMSO or MG132. SirT6 protein levels were measured via Western blotting. The accumulation of WT SirT6 or K170R SirT6 caused by MG132 for each cell condition was quantified and expressed relative to DMSO treatment (below; n = 3). The results represent means and SEM; *, P < 0.05. Graphed results represent SirT6 protein relative to β-tubulin and normalized to control in arbitrary units.
Our data indicate that, in the absence of CHIP, SirT6 is canonically ubiquitinated (Fig. 3A) and degraded by the proteasome (Fig. 1D and E), which reduces WT SirT6 protein stability (Fig. 4A and B). To confirm whether K170R SirT6 is less susceptible to proteasomal degradation, we compared the effects of MG132 treatment on accumulation of WT SirT6 and K170R SirT6 in shControl and shCHIP cells. If K170 is the main site for canonical ubiquitination by a ubiquitin ligase competing with CHIP, it is assumed that mutation of this lysine would prevent MG132-induced SirT6 accumulation in CHIP-depleted cells. To control for differences in protein accumulation due to differences in initial protein levels, we employed a transient-transfection strategy so that the levels of FLAG-SirT6 expression were similar across all conditions and then split the cells and added vehicle or MG132 for 16 h. In shControl cells, proteasome inhibition did not lead to a statistically significant accumulation of either WT or K170R SirT6 (Fig. 4C), consistent with our data demonstrating a long half-life of SirT6 in the presence of CHIP. In contrast, proteasome inhibition in shCHIP cells resulted in a dramatic increase in WT SirT6 levels, similar to data in Fig. 1D and E. However, no MG132-induced accumulation of K170R SirT6 was observed in shCHIP cells, suggesting that the K170 residue is the predominant lysine involved in the proteasome-mediated degradation of SirT6.
Our data in Fig. 1 to 4 demonstrate that CHIP promotes SirT6 stability, that CHIP directly interacts with SirT6 through CHIP's U-box domain, that reintroduction of ubiquitination-competent CHIP increases SirT6 protein expression, that SirT6 is noncanonically ubiquitinated in the presence of CHIP, and that CHIP can directly ubiquitinate SirT6 with noncanonical chains at K170. In the absence of CHIP, WT SirT6 (but not K170R SirT6) is degraded in a proteasome-dependent manner, suggesting that K170 is decorated with canonical ubiquitin chains when CHIP is depleted. Taken together, these data suggest a model in which CHIP competes with a second ubiquitin ligase to noncanonically ubiquitinate SirT6 at K170, preventing canonical ubiquitination at K170 and promoting SirT6 stabilization. Although the overwhelming majority of CHIP substrates are degraded by the proteasome following canonical ubiquitination, the data presented here suggest a new role for CHIP in protein stabilization.
CHIP's stabilization of SirT6 results in decreased H3K9Ac levels and increased transcription of target genes.
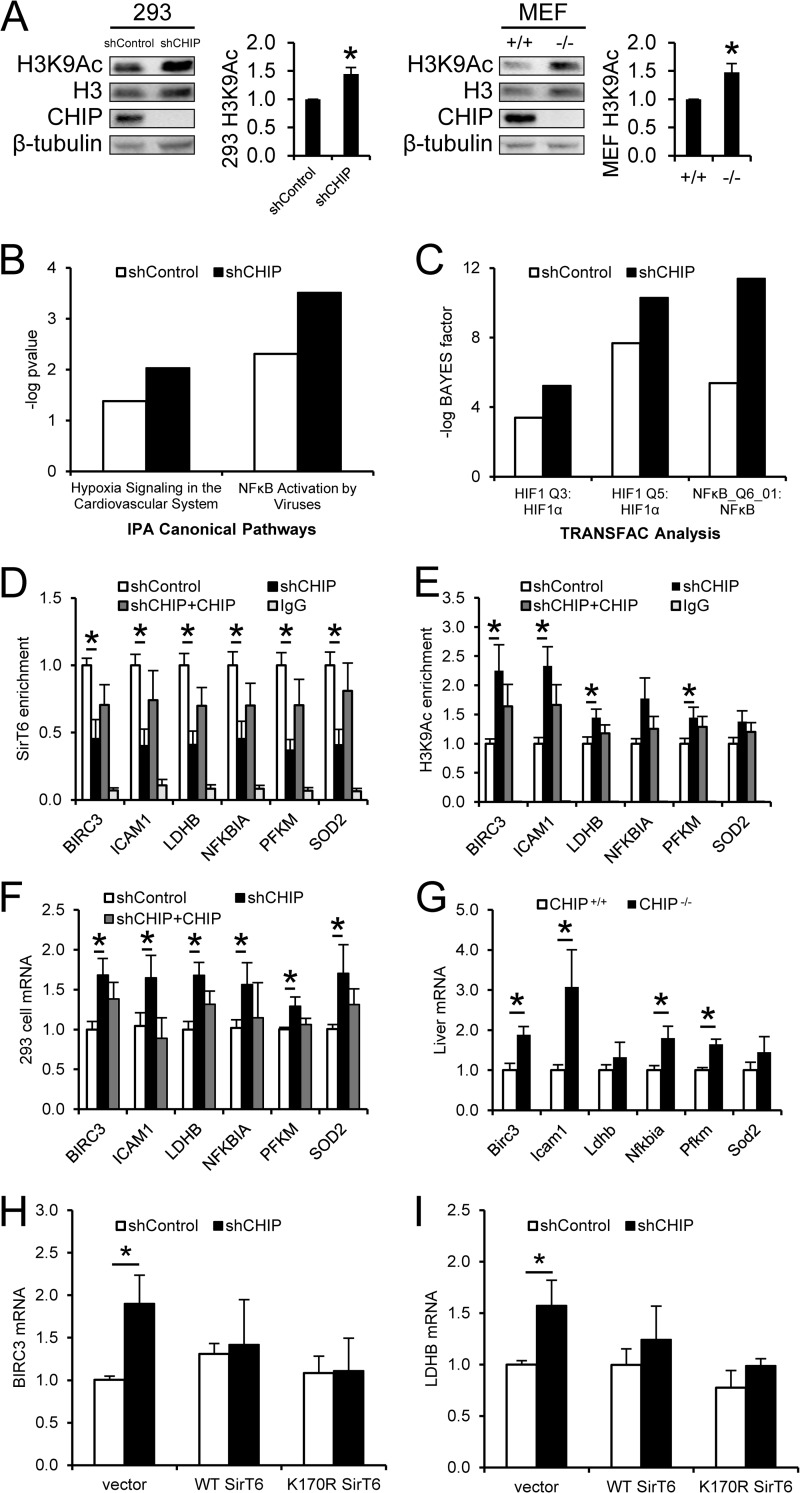
In order to understand the biochemical relevance of CHIP's effect on SirT6 stability, we examined the levels of acetylated histone H3K9 (H3K9Ac), a target of SirT6 KDAC activity (30), in control and CHIP-depleted cells. We hypothesized that, in the absence of CHIP, decreased SirT6 steady-state levels (Fig. 1B) should lead to accumulation of H3K9Ac. Using 293 cells and MEFs, we found that cells lacking CHIP also demonstrated increased H3K9Ac relative to total H3 (Fig. 5A).
Fig 5.
CHIP depletion decreases SirT6 and increases H3K9Ac promoter occupancy. (A) H3K9Ac was measured and expressed relative to total H3 via Western blot analysis in shControl and shCHIP 293 cells and in CHIP+/+ and CHIP−/− MEFs. Quantification is shown on the right of each Western blot (n = 3). (B and C) Analysis of H3K9Ac ChIP-seq data from shControl and shCHIP cells was performed by Ingenuity pathway analysis (B) and TRANSFAC (C), using peaks associated with the 5′ regions of annotated genes. (D and E) ChIP-qPCR for endogenous SirT6 (D) and H3K9Ac (E) was performed in shControl cells, shCHIP cells, or shCHIP cells overexpressing WT CHIP (shCHIP+CHIP). Protein enrichment within the promoter regions of the indicated genes was calculated relative to input and normalized to shControl levels (n = 3). (F and G) Real-time PCR was used to measure mRNA in 293 cells used in ChIP studies (F) and livers from CHIP+/+ and CHIP−/− mice (n = 3) (G). (H and I) shControl and shCHIP cells were transfected with vector, WT SirT6, or K170R SirT6 constructs; 48 h later, RNA was isolated. BIRC3 mRNA (H) and LDHB mRNA (I) were measured by qPCR and calculated relative to 18S (n = 3). The results represent means and SEM; *, P < 0.05.
SirT6 binds to the promoters of NF-κB-dependent inflammatory genes (31), hypoxia-inducible factor 1α (HIF-1α)-dependent glycolytic genes (32), and c-Jun-dependent IGF-related genes (26, 33), where SirT6 deacetylates H3K9Ac at these positions to inhibit transcription of downstream gene products. Conversely, SirT6 deletion increases H3K9Ac promoter enrichment and enhances transcription of these genes. Therefore, we hypothesized that the decreased WT SirT6 protein half-life (Fig. 4B) and increased H3K9Ac (Fig. 5A) measured in CHIP-deficient cells would correspond to increased H3K9Ac at the promoters of SirT6-regulated NF-κB- and HIF-1α-dependent genes. Furthermore, because overexpression of WT CHIP in shCHIP cells increases SirT6 protein stability (Fig. 2E and F), we hypothesized that CHIP reintroduction in shCHIP cells would ameliorate the effects of CHIP depletion. To obtain an unbiased understanding of how SirT6 depletion might affect H3K9Ac levels across the genome, we first performed ChIP with next-generation sequencing analysis (ChIP-seq) of H3K9Ac-enriched genomic regions. in silico analysis of H3K9Ac-enriched genomic regions was performed using. Ingenuity pathway analysis and TRANSFAC to identify representation in biological pathways and transcription factor binding sites, respectively. Consistent with the hypothesis that decreased SirT6 stability in shCHIP cells would lead to increased H3K9Ac at the promoters of SirT6 target genes, H3K9Ac enrichment was increased in transcriptional networks associated with HIF-1α and NF-κB in shCHIP cells (Fig. 5B and C).
To directly demonstrate whether decreased SirT6 protein stability in shCHIP cells affects SirT6 and H3K9Ac enrichment at promoters previously determined to demonstrate SirT6 occupancy, we performed ChIP with real-time PCR (ChIP-qPCR) for endogenous SirT6 and H3K9Ac at the promoters of several previously identified SirT6-regulated genes (31, 32) in shControl or shCHIP cells or shCHIP cells transiently overexpressing WT CHIP. In accordance with our data demonstrating an overall decrease in SirT6 and increase in H3K9Ac in CHIP-depleted cells, SirT6 enrichment was dramatically decreased at promoter regions in shCHIP cells (Fig. 5D), resulting in increased H3K9Ac enrichment at these positions (Fig. 5E). However, compared with shCHIP cells, CHIP overexpression in shCHIP cells slightly increased SirT6 enrichment and, conversely, decreased H3K9Ac enrichment. Since H3K9Ac promotes gene transcription, we expected that the mRNA of the genes controlled by these promoters would also be increased in CHIP-deficient cells, as has been observed in SirT6-depleted cells (31, 32). We found that mRNA levels of several gene products of the promoters measured in Fig. 5D and E were increased in shCHIP cells (Fig. 5F) and CHIP−/− mouse livers (Fig. 5G); however, CHIP overexpression blunted the effect of CHIP depletion on mRNA (Fig. 5F). The increase in SirT6-regulated mRNAs in CHIP-depleted cells was directly dependent on SirT6, as transient overexpression of either WT SirT6 or K170 SirT6 in shCHIP cells reduced mRNA levels to normal (Fig. 5H and I). Taken together, these data indicate that CHIP increases SirT6 stability, but in the absence of CHIP, the reduction in SirT6 protein half-life leads to decreased SirT6 promoter occupancy, causing increased H3K9Ac promoter occupancy and increased transcription of inflammatory and glycolytic genes. Thus, CHIP prevents aberrant NF-κB- and HIF-1α-dependent gene transcription by stabilizing SirT6, a negative regulator of these transcription factors.
CHIP and SirT6 are required for resistance to cellular stress.
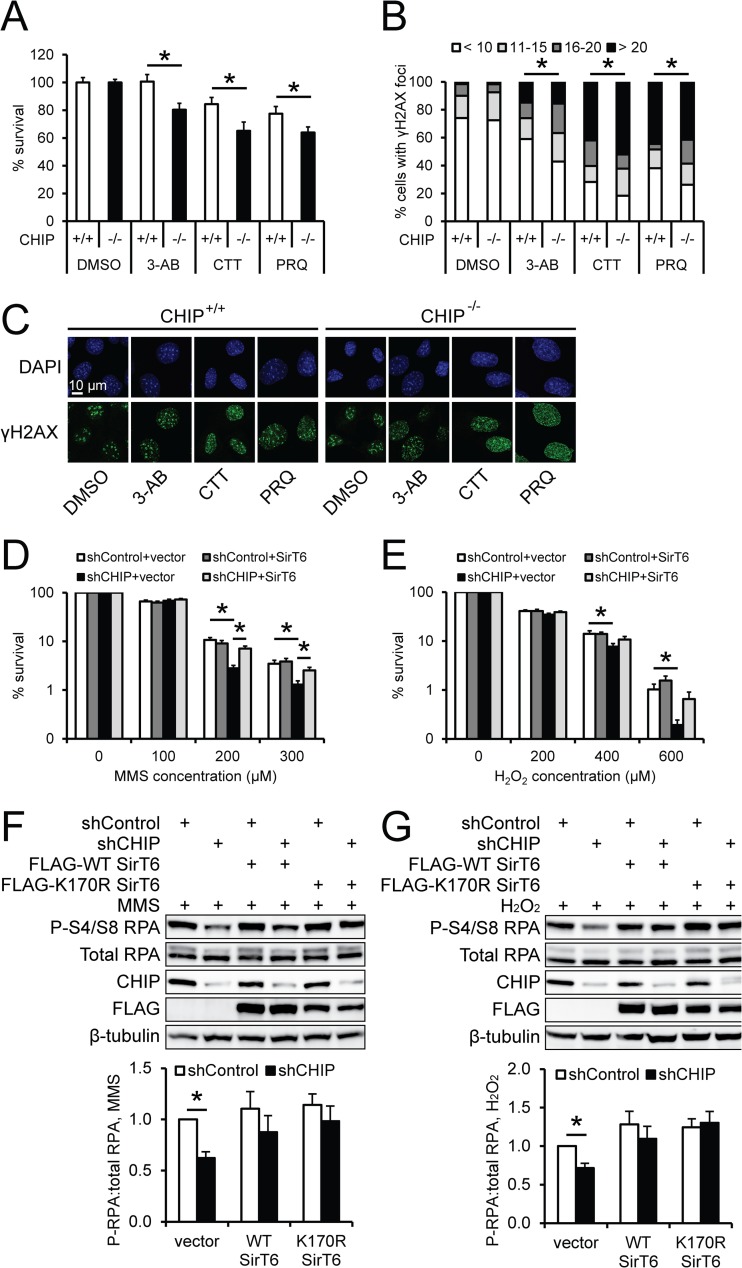
In addition to SirT6's role in transcriptional regulation, SirT6 also enhances base excision repair and double-strand break (DSB) repair by activating poly(ADP-ribose) polymerase 1, stabilizing DNA-PK, and deacetylating CtIP (34–36). Therefore, we explored whether CHIP depletion and subsequent effects on SirT6 protein stability also affect DNA repair mediated through SirT6. We first measured the effects of multiple DNA-damaging agents on viability and DSBs in CHIP+/+ and CHIP−/− MEFs. As shown in Fig. 6A, CHIP−/− cells demonstrated higher toxicity when exposed to 3-aminobenzamide, camptothecin, or paraquat. Quantification of γH2AX foci (an indicator of DSBs) showed that significantly more CHIP−/− cells had increased numbers of foci per cell than CHIP+/+ cells (Fig. 6B and C). To further test whether the loss of CHIP is associated with reduced DNA repair capacity, we exposed 293 cells to the DNA-damaging agent MMS or H2O2 and found that shCHIP 293 cells demonstrated less viability (Fig. 6D and E). However, this decrease in viability was directly linked to a decrease in SirT6 in these cells, as SirT6 overexpression in shCHIP-expressing cells (shCHIP plus SirT6) increased viability (Fig. 6D) or caused a trend toward increased viability (Fig. 6E). SirT6 depletion is associated with reduced phosphorylation of RPA, which is recruited to DNA repair sites and is a marker of successful homologous recombination (34). In accordance with our cell viability data, we found that RPA phosphorylation was decreased in shCHIP cells treated with MMS (Fig. 6F) or H2O2 (Fig. 6G). However, ectopic expression of WT SirT6 or K170R SirT6 in CHIP-depleted cells restored RPA phosphorylation levels. Although a plethora of proteins are involved in DNA damage response pathways, the viability and DNA damage phenotype produced in the absence of CHIP is strikingly similar to the phenotype reported for SirT6-depleted cells (34, 35) and indicates that CHIP is required for maximum SirT6-mediated DNA repair activity. The sum total of the data presented here demonstrate that the convergence of CHIP- and SirT6-related functions represents an intersection between PQC and epigenetics that supports the foundation for signaling associated with longevity.
Fig 6.
Decreased DNA repair in the absence of CHIP is rescued with SirT6 overexpression. (A) CHIP+/+ (white bars) and CHIP−/− (black bars) MEFs were cultured in DMSO, 3-aminobenzamide (3-AB), camptothecin (CTT), or paraquat (PRQ), and viability was assessed using an MTS assay (n = 3). (B) Following drug treatment, MEFs were fixed and stained for γH2AX and DAPI, and foci of >90 nuclei/condition were quantified. The results were clustered by the number of foci per cell and expressed as percentages of the total distribution. (C) Representative images from γH2AX (green) staining. DAPI (blue) is included as a nuclear stain. (D and E) shControl and shCHIP cells stably expressing empty vector or FLAG-SirT6 were treated with MMS (D) or H2O2 (E) for 6 days. The cells were stained with trypan blue and counted, and the percentage of viable cells was calculated (n = 3). (F and G) shControl and shCHIP cells were transiently transfected with vector, WT SirT6, or K170R SirT6. Forty-eight hours later, the cells were exposed to 200 μM MMS (F) or 400 μM H2O2 (G) for 1 h and then allowed to recover for 30 min. Cells were harvested, and the levels of total RPA and RPA phosphorylated at serines 4 and 8 were measured via Western blotting. Quantification of phosphorylated RPA normalized to total RPA is shown below the representative blots (n = 4 for each). The results represent means and SEM; *, P < 0.05.
DISCUSSION
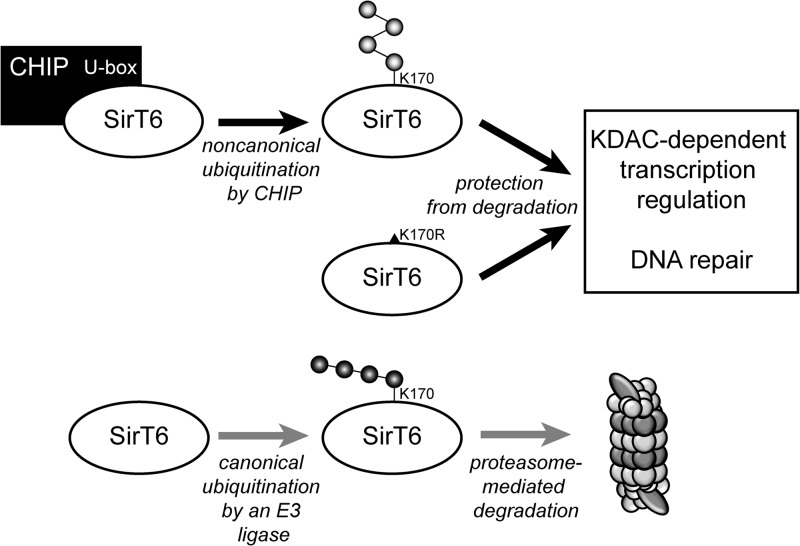
Cells possess complicated networks of quality control systems that facilitate the production and repair of functional nucleic acids and proteins. In the later stages of an organism's life span, by-products of normal molecular processes and damage from environmental insults gradually overwhelm the cell's ability to effectively counteract these stresses, which can lead to early senescence and the eventual deterioration of entire organ systems. The data presented here reveal a previously unknown intersection between two quality control systems that work in unison to facilitate the surveillance and repair of damage that would otherwise compromise cellular integrity. Our observations suggest that the ubiquitin ligase CHIP noncanonically ubiquitinates the sirtuin family member SirT6 at K170 and, by doing so, stabilizes and protects SirT6 from proteasome-dependent degradation mediated by a second, as yet undetermined ubiquitin ligase. This allows SirT6 to participate in histone deacetylation and DNA repair pathways (Fig. 7). Although the stable interaction between CHIP and SirT6 and evidence of ubiquitination from in vitro assays would indicate a direct mechanism for CHIP's effects on SirT6 stability, it is possible that CHIP stabilizes SirT6 through an indirect mechanism, such as degrading an E3 ligase that ubiquitinates SirT6 in the absence of CHIP. Future work will focus on identifying other factors involved in SirT6 stability and their potential regulation by CHIP. In addition, it is important that we use mass spectrometry methods to directly confirm ubiquitination of SirT6 at K170 and solidify evidence that the presence of CHIP affects ubiquitin chain linkages associated with SirT6.
Fig 7.
Model of SirT6 protein stability in the presence and absence of CHIP. Our data demonstrate that CHIP stably interacts with SirT6 in a manner dependent on CHIP's U-box domain. SirT6 is noncanonically ubiquitinated at K170 by CHIP, which prevents canonical ubiquitination at K170 and stabilizes SirT6. This allows SirT6 to exert KDAC and DNA repair activities. In the absence of CHIP, the availability of K170 allows canonical ubiquitination by another E3 ligase, leading to proteasomal degradation.
Aging cells experience reduced DNA repair activity and increased DNA lesions, and several progeria syndromes are characterized by major deficiencies in DNA repair proteins (37). SirT6 augments DNA repair processes and preserves genomic integrity by interacting with multiple DNA repair proteins (34–36), including the telomere-associated protein WRN, which specifically prevents telomere shortening and early cell senescence (30). Additionally, SirT6 KDAC activity targeting H3K9Ac blocks the transcription of genes regulated by the NF-κB subunit p65, as well as genes associated with glycolysis, glucose transport, and IGF signaling (26, 27, 31, 32), which are upregulated during aging (38, 39). Given these roles for SirT6, it is not surprising that a large portion of SirT6−/− mice demonstrate perinatal lethality due to dysregulation of glucose homeostasis (27). Similar to our observations of CHIP−/− mice (5), SirT6−/− mice that survive the perinatal period are smaller than WT littermates and exhibit a premature-aging phenotype characterized by loss of subcutaneous fat, decreased bone mineral density, lordokyphosis, and increased senescence-associated β-galactosidase staining (25). Although the average life span of SirT6−/− mice is substantially shorter than that of CHIP−/− mice, the similarity in the phenotypes is striking. Given the data demonstrating that SirT6 protein is dramatically decreased in CHIP−/− cells and tissues, it is likely that the loss of SirT6 in CHIP−/− mice is partially responsible for the dramatic aging phenotype observed in CHIP−/− mice.
The original discovery of CHIP has yielded many studies identifying substrates that are canonically ubiquitinated by CHIP and subsequently targeted to the proteasome for degradation (10, 13, 14). However, there is ample evidence demonstrating CHIP's capability to produce a variety of ubiquitin chains, both canonical and noncanonical in nature, and proteasomal degradation is not necessarily the result of CHIP-mediated ubiquitination (15, 40). The mechanics and biology of noncanonical ubiquitin chains are not fully understood, but our current understanding suggests that noncanonical chains have diverse effects on substrate protein localization, activity, and interactions with other proteins (16). Our recent work demonstrating that CHIP noncanonically ubiquitinates and sequesters Daxx describes a noncanonical ubiquitination event but is an example of how CHIP inhibits the activity of a ubiquitinated substrate (17). The data presented here also demonstrate CHIP's ability to noncanonically ubiquitinate the target substrate SirT6. However, in contrast to the biology of Daxx, CHIP-mediated SirT6 ubiquitination increases SirT6 stability and supports catalytic activity. Given that this is the second demonstration of CHIP-mediated noncanonical ubiquitination, it is possible that CHIP modifies other substrates in a similar manner. It will be exciting to identify other CHIP ubiquitination substrates and to determine whether their fate is degradation or stabilization. In summary, we have identified SirT6 as a substrate for CHIP-dependent noncanonical ubiquitination, which improves SirT6 stability and enhances SirT6 activity in supporting DNA repair and reducing transcription of inflammatory and glycolytic genes. The discovery of the CHIP-SirT6 interaction provides a bridge between protein homeostasis and epigenetic regulation, two networks essential to understanding the molecular biology of aging.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hemant Kelkar and Xiaojun Guan at the UNC Center for Bioinformatics for analysis of ChIP-seq data and Will Thompson at the Duke University Proteomics Core for identification of the ubiquitinated SirT6 residue. We also thank Andrea Portbury for helpful review of the manuscript.
This work was supported in part by NIH grants R01-GM061728 and R37-HL65619 and by the Fondation Leducq (to C.P.) and by NIH grants T32-ES007126 and F32-AG038061 (to S.M.R.). This research was conducted while S.M.R. was an Ellison Medical Foundation/AFAR Postdoctoral Fellow.
S.M.R., Y.W., and H.M. performed the experiments. S.M.R. performed statistical analysis and made the figures. S.M.R. and C.P. designed experiments and wrote the manuscript.
We have no conflicts of interest to report.
Footnotes
Published ahead of print 16 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00480-13.
REFERENCES
- 1.Fontana L, Partridge L, Longo VD. 2010. Extending healthy life span—from yeast to humans. Science 328:321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson FB, Sinclair DA, Guarente L. 1999. Molecular biology of aging. Cell 96:291–302 [DOI] [PubMed] [Google Scholar]
- 3.Kenyon CJ. 2010. The genetics of ageing. Nature 464:504–512 [DOI] [PubMed] [Google Scholar]
- 4.McDonough H, Patterson C. 2003. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones 8:303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Min JN, Whaley RA, Sharpless NE, Lockyer P, Portbury AL, Patterson C. 2008. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol. Cell. Biol. 28:4018–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morimoto RI. 2008. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22:1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai Q, Zhang C, Wu Y, McDonough H, Whaley RA, Godfrey V, Li HH, Madamanchi N, Xu W, Neckers L, Cyr D, Patterson C. 2003. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 22:5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. 2001. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276:42938–42944 [DOI] [PubMed] [Google Scholar]
- 9.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. 2006. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 440:551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3:100–105 [DOI] [PubMed] [Google Scholar]
- 11.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. 2002. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. U. S. A. 99:12847–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Xin H, Xu X, Huang M, Zhang X, Chen Y, Zhang S, Fu XY, Chang Z. 2004. CHIP mediates degradation of Smad proteins and potentially regulates Smad-induced transcription. Mol. Cell. Biol. 24:856–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. 2005. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J. Biol. Chem. 280:23727–23734 [DOI] [PubMed] [Google Scholar]
- 14.Dickey CA, Kamal A, Lundgren K, Klosak N, Bailey RM, Dunmore J, Ash P, Shoraka S, Zlatkovic J, Eckman CB, Patterson C, Dickson DW, Nahman NS, Jr, Hutton M, Burrows F, Petrucelli L. 2007. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 117:648–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. 2007. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282:17375–17386 [DOI] [PubMed] [Google Scholar]
- 16.Komander D, Rape M. 2012. The ubiquitin code. Annu. Rev. Biochem. 81:203–229 [DOI] [PubMed] [Google Scholar]
- 17.McDonough H, Charles PC, Hilliard EG, Qian SB, Min JN, Portbury A, Cyr DM, Patterson C. 2009. Stress-dependent Daxx-CHIP interaction suppresses the p53 apoptotic program. J. Biol. Chem. 284:20649–20659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11:437–444 [DOI] [PubMed] [Google Scholar]
- 19.Hallows WC, Lee S, Denu JM. 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U. S. A. 103:10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai Q, Qian SB, Li HH, McDonough H, Borchers C, Huang D, Takayama S, Younger JM, Ren HY, Cyr DM, Patterson C. 2005. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J. Biol. Chem. 280:38673–38681 [DOI] [PubMed] [Google Scholar]
- 21.Zhou P. 2004. Determining protein half-lives. Methods Mol. Biol. 284:67–77 [DOI] [PubMed] [Google Scholar]
- 22.Homma S, Jin X, Wang G, Tu N, Min J, Yanasak N, Mivechi NF. 2007. Demyelination, astrogliosis, and accumulation of ubiquitinated proteins, hallmarks of CNS disease in hsf1-deficient mice. J. Neurosci. 27:7974–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ. 1999. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 18:5943–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan LJ, Christians ES, Liu L, Xiao X, Sohal RS, Benjamin IJ. 2002. Mouse heat shock transcription factor 1 deficiency alters cardiac redox homeostasis and increases mitochondrial oxidative damage. EMBO J. 21:5164–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. 2006. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124:315–329 [DOI] [PubMed] [Google Scholar]
- 26.Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V, Pillai VB, Ravindra PV, Gupta M, Jeevanandam V, Cunningham JM, Deng CX, Lombard DB, Mostoslavsky R, Gupta MP. 2012. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat. Med. 18:1643–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao C, Kim HS, Lahusen T, Wang RH, Xu X, Gavrilova O, Jou W, Gius D, Deng CX. 2010. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J. Biol. Chem. 285:36776–36784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haigis MC, Sinclair DA. 2010. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5:253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tennen RI, Berber E, Chua KF. 2010. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech. Ageing Dev. 131:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. 2008. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452:492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. 2009. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 136:62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. 2010. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140:280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao C, Wang RH, Lahusen TJ, Park O, Bertola A, Maruyama T, Reynolds D, Chen Q, Xu X, Young HA, Chen WJ, Gao B, Deng CX. 2012. Progression of chronic liver inflammation and fibrosis driven by activation of c-JUN signaling in Sirt6 mutant mice. J. Biol. Chem. 287:41903–41913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaidi A, Weinert BT, Choudhary C, Jackson SP. 2010. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 329:1348–1353 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. 2011. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332:1443–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, Bohr VA, Chua KF. 2009. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging 1:109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. 2005. DNA repair, genome stability, and aging. Cell 120:497–512 [DOI] [PubMed] [Google Scholar]
- 38.Adler AS, Sinha S, Kawahara TL, Zhang JY, Segal E, Chang HY. 2007. Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev. 21:3244–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holt SJ, Riddle DL. 2003. SAGE surveys C. elegans carbohydrate metabolism: evidence for an anaerobic shift in the long-lived dauer larva. Mech. Ageing Dev. 124:779–800 [DOI] [PubMed] [Google Scholar]
- 40.Alberti S, Demand J, Esser C, Emmerich N, Schild H, Hohfeld J. 2002. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J. Biol. Chem. 277:45920–45927 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.