Abstract
Three interlocking problems in gene regulation are: how to explain genome-wide targeting of transcription factors in different cell types, how prior transcription factor action can establish an ‘epigenetic state’ that changes the options for future transcription factor action, and how directly a sequence of developmental decisions can be memorialized in a hierarchy of repression structures applied to key genes of the ‘paths not taken’. This review uses the finely staged process of T-cell lineage commitment as a test case in which to examine how changes in developmental status are reflected in changes in transcription factor expression, transcription factor binding distribution across genomic sites, and chromatin modification. These are evaluated in a framework of reciprocal effects of previous chromatin structure features on transcription factor access and of transcription factor binding on other factors and on future chromatin structure.
Keywords: DNA binding, histone methylation, DNA methylation, lineage commitment, repression, hematopoiesis
THE PROBLEM OF TRANSCRIPTION FACTOR TARGETING
Multicellular biological systems frequently use the same transcription factors in different tissues to participate in control of completely different gene sets. A central question for many years has been how transcription factors are guided to these distinct assignments. The binding specificities of factors as measured in purified systems are frankly inadequate to account for the small number of sites of occupancy observed in a given cell type by genome-wide mapping analysis (chromatin immune precipitation analyzed by deep sequencing, ‘ChIP-seq’, or by hybridization to unique sequence arrays, ‘ChIP-chip’), relative to the number of potentially equivalent motifs in a mammalian genome [1–7]. A common finding in ChIP-seq analyses is that 103–104 sites are generally seen to be bound; although this seems like a large number of potential target sites, it is a small fraction out of ∼106 apparently equivalent motifs per genome. In fact, the problem is even harder. Genome-wide protein binding data have confirmed that the same factor actually binds selectively to different genomic sites in different cell types or at different stages in a developmental process [3, 5, 8–10]. Thus, not only does the factor fail to engage potentially high-affinity sites in any one context, it also rejects in one context the same sites that it does engage with high affinity in another. These results mean that to understand how a transcription factor works to control gene expression, one must first understand how it discriminates between binding sites it may and may not occupy in a particular cell type.
In general, there are two nonexclusive ways to answer this question [11]. One is to invoke combinatorial transcription factor action: genomic regions with multiple transcription factors binding in a given cell type are particularly likely to be active cis-regulatory elements in that cell type [2, 11–16]. Indeed, transcription factor binding to a given site may be unstable unless facilitated by neighboring occupancy by another transcription factor or factors [17]. The most dramatic versions of this possibility are those where two transcription factors directly interact at the protein level, either to create a heterodimer with a distinctive specificity [1, 18–20] or to induce conformational changes that mutually increase DNA binding affinity at combined sites. Well-documented cases include the Ets1–Runx1 [21, 22] and Ets1–Pax5 [23] interactions, which relieve autoinhibitory structures in the two factors. However, combinatoriality need not be as specific or direct as this in all cases. Many transcription factor quorum sensing phenomena, like those that apparently mediate function of the IFNβ and IL-2 enhancers [24, 25], could involve stabilization of varied combinations of factors once a threshold has been crossed to recruit p300 or CBP co-activators. This could enhance binding of all participating factors because the engagement of the large co-activator complex creates another affinity trap at the site besides affinity for the DNA itself. Many variations of co-occupancy that could result in such ‘AND logic’ for enhancer activation can be envisioned.
The other way to address the question of transcription factor binding selectivity is to invoke selective masking of large parts of the genome from accessibility, and to posit that different regions of the genome are masked in different cell types [5, 11]. This is generally considered to be mediated through ‘epigenetic modifications’ of nucleosomal structure that divide the genome into open and closed chromatin, with the partitioning different in different cell types. Indeed, evidence shows that many transcription factors can be selectively recruited to regions that are marked with particular histone modifications correlated with open chromatin, e.g. histone H3K4me1 and H3K4me2 rich regions which are free of H3K27me3 or H3K9me3 marks (e.g. [3, 8, 13, 26–28]). However, this mechanism raises two substantial questions. First is how these marks come to be set so selectively. The second is under what conditions these modifications are not just symptoms of local transcription factor activity, but in fact act as a constraint on future access of transcription factors to particular binding sites.
Transcription factor action frequently results in local alteration of histone modifications, nucleosome spacing and DNA methylation, all aspects of chromatin that have been termed ‘epigenetic’marks. The mechanisms clearly involve recruitment of histone modification enzymes to local sites by complexing with transcription factors on the DNA (e.g. [3, 13, 29]). Although mechanisms exist to propagate such epigenetic marks through DNA replication once they are in place, it is clear that any changes in epigenetic marks follow the activity of the transcription factors at these sites. The prior transcription factor binding that has recruited the chromatin marking complexes to generate a pattern of more and less accessible sites may help to solve the specificity problem for future transcription factor binding (e.g. for Runx1 sites in hematopoietic cells, [30]). However, the rules that relate transcription factor binding to chromatin modification are not yet general enough for function to be inferred simply from occupancy.
MODES OF TRANSCRIPTION FACTOR INTERACTION WITH CHROMATIN STATES: IMPLICATIONS OF COMBINATORIALITY AND COMBINATORIALITY ACROSS TIME
One striking result emerging from genome-wide transcription factor occupancy analysis is that factors are commonly found binding to cis-regulatory elements of many genes that do not seem to be functional targets, in addition to those of genes that they do regulate (for hematopoietic examples, see [5, 8, 28, 31]). The ‘nonfunctional’ binding is selective, in the sense that it is occurring at accessible sites while other potential sites are not bound and this binding can be quite strong, with steady-state occupancy at these sites sometimes ranking among the highest peaks in the genome for that transcription factor. Thus, most excess binding is not simply an analytical error of setting a threshold for peak calling at too permissive a level. Nevertheless, the genes associated with many binding sites may be uncorrelated in expression with the transcription factor gene and unaffected by perturbation of the transcription factor’s activity. On a case-by-case basis, some of these enigmatic occupancy sites can be explained by looping to some distant gene that is the true functional target; alternatively, if the neighboring gene is not affected by deletion of the transcription factor gene, then its continued expression may be explained by complementation of the factor’s role with a co-expressed related factor. But this is not a minor aspect of transcription factors’ engagements with the genome: for a factor that appears to regulate no more than ∼102–103 genes, there may be as many as 10 times more genes that have linked, robustly occupied binding sites [5, 8, 28, 31].
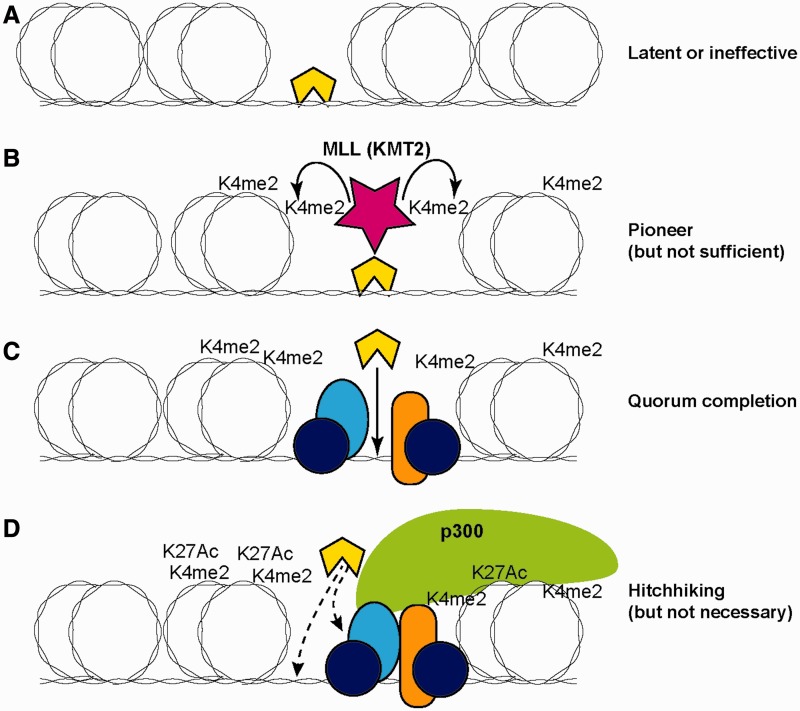
Although this is a problem for predicting the target genes of a given transcription factor, it yields important clues about the ways that combinatorial transcription factor action works. From the transcription factor’s ‘perspective’, the important features of the genome are not genes as such, but target sequences of greater and lesser accessibility and with more or fewer potentially collaborative transcription factors already bound [11]. Transcription factor effects on promoter activity depend on binding at cis-regulatory elements which can loop to the promoter to deliver or release RNA polymerase. But, binding can also cause effects on other transcription factors’ binding and future chromatin state, even if these do not reach the endpoint of immediately turning on or off transcription. Figure 1 illustrates how the same level of transcription factor occupancy can reflect four different functional cases.
Figure 1:
Functional modes of transcription factor engagement in a chromatin context. The transcription factor of interest is represented by a yellow (light color) polygon. Double coils around an oval represent DNA wrapped around nucleosomes. (A) Binding alone to a temporarily accessible site, without activation of histone modifications or redistribution. (B) Binding stably enough to recruit local histone modifying enzymes (MLL = KMT2, histone H3 Lys 4 methyltransferase) and enhance access to DNA, thus providing a local pioneering effect even if not conferring immediate transcriptional regulatory function. K4me2 = H3K4me2. (C) Binding to complete a ‘regulatory quorum’. Factor of interest is selectively recruited to join a pre-established assemblage of other transcription factors at a cis-regulatory site that was poised but not functionally active until the factor joined. Recruitment in this case is shown by protein–DNA binding, but may also include protein–protein binding. (D) ‘Hitchhiking’: factor binding adventitiously to a cis-regulatory element at which function is already established independently of the factor of interest, and unaffected by the presence of the factor of interest. Factor may bind to exposed DNA sites or through interaction with other recruited proteins or histone marks. Element diagrammed is an activating enhancer element where H3K27Ac (H3K27Ac) modifications have occurred and the p300 co-activator has been recruited already; however, hitchhiking can also occur at active promoters where the marks are different.
If transcriptional activators or repressors simply worked alone, as is often assumed by using terms like ‘master regulator’, then occupancies should indeed be neatly separable into categories of functional versus nonfunctional. However, even in biological contexts where a factor is rate limiting, it is likely not to be working alone. Careful dissections of developmentally controlled cis-regulatory elements have repeatedly emphasized that combinations of distinct transcription factors need to work together at given cis-regulatory modules to make a gene turn off or on correctly [32–34]. Combinatorial transcription factor action makes the effects of a factor conditional on other factors and this condition can affect binding; in addition, it affects the impact of any binding that occurs on the target gene’s transcription (activation or repression).
In a case where a factor is binding to its target site, but a needed co-factor is not present, then a neighboring transcription unit may not respond. The factor’s occupancy may be entirely nonfunctional (Figure 1A), but may alternatively serve as a pioneer (Figure 1B), to establish a beachhead for future transcription factor recruitment. Precedents for this kind of pioneering occupancy include PU.1 at many sites in myeloid precursors and macrophages and Runx1 in hematopoietic progenitors [3, 30, 35]. Often, pioneering function is seen when the bound transcription factor recruits histone-modifying enzymes to initiate a change in chromatin state [3, 13], even when no transcriptional regulatory effect on the target gene is seen. This sets the conditions to enhance targeting of later activated factors to the same site, when a transcriptional effect finally results.
Conversely, a transcription factor can be found selectively binding to cis-regulatory elements that have active histone marks and seem to be engaged in transcriptional regulatory function. This can indeed be the result of coordinated transcription factor binding that establishes an active cis-regulatory module. But in some cases, the transcription factor itself may be unable to initiate chromatin opening at the cis-regulatory module, or even to find the right site to occupy on its own; it may depend on other, pre-bound factors to do this (Figure 1C and D), something that is only detected if the regulatory steps leading up to the transcription factor’s occupancy can be tracked. This kind of binding is readily assumed to be functional, but again it may reflect two rather different roles. If the factor is functional, it may be because it provides the final member of the transcription factor combination that is needed to make a ‘regulatory quorum’ to activate or repress the target gene (Figure 1C). For example, Foxp3 in T cells becoming T-regs is recruited only to sites that already have Runx1, Ets family factor, FoxO1 or activation-dependent transcription factors already bound, although Foxp3 then radically changes the impact of activation [36]. Similarly, binding of Signal Transducer and Activator of Transcription (STAT) factor STAT4 or STAT6 occurs at sites that were already marked for accessibility, in Th1 and Th2 cells, respectively, but the co-activator p300 is only recruited when the STAT factor is engaged [37]. Some of the later-activated macrophage factors that are recruited to sites where PU.1 is a pioneer are also rate limiting for transcriptional responses [3, 35]. Yet, transcription factors can also be recruited to active sites as ‘hitchhikers’, simply because those sites are highly accessible and rich in interaction surfaces (Figure 1D), even when the factors stick there without being essential for transcriptional regulation at all. Co-recruitment of multiple factors to the same open and active cis-regulatory elements is common, whether or not they are coordinately exerting function there [12, 16, 38]. The hitchhiking phenomenon is a vexing problem for interpretation of genome-wide binding analyses, but it sheds light on the need to consider functionality in terms of transcription factor complexes rather than single transcription factor binding events.
The roles that a transcription factor can play in gene regulation are thus complicated, but also expanded by the ability of sub-quorum transcription factor binding to alter chromatin state. Not only can this contribute a necessary function even when it is not sufficient; more interestingly, it can contribute a combinatorial function that works across time. For example, active chromatin marks are already established at multiple B-cell and T-cell genes before they are expressed, even in a hematopoietic stem cell [27, 39, 40]. In the case of hematopoietic cells, this pattern of accessibility may be partly due to pioneering of factors like Runx and Ets family members that are consistently present throughout hematopoiesis and have their target sites over-represented among cis-regulatory elements that will later be bound by most lymphocyte transcription factors. Although immediate changes in transcription of target genes in response to a factor’s activation or removal are still the clearest evidence for function, it is likely that local changes in histone modification in response to the factor’s binding or removal can also be indicators of potentially important biological effects.
THE MISSING FUNCTIONS: REPRESSION AND LAYERS OF SILENCING
Although most experimental evidence for mechanisms to date has been focused on transcription factor effects in gene activation, a most important aspect of regulation across time is the establishment of repression. Taking the longest view, tissue-specific genes are generally not expressed in the initial fertilized egg [41], and it could be imagined that repression generally is just a default continuation of an initial silent state when it is not directly overcome by transcriptional activators. However, there are many clear developmental paths through which genes become activated before they are silenced, and these obviously require a cause of repression. Furthermore, there are hierarchies of gene silencing which are distinct from simple lack of activation and may involve different levels of irreversibility [42].
An example is the state of genes involved in particular T-cell effector functions, genes involved in Th1 (e.g. Ifng, Tnf), Th2 (e.g. Il4, Il13) and Th17 (e.g. Il17a, Il17f) responses to antigen (reviewed by [43]). None of these effector genes is transcriptionally active in the resting state in which most naïve CD4+ T cells exist in the body for weeks or months. Yet, they may all be accessible to activation in these cells, if the cells encounter antigenic stimulation in appropriate conditions. Even so, under continued stimulation, the activated T cells differentiate into one of these subtypes, and when that happens the genes associated with the alternative options become progressively more difficult to activate than they were in the naïve state. The DNA around the unused genes becomes more methylated and repressive histone marks are deposited. This is clearly a ‘more silent’ state than the initial one.
Even more functionally silent are the genes that would have been used by the hematopoietic precursors of the T cells, if they had differentiated into nonlymphoid lineages instead of T cells. These genes become permanently inaccessible during the course of T-cell development. These are clearly some form of transcription factor effects exerted on target gene accessibility across long time scales. As we will discuss, however, our existing knowledge of chromatin accessibility mechanisms leaves some major questions about how different levels of silence are established and maintained.
DEVELOPMENTAL SYSTEMS AS A TEST OF THE IMPACT OF CHROMATIN MARKING
The best biological systems in which to consider how chromatin modification affects gene expression are developmental processes in which cells traverse a hierarchy of sequential, irreversible gene expression states. These processes result in gene regulatory state ‘decisions’ that persist later even under highly varied environmental conditions. Not only do certain genes turn on and remain on stably, but also other genes are turned off permanently. The long-term silencing of some genes is an important point in these developmental processes, because such genes are not only not transcribed, but prohibited from future expression. Knowing the genes involved in these cases, it is possible to test how known chromatin modifications may play a role in the irreversibility of the process and in the reasons why a factor that may be involved in multiple steps could play different roles in the earlier steps than in the later ones.
Hematopoiesis in mammals provides excellent examples of a hierarchical developmental process that installs diverse, lasting gene regulatory states in descendants of a multipotent precursor. Lineage commitment occurs through a sequence of partial restriction events that eliminate particular developmental options for the cells, until only one possible fate is left. This implies that commitment has repression at its core. Each fate is based on establishing stable expression of a distinct combination of transcription factors, although individual transcription factors often play roles in more than one end state. Thus, it is very informative to relate the binding patterns of individual transcription factors in vivo to their shifting regulatory contexts, in different hematopoietic developmental intermediates and end states.
T cell development is a particularly useful model for this, because the intermediates between stem cells and committed pre-T cells are so well defined [44–46]. Throughout early life and young adulthood, multiple waves of T-cell precursors need to migrate to the thymus to begin their developmental program, entering this specialized organ while they are still in a highly uncommitted state. The cells that enter can still generate other kinds of lymphocytes and also several different kinds of myeloid and dendritic cells, depending on environmental conditions. These options then get eliminated, one by one, as the cells begin to differentiate under the influence of Notch-pathway signaling from the thymic microenvironment. Commitment to the T-cell pathway is complete even before the cells finish the gene rearrangement events that will establish their later antigen recognition specificity. However, the cells require multiple cell cycles to undergo full commitment, ranging up to >12 cell generations in the steady-state young adult mouse thymus [47, 48]. In the process, they change their cell surface phenotypes in a way coordinated with their changes in regulatory state, and this has enabled a well-defined sequence of intermediates to be defined. Thus, cells at specific intermediate states in the developmental commitment process can be prospectively isolated based on surface markers and analyzed for the gene expression, transcription factor binding patterns and functional roles of those transcription factors that are responsible for these developmental properties.
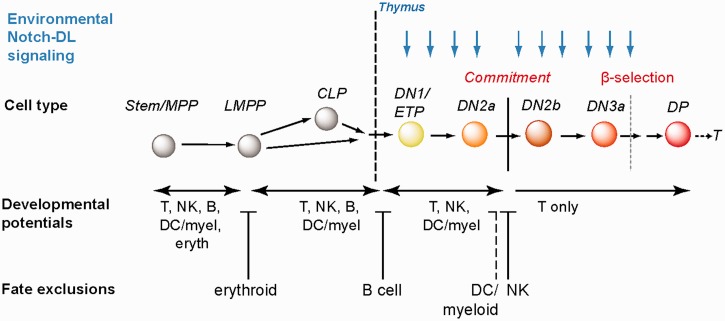
T-lineage commitment involves both positive and negative regulation events (Figure 2). Negative regulation is the basis for the loss of access to particular alternative fates and because particular fates are irreversibly blocked off at different stages, negative regulatory mechanisms must act sequentially. Does sequentiality mean that there are different ‘depths’ of repression applied to genes involved in the earliest fates to be excluded than in the last ones to be excluded? The genes that are permanently decommissioned from use during T-lineage commitment provide a good test case to see whether there is a hierarchy of repressive mechanisms defined by our knowledge that reflect the order of the exclusion of different developmental fates.
Figure 2:
Schematic of T-cell development with emphasis on stages when multipotency is narrowed and then relinquished. Blue vertical arrows indicate the stages at which Notch pathway signaling within the thymus is required for progression. Labels above the cell cartoons depict common names for the stages. For prethymic cells: MPP, multipotent progenitor; LMPP, lymphoid-primed multipotent progenitor; CLP, common lymphoid progenitor. For intrathymic cells, DN = CD4-CD8-TCR-stages; substages (DN1, 2a, 2b, etc.) are distinguished by expression of Kit, CD25, CD44, and other markers including CD27 and CD28. ETP, early T-cell precursor, same as Kit-high DN1. β-Selection indicates first stage when proliferation and survival depend on expression of a part of the T-cell receptor complex, a phase of rapid proliferation and differentiation triggered by the successful rearrangement of the TCRβ chain gene. DP = CD4+ CD8+ TCRβ+ cells. DC = dendritic cell potential; myel = myeloid (macrophage and/or granulocyte) potential; eryth = erythrocyte potential; NK = natural killer potential. For explanation, see text.
A MOLECULAR TARGET DEFINITION OF T-CELL LINEAGE COMMITMENT
In hematopoiesis, the crucial transcription factors that direct gene expression in a particular cell type and the crucial growth factor receptors that sustain viability in cells adopting that fate are well defined for most, if not all hematopoietic fates. Thus, each lineage exclusion step of the commitment process can be understood as permanent loss of access to at least one critical transcription factor and/or growth factor receptor that would have been needed for that lineage.
The order in which T-cell precursors lose access to these alternatives is shown in Figure 2 (reviewed in [44]): (1) the cells first lose access to the erythroid and megakaryocytic (platelet-forming) fates, before they enter the thymus; (2) many of the precursors then lose some efficiency at generating myeloid cells before entering the thymus, but this is optional and not important for their ultimate ability to generate T cells; (3) soon after entering the thymus (probably earlier in fetal T cells), they lose the ability to generate B cells; (4) after multiple cell divisions in the thymus, they fully lose the ability to generate macrophages, granulocytes and dendritic cells; and (5) in parallel or soon afterwards, they lose the ability to generate full-fledged natural killer (NK) cells. However, some NK-like effector gene expression remains within the repertoire of T-cell responses, and so access to aspects of this fate may never be fully eliminated by the T-lineage. Can the repression mechanisms that block NK-cell differentiation in early T cells be distinguished from those that much earlier eliminate the erythroid option?
T-cell-specific regulatory factors are turned on in early stages of T-cell development before lineage commitment and two of the most important are GATA-3 and TCF-1 (encoded by Tcf7). These factors are induced in the context of a set of broadly used transcription factors: Myb, Ikaros (Ikzf1), E2A (Tcf3 = Tcfe2a) and Runx1/CBFβ, which play numerous roles in pioneering and direct transcriptional activation in several blood lineages. Initially, early T cells also share with stem and progenitor cells expression of a suite of progenitor-associated regulatory genes, e.g. Gata2, Tal1, Lyl1, Gfi1b, Sfpi1 (encoding PU.1) and the thrombopoietin receptor gene Mpl. These stem and progenitor-specific genes become silenced in the course of T-cell lineage commitment and provide a valuable window on the dynamics and mechanisms of the commitment-related silencing process [8, 49–52]. The mechanism of exclusion of developmental alternatives can be tracked at the level of the key genes they depend on (reviewed by [53]) that are not shared with the T-cell program.
B-cell differentiation is based on a core mutual activation circuit involving the two lineage-specific transcription factors, EBF1 and Pax5, and it also is supported by the multilineage factor PU.1, in addition to transcription factors shared with early T cells. Early B-cell precursors receive growth signals through the IL-7R, shared with early T cells, and through the TSLPR (Tpte2). Especially because of the large number of shared factors, B lineage exclusion in T cells depends on permanent inactivation of Ebf1 and Pax5.
Erythroid cell differentiation depends on the transcription factors GATA-1 and FOG-1 (Zfpm1 gene), and EKLF (Klf1). The erythropoietin receptor (EpoR) provides crucial viability support for erythroid development. Additional regulatory factors involved are SCL (Tal1) and Gfi1b which are also shared with stem and progenitor cells. FOG-1 is also needed in T-cell precursors, so the most likely genes to be serving as specific switches for access to erythroid development are Gata1, Klf1 and Epor.
Dendritic cells depend on the transcription factor PU.1 (Sfpi1) and on growth signals transduced through the Flt3 or GM-CSF-R (Csf2ra/Csf2rb) growth factor receptors.
Granulocyte and macrophage development is driven by the combination of PU.1 with one of the C/EBP family members (Cebpa or Cebpb). Crucial for survival, proliferation and even instruction of the macrophage versus granulocyte fates are signals through at least one of the myeloid cytokine receptors, M-CSF-R (Csf1r), GM-CSF-R (Csf2ra/Csf2rb) and/or G-CSF-R (Csf3r).
NK-cell activity and programming depend on many factors shared with T cells, but also particularly on the T-box factors, T-bet (Tbx21) and Eomesodermin (Eomes), and a recently discovered zinc finger protein, Zfp105. Growth and activation depend on the IL-2/IL-15 receptor, product of the Il2rb gene, which is not normally expressed in the early stages of T-cell specification. However, all these genes except Zfp105 can be expressed conditionally later in some subsets of T cells.
At the time the cells finally relinquish the dendritic cell option, it is likely to have been at least 10–12 cell divisions since the time they lost the erythroid option and at least 5–7 cell divisions since they gave up the B cell option. If the epigenetic marks correlated with repression were the main mechanism for ensuring permanence of silencing, then in newly committed T-cell precursors, these marks might be most heavily and consistently applied to the genes for the erythroid fate and most shallowly applied to the genes for the NK-cell fate.
EPIGENETIC MODIFICATIONS AND NEGATIVE REGULATION OF ALTERNATIVE FATES IN T-CELL LINEAGE COMMITMENT
These predictions could be tested as stages of the commitment process have been tracked by ChIP-seq and RNA-seq analyses of early T-cell development [8], spanning stages from cells corresponding to the earliest intrathymic stage (DN1) through to cells that have completed commitment and undergone some successful TCR gene rearrangement (DP cells; i.e. CD4+ CD8+ cells). Other published results have enabled the comparisons to be extended to putative prethymic stages and much later mature T cells [13, 54]. To summarize, developing T cells relinquish access to alternative fates by a mosaic of diverse, gene-specific silencing mechanisms that emphasize the unique regulatory requirements of each of the genes being repressed.
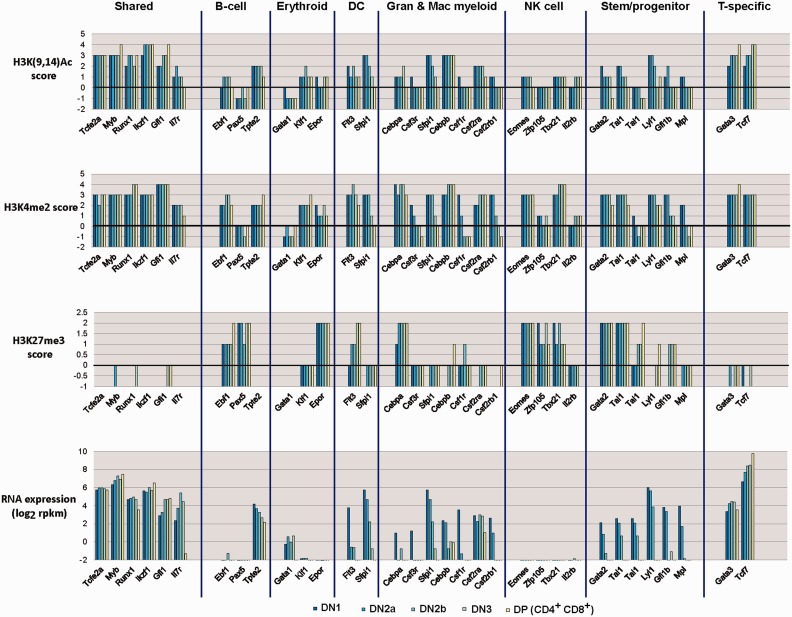
Figure 3 summarizes take-home messages from our own recent study of chromatin modifications in cells spanning the stages of commitment, results which bridge and confirm previous results from other groups [27, 39, 55]. RNA expression patterns (bottom graphs) and the levels of promoter-associated histone modifications that accompany them (upper three graphs) are shown for key genes according to the developmental programs in which they participate. At a crude level, the relationships are not surprising: genes that have active histone marks [H3K(9,14)Ac] at their promoters are generally active in these early T-lineage samples; genes that have repressive histone marks (H3K27me3) at their promoters are generally silent; the ‘accessibility’ mark (H3K4me2) is associated with many promoters in various activity states.
Figure 3:
Expression and histone modification changes in genes associated with non-T developmental potentials from precommitment to postcommitment stages of T-cell development. Data summarized are from reference [8], and show histone modification scores around transcriptional start sites of the genes correlated with their RNA expression levels as early T-lineage cells progress from multipotency to commitment (Figure 2). Genes for which data are displayed are grouped according to the developmental program(s) in which they play an important role. Colored bars show values for the indicated parameters in cells from precommitment (DN1, DN2a) to postcommitment (DN2b, DN3, DP) stages of early T-cell development (dark colors: earlier stages; light colors: later stages). For histone marks shown, any score of 0 or less is insignificant. Log2 RNA expression values above a score of 0–1 (1–2 rpkm) are significant. See text for additional discussion.
The interesting aspect of the results is that the pattern of modifications does not follow a lineage-specific hierarchy reflecting the known developmental exclusion sequence. Consider the earliest fate to be excluded, the erythroid fate. Erythroid genes indeed are poorly expressed or not expressed in all these early T-cell stages. But the silent Klf1 gene is in ‘open’ chromatin, there is slight, but consistent transcription from the ‘master regulator’ Gata1 locus, and only the Epor gene has repression marks. The B-cell fate, which is much more closely related to the T-cell fate and repressed much later, shows much more convincing silencing applied to the Ebf1 and Pax5 loci.
Myeloid genes and dendritic genes again show heterogeneity in their use of H3K27me3 marking. Ironically, the fate for which regulatory genes are most obviously subject to repressive marking is the fate that is closest to T cells—namely the NK-cell fate. Not only are Eomes, Zfp105, Tbx21 and Il2rb transcriptionally silent in the early T-cell stages, but also the promoters of all three transcription factor loci are heavily modified with H3K27me3. Yet, T-cell precursors retain until the very last step of commitment (until DN2b) the ready ability to switch into the NK lineage and remain able to use most of these genes later in development, if they pick the CD8+ cytolytic T-cell functional speciality.
Thus, clearly H3K27me3 is not a simple index of gene silencing. H3K27me3 intensity does not correlate with permanence of repression, and at least half of the silent and silenced genes in the genome of these early T cells lack these marks, agreeing with results found in other developmental systems [56, 57]. These examples of silent genes without H3K27me3 marks also accord well with evidence that the ‘need’ to use polycomb repressive complex 2 to trimethylate H3K27 is greater to silence genes with high CpG promoters than for genes with low CpG promoters. However, this gene specificity begs the question of how many different kinds of repression exist, and whether chromatin modification state really does reflect the machinery needed to block access to a particular fate forever.
DYNAMICS OF REPRESSION AND DEREPRESSION: EPIGENETIC MARKING AS A QUANTITATIVE THRESHOLD SETTER
For genes that undergo repression only after T-cell development has begun, it has been possible in our study to catch the stages through which the H3K27me3 mark is deposited [8]. Here as in other developmental systems, for most of these acutely repressed genes, the substantial drops in transcriptional activity commonly occur several stages before the appearance of H3K27me3 at any significant fraction of the promoters of the repressed genes. In other words, either through removal of positive regulators or through interference from negative regulators, the genes are being actively turned off before the H3K27 methylation by polycomb repressive complex 2 is called in. If H3K27me3 is needed, it seems to play a repression stabilizing role.
However, what is it that makes the repression stable? The example of the NK-cell genes shows that even heavy H3K27me3 marking must be conditionally removable. However, the silencing of the B-cell regulatory genes Ebf1 and Pax5 is robust and functionally irreversible. Thus, not only is the negative regulatory machinery involved in commitment highly complex, it is also functionally multilayered in a way that current histone mark detection does not fully capture. This is in good agreement with studies from Hoogenkamp et al. [58], who have described a localized chromatin decondensation step that can be needed for gene activation under separate control from the mechanisms that control histone marking.
The example of an H3K27me3+ repressed gene that turns on compared with another such gene that does not turn on sheds light on the interplay between histone modification state and transcription factor regulatory activity. One of the most T-cell-specific of all regulatory genes, Bcl11b, is first turned on dramatically just before the transition from uncommitted to committed (DN2a to DN2b transition) and fully activated only by the DN3 stage. As for all T-cell program genes, Bcl11b depends on Notch pathway signaling to be activated [59]. However, it does not respond to the Notch signals that are available in the earliest stages of T-cell development in the thymus. It depends on Notch signals and GATA-3 [60], and Runx factors [61], and probably also TCF-1 (Tcf7 gene product)[62], two of which, GATA-3 and TCF-1, also need to be induced during T-cell specification. A strong likelihood is that epigenetic modifications are used to enforce this strict combinatorial requirement.
The 5′ sequence of the Bcl11b gene is not only invested with CpG methylation in the early T-cell stages [63], but also coated with H3K27me3-modified nucleosomes [8]. We have recently obtained evidence for a distal enhancer of the Bcl11b gene that is needed to complement promoter and intragenic regulatory sequences to promote expression in immature T-lineage cells; this enhancer is also initially covered with H3K27me3-modified histones [64]. However, the repressive histone mark and the DNA methylation both are removed sharply as the gene turns on, and this becomes a site then where at least some of the essential T-cell regulatory inputs (TCF-1 and Runx1; [61, 62, 64]) are delivered. The barrier can fall.
Although Bcl11b has repressive marks, key parts of its structure appear to be functionally accessible before it is turned on by criterion of transcription factor binding. Despite the H3K27me3 marking the distal enhancer, the PU.1 transcription factor is already able to bind to that element. PU.1 is one of the factors that binds to a large excess of sites where it does not apparently exert a regulatory influence, and evidence currently in hand suggests that PU.1 is acting as a ‘joyrider’ or binding without function, not actually participating in regulating enhancer activity. However, its binding there is very interesting because, genome-wide, as a rule, PU.1 is very strongly excluded from most H3K27me3-modified regions. It is indeed excluded from binding at this same element in early B-lineage cells, where there is also H3K27me3 deposited [64], and in mature cells of myeloid lineages, despite their higher levels of PU.1 per cell (S. Damle, unpublished results). In this earliest T-lineage context, then, PU.1 is acting as a highly useful probe that detects chinks in the armor associated with this element in at least a substantial fraction of nuclei, long before the gene goes on. When sufficient levels of positive regulatory factors needed for Bcl11b upregulation are induced, these chinks allow the whole chromatin opening cascade to begin and the assembly of active enhancer complexes can take off.
In contrast, the Pax5 gene which is also coated with H3K27me3-modified nucleosomes in early T-lineage cells is in a much less accessible state. Not only its promoter, but also a number of intragenic enhancers bear H3K27me3-modified nucleosomes. Peaks of H3K4me2 still mark the positions of hematopoietic cis-regulatory elements, but the gene is functionally closed: a major PU.1 binding site that is occupied in early B cells is not engaged by the PU.1 in the early T cells [8]. This enhancer is a major site of action for B-lineage promoting E2A and EBF1 transcription factors in early B cells [65]. As we saw, early T cells do not express EBF1, but at the very early stage when B-cell lineage exclusion actually occurs, their precursors may still be competent to use the shared factor E2A and other factors to activate Ebf1. Thus, once the early T-lineage cells impose inaccessibility on this crucial Pax5 cis-regulatory element, it may be one of the things that make the exclusion of B-cell fate irreversible.
CONCLUDING REMARKS
The developmental process of commitment is best modeled as a stepwise process of transcription factor action on available sites to induce or silence other regulatory genes that alter the accessibility landscape for the next stage. Analysis of the actual steps involved in the early T-cell lineage decisions indicate that histone modifications (and apparently DNA methylation as well) are regulated in a gene-by-gene manner, i.e. following the transcriptional requirements of each specific gene, not in a groupwise lineage-specific manner. This means that chromatin marks are indices of transcription factor action and impedance setters for specific genes with their own regulatory requirements, i.e. with roles that depend on their context within specific gene regulatory network nodes. Epigenetic modifications that we know how to detect offer clues to quantitatively preferred or disfavored sites, but both DNA methylation and ‘repressed chromatin’ are both reversible over fairly rapid developmental time frames, probably on the order of just a small number of cell cycles. Much remains to be learned about repression, and we still lack a clear mechanistic picture of the molecular controls imposing ‘deep repression’. Recent evidence suggests that relocation of genes to complexes in different nuclear compartments can be associated with major changes in expression status [66], but whether this is a primary causal factor or a consequence is still to be determined. However, the chromatin modifications that affect developmental gene regulation are acting not as separate constraints, but as links across time within the network of transcription factor regulatory actions.
Key points.
Transcription factor binding site accessibility in the genome is modulated by previous developmental history of the cell and the previous regulatory states through which a cell has passed.
Transcription factor engagement can modify local histone markings and DNA accessibility to affect future access to additional factors.
Developmental progression from multipotency to lineage commitment of early T cells involves multiple local changes in chromatin states under the influence of new combinations of developmentally regulated transcription factors.
Lineage choices involve repression or permanent silencing of regulators for rejected fate alternatives, but the biochemical markers distinguishing functionally ‘deeper’ from ‘shallower’ silencing still needs to be defined.
Silencing and activation mechanisms for regulatory genes that control T cells are gene-specific, according to the unique gene network inputs required for individual regulatory genes involved in the alternative fates.
Acknowledgements
I thank current and former members of my laboratory for stimulating discussions and for the chance to discuss our unpublished work.
Biography
Ellen V. Rothenberg received her A.B. from Harvard University (A.B.) and Ph.D. from MIT. She was a Jane Coffin Childs fellow at Sloan-Kettering Institute and a junior faculty member at the Salk Institute before coming to Caltech, where she is Albert Billings Ruddock Professor of Biology.
FUNDING
The US Public Health Service (National Institutes of Health grants RC2CA148248, R33HL089123, R01CA90233); the Albert Billings Ruddock Professorship of Biology.
References
- 1.Fong AP, Yao Z, Zhong JW, et al. Genetic and epigenetic determinants of neurogenesis and myogenesis. Dev Cell. 2012;22:721–35. doi: 10.1016/j.devcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garber M, Yosef N, Goren A, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47:810–22. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badis G, Berger MF, Philippakis AA, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–3. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Treiber T, Mandel EM, Pott S, et al. Early B cell Factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–25. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Hollenhorst PC, Chandler KJ, Poulsen RL, et al. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaeger SA, Chan ET, Berger MF, et al. Conservation and regulatory associations of a wide affinity range of mouse transcription factor binding sites. Genomics. 2010;95:185–95. doi: 10.1016/j.ygeno.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang JA, Mortazavi A, Williams BA, et al. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–82. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palii CG, Perez-Iratxeta C, Yao Z, et al. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. EMBO J. 2011;30:494–509. doi: 10.1038/emboj.2010.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonn S, Furlong EE. cis-Regulatory networks during development: a view of Drosophila. Curr Opin Genet Dev. 2008;18:513–20. doi: 10.1016/j.gde.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Arvey A, Agius P, Noble WS, et al. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res. 2012;22:1723–34. doi: 10.1101/gr.127712.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tijssen MR, Cvejic A, Joshi A, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin YC, Jhunjhunwala S, Benner C, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–43. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J, Shao Z, Glass K, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23:796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Natoli G, Ghisletti S, Barozzi I. The genomic landscapes of inflammation. Genes Dev. 2011;25:101–6. doi: 10.1101/gad.2018811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson NK, Foster SD, Wang X, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–44. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 17.Ogata K, Sato K, Tahirov TH. Eukaryotic transcriptional regulatory complexes: cooperativity from near and afar. Curr Opin Struct Biol. 2003;13:40–8. doi: 10.1016/s0959-440x(03)00012-5. [DOI] [PubMed] [Google Scholar]
- 18.Chlon TM, Dore LC, Crispino JD. Cofactor-mediated restriction of GATA-1 chromatin occupancy coordinates lineage-specific gene expression. Mol Cell. 2012;47:608–21. doi: 10.1016/j.molcel.2012.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P, Spolski R, Liao W, et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490:543–6. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glasmacher E, Agrawal S, Chang AB, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–80. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goetz TL, Gu TL, Speck NA, et al. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor α2. Mol Cell Biol. 2000;20:81–90. doi: 10.1128/mcb.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu TL, Goetz TL, Graves BJ, et al. Auto-inhibition and partner proteins, core-binding factor β (CBFβ) and Ets-1, modulate DNA binding by CBFα2 (AML1) Mol Cell Biol. 2000;20:91–103. doi: 10.1128/mcb.20.1.91-103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol Cell. 2001;8:1267–76. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 24.Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 25.Rothenberg EV, Ward SB. A dynamic assembly of diverse transcription factors integrates activation and cell-type information for interleukin-2 gene regulation. Proc Natl Acad Sci USA. 1996;93:9358–65. doi: 10.1073/pnas.93.18.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet. 2012;13:469–83. doi: 10.1038/nrg3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer EM, Lin YC, Benner C, et al. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 2011;35:413–25. doi: 10.1016/j.immuni.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revilla-i-Domingo R, Bilic I, Vilagos B, et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31:3130–46. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller SA, Huang AC, Miazgowicz MM, et al. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–93. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lichtinger M, Ingram R, Hannah R, et al. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 2012;31:4318–33. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka Y, Joshi A, Wilson NK, et al. The transcriptional programme controlled by Runx1 during early embryonic blood development. Dev Biol. 2012;366:404–19. doi: 10.1016/j.ydbio.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 33.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–52. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 34.Clyde DE, Corado MS, Wu X, et al. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426:849–53. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- 35.Ghisletti S, Barozzi I, Mietton F, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–28. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Samstein RM, Arvey A, Josefowicz SZ, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–66. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vahedi G, Takahashi H, Nakayamada S, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–93. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher WW, Li JJ, Hammonds AS, et al. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc Natl Acad Sci USA. 2012;109:21330–5. doi: 10.1073/pnas.1209589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weishaupt H, Sigvardsson M, Attema JL. Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood. 2010;115:247–56. doi: 10.1182/blood-2009-07-235176. [DOI] [PubMed] [Google Scholar]
- 40.Maës J, Maleszewska M, Guillemin C, et al. Lymphoid-affiliated genes are associated with active histone modifications in human hematopoietic stem cells. Blood. 2008;112:2722–9. doi: 10.1182/blood-2008-02-140806. [DOI] [PubMed] [Google Scholar]
- 41.Hawkins RD, Hon GC, Lee LK, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–91. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–35. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 43.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 44.Rothenberg EV. T cell lineage commitment: Identity and renunciation. J Immunol. 2011;186:6649–55. doi: 10.4049/jimmunol.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Q, Bell JJ, Bhandoola A. T-cell lineage determination. Immunol Rev. 2010;238:12–22. doi: 10.1111/j.1600-065X.2010.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Pooter RF, Kee BL. E proteins and the regulation of early lymphocyte development. Immunol Rev. 2010;238:93–109. doi: 10.1111/j.1600-065X.2010.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–79. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 48.Manesso E, Chickarmane V, Kueh HY, et al. Computational modelling of T-cell formation kinetics: output regulated by initial proliferation-linked deferral of developmental competence. J R Soc Interface. 2013;10:20120774. doi: 10.1098/rsif.2012.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. 2010;185:284–93. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mingueneau M, Kreslavsky T, Gray D, et al. The transcriptional landscape of αβ T cell differentiation. Nat Immunol. 2013;14:619–32. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawazu M, Yamamoto G, Yoshimi M, et al. Expression profiling of immature thymocytes revealed a novel homeobox gene that regulates double-negative thymocyte development. J Immunol. 2007;179:5335–45. doi: 10.4049/jimmunol.179.8.5335. [DOI] [PubMed] [Google Scholar]
- 52.Ikawa T, Hirose S, Masuda K, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–6. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 53.Mercer EM, Lin YC, Murre C. Factors and networks that underpin early hematopoiesis. Semin Immunol. 2011;23:317–25. doi: 10.1016/j.smim.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei G, Wei L, Zhu J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–67. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui K, Zang C, Roh TY, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Filion GJ, van Bemmel JG, Braunschweig U, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–24. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mendenhall EM, Koche RP, Truong T, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoogenkamp M, Lichtinger M, Krysinska H, et al. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood. 2009;114:299–309. doi: 10.1182/blood-2008-11-191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tydell CC, David-Fung ES, Moore JE, et al. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–38. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- 60.García-Ojeda ME, Klein Wolterink RG, Lemaître F, et al. GATA-3 promotes T cell specification by repressing B cell potential in pro-T cells. Blood. 2013;121:1749–59. doi: 10.1182/blood-2012-06-440065. [DOI] [PubMed] [Google Scholar]
- 61.Guo Y, Maillard I, Chakraborti S, et al. Core binding factors are necessary for natural killer cell development, and cooperate with Notch signaling during T cell specification. Blood. 2008;112:480–92. doi: 10.1182/blood-2007-10-120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber BN, Chi AW, Chavez A, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–8. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ji H, Ehrlich LI, Seita J, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–42. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L, Zhang JA, Dose M, et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013 doi: 10.1182/blood-2012-08-447839. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Decker T, Pasca dM, McManus S, et al. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–20. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 66.Lin YC, Benner C, Mansson R, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13:1196–204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]