Abstract
The continual public health threat posed by the emergence of novel influenza viruses necessitates the ability to rapidly monitor infection and spread in experimental systems. To analyze real-time infection dynamics, we have created a replication-competent influenza reporter virus suitable for in vivo imaging. The reporter virus encodes the small and bright NanoLuc luciferase whose activity serves as an extremely sensitive readout of viral infection. This virus stably maintains the reporter construct and replicates in culture and in mice with near-native properties. Bioluminescent imaging of the reporter virus permits serial observations of viral load and dissemination in infected animals, even following clearance of a sublethal challenge. We further show that the reporter virus recapitulates known restrictions due to host range and antiviral treatment, suggesting that this technology can be applied to studying emerging influenza viruses and the impact of antiviral interventions on infections in vivo. These results describe a generalizable method to quickly determine the replication and pathogenicity potential of diverse influenza strains in animals.
INTRODUCTION
Influenza A virus is a serious public health threat. Each year, 3 to 5 million people are infected with influenza virus, resulting in 250,000 to 500,000 deaths (1). Infections from seasonal influenza viruses cause upwards of ∼24,000 to 36,000 deaths per year in the United States alone (2). Pandemic viruses have the potential for significantly higher mortality rates, with the 1918 outbreak estimated to have caused upwards of 50 million deaths (3). Reinforcing the immediacy of these concerns, the reassortant H7N9 virus recently isolated from patients in China currently displays a case fatality rate approaching 25% (4–6).
Animal models of infection are useful for determining the pathogenicity and transmission potential of emerging influenza viruses (7–9). Mice and ferrets are routinely used to study influenza virus infections, host defenses, and responses to antiviral therapies or vaccines. However, animal models of influenza virus infections are constrained by the inability to monitor viral dynamics in real time. Additionally, in mouse models viral load determination typically requires sacrificing the animal, precluding longitudinal assessment. In vivo imaging of bioluminescent reporter viruses is a powerful alternative that allows real-time detection of viral load and spread in the same animal over time (10). This strategy has been exploited in animal models for multiple viruses, including dengue virus (11, 12), herpes simplex virus 1 (HSV-1) (13), Sindbis virus (14), vaccinia virus (15), and Sendai virus (16).
Generation of a replication-competent influenza reporter virus is confounded by the complex architecture of the segmented genome: (i) all of the viral genes are critical in vivo, precluding simple replacement with the reporter; (ii) the compact genome does not tolerate large insertions, which can destabilize the genome and are lost over time (17–19); (iii) most insertions severely attenuate replication (17, 20, 21); and (iv) insertion of reporter gene sequences at either end of the coding region disrupts packaging signals essential for virion assembly that are present in both the untranslated region (UTR) and the viral open reading frame (ORF) (22, 23). Some of these limitations have been addressed by inserting foreign sequences into viral genes to create protein fusions or polyproteins (17, 20, 21, 24, 25). Alternatively, entire viral genes have been replaced with foreign sequence and reporter viruses were grown in specialized complementing cell lines (18, 19, 26, 27). In extreme cases, an additional genome segment carrying extra sequence was inserted into virions by re-engineering viruses to package nine viral RNAs, as opposed to the normal complement of eight (28). Whereas these reporter virus have been used successfully for in vitro studies and most recently with limited sensitivity in animal infections (17, 20), the genomes containing the reporter are generally unstable and/or the reporter viruses are highly attenuated. As such, no reporter has yet been developed that closely mimics the native virus, especially in animal infections.
We have overcome these limitations by using the very small and extremely bright luciferase variant NanoLuc (NLuc) to develop a replication-competent reporter virus suitable for in vivo imaging. NLuc is a 19-kDa engineered luciferase that possesses ∼150-fold-greater specific activity (i.e., light output) than both Renilla and firefly luciferases (29). We show that an NLuc reporter virus replicates with near-native properties in culture and in vivo. Moreover, the NLuc reporter virus possesses pathogenicity and lethality in mice that is indistinguishable from those of the parental virus. We used this virus to perform in vivo imaging and to track viral load and dissemination of influenza virus infections in the lungs of mice. We further show that the reporter virus recapitulates known restrictions due to host range and antiviral treatment, suggesting that this technology can be applied to studying emerging influenza viruses and the impact of novel antiviral interventions on infections in vivo. These findings suggest that bioluminescent imaging of NLuc reporter viruses can be used to rapidly assess the properties of emerging viruses and screen the efficacy of candidate vaccines or antiviral therapies.
MATERIALS AND METHODS
Plasmids, cells, and antibodies.
The virus rescue plasmids pTM-polI-WSN-All (a kind gift from Y. Kawaoka [30]) and pTM-ΔRNP (31) express viral RNAs (vRNAs) from a genomic sequence of an influenza virus (A/WSN/33 [WSN]) flanked by a polymerase (pol) I promoter and terminator. pBD-PB1, -PB2, -PA, and -NP contain bidirectional pol I and pol II cassettes to express both vRNA and mRNA (32, 33). NanoLuc (NLuc) sequences were amplified from pNL1.1 (Promega). p3X-1T expresses PB1 with a tandem affinity purification (TAP) tag, PB2, and PA. NP was expressed from pCAGGS-WSN-NP (30). The vRNA-luciferase reporter plasmid pHH21-vNA-luc contains the firefly luciferase-coding sequence in the antisense orientation flanked by untranslated regions of the vNA gene segment from the A/WSN/33 isolate and was previously described (34).
A549, 293T, MDBK, and MDCK cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Calu-3 cells were grown in DMEM/F12 supplemented with 20% FBS (HyClone). Infections were performed in media supplemented with 0.3% bovine serum albumin (BSA), 25 mM HEPES, 1× penicillin/streptomycin, and 0.25 to 0.5 μg/ml N-tosyl-l-phenylalanine chloromethyl ketone (TPCK)–trypsin. All cells were grown at 37°C in 5% CO2. PA antibodies were raised in rabbits (kindly provided by T. Parlsow [34]).
Creation of PA-NLuc constructs.
PA fusions were created in pBD-PA by Gibson Assembly (NEB) as diagrammed (Fig. 1A). Briefly, the NLuc coding sequence and stop codon were placed downstream of the PA sequence to create a contiguous ORF. Translation of this sequence appends NLuc to the C terminus of PA. Native packaging sequences were restored by repeating the terminal 30, 40, or 50 nucleotides (nt) of the PA ORF (including the stop codon) after the NLuc stop codon adjacent the native UTR. Where indicated, sequence encoding the “self-cleaving” 2A peptide from porcine teschovirus was placed between the PA sequence and the NLuc sequence (35). Direct repeats were removed from the reporter gene by introducing 18 silent mutations in the wobble positions at the 3′ end of the PA ORF.
Fig 1.
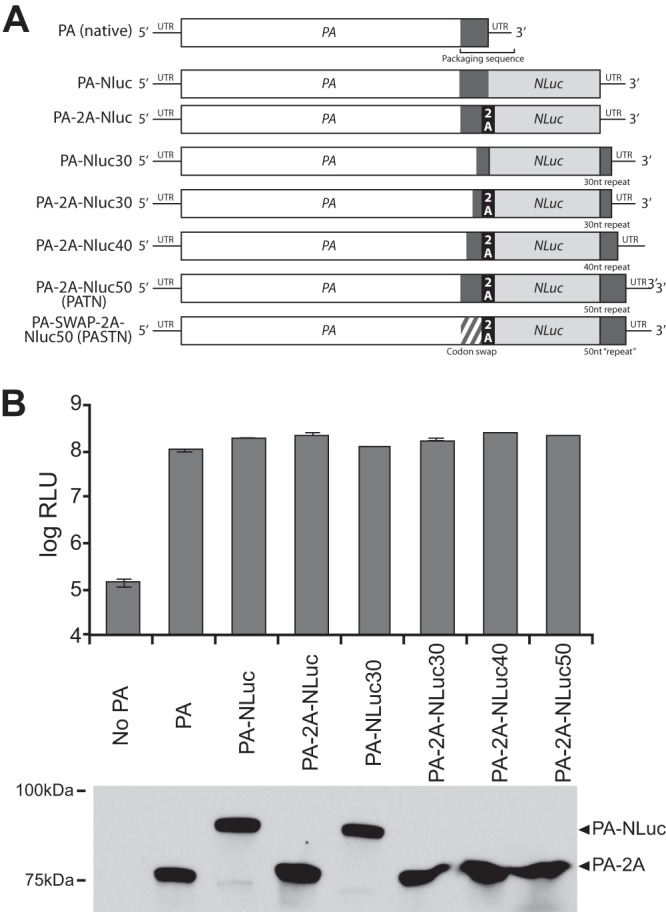
Generation of an influenza reporter virus encoding a PA-NLuc fusion. (A) Schematic of the PA-NLuc gene fusions encoding the polyprotein containing PA, the self-cleaving 2A peptide where indicated, and NLuc. Repeated regions of the PA ORF that contain packaging signals and are repeated downstream of NLuc are depicted as dark gray boxes. Silent mutations were introduced into the terminal 47 nt of the PA coding sequence in the PA-SWAP-2A-NLuc50 (PASTN) gene. (B) Polymerase activity assay of PA fusions with or without the 2A peptide and with packaging sequence repeats of increasing length. Polymerase activity assays were performed with human 293T cells by transfecting vectors expressing PB1, PB2, the indicated PA proteins, NP, and a vNA-based firefly luciferase reporter. PA proteins were detected by Western blotting. Data are presented as means ± standard deviations (sd) (n = 3).
Polymerase activity assays.
293T cells were reverse transfected in triplicate with TransIT-2020 (Mirus) with vectors expressing PB1, PB2, NP, and the indicated PA proteins in addition to the reporter pHH21-vNA-luc. Cells were lysed 24 to 36 h posttransfection, and luciferase activity was measured with a luciferase assay system (Promega). Western blotting was performed on lysates to confirm equivalent levels of transfection and expression.
Virus rescue, titration, and multicycle replication.
Virus was rescued by reverse transfection of a 293T/MDBK coculture using TransIT-2020 (Mirus). To obtain native WSN, cells were reverse transfected with pTM-polI-WSN-All, p3X-1T, and pCAGGS-NP. PA fusion viruses were obtained by transfection with pTM-ΔRNP, pBD-PB1, -NP, -PB2 (or PB2 K627E), and the desired pBD-PA-NLuc fusion. Wild-type (WT) and PB2 K627E PA-2A-NLuc50 viruses were plaque purified. Viruses were subsequently amplified in MDBK cells or embryonated eggs, and titers were determined by a plaque assay performed on MDCK cells. Plaque sizes were determined in a single-blind analysis of scanned images. Diameters were measured using ImageJ. Multicycle replication assays were performed in triplicate in MDBK and Calu-3 cells infected at multiplicities of infection (MOI) of 0.01 and 0.001, respectively. Aliquots were removed at the indicated time points, and viral titers were determined by a plaque assay (36). Titers of NLuc viruses were also determined with a Nano-Glo assay (see below). Blind serial passaging was performed in MDBK and Calu-3 cells by transfer of a small aliquot of virus from one culture to the next without plaque purification or amplification.
Nano-Glo viral titer assay.
A Nano-Glo viral titer assay was developed to establish NLuc activity as a proxy for traditional forms of viral titer (e.g., PFU/ml or tissue culture infective dose [TCID50]/ml). MDCK cells were grown to confluence in a white 96-well plate and infected in at least triplicate with 20 μl of viral supernatant for 1 h. Cells were washed to remove the inoculum (and any residual NLuc produced in previous infections) and incubated for 8 or 16 h. NLuc activity was measured in situ with the Nano-Glo assay system (Promega), and luminescence was detected with a plate reader (Tecan). For antiviral treatment, cells were incubated with 100 μg/ml ribavirin for 2 h prior to infection; ribavirin was then present throughout the infection.
Mouse infections.
Female BALB/c mice (Jackson Laboratory) (4 to 6 weeks old) were inoculated intranasally with the indicated amount of virus in 25 μl under light isoflurane anesthesia. Body weight was monitored daily. Mice losing >20% of their original body weight were humanely euthanized. Viral load in lung homogenates was determined by TCID50 and the Nano-Glo viral titer assay. In vivo imaging was performed with an IVIS 200 imaging system on anesthetized mice. Nano-Glo reagent was diluted 1:20 in PBS, and 100 μl was injected retro-orbitally. Image acquisition and analysis were performed with Living Image software (PerkinElmer). Flux measurements were acquired from regions of interest automatically gated to the signal contours. All data in composite images utilized the same scale. Mouse experiments were performed under protocols approved by the University of Wisconsin—Madison Institutional Animal Care and Use Committees.
Statistics.
Data represent means ± standard deviations (n ≥ 3) unless otherwise noted. Curve fitting was performed in Excel, and goodness of fit is depicted by R2. Where indicated, a comparison between groups was performed by a two-tailed Student's t test. Significance is defined as P < 0.05 and is indicated when necessary with an asterisk (*).
RESULTS
Development of a bioluminescent influenza reporter virus.
Creation of an influenza reporter virus requires that the inserted reporter gene neither interfere with functions of the native viral proteins nor disrupt the packaging signals of the native gene segment. The polymerase subunit PA has previously been shown to tolerate fusion of exogenous protein sequence to its C terminus without disrupting polymerase function (37). In addition, a minimal packaging sequence has been identified in the PA gene (23). Based on this information, we attempted to make a replication-competent reporter virus by creating a series of fusion proteins with NanoLuc (NLuc) appended to the C terminus of PA. Reporter constructs were created in a WSN background (Fig. 1A). Where indicated, the PA fusions were further modified by inserting the self-cleaving 2A peptide from porcine teschovirus to create discrete PA and NLuc proteins from a polyprotein precursor (35). As packaging sequences have been identified in the end of the PA ORF (23, 38, 39), we also duplicated increasing portions of the PA ORF downstream of the NLuc stop codon.
To confirm the functionality of PA in the fusion proteins, polymerase activity assays were performed in 293T cells. PA-NLuc fusions were expressed with the remaining polymerase subunits PB1 and PB2, the viral nucleoprotein (NP), and a firefly-based luciferase reporter construct. Firefly luciferase and NLuc utilize different substrates and are thus suitable for multiplexing (29). All PA fusions, regardless of the inclusion of the 2A peptide or repeated packaging sequences, supported robust polymerase activity within ∼2-fold of the native PA level (Fig. 1B). Western blotting confirmed relatively equivalent levels of expression of all PA proteins (Fig. 1B). Blotting also showed effectively complete separation of PA and NLuc polypeptides when the 2A peptide was present. PA-NLuc fusions without the 2A peptide also produce small amounts of protein with a molecular mass similar to that of native PA, suggesting the presence of breakdown products or premature terminations. Based on these results, PA-NLuc genes containing the 2A peptide were used in subsequent experiments to minimize the possibility that the fusion would interfere with other PA functions, such as trafficking and assembly.
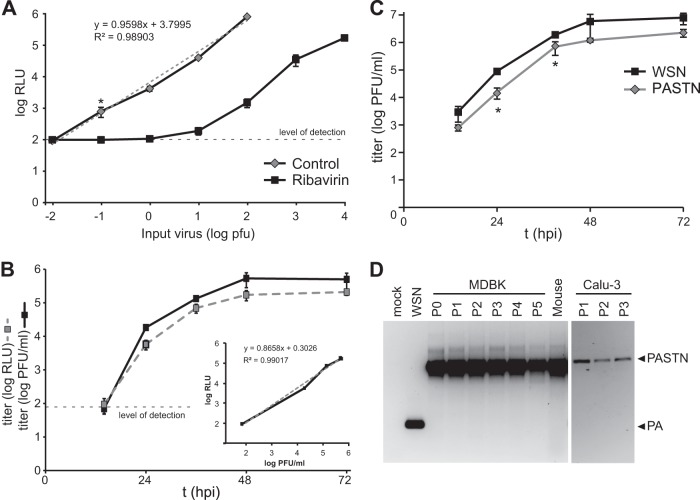
WSN-based reporter viruses were rescued with PA-2A-NLuc genes containing restored packaging signals (PA-2A-NLuc30, -40, and -50). Our initial experiments demonstrated that viruses with only 30 or 40 nt of repeated packaging signals displayed obvious attenuation compared to strain WSN (data not shown); therefore, we focused on virus with 50 nt of repeated packaging sequence, which we termed PA-2A-NLuc50 (PATN). A high-throughput Nano-Glo viral titer assay was developed to rapidly detect luciferase activity produced as a consequence of viral infection. The sensitivity and dynamic range of the assay were determined by performing infections with known amounts of input PATN virus. MDCK cells were infected in a 96-well format with reporter virus for 16 h and subjected to a Nano-Glo viral titer assay. The Nano-Glo viral titer assay detected robust luciferase activity, with a linear range over 4 logs of input virus (Fig. 2A). The assay was extremely sensitive, as infection with 0.1 PFU of virus routinely produced a signal greater than 5-fold above the background (P < 0.005). This raises the possibility that the Nano-Glo viral titer assay may be more sensitive than plaque assays. Additionally, the Nano-Glo viral titer assay may detect infection by the recently proposed “semi-infectious” particles that express viral proteins but cannot otherwise form a plaque (40), thus increasing sensitivity relative to that of plaque assays. Further experimentation is required to make a definitive determination. In addition, while plaque formation may require 48 to 72 h, the Nano-Glo titer assay quantitatively measures viral levels in as little as 8 h (data not shown). The Nano-Glo titer assay was repeated in the presence of ribavirin to confirm that the luciferase output was a product of viral gene expression and not the result of NLuc carried forward in the viral supernatant or fortuitously packaged into virions. Ribavirin treatment reduced NLuc activity to below the level of detection at low doses of input virus and reduced activity by 2 to 3 logs compared to that seen with untreated samples at higher doses (Fig. 2A). Ribavirin does not directly inhibit NLuc activity (data not shown). Thus, the Nano-Glo titer assay is a sensitive, replication-dependent measure of input virus levels. Our results with ribavirin further show that this assay is highly responsive to therapeutic interventions and thus may be suitable for rapid screening of antiviral compounds or antibodies.
Fig 2.
PA-2A-NLuc permits exquisitely sensitive detection of an influenza reporter virus. (A) Nano-Glo viral titer assay performed with defined amounts of input virus. Where indicated, cells were pretreated with 100 μg/ml ribavirin, which was then present throughout the assay. A trend line (gray dashed line) was fitted to untreated samples. *, P < 0.005; n = 4 ± sd. (B) Multicycle replication in human Calu-3 lung cells infected at MOI = 0.001. Viral titers were determined by both a plaque assay and a Nano-Glo viral titer assay (n = 3 ± sd). The limit of detection for luciferase activity is indicated as a black dashed line in panels A and B. To demonstrate the relationship between the two titer measurements, RLU was plotted as a function of PFU (inset). (C) Multicycle replication kinetics of native and PASTN viruses in MDBK cells infected at MOI = 0.01. Viral titers were determined by a plaque assay. Titers of WSN and PASTN at 24 and 39 h were significantly different (*, P < 0.05; n = 3 ± sd). (D) The PASTN gene is stable in vivo and through multiple passages in culture. RT-PCR was performed on RNA recovered from viral stocks (passage 0 [P0]), mouse lung homogenate from 5 dpi, or virus following serial passage in MDBK or Calu-3 cells. The PCR product was analyzed by agarose gel electrophoresis. Expected product sizes for native PA and the full-length reporter gene are indicated.
NLuc activity in the Nano-Glo titer assay was directly related to the amount of input virus, suggesting that NLuc activity could be used to measure titers in samples with unknown amounts of virus. To determine the utility of the reporter gene as a readout of viral level, PATN was used to initiate a multicycle infection in human Calu-3 lung cells. Titers were determined by both plaque and Nano-Glo assays. Viral titers determined by standard plaque assay paralleled the NLuc activity detected in our Nano-Glo titer assay (Fig. 2B). These data also established a linear relationship between PFU and relative light unit (RLU) measurements throughout the time course (R2 = 0.990; Fig. 2B, inset). These results demonstrate that the Nano-Glo titer assay serves as a direct measure of viral titer similar to plaque assay results.
Previous influenza reporter viruses were unstable and rapidly lost the reporter coding sequence, possibly due to the presence of direct repeats in the reporter gene (17, 19). Consequently, 18 silent mutations were introduced into the 3′ end of the PA ORF to remove the direct repeat arising from duplication of the packaging signal (Fig. 1A). This construct with codon-swapped mutations was used to create the PA-SWAP-2A-NLuc50 (PASTN) virus. The properties of the PASTN reporter virus in culture relative to the parental virus were determined in multicycle replication assays. MDBK cells were infected with either WSN or PASTN, and viral titers were determined by plaque assay (Fig. 2C). PASTN reached high titers, replicating with kinetics similar to native WSN kinetics. PASTN titers were ∼0.5 log lower than the parental strain titers throughout the time course, although the difference in titers was statistically significant only at 24 and 39 h postinfection (hpi) (P < 0.05) (Fig. 2C). Similar patterns of replication were observed during WSN and PASTN infections in human lung A549 cells (data not shown). We also amplified PASTN in embryonated eggs as this is a common source of viral stocks. PASTN grew to average titers of 1.1 × 107 PFU/ml, which was slightly less than but not significantly different from those determined for WSN, which grew to average titers of 6.1 × 107 PFU/ml (n = 3 eggs, P = 0.32). Plaque size was measured during the course of these experiments. Plaques for PASTN were less sharply defined and ∼80% the diameter of WSN plaques, with sizes of 2.85 mm ± 0.63 (n = 137) and 3.53 mm ± 0.98 (n = 76), respectively.
The stability of the PASTN reporter gene was tested following amplification in MDBK cells (passage 0 [P0]) or serial passage through MDBK cells (P1 to P5) or Calu-3 cells (P1 to P3) or 5 days postinfection (dpi) in a mouse lung. RT-PCR was performed on viral RNA with primers capable of simultaneously detecting native PA, the full-length PA-2A-NLuc fusion, or deletion products that might arise. Only the full-length PASTN gene was detectable in all of our reporter virus stocks (Fig. 2D). Moreover, only the full-length reporter was amplified from RNA extracted from mouse lungs, indicating the absence of detectable deletion products in both cell-free virions and infected tissue. These results show that the PASTN reporter virus replicates in culture with near-native properties while stably maintaining the reporter gene. Combined, these experiments validate the PASTN virus and titer assay as a robust reporter system that accurately measures influenza virus replication. In an advance over previous reporter viruses (17–21, 24–28), PASTN displays minimal attenuation and is not restricted to replication in select complementing cell lines.
Real-time in vivo imaging of influenza virus infection.
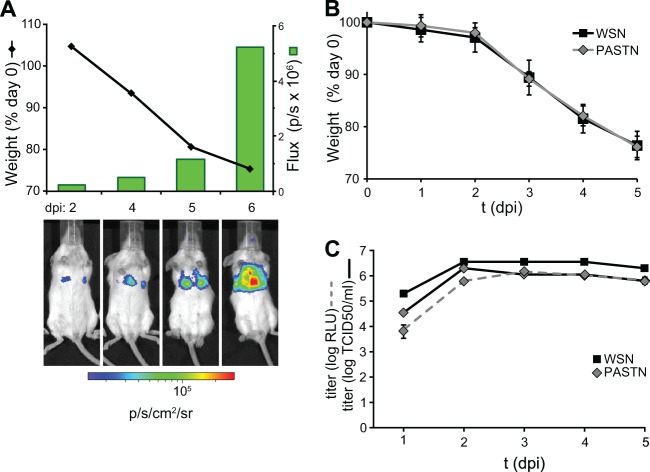
In vivo imaging of reporter viruses has proven useful in the study of replication dynamics and spread for many viral systems, including Sindbis virus (14), dengue virus (11, 12), HSV-1 (13), and vaccinia virus (15). To apply this approach to influenza virus, we used PASTN in an attempt to visualize influenza virus replication in the mouse model. BALB/c mice were infected with reporter virus and subjected to longitudinal weighing and in vivo imaging (Fig. 3A). Bilateral multifocal bioluminescence was detected as early as 2 dpi, indicating the initiation of infections in both the right and left lobes of the lung. Light output, measured as flux, increased over the course of infection and peaked on day 6 when the animal reached the experimental endpoint. The increase in flux accompanied spread of virus throughout the lungs and weight loss in the mouse, demonstrating that the NLuc virus was capable of supporting a pathogenic infection. Whole-body imaging has the potential to detect viral spread without a priori knowledge of tissue tropism. While WSN has the potential to initiate neurotropic infections, primarily by intracerebral inoculation (41), bioluminescence was not detected in the brain and virus was not recovered from the brain in any of our experiments. Whether bioluminescent imaging of this reporter virus will quantitatively detect extrapulmonary replication remains to be determined.
Fig 3.
In vivo imaging of an influenza reporter virus with near-native replication properties. Noninvasive imaging detects robust bioluminescence in mice infected with PASTN. (A) Mice were infected with 104 PFU of PASTN virus and analyzed for weight loss and bioluminescence. Representative data from serial imaging of one mouse are shown. (B) In a separate experiment, weight loss was measured for cohorts of mice infected with 104 PFU of either WSN or PASTN virus (n = 3 ± sd). (C) Cohorts of mice were infected as described for panel B and sacrificed at the indicated times. Viral titers in lung homogenates were determined by TCID50 and a Nano-Glo titer assay (n = 3 ± sd).
For the reporter virus to be most useful in animal models, it must replicate with properties similar to those of the parental strain. The replicative capacity and pathogenicity of PASTN in mice were thus directly compared to those of WSN, the parental virus from which it was derived. Cohorts of mice were infected with 104 PFU of PASTN or WSN and weighed daily. In addition, one set of mice infected with PASTN was imaged each day and euthanized to determine viral titers in the lung (Fig. 3C). Viral lung titers were also determined each day for a set of mice infected with WSN. Weight loss results were indistinguishable between PASTN and WSN throughout the infection time course (Fig. 3B), revealing that pathogenicity levels determined by this single measure were similar between the reporter and parental virus. Viral titers from lung homogenate showed that WSN and PASTN replicated to high levels in the mouse (Fig. 3C). Increases in viral titer were accompanied by increased bioluminescence. PASTN titers were marginally lower than WSN titers but were on average within ∼3-fold of the WSN titers. This subtle difference is consistent with our previous results showing that PASTN replicates to only slightly lower levels than WSN in culture (Fig. 2C). The PASTN viral load in the mouse lung as determined using the Nano-Glo titer assay was in close agreement with that determined by TCID50 (Fig. 3C) and was consistent with our analysis in culture (Fig. 2B).
To determine if our reporter virus could be used to rapidly assess the ability of influenza viruses to replicate and/or transmit to a new host species, we performed infections in mice with human strain- and avian strain-like viruses. The viral polymerase has long been known to facilitate cross-species transmission from avian to mammalian hosts, with major adaptive changes occurring in the PB2 subunit (42–44). In general, polymerases encoding the avian-signature glutamic acid at position 627 in PB2 function efficiently in avian hosts but are highly restricted in mammals; conversely, evolution of the human-signature lysine at PB2 position 627 dramatically rescues polymerase activity in mammalian hosts (33, 45–50). PATN viruses encoding either WT K627 PB2 or the avian strain-like PB2 K627E were rescued and used in in vivo imaging experiments to assess host range in real time. As expected, mice infected with WT PATN lost weight and displayed robust bioluminescence that increased over time (Fig. 4 and data not shown). In contrast, virus encoding the avian strain-like PB2 K627E was severely restricted, as infected mice showed little weight loss and bioluminescence was near background levels throughout the experiment. Nonspecific bioluminescence was occasionally detected in the eye at the site of substrate injection. Thus, the reporter virus faithfully recapitulates the known polymerase-mediated restriction due to host range and could therefore also be used with newly isolated influenza virus strains to determine their host range and the role of species-specific adaptive mutations.
Fig 4.
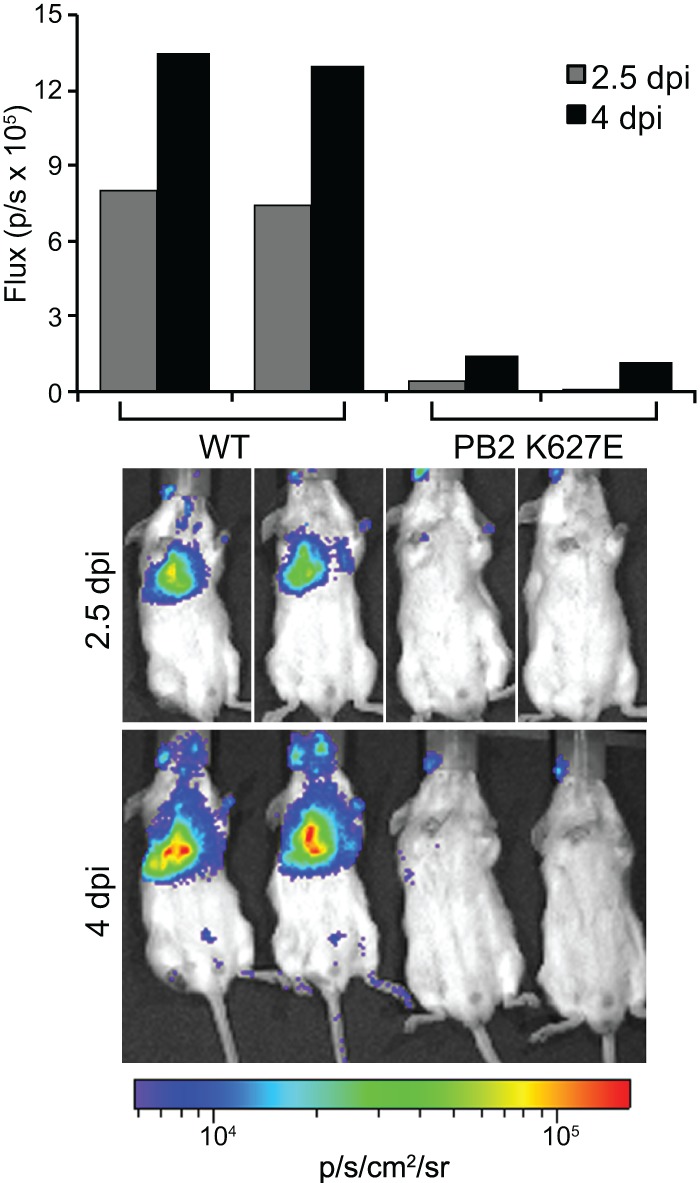
The NLuc influenza reporter virus recapitulates viral attenuation due to species-specific restriction of polymerase function. Mice were infected with 104 PFU of PATN virus encoding either WT PB2 or PB2 with the avian-signature K627E mutation. Flux was determined for mice imaged individually (2.5 dpi) or concurrently (4 dpi).
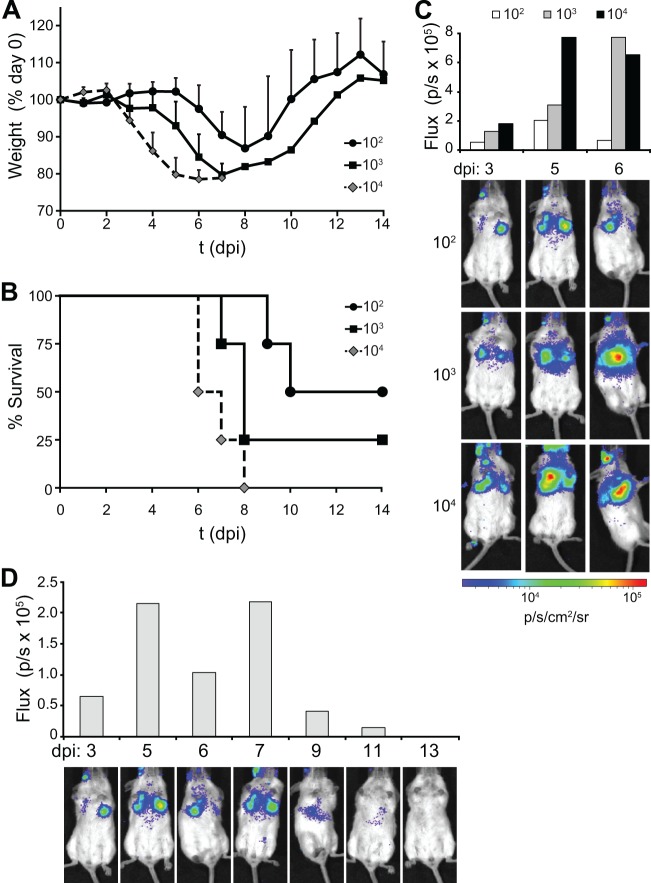
In a final series of experiments, mice were infected with serial dilutions of PASTN to simultaneously measure the lethality and sensitivity of the reporter virus. Groups of four mice were infected with 102, 103, or 104 PFU of PASTN and monitored daily for weight loss. Imaging was performed throughout. Weight loss and viability displayed a strong dose-dependent effect (Fig. 5A and B). All of the mice inoculated with 104 PFU and a majority of those infected with 103 PFU succumbed to infection, whereas half of the mice infected with 102 PFU survived. These initial experiments suggest that the minimal lethal dose (MLD50) of PASTN in BALB/c is ∼103 PFU, in remarkable agreement with the reported MLD50 for WSN of 103 to 103.4 PFU (27, 51–53). This is a significant improvement over other reporter viruses that show an MLD50 50 to 100 times higher than the parental MLD50 (17, 20).
Fig 5.
The PASTN reporter virus retains near-native pathogenicity in vivo. (A) BALB/c mice were infected intranasally with 102, 103, or 104 PFU of PASTN virus, and body weights were measured for 14 days (n = 4 + sd). (B) Survival rates of mice infected as described for panel A. (C) The relationship between bioluminescent signal and viral dose was examined by longitudinal observation of mice infected with increasing doses of PASTN virus. (D) Serial observations over the course of a sublethal infection of a single mouse from the experiment described for panel C with 102 PFU of PASTN virus permitted real-time observation of viral clearance. The same scale was used for all mouse images.
Longitudinal imaging of the same mouse from each group revealed that viral levels and dissemination in the lungs increased faster with higher doses, concomitant with weight loss (Fig. 5C). Notably, imaging of PASTN is sensitive enough to detect viral clearance of a sublethal infection (Fig. 5D). Combined, these data suggest that the PASTN reporter virus replicates with properties very close to those of the parental strain and, unlike previous influenza reporter viruses, shows very little attenuation in culture or in vivo. Further, bioluminescence detected by in vivo imaging of this extremely sensitive reporter virus measures viral localization, spread in the lungs, and levels in the infected animal.
DISCUSSION
The continual emergence of novel influenza viruses requires the ability to quickly assess their pathogenicity, pandemic potential, and susceptibility to antiviral interventions. Public health responses to outbreaks rely heavily upon experimental determination of viral pathogenicity and transmission capabilities. The rapid acquisition of these data has been hampered by the inability to monitor influenza virus replication in animals in real time. Here we describe a novel influenza reporter virus capable of simultaneously measuring viral load and dissemination in vivo. This virus permits longitudinal measurements in the same animal throughout the course of infection. The virus was engineered to maintain native protein expression and packaging signals. As such, the replication characteristics of the reporter virus are nearly identical to those of the native virus. Furthermore, the reporter virus enabled rapid detection of viral replication and titers in culture. Thus, our influenza reporter virus is a powerful tool to visualize infection in animal models and quantitatively measure replication in culture.
The NLuc reporter virus permits exquisitely sensitive detection of viral replication. The sensitivity and noninvasive longitudinal measurements made possible the imaging of infection and clearance of a sublethal dose of PASTN in BALB/c mice, a mouse background that is relatively resistant to influenza virus (54) (Fig. 5). In a mouse inoculated with the low dose of 102 PFU, infection began primarily in the left lung. By 5 dpi, two foci appeared on the right side, indicating spread of virus to the lobes in the right lung. Flux decreased in the left lung by 6 dpi, suggesting that the infection had begun to resolve. Unexpectedly, flux measurements showed that large amounts of virus were again present in the left lung at 7 dpi, accompanied by an 8% drop in body weight. The infection then continued to clear and was undetectable at 13 dpi. Viral lung titers were determined at 14 dpi, and no virus was detected. While only a single example, this time course alludes to the influenza viral dynamics that in vivo imaging can detect, with the potential to elucidate mechanisms of dissemination in the lungs, immune escape, or emergence of antiviral resistance. It will be important to determine if this approach is viable in ferrets, the currently accepted best animal model of human influenza virus infection.
Our NanoLuc reporter virus complements two other recently described Gaussia luciferase (GLuc) viruses. GLuc was incorporated into the NS gene of a rearranged influenza virus genome engineered to create vaccine candidate viruses (21). Whereas versions of the rearranged virus stably expressed GLuc, the rearranged virus was severely attenuated in culture and displayed a complete loss of pathology in mice. While beneficial for vaccine safety, the dramatic reduction in viral growth suggests a limited utility of this rearranged genome as a reporter virus. GLuc was also incorporated into the viral genome of PR8 (20). Using an approach similar to the one described here, GLuc was encoded as a polyprotein on the PB2 segment. The PR8-GLuc virus supported in vivo imaging, although high doses of virus (15 to 1,500 MLD50) were used in the DBA.2 mouse, a background that is highly sensitive to influenza virus infection (54). In addition, the PR8-GLuc virus was highly attenuated, with an MLD50 50- to 100-fold higher than the parental PR8 MLD50. This contrasts with the PASTN strain described here, which displays robust bioluminescence during even sublethal infections with 0.1 MLD50 as well as pathology and mortality that are indistinguishable from those of the parental WSN. Nevertheless, both PR8-GLuc and PASTN have the potential to make significant contributions to understanding the spatiotemporal dynamics of influenza virus replication in vivo. Given the modular nature of the NLuc reporter construct, this approach should be broadly applicable to any influenza virus isolate and may also be amenable to other small reporters. To that end, we have already rescued a NLuc reporter virus encoding the RNP genes from the patient isolate A/Utah/01/2009 from the 2009 pandemic (data not shown). Thus, reporter viruses will be useful in monitoring replication in existing strains, and their ease of construction will make them particularly useful in rapidly examining the replication characteristics of emerging strains as well.
Positron emission tomography (PET), fluorescence, and bioluminescence elicited by firefly luciferase are all compatible with NanoLuc imaging. Thus, multiplexing influenza luciferase viruses with other in vivo imaging modalities could simultaneously measure viral replication and the host response. For example, previous experiments monitoring inflammatory responses to influenza virus infection via PET imaging could be coupled with PASTN to determine the impact of the viral load and the site of replication on the severity of the response (55). Similarly, recruitment of discrete immune cell populations to sites of infection can be monitored by infecting mice with PASTN and detecting immune cells that have been labeled ex vivo with tracking dyes (56) or by detecting fluorogenic probes that are selectively activated by specific immune cell populations (57). Alternatively, innate immune responses and viral replication could be measured concurrently by using PASTN to infect reporter mice that drive firefly luciferase from an NF-κB or IFN-β promoter(s), as NLuc and firefly luciferase exclusively use different substrates (29, 58, 59). Multiplexing could also be used to study the interplay between primary viral infections and the development of secondary bacterial pneumonia by performing coinfections with PASTN and bioluminescent Streptococcus (60). Thus, with near-native replication properties, a stable genome, and a very strong bioluminescent output, our influenza reporter virus provides a noninvasive spatiotemporal measure of viral replication that now enables in vivo studies of the interplay between infection, host response, and disease severity.
ACKNOWLEDGMENTS
This work was supported in part by the National Institute of General Medical Sciences (R00GM088484), a Shaw Scientist Award, a Mirus Research Award, and a Wisconsin Partnership Education and Research Committee New Investigator Program grant to A.M. and an NIH National Research Service Award (T32 GM07215) to V.T.
We thank L. Moncla, T. Wesely, J. D. Sauer, L. Knoll, and members of the Mehle and Knoll laboratories for valuable contributions and Y. Kawaoka, T. Parslow, and Promega for sharing reagents.
Footnotes
Published ahead of print 2 October 2013
REFERENCES
- 1.WHO 2009. 2009 Influenza (seasonal) fact sheet. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs211/en/ [Google Scholar]
- 2.Thompson MG, Shay DK, Zhou H, Bridges CB, Cheng PY, Burns E, Bresee JS, Cox NJ. 2010. Estimates of deaths associated with seasonal influenza—United States, 1976–2007. MMWR Morb. Mortal. Wkly. Rep. 59:1057–1062 [PubMed] [Google Scholar]
- 3.Taubenberger JK, Morens DM. 2006. 1918 influenza: the mother of all pandemics. Emerg. Infect. Dis. 12:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368:1888–1897 [DOI] [PubMed] [Google Scholar]
- 5.Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, Meng L, Hong Z, Tu W, Cao Y, Li L, Ding F, Liu B, Wang M, Xie R, Gao R, Li X, Bai T, Zou S, He J, Hu J, Xu Y, Chai C, Wang S, Gao Y, Jin L, Zhang Y, Luo H, Yu H, Gao L, Pang X, Liu G, Shu Y, Yang W, Uyeki TM, Wang Y, Wu F, Feng Z. 24 April 2013. Preliminary report: epidemiology of the avian influenza A (H7N9) outbreak in China. N. Engl. J. Med. [Epub ahead of print.] 10.1056/NEJMoa1304617 [DOI] [Google Scholar]
- 6.WHO 2013. Overview of the emergence and characteristics of the avian influenza A(H7N9) virus. WHO, Geneva, Switzerland: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/WHO_H7N9_review_31May13.pdf [Google Scholar]
- 7.Barnard DL. 2009. Animal models for the study of influenza pathogenesis and therapy. Antiviral Res. 82:A110–A122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belser JA, Katz JM, Tumpey TM. 2011. The ferret as a model organism to study influenza A virus infection. Dis. Model Mech. 4:575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Donnell CD, Subbarao K. 2011. The contribution of animal models to the understanding of the host range and virulence of influenza A viruses. Microbes Infect. 13:502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luker KE, Luker GD. 2008. Applications of bioluminescence imaging to antiviral research and therapy: multiple luciferase enzymes and quantitation. Antiviral Res. 78:179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mondotte JA, Lozach PY, Amara A, Gamarnik AV. 2007. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J. Virol. 81:7136–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoggins JW, Dorner M, Feulner M, Imanaka N, Murphy MY, Ploss A, Rice CM. 2012. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proc. Natl. Acad. Sci. U. S. A. 109:14610–14615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luker GD, Bardill JP, Prior JL, Pica CM, Piwnica-Worms D, Leib DA. 2002. Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. J. Virol. 76:12149–12161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook SH, Griffin DE. 2003. Luciferase imaging of a neurotropic viral infection in intact animals. J. Virol. 77:5333–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez JF, Rodriguez D, Rodriguez JR, McGowan EB, Esteban M. 1988. Expression of the firefly luciferase gene in vaccinia virus: a highly sensitive gene marker to follow virus dissemination in tissues of infected animals. Proc. Natl. Acad. Sci. U. S. A. 85:1667–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke CW, Mason JN, Surman SL, Jones BG, Dalloneau E, Hurwitz JL, Russell CJ. 2011. Illumination of parainfluenza virus infection and transmission in living animals reveals a tissue-specific dichotomy. PLoS Pathog. 7:e1002134. 10.1371/journal.ppat.1002134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. U. S. A. 107:11531–11536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Sobrido L, Cadagan R, Steel J, Basler CF, Palese P, Moran TM, García-Sastre A. 2010. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 84:2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozawa M, Victor ST, Taft AS, Yamada S, Li C, Hatta M, Das SC, Takashita E, Kakugawa S, Maher EA, Neumann G, Kawaoka Y. 2011. Replication-incompetent influenza A viruses that stably express a foreign gene. J. Gen. Virol. 92:2879–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heaton NS, Leyva-Grado VH, Tan GS, Eggink D, Hai R, Palese P. 22 May 2013. In vivo bioluminescent imaging of influenza A virus infection and characterization of novel cross-protective monoclonal antibodies. J. Virol. 10.1128/JVI.00969-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pena L, Sutton T, Chockalingam A, Kumar S, Angel M, Shao H, Chen H, Li W, Perez DR. 2013. Influenza viruses with rearranged genomes as live-attenuated vaccines. J. Virol. 87:5118–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dos Santos Afonso E, Escriou N, Leclercq I, van der Werf S, Naffakh N. 2005. The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the PB2 segment. Virology 341:34–46 [DOI] [PubMed] [Google Scholar]
- 23.Liang Y, Hong Y, Parslow TG. 2005. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 79:10348–10355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F, Feng L, Pan W, Dong Z, Li C, Sun C, Chen L. 2010. Generation of replication-competent recombinant influenza A viruses carrying a reporter gene harbored in the neuraminidase segment. J. Virol. 84:12075–12081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kittel C, Sereinig S, Ferko B, Stasakova J, Romanova J, Wolkerstorfer A, Katinger H, Egorov A. 2004. Rescue of influenza virus expressing GFP from the NS1 reading frame. Virology 324:67–73 [DOI] [PubMed] [Google Scholar]
- 26.Avilov SV, Moisy D, Munier S, Schraidt O, Naffakh N, Cusack S. 2012. Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. J. Virol. 86:1433–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinya K, Fujii Y, Ito H, Ito T, Kawaoka Y. 2004. Characterization of a neuraminidase-deficient influenza a virus as a potential gene delivery vector and a live vaccine. J. Virol. 78:3083–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Q, Lowen AC, Wang TT, Palese P. 2010. A nine-segment influenza A virus carrying subtype H1 and H3 hemagglutinins. J. Virol. 84:8062–8071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. 2012. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 7:1848–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann G, Fujii K, Kino Y, Kawaoka Y. 2005. An improved reverse genetics system for influenza A virus generation and its implications for vaccine production. Proc. Natl. Acad. Sci. U. S. A. 102:16825–16829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mehle A, Doudna JA. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. U. S. A. 106:21312–21316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehle A, Doudna JA. 2008. An inhibitory activity in human cells restricts the function of an avian-like influenza virus polymerase. Cell Host Microbe 4:111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regan JF, Liang Y, Parslow TG. 2006. Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J. Virol. 80:252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donnelly MLL, Hughes LE, Luke G, Mendoza H, ten Dam E, Gani D, Ryan MD. 2001. The ‘cleavage' activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like' sequences. J. Gen. Virol. 82:1027–1041 [DOI] [PubMed] [Google Scholar]
- 36.Matrosovich M, Matrosovich T, Garten W, Klenk HD. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. 10.1186/1743-422X-3-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fodor E, Smith M. 2004. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 78:9144–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Y, Huang T, Ly H, Parslow TG, Liang Y. 2008. Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J. Virol. 82:229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y. 2006. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 80:2318–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooke CB, Ince WL, Wrammert J, Ahmed R, Wilson PC, Bennink JR, Yewdell JW. 2013. Most influenza a virions fail to express at least one essential viral protein. J. Virol. 87:3155–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Francis T, Moore AE. 1940. A study of the neurotropic tendency in strains of the virus of epidemic influenza. J. Exp. Med. 72:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Almond JW. 1977. A single gene determines the host range of influenza virus. Nature 270:617–618 [DOI] [PubMed] [Google Scholar]
- 43.Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naffakh N, Tomoiu A, Rameix-Welti MA, van der Werf S. 2008. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu. Rev. Microbiol. 62:403–424 [DOI] [PubMed] [Google Scholar]
- 45.Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842 [DOI] [PubMed] [Google Scholar]
- 46.Labadie K, Dos Santos Afonso E, Rameix-Welti MA, van der Werf S, Naffakh N. 2007. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 362:271–282 [DOI] [PubMed] [Google Scholar]
- 47.Munster VJ, de Wit E, van Riel D, Beyer WE, Rimmelzwaan GF, Osterhaus AD, Kuiken T, Fouchier RA. 2007. The molecular basis of the pathogenicity of the Dutch highly pathogenic human influenza A H7N7 viruses. J. Infect. Dis. 196:258–265 [DOI] [PubMed] [Google Scholar]
- 48.Naffakh N, Massin P, Escriou N, Crescenzo-Chaigne B, van der Werf S. 2000. Genetic analysis of the compatibility between polymerase proteins from human and avian strains of influenza A viruses. J. Gen. Virol. 81:1283–1291 [DOI] [PubMed] [Google Scholar]
- 49.Salomon R, Franks J, Govorkova EA, Ilyushina NA, Yen HL, Hulse-Post DJ, Humberd J, Trichet M, Rehg JE, Webby RJ, Webster RG, Hoffmann E. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203:689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinya K, Hamm S, Hatta M, Ito H, Ito T, Kawaoka Y. 2004. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology 320:258–266 [DOI] [PubMed] [Google Scholar]
- 51.Goto H, Wells K, Takada A, Kawaoka Y. 2001. Plasminogen-binding activity of neuraminidase determines the pathogenicity of influenza A virus. J. Virol. 75:9297–9301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. 2008. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. U. S. A. 105:4381–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobasa D, Takada A, Shinya K, Hatta M, Halfmann P, Theriault S, Suzuki H, Nishimura H, Mitamura K, Sugaya N, Usui T, Murata T, Maeda Y, Watanabe S, Suresh M, Suzuki T, Suzuki Y, Feldmann H, Kawaoka Y. 2004. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431:703–707 [DOI] [PubMed] [Google Scholar]
- 54.Srivastava B, Błażejewska P, Heßmann M, Bruder D, Geffers R, Mauel S, Gruber AD, Schughart K. 2009. Host genetic background strongly influences the response to influenza A virus infections. PLoS One 4:e4857. 10.1371/journal.pone.0004857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jonsson CB, Camp JV, Wu A, Zheng H, Kraenzle JL, Biller AE, Vanover CD, Chu Y-K, Ng CK, Proctor M, Sherwood L, Steffen MC, Mollura DJ. 2012. Molecular imaging reveals a progressive pulmonary inflammation in lower airways in ferrets infected with 2009 H1N1 pandemic influenza virus. PLoS One 7:e40094. 10.1371/journal.pone.0040094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tario JD, Jr, Gray BD, Wallace SS, Muirhead KA, Ohlsson-Wilhelm BM, Wallace PK. 2007. Novel lipophilic tracking dyes for monitoring cell proliferation. Immunol. Invest. 36:861–885 [DOI] [PubMed] [Google Scholar]
- 57.Kossodo S, Zhang J, Groves K, Cuneo GJ, Handy E, Morin J, Delaney J, Yared W, Rajopadhye M, Peterson JD. 2011. Noninvasive in vivo quantification of neutrophil elastase activity in acute experimental mouse lung injury. Int. J. Mol. Imaging 2011:581406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carlsen H, Moskaug JO, Fromm SH, Blomhoff R. 2002. In vivo imaging of NF-kappa B activity. J. Immunol. 168:1441–1446 [DOI] [PubMed] [Google Scholar]
- 59.Lienenklaus S, Cornitescu M, Zietara N, Lyszkiewicz M, Gekara N, Jablonska J, Edenhofer F, Rajewsky K, Bruder D, Hafner M, Staeheli P, Weiss S. 2009. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J. Immunol. 183:3229–3236 [DOI] [PubMed] [Google Scholar]
- 60.McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, Bennink J, Yewdell JW, McCullers JA. 2007. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]