Abstract
The high-risk human papillomavirus (HR HPV) E6 oncoprotein binds host cell proteins to dysregulate multiple regulatory pathways, including apoptosis and senescence. HR HPV16 E6 (16E6) interacts with the cellular protein NFX1-123, and together they posttranscriptionally increase hTERT expression, the catalytic subunit of telomerase. NFX1-123 interacts with hTERT mRNA and stabilizes it, leading to greater telomerase activity and the avoidance of cellular senescence. Little is known regarding what other transcripts are dependent on or augmented by the association of NFX1-123 with 16E6. Microarray analysis revealed enhanced expression of Notch1 mRNA in 16E6-expressing keratinocytes when NFX1-123 was overexpressed. A moderate increase in Notch1 mRNA was seen with overexpression of NFX1-123 alone, but with 16E6 coexpression the increase in Notch1 was enhanced. The PAM2 motif and R3H protein domains in NFX1-123, which were important for increased hTERT expression, were also important in the augmentation of Notch1 expression by 16E6. These findings identify a second gene coregulated by 16E6 and NFX1-123 and the protein motifs in NFX1-123 that are important for this effect.
INTRODUCTION
Human papillomaviruses (HPVs) are nonenveloped double-stranded DNA viruses. Of the more than 150 HPVs that have been identified to date, over 30 types are capable of infecting the genital and oral mucosa. HPVs that infect mucosa are divided into low-risk (LR) and high-risk (HR) categories based on their associated risk for anogenital and, increasingly, head and neck cancers (1–11). As these HR HPV infections are linked to malignancies, much focus has been placed on studying the cellular changes these viruses trigger that lead to cancer and on studying the functions of two oncogenes expressed by HR HPV, E6 and E7.
HR E6 induces oncogenic changes primarily through its interactions with host cell proteins, and those interactions block apoptotic signaling and drive cellular immortalization (for a review, see reference 12). E6-associated protein (E6AP) is an E3 ubiquitin ligase that is the most well-studied protein partner of HR E6 (13–19). However, HR E6 interacts with other cellular proteins, such as NFX1 splice variants, and these are important in oncogenesis as well (20, 21).
In human epithelial cells, NFX1 is a gene expressed as two splice variants, NFX1-91 and NFX1-123 (22). NFX1-91 has been shown to transcriptionally repress telomerase expression in epithelial cells, and HR HPV 16E6 with E6AP targets NFX1-91 for degradation (22, 23). Our work has focused on NFX1-123, the longer splice variant of NFX1, and on its role with 16E6 in oncogenesis. Previously we had shown that 16E6 bound NFX1-123 in its central domain, but unlike NFX1-91 it was not degraded; together, 16E6 and NFX1-123 increased hTERT expression and telomerase activity in human foreskin keratinocytes (HFKs) through a posttranscriptional mechanism (20). 16E6 and NFX1-123 increased hTERT by utilizing both the PAM2 motif and R3H domain of the NFX1-123 protein. The N-terminal PAM2 motif is required to interact with cytoplasmic poly(A) binding proteins, and the C-terminal R3H domain has putative nucleic acid binding capabilities (20, 21). With these motifs, NFX1-123 interacted with and stabilized the hTERT mRNA, posttranscriptionally increased its expression, and increased telomerase activity (21), which is important in HPV-associated cancer development (24, 25).
Because 16E6 interacts with NFX1-123 to direct posttranscriptional gene regulation of an oncogene, we hypothesized that other critical cellular transcripts are coregulated by 16E6 and NFX1-123. Using microarray analysis of mRNA transcripts in HFKs, we identified and confirmed that Notch1 was increased by NFX1-123 and 16E6. Notch1 is an important regulator of cell growth and differentiation in multiple cell types, including epithelial cells. While Notch and its signaling pathway can lead to either oncogenic or tumor suppressor effects, the normal control of Notch1 expression and its derangement in cancer is not fully understood (26). In cervical cancer specifically, investigators have found decreased Notch1 expression (27–29). Conversely, others have found increased Notch1 expression during cervical cancer development (30–32) and increased Notch1 expression and activation by the HR E6 and E7 oncogenes (33), which leads to improved cell growth (34, 35). Our findings support the positive regulation of Notch1 by 16E6 and NFX1-123. Similar to the regulation of hTERT, the PAM2 motif and R3H domain of NFX1-123 were important in increasing Notch1 expression when 16E6 was coexpressed. The coregulation of Notch1 by 16E6 and NFX1-123 reveals additional roles that NFX1-123 plays in HPV-associated cancer development (36).
MATERIALS AND METHODS
Plasmids.
The FLAG-tagged NFX1-123WT, FLAG-tagged NFX1-123 PAM2-deleted, FLAG-tagged NFX1-123 R3H-deleted, pBabe-puro 16E6, LXSN vector control, scramble, and short hairpin 1 RNA to NFX1-123 (sh1) plasmids have been described previously (20). The short hairpin 2 RNA to NFX1-123 (sh2) was made with phosphorylated and annealed oligonucleotides 5′ GATCCGCGTGAATAAGGGAAAGAATTTCAAGAGAATTCTTTCCCTTATTCACGTTTTTTGG 3′ (forward) and 5′ AATTCCAAAAAACGTGAATAAGGGAAAGAATTCTCTTGAAATTCTTTCCCTTATTCACGCG 3′ (reverse). These annealed oligonucleotides were ligated into the C-FUGW vector using BamHI and EcoRI restriction sites. BLAST was used to confirm that there were no off-target effects of short hairpin RNA (shRNA) and scramble sequences.
Tissue culture.
Primary human foreskin keratinocytes (HFKs) were cultured as described previously (22). Briefly, HFKs were derived from neonatal foreskins and grown in EpiLife medium supplemented with calcium chloride (60 μM), human keratinocyte growth supplement (Life Technologies, Carlsbad, CA), and penicillin-streptomycin. 293T cells were grown in Dulbecco's modified Eagle's medium (GIBCO-BRL, Carlsbad, CA) containing 10% fetal calf serum and penicillin-streptomycin.
Retrovirus production and infection.
Retrovirus was produced either in established viral producer cell lines (PA317 and PG13) or in 293T cells by a transient vesicular stomatitis virus G-pseudotyped virus (VSV-G) production protocol as previously described (37). Lentivirus was also produced as previously described (20). Briefly, short hairpin NFX1-123 (sh1 or sh2) c-FUGW or scramble c-FUGW constructs were cotransfected with cytomegalovirus-vesicular stomatitis virus G and Δ8.9 plasmids into 293T cells using FuGENE6 (Roche, Alameda, CA), and lentiviral retrovirus was serially collected. Retrovirus was concentrated by ultracentrifugation, mixed with Polybrene (8 μg/ml) (EMD Millipore, Billerica, MA), and incubated with 50 to 60% confluent HFKs. Three hours later, the Epilife medium was replaced. For VSV-G-pseudotyped virus infections (FNFX1-123WT, LXSN vector control, and pBabe-puro 16E6), the cells were expanded 24 h posttransduction, and after 48 h the cells were placed under neomycin/G418 selection (50 μg/ml) or puromycin selection (0.5 μg/ml). All lentivirus infections (scramble, sh1, and sh2) were confirmed by green fluorescent protein expression.
Microarray.
Total RNA was isolated with TRIzol reagent (Life Technologies, Carlsbad, CA) and further purified using the RNeasy kit (Qiagen, Valencia, CA). Samples were converted to cDNA, labeled using the Illumina TotalPrep RNA amplification kit (Life Technologies, Carlsbad, CA), and hybridized to the HumanHT-12 v4 expression BeadChip array (Illumina, San Diego, CA). All labeling, hybridizations, and quality control were performed at the Fred Hutchinson Cancer Research Center Genomics Core (Seattle, WA). Array data were initially analyzed using GenomeStudio (Illumina, San Diego, CA). Subsequent analyses were performed using GeneSpring GX11.5.1 (Agilent Technologies, Santa Clara, CA), Microsoft Excel (Redmond, WA), and ingenuity pathway analysis (Redwood City, CA). Data were log2 transformed, followed by quantile normalization. Data were filtered on the detection P value, and only those probes with a detection P value of <0.01 across all samples were carried forward for analysis. Comparative analyses were performed to determine fold change values between the normalized probe intensities using the isogenic scramble control as the baseline.
qPCR and TaqMan array.
RNA was isolated as described above. Total RNA (1 μg) was DNase treated and used to generate cDNA with random hexamer primers and SuperScript II reverse transcriptase (Life Technologies, Carlsbad, CA). Quantitative real-time PCR (qPCR) was performed using an ABI StepOne Plus system (Applied Biosystems, Foster City, CA). Amplification was carried out using TaqMan master mix and the following predesigned TaqMan probes according to the manufacturer's instructions: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (4333764F), Notch1 (HS01062014_m1), HES1 (Hs00172878_m1), and HES5 (Hs01387463_g1) (Applied Biosystems, Foster City, CA). Predesigned TaqMan array human notch signaling (4418810) was also performed according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). Reactions were performed in triplicate. Data analysis was performed using the comparative threshold cycle method (ΔΔCT; Applied Biosystems, Foster City, CA) to determine relative expression levels, with GAPDH to normalize mRNA levels within each sample. Values graphed are the mean fold change in each sample compared to the control, and error bars graphed represent the standard deviations for each sample (n = 3).
Primer sequences for NFX1-123 and 36B4 were described previously (20), and amplification was carried out as previously published using Power Sybr green master mix (Applied Biosystems, Foster City, CA) (20). Error bars using NFX1-123 and 36B4 primers represent 95% confidence intervals.
Immunoblotting.
Whole-cell protein extracts were electrophoresed on SDS-polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF; Millipore, Billerica, MA). Blots were probed with anti-GAPDH (1:100,000; Abcam, Cambridge, MA) and Notch1 (5B5) (1:1,000; Cell Signaling, Danvers, MA) antibodies. The rabbit polyclonal anti-NFX1-123 antibody was generously provided by Ann Roman. All films were scanned using Adobe Photoshop, and densitometry was calculated using Quantity One 4.5.0.
Luciferase assays.
Luciferase assays were performed as described previously, with the following modifications (20, 27, 28). HFKs were grown to 60% confluence in six-well plates and transiently transfected with a total of 2 μg of DNA per well using a 1:3 DNA/TransIT keratinocyte transfection reagent (Mirus Bio, Madison, WI) ratio. All cells were harvested 24 h after transfection. Cellular debris was pelleted, and luminescence was quantified with 20 μl of lysate mixed with luciferase assay buffer (Promega, Madison, WI) in a Victor3 1420 luminometer. Each experiment was done in triplicate and normalized for total protein concentration. Error bars graphed represent 95% confidence intervals. P values were calculated using the two-tailed student t test. Notch1 promoter reporter N1PR2-Luc (2-kb promoter) and Prless-Luc (promoterless reporter) were gifts from Tohru Kiyono (27, 28).
Microarray data accession number.
The data discussed in this article have been deposited in the NCBI Gene Expression Omnibus and are accessible under GEO series accession number GSE43082.
RESULTS
Microarray screening for genes coregulated by NFX1-123 and 16E6.
While we have shown that NFX1-123 and 16E6 regulate hTERT expression (20, 21), little is known regarding what other transcripts and downstream signaling pathways depend on NFX1-123 and 16E6. To identify novel cellular transcripts affected by NFX1-123 and 16E6, global gene expression changes were assessed using a microarray screen in three independently derived HFK cell lines transduced with a 16E6 retrovirus (Fig. 1A). 16E6 expression was confirmed by p53 protein degradation (data not shown). Each of the three 16E6 HFK cell lines was then expanded and transduced with a second retrovirus containing either a FLAG-tagged NFX1-123 overexpression construct (FNFX1-123WT) or an isogenic scramble short hairpin RNA control construct (scramble) (Fig. 1A). On average, FNFX1-123WT expression led to three-and-a-half times the mRNA of endogenous NFX1-123, placing its overexpression within the range of NFX1-123 mRNA and protein amounts detected in SiHa and HeLa cervical cancer cell lines (data not shown). The scramble control was chosen for the reference value in order to validate our microarray data using short hairpin RNA constructs to knock down NFX1-123 expression.
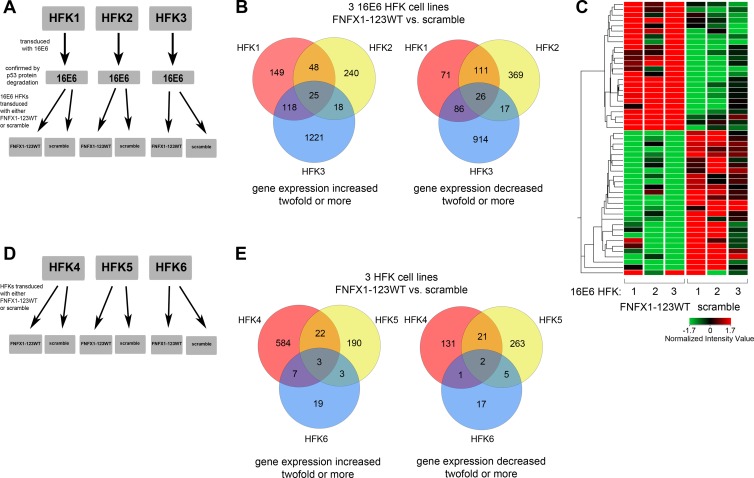
Fig 1.
Gene expression changes in HFKs with increased NFX1-123 by microarray. (A) Diagram of 16E6 HFK cell lines used in the microarray. Three independent HFK cell lines (HFK1, HFK2, and HFK3) were transduced with 16E6. These three 16E6 HFK cell lines were then divided and transduced with FLAG-tagged NFX1-123 in its wild-type form of overexpression (FNFX1-123WT) or with an isogenic scramble short hairpin RNA control (scramble). (B) Venn diagrams of genes identified as increased or decreased by at least 2-fold in FNFX1-123WT cells. Fold changes were calculated by using the isogenic scramble shRNA control as the baseline for each independent 16E6 HFK cell line. (C) Hierarchical clustering of 51 genes identified in panel B as increased or decreased at least 2-fold or more with FNFX1-123WT in all 16E6 HFK cell lines. Red, high expression; green, low expression. (D) Diagram of HFK cell lines (HFK4, HFK5, and HFK6) used in the microarray. Three independent HFK cell lines were divided and transduced with either FNFX1-123WT or scramble. (E) Venn diagrams of genes identified as increased or decreased by at least 2-fold in FNFX1-123WT cells. Fold changes were calculated as described for panel B for each independent HFK cell line.
For the three independent 16E6 HFK cell lines, 398 genes were up- or downregulated 2-fold or more in two out of three cell lines overexpressing NFX1-123 (FNFX1-123WT) compared to isogenic scramble shRNA control, and an additional 51 genes were up- or downregulated across all three cell lines (Fig. 1B). The 51 up- or downregulated genes are represented by hierarchical clustering in Fig. 1C, and the 25 genes increased 2-fold or more across all three 16E6 HFK cell lines are listed in Table 1. Of particular interest was the gene Notch1, which was found to be significantly increased by microarray in all three overexpressing NFX1-123 HFK cell lines when 16E6 was coexpressed.
Table 1.
Gene with expression increased at least 2-fold in 16E6 HFKs with overexpressed NFX1-123
| Name | Entrez gene ID | Definition | Mean fold change in expression |
|---|---|---|---|
| RAB7B | 338382 | RAB7B, member of the RAS oncogene family | 2.0 |
| LOC643031 | 643031 | Similar to NADH dehydrogenase subunit 5 (predicted) | 2.1 |
| TGM1 | 7051 | Transglutaminase 1 | 2.1 |
| FBN2 | 2201 | Fibrillin 2 | 2.2 |
| HSPBL2 | 653553 | Heat shock 27-kDa protein-like 2 pseudogene | 2.3 |
| PPL | 5493 | Periplakin | 2.5 |
| SLPI | 6590 | Secretory leukocyte peptidase inhibitor | 2.5 |
| FOXA2 | 3170 | Forkhead box A2, transcript variant 2 | 2.6 |
| BNIPL | 149428 | BCL2/adenovirus E1B 19-kDa interacting protein-like | 2.6 |
| CCNB1IP1 | 57820 | Cyclin B1 interacting protein 1, transcript variant 3 | 2.7 |
| IMPA2 | 3613 | Inositol(myo)-1(or 4)-monophosphatase 2 | 2.8 |
| RPS29 | 6235 | Ribosomal protein S29, transcript variant 2 | 2.8 |
| KRT16 | 3868 | Keratin 16 (focal nonepidermolytic palmoplantar keratoderma) | 2.9 |
| RAET1G | 353091 | Retinoic acid early transcript 1G | 2.9 |
| LCE2B | 26239 | Late cornified envelope 2B | 3.0 |
| LCE1B | 353132 | Late cornified envelope 1B | 3.0 |
| ALDH3B2 | 222 | Aldehyde dehydrogenase 3 family, member B2, transcript variant 2 | 3.1 |
| MGC102966 | 644945 | Similar to keratin, type I cytoskeletal 16 (predicted) | 3.2 |
| LOC400578 | 400578 | Similar to keratin, type I cytoskeletal 14 (predicted) | 3.2 |
| NOTCH1 | 4851 | Notch homolog 1, translocation associated (Drosophila) | 3.5 |
| SPRR2G | 6706 | Small proline-rich protein 2G | 3.5 |
| LOC400578 | 400578 | Similar to keratin, type I cytoskeletal 14 (predicted) | 3.7 |
| LOC729252 | 729252 | Similar to keratin, type I cytoskeletal 14 (predicted) | 3.7 |
| CEBPD | 1052 | CCAAT/enhancer binding protein (C/EBP), delta | 5.5 |
| LOR | 4014 | Loricrin | 6.0 |
Whole-genome expression microarray analysis was also conducted in three independent non-16E6-expressing HFKs that were expanded and transduced with retrovirus containing either a FLAG-tagged NFX1-123 overexpression construct (FNFX1-123WT) or an isogenic scramble shRNA control construct (scramble) (Fig. 1D). This second microarray study permitted a comparison of gene expression changes with or without 16E6 coexpression to then determine whether they depended on 16E6 or occurred regardless of it.
Interestingly, fewer genes were found to be increased or decreased by NFX1-123 overexpression in non-16E6-expressing HFKs. For the three independent HFK cell lines, 59 genes were up- or downregulated 2-fold or more in two out of three FNFX1-123WT cell lines compared to isogenic scramble shRNA control, and an additional five genes were up- or downregulated across all three cell lines (Fig. 1E). The three genes found to be upregulated 2-fold or more in FNFX1-123WT HFKs compared to the scramble control were the following (gene symbol, mean fold change): HSP90AA1, 3.0; SPRR3, 3.7; and LOR, 5.4.
16E6 and NFX1-123 increased Notch1 expression.
Microarray analysis results for Notch1 were validated at the RNA and protein levels by qPCR and immunoblot assays. Analysis was done using additional HFK cell lines where NFX1-123 expression was modulated. The impact of 16E6 expression was also evaluated. Similar to the array-based whole-genome expression analysis (Fig. 1A and D), three independent 16E6-expressing HFKs and three independent non-16E6-expressing HFKs were expanded and transduced with FNFX1-123WT or scramble or with one of two shRNA constructs for NFX1-123 (sh1 or sh2). Background expression of either NFX1-123 or Notch1 in HFKs with either the LXSN vector control or the scramble shRNA isogenic control was found to be equivalent (data not shown).
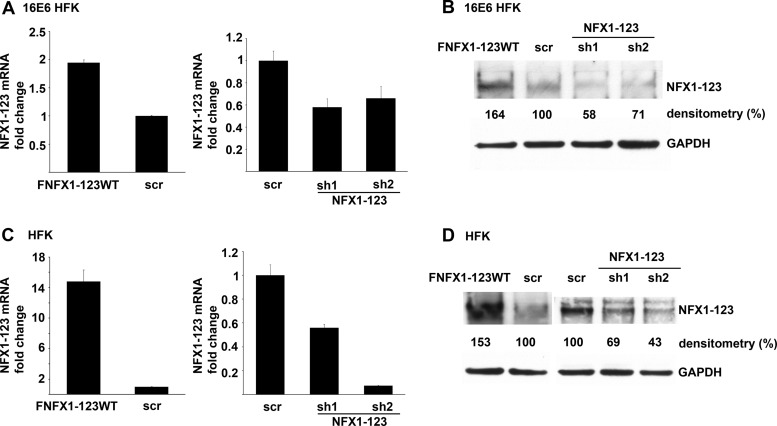
Across three 16E6 HFK cell lines, overexpression of FLAG-tagged NFX1-123 led to a 2-fold increase in NFX1-123 mRNA, and sh1 or sh2 knocked down NFX1-123 mRNA to approximately half the endogenous level in control cells (scr) (Fig. 2A). NFX1-123 protein levels paralleled the mRNA changes due to overexpression or knock down (Fig. 2B). In three non-16E6 HFK cell lines, NFX1-123 mRNA expression increased by 15-fold on average using the overexpression FLAG-tagged construct, and NFX1-123 mRNA expression was knocked down by at least half using either sh1 or sh2 (Fig. 2C). Again, NFX1-123 protein levels followed mRNA changes, although less dramatically (Fig. 2D).
Fig 2.
Modulation of NFX1-123 expression in keratinocytes. (A) NFX1-123 mRNA expression by qPCR in 16E6 HFKs transduced with overexpressed wild-type FLAG-tagged NFX1-123 (FNFX1-123WT), isogenic scramble short hairpin RNA control (scr), short hairpin 1 RNA to NFX1-123 (sh1), or short hairpin 2 RNA to NFX1-123 (sh2). Relative levels of NFX1-123 mRNA were normalized to the mRNA levels of housekeeping gene 36B4. Values shown were the mean fold change in each sample compared to the scr vector control. Error bars represent 95% confidence intervals from the technical replicates shown (n = 3). (B) Representative immunoblot showing NFX1-123 protein expression in 16E6 HFKs. Cells were transduced as described for panel A, and densitometry was calculated using Quantity One. GAPDH is shown as a loading control. (C) NFX1-123 mRNA levels by qPCR of HFKs transduced with FNFX1-123WT, scr, sh1, or sh2. Relative levels of NFX1-123 mRNA were normalized to 36B4 mRNA levels. Values shown were the mean fold changes in each sample compared to the scr vector control. Error bars represent 95% confidence intervals from the technical replicates shown (n = 3). (D) Representative immunoblot showing NFX1-123 protein expression in HFKs. Cells were transduced as described for panel C, and densitometry was calculated using Quantity One. GAPDH is shown as a loading control.
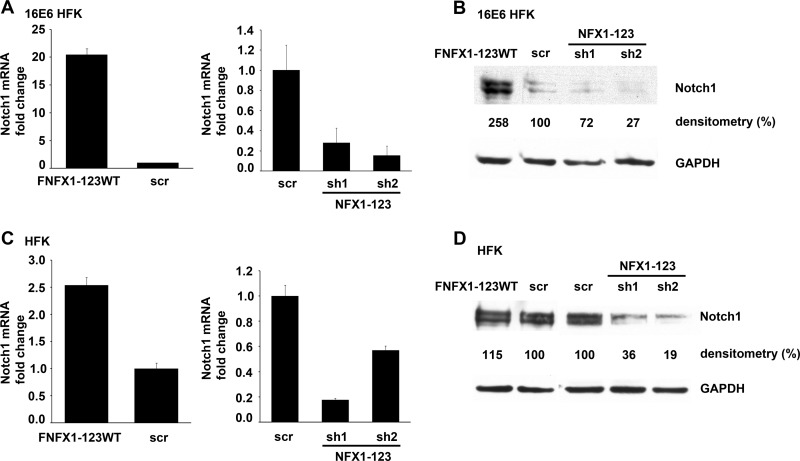
In 16E6 HFKs, Notch1 mRNA was increased up to 20-fold with NFX1-123 overexpression and was reduced at least 70% with knockdown of NFX1-123 by sh1 and sh2 (Fig. 3A). Notch1 protein also increased with more NFX1-123 and decreased with knockdown of NFX1-123 (Fig. 3B). In the absence of 16E6, Notch1 mRNA expression was moderately increased (2-fold) in HFKs with NFX1-123 overexpression and decreased by 40 to 80% when NFX1-123 was reduced by sh1 or sh2 (Fig. 3C). Without 16E6 coexpression, Notch1 protein did not increase with overexpression of NFX1-123 as robustly as with 16E6 (Fig. 3D). However, the endogenous protein expression of Notch1 in HFKs was decreased by at least two-thirds by knockdown of NFX1-123 in the absence of 16E6 coexpression (Fig. 3D). These data link Notch1 mRNA and protein expression to changes in NFX1-123 expression and demonstrate that 16E6 synergized with NFX1-123 to increase Notch1 expression in HFKs.
Fig 3.
Expression level of NFX1-123 affected Notch1 expression. (A) Notch1 mRNA levels in 16E6 HFKs transduced with overexpressed FLAG-tagged NFX1-123 wild type (FNFX1-123WT), isogenic scramble short hairpin RNA control (scr), short hairpin 1 RNA to NFX1-123 (sh1), or short hairpin 2 RNA to NFX1-123 (sh2). Relative levels of Notch1 mRNA were calculated using the ΔΔCT method, normalizing mRNA levels to GAPDH within each sample. Values shown are the mean fold change in each sample compared to the scr vector control. Error bars represent the standard deviations for each sample (n = 3). (B) Representative immunoblot showing the Notch1 protein expression in 16E6 HFKs transduced as described for panel A. Densitometry was calculated using Quantity One. GAPDH is shown as a loading control. (C) Notch1 mRNA levels in HFKs transduced with FNFX1-123WT, scr, sh1, or sh2. Relative levels of Notch1 mRNA were calculated using the ΔΔCT method, normalizing mRNA levels to GAPDH within each sample. Values shown are the mean fold changes in each sample compared to the scr vector control. Error bars represent the standard deviations for each sample (n = 3). (D) Representative immunoblot showing Notch1 protein expression in HFKs transduced as described for panel C. Densitometry was calculated using Quantity One. GAPDH is shown as a loading control.
Notch pathway components increased with NFX1-123 in 16E6 HFKs.
As Notch1 mRNA and protein expression rose with increased expression of NFX1-123 in 16E6 HFKs, a TaqMan quantitative reverse transcription-PCR-based Notch pathway array was used to determine expression changes in Notch pathway components. Three independent 16E6 HFK cell lines were compared where either NFX1-123 was overexpressed (FNFX1-123WT) or an LXSN vector control was used. The array included 44 genes associated with Notch signaling, including Notch pathway ligands, receptors, and downstream effectors. Nineteen of these components were increased 2-fold or more in at least two of the three 16E6 HFK cell lines in which NFX1-123 was overexpressed (Table 2), and no genes were found to be decreased 2-fold or more. Again, Notch1 itself was increased (7-fold), as were Notch2 (2-fold) and Notch3 (8-fold). Additionally, there were increases in RBPJ, a transcriptional regulator that plays a key role in Notch signaling; JAG1, a Notch ligand; and ADAM17, APH1B, PSEN1, and NCSTN, four proteases that activate Notch. Although increases in several negative regulators of Notch were also detected, we hypothesized that these were the result of the complex feedback loop involved in Notch signaling. Therefore, not only was Notch1 expression increased by NFX1-123 with 16E6 coexpression, but the Notch downstream signaling pathway also was activated and upregulated.
Table 2.
Notch pathway genes increased in 16E6 HFKs with overexpressed NFX1-123
| Name | Mean fold changea |
|---|---|
| ADAM17 | 6.4 |
| APH1B | 2.2 |
| DLK2 | 2.2 |
| FBXW7 | 4.7 |
| HAT1 | 7.1 |
| HDAC2 | 8.4 |
| HDAC4 | 2.9 |
| HDAC8 | 2.0 |
| HDAC9 | 2.6 |
| JAG1 | 2.8 |
| NCOR2 | 2.3 |
| NCSTN | 2.2 |
| NOTCH1 | 6.8 |
| NOTCH2 | 2.2 |
| NOTCH3 | 8.4 |
| PSEN1 | 7.0 |
| RBPJ | 23.3 |
| RFNG | 2.4 |
| SNW1 | 7.7 |
Notch pathway transcript levels in 16E6 FNFX1-123 HFKs were compared to transcript levels from 16E6 LXSN HFKs (n = 3).
Notch reduction by NFX1-123 knockdown led to decreased Hes1 and Hes5.
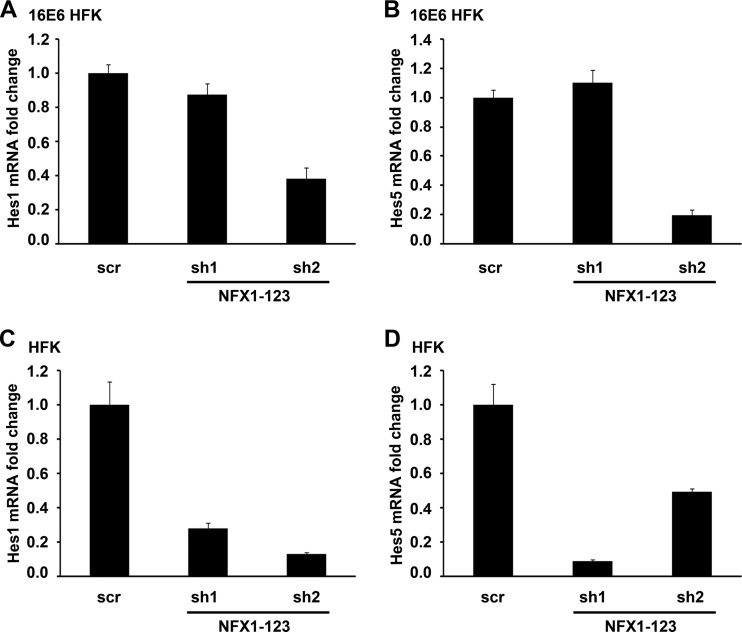
Since Notch1 and multiple factors in its pathway were increased with overexpression of NFX1-123 in 16E6 HFKs, knockdown of NFX1-123 likely would lead to decreased signaling within the Notch pathway. Hes family members are direct targets of Notch activation (38); thus, a reduction in Notch1 expression and activity likely would lead to a decrease in their expression and activity. Indeed, when NFX1-123 was reduced in three independent 16E6 HFK cell lines by 80% using sh2 and Notch1 was reduced nearly three-quarters in parallel, there was a 3-fold reduction in Hes1 mRNA (Fig. 4A) and a 5-fold reduction in Hes5 mRNA (Fig. 4B). Although there was no decrease in Hes1 or Hes5 mRNA by sh1 (Fig. 4A and B), these samples had only a 30% decrease in Notch1 protein (Fig. 3B). In HFKs without 16E6, where knockdown of NFX1-123 by both sh1 and sh2 led to decreased Notch1 expression (Fig. 3C and D), Hes1 mRNA was reduced 4-fold by sh1 and nearly 8-fold by sh2, and Hes5 mRNA was reduced 11-fold by sh1 and 2-fold by sh2 (Fig. 4C and D).
Fig 4.
Notch downstream targets Hes1 and Hes5 decreased with NFX1-123 protein knockdown. (A) Hes1 mRNA levels in 16E6 HFKs transduced with either scramble short hairpin RNA control (scr), sh1RNA to NFX1-123 (sh1), or sh2RNA to NFX1-123 (sh2). Relative levels of Hes1 mRNA were calculated using the ΔΔCT method, normalizing mRNA levels to GAPDH within each sample. Values shown were the mean fold change in each sample compared to the scr vector control. Error bars represent the standard deviations for each sample (n = 3). (B) Hes5 mRNA levels in 16E6 HFKs transduced with either scr, sh1, or sh2. Relative levels of Hes5 mRNA were calculated using the ΔΔCT method, normalizing mRNA levels to GAPDH within each sample. Values shown were the mean fold change in each sample compared to the scr vector control. Error bars represent the standard deviations for each sample (n = 3). (C) Hes1 mRNA levels in HFKs transduced with either scr, sh1, or sh2. Relative levels of Hes1 mRNA were calculated using the ΔΔCT method, normalizing mRNA levels to GAPDH within each sample. Values shown are the mean fold change in each sample compared to the scr vector control. Error bars represent the standard deviations for each sample (n = 3). (D) Hes5 mRNA levels in HFKs transduced with either scr, sh1, or sh2. Relative levels of Hes5 mRNA were calculated using the ΔΔCT method, normalizing mRNA levels to GAPDH within each sample. Values shown are the mean fold change in each sample compared to the scr vector control. Error bars represent the standard deviations for each sample (n = 3).
NFX1-123-mediated Notch1 gene upregulation.
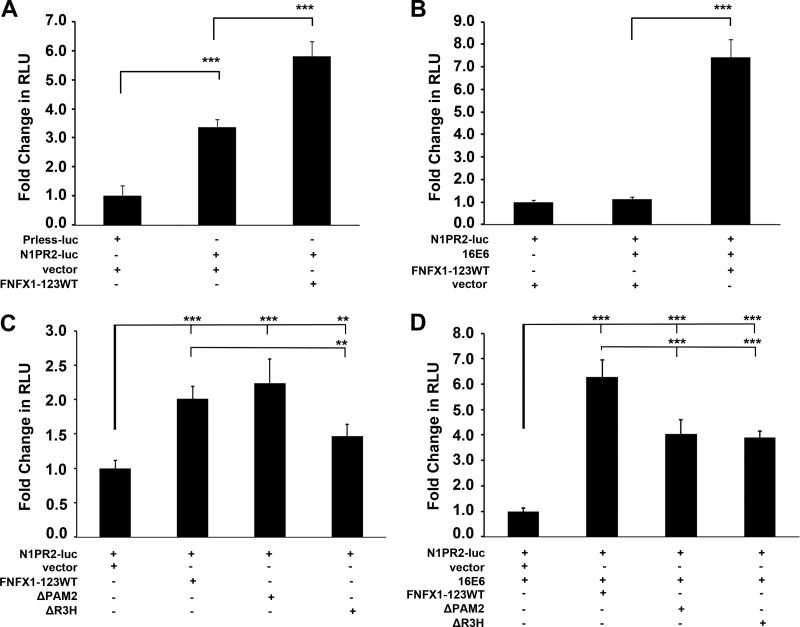
To clarify the role of increased NFX1-123 and 16E6 on Notch1 expression, luciferase assays were used to quantify changes in Notch1 promoter-driven expression. First, in HFKs a promoterless luciferase construct (Prless-luc) was shown to have no activity above that of buffer alone (data not shown). Second, a luciferase construct that included 2 kb of the Notch1 promoter and its entire 5′ untranslated region (N1PR2-luc) (27–29) was transfected into HFKs. Compared to the Prless-luc construct, the N1PR2-luc construct expression level was three times higher than the background expression level (Fig. 5A). This level of expression demonstrated that there was endogenous activation of the Notch1 promoter in HFKs, and it allowed for confirmation of adequate transfection efficiency with each experimental protocol. In HFKs transfected with FNFX1-123WT versus its empty vector control (vector), the Notch1 promoter-driven luciferase activity doubled with increased NFX1-123 expression (Fig. 5A). Thus, overexpression of NFX1-123 increased the Notch1 promoter-driven expression compared to that of HFKs with endogenous levels of NFX1-123.
Fig 5.
Notch1 promoter-driven expression increased by coexpression of FNFX1-123WT and 16E6. (A) HFKs were transfected with either a promoterless luciferase construct (Prless-luc) or a 2-kb Notch1 promoter luciferase construct (N1PR2) and cotransfected with the FLAG-tagged NFX1-123 wild-type construct (FNFX1-123WT) or the pDEST12.2 vector (vector). N1PR2 produced significantly more baseline luciferase activity (RLU, relative light units) than Prless-luc. ***, P < 0.001. HFKs cotransfected with N1PR2 and FNFX1-123WT expressed more luciferase than N1PR2 and vector. ***, P < 0.001. (B) HFKs were cotransfected with N1PR2 and either vector, 16E6, or 16E6 and FNFX1-123WT. Eightfold more luciferase was produced when HFKs were cotransfected with N1PR2, 16E6, and FNFX1-123WT than with N1PR2, 16E6, and vector. ***, P < 0.001. (C) HFKs were cotransfected with N1PR2 and either the FNFX1-123WT construct, the FLAG-tagged NFX1-123 PAM2-deleted construct (ΔPAM2), the FLAG-tagged NFX1-123 R3H-deleted construct (ΔR3H), or vector. FNFX1-123WT had a moderate increase in luciferase compared to vector alone. ***, P < 0.001. A reduction in luciferase was detected when the R3H domain was removed from FNFX1-123WT. **, P < 0.01. (D) HFKs were cotransfected with N1PR2 and 16E6 and either FNFX1-123WT, ΔPAM2, ΔR3H, or vector. Sixfold more luciferase was produced when HFKs were cotransfected with N1PR2, 16E6, and FNFX1-123WT than with N1PR2, 16E6, and vector. ***, P < 0.001. Luciferase was significantly decreased when the PAM2 motif or R3H domain was removed from FNFX1-123WT in 16E6-cotransfected HFKs. ***, P < 0.001. All graphs are of at least three combined experiments, each conducted in triplicate. Error bars graphed represent 95% confidence intervals, and P values were calculated by two-tailed student t test with vector as the reference.
In HFKs transfected with 16E6, there was no increase in the background luciferase activity driven by the 2-kb Notch1 promoter over vector alone (Fig. 5B). However, when 16E6 and FNFX1-123WT were cotransfected, an 8-fold increase in activity was detected (Fig. 5B). Therefore, 16E6 itself had no effect on Notch1 promoter-driven expression, but with increased NFX1-123, 16E6 augmented the increase in luciferase activity.
Because NFX1-123 required its PAM2 motif (20, 39) and its R3H domain (40) to increase 16E6-driven hTERT expression, FNFX1-123-WT was compared to FLAG-tagged NFX1-123 with the PAM2 motif deleted (ΔPAM2), the R3H domain deleted (ΔR3H), or a vector control. These mutant constructs and their expression have been previously published (20, 21). The 2-fold increase in Notch1-driven luciferase expression with FNFX1-123WT versus vector was unaffected by deletion of the PAM2 motif but was significantly reduced with deletion of the R3H domain (Fig. 5C).
The role of 16E6 coexpression on Notch1 promoter-driven luciferase activity with FNFX1-123WT, mutant, or vector control constructs was also compared. When 16E6 was cotransfected with FNFX1-123WT, there was a 6-fold increase in luciferase activity over the vector (Fig. 5D). However, when 16E6 was cotransfected with either the ΔPAM2 or the ΔR3H construct, the increase in Notch1 promoter-driven expression seen with FNFX1-123WT was reduced by one-third (Fig. 5D). Therefore, in HFKs, the basal luciferase activity from the N1PR2-luc construct was increased with more NFX1-123, this activity was augmented by 16E6 coexpression, and both the PAM2 and R3H protein motifs were important for full luciferase activity.
DISCUSSION
Our data reveal Notch1 as a second gene whose expression was regulated by NFX1-123 and 16E6 in HFKs. Similar to the upregulation of hTERT expression by NFX1-123 and 16E6 (20), Notch1 levels increased with more NFX1-123 and decreased with less NFX1-123 in 16E6 HFKs (Fig. 3A and B). Moreover, like the expression of hTERT, Notch1 promoter-driven luciferase expression depended on the PAM2 motif and R3H domains of NFX1-123 to fully increase expression and to be augmented by 16E6 coexpression (Fig. 5D).
However, the role of NFX1-123 in hTERT regulation does differ from that of Notch1. Both Notch1 and NFX1-123 are endogenously expressed in HFKs, while hTERT and 16E6 are not. We previously found that hTERT expression only occurred when 16E6 activated the hTERT promoter, and an increase in NFX1-123 expression itself had no effect (20). With 16E6 expression, a decrease in NFX1-123 blocked the full activation of the hTERT promoter and telomerase activity by 16E6 (20). We now report that regardless of 16E6 coexpression, a knockdown in NFX1-123 expression led to a reduction in endogenous Notch1 mRNA and protein levels (Fig. 3C and D). This effect on the level of Notch1 expression by NFX1-123 without requiring 16E6 (Fig. 5A and C) denotes that NFX1-123 is a regulator of Notch1. This gene regulation by NFX1-123 in turn is augmented by the coexpression of 16E6, but alone 16E6 had no effect (Fig. 5B).
The NFX1 gene is conserved in yeast, Drosophila, Caenorhabditis elegans, Arabidopsis, mice, and humans (41–47). Studies of NFX1 homologues demonstrate its importance in normal cell growth, function, and homeostasis across species, as mutations in the Drosophila melanogaster NFX1 homologue (shuttle craft) are embryonic lethal (42, 48), and gain-of-function mutations in the Arabidopsis thaliana homologue (AtNFXL1) lead to improved growth during stress (41, 47, 49). Our studies in HFKs support the importance of NFX1 in cell growth and longevity, as increases in the NFX1 isoform NFX1-123 improves expression of two important cellular genes, hTERT and Notch1. These two genes are each important in HPV-associated cancers, and their regulation by NFX1-123 is strongly dictated through 16E6.
NFX1-123 binds cytoplasmic poly(A) binding proteins and posttranscriptionally increases the expression of hTERT through RNA binding and stabilization (20, 21). The 5′ untranslated region (UTR) of hTERT is critical for this increase in hTERT expression by NFX1-123 with 16E6 (21). Therefore, it is interesting to hypothesize that the 5′UTR sequence of Notch1 is also important for an increase in expression by NFX1-123 and 16E6. The Notch1 5′UTR sequence was included in the 2-kb Notch1 luciferase reporter construct used in our assays (26). Therefore, when a transcript was produced from this construct, the 5′UTR of Notch1 was synthesized in frame with the luciferase coding sequence. If NFX1-123 posttranscriptionally upregulated the endogenous expression of Notch1 through its 5′UTR, an increase in luciferase activity would be detected. Indeed, background expression from the 2-kb Notch1 luciferase reporter construct was increased 2-fold by NFX1-123 overexpression (Fig. 5A). This increase may have been due to posttranscriptional upregulation by NFX1-123 of the fused Notch1/luciferase RNA. Additionally, the R3H domain of NFX1-123 was required for increased expression, as were both the R3H domain and PAM2 motif for the increased expression with 16E6 (Fig. 5C and D). These data underscore that NFX1-123 increased baseline expression of Notch1 from the 2-kb Notch1 luciferase reporter plasmid, 16E6 synergized this increase, and the two motifs in NFX1-123 that were important in posttranscriptional regulation of hTERT with 16E6 were also important for Notch1 regulation. As 16E6 did not increase Notch1 luciferase expression by itself but augmented the increase with overexpressed NFX1-123, we hypothesize that 16E6 requires a greater amount of available NFX1-123 protein in the cell to affect a significant change in Notch1 expression.
Increased NFX1-123 in 16E6 HFKs led to higher expression of Notch1 and also multiple components of the Notch pathway (Table 2). If Notch1 levels rose without changes to its cleavage or movement into the nucleus, there would be no changes to downstream target genes, cell growth, or differentiation due to NFX1-123 and 16E6. However, we found that numerous genes associated with Notch signaling were increased by NFX1-123 and 16E6, including Notch ligand; Notch1, Notch2, and Notch3 receptors; proteases involved in Notch receptor/ligand processing; and transcriptional coregulators that play a key role in Notch signaling (Table 2). This suggested that there would be changes in Notch1 downstream targets. Indeed, we found that Notch1 target genes Hes1 and Hes5 were decreased when Notch1 expression was decreased by knockdown of NFX1-123 (Fig. 4). These novel findings indicate consistent activation of the Notch signaling pathway, with increases in downstream targets driven by increased NFX1-123 that was reduced in parallel when NFX1-123 and Notch1 were decreased. Interestingly, the Notch1 transcription protein partner MAML is bound and targeted for degradation by beta-HPV E6 types to inhibit Notch signaling (50–53); however, alpha-HPV E6 family members do not bind MAML (50, 51, 53). Our data support these findings, as we found no changes in MAML expression due to NFX1-123 in 16E6 HFKs by the TaqMan Notch pathway array (data not shown).
Notch1 is a master regulator of cell growth and differentiation across many cell types, and perturbations in Notch signaling can confer either oncogenic or tumor suppressor effects on a cell, depending on its context (36). The normal regulation of Notch1 expression in human keratinocytes has not been studied in great detail (26); however, our data identified NFX1-123 as a regulator of Notch1 expression. Several studies have looked at expression levels of Notch1 in cervical cancer development and disease with conflicting results, as modulation of Notch1 expression by HR HPV varies based on experimental timing and design and the chronologies of samples collected from patients. However, moderate increases in Notch1 appear to aid HPV-positive cell growth (34), and our findings also support that 16E6 augments increased Notch1 expression driven by NFX1-123.
ACKNOWLEDGMENTS
We are grateful to D. A. Galloway and members of her laboratory for helpful discussions and suggestions. We appreciate the gifts of the Notch1 promoter luciferase constructs from T. Kiyono and T. Yugawa, NFX1-123 antibody from A. Roman, computational support from H. L. Howie, and manuscript review from A. Geballe and A. Roman.
This work was supported by NIH grant K08 CA131171 to R.A.K.
The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print 9 October 2013
REFERENCES
- 1.Munoz N, Bosch FX, de Tafur SSL, Izarzugaza I, Gili M, Viladiu P, Navarro C, Martos C, Ascunce N, Gonzalez LC, Kaldor JM, Guerrero E, Lorincz A, Santamaria M, Alonso De Ruiz P, Aristizabal N, Shah K. 1992. The causal link between human papillomavirus and invasive cervical cancer: a population-based case-control study in Colombia and Spain. Int. J. Cancer 52:743–749 [DOI] [PubMed] [Google Scholar]
- 2.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12–19 [DOI] [PubMed] [Google Scholar]
- 3.de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin HR, Vallejos CS, de Ruiz PA, Lima MA, Guimera N, Clavero O, Alejo M, Llombart-Bosch A, Cheng-Yang C, Tatti SA, Kasamatsu E, Iljazovic E, Odida M, Prado R, Seoud M, Grce M, Usubutun A, Jain A, Suarez GA, Lombardi LE, Banjo A, Menendez C, Domingo EJ, Velasco J, Nessa A, Chichareon SC, Qiao YL, Lerma E, Garland SM, Sasagawa T, Ferrera A, Hammouda D, Mariani L, Pelayo A, Steiner I, Oliva E, Meijer CJ, Al-Jassar WF, Cruz E, Wright TC, Puras A, Llave CL, Tzardi M, Agorastos T, Garcia-Barriola V, Clavel C, Ordi J, Andujar M, Castellsague X, Sanchez GI, Nowakowski AM, Bornstein J, Munoz N, Bosch FX. 2010. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 11:1048–1056 [DOI] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML. 2011. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 29:4294–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, Shah KV, Sidransky D. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92:709–720 [DOI] [PubMed] [Google Scholar]
- 6.D'Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, Westra WH, Gillison ML. 2007. Case-control study of human papillomavirus and oropharyngeal cancer. N. Engl. J. Med. 356:1944–1956 [DOI] [PubMed] [Google Scholar]
- 7.Carter JJ, Madeleine MM, Shera K, Schwartz SM, Cushing-Haugen KL, Wipf GC, Porter P, Daling JR, McDougall JK, Galloway DA. 2001. Human papillomavirus 16 and 18 L1 serology compared across anogenital cancer sites. Cancer Res. 61:1934–1940 [PubMed] [Google Scholar]
- 8.Daling JR, Madeleine MM, Johnson LG, Schwartz SM, Shera KA, Wurscher MA, Carter JJ, Porter PL, Galloway DA, McDougall JK, Krieger JN. 2005. Penile cancer: importance of circumcision, human papillomavirus and smoking in in situ and invasive disease. Int. J. Cancer 116:606–616 [DOI] [PubMed] [Google Scholar]
- 9.Daling JR, Madeleine MM, Schwartz SM, Shera KA, Carter JJ, McKnight B, Porter PL, Galloway DA, McDougall JK, Tamimi H. 2002. A population-based study of squamous cell vaginal cancer: HPV and cofactors. Gynecol. Oncol. 84:263–270 [DOI] [PubMed] [Google Scholar]
- 10.Gross G, Pfister H. 2004. Role of human papillomavirus in penile cancer, penile intraepithelial squamous cell neoplasias and in genital warts. Med. Microbiol. Immunol. 193:35–44 [DOI] [PubMed] [Google Scholar]
- 11.Salit IE, Tinmouth J, Chong S, Raboud J, Diong C, Su D, Sano M, Lytwyn A, Chapman W, Mahony J. 2009. Screening for HIV-associated anal cancer: correlation of HPV genotypes, p16, and E6 transcripts with anal pathology. Cancer Epidemiol. Biomarkers Prev. 18:1986–1992 [DOI] [PubMed] [Google Scholar]
- 12.Howie HL, Katzenellenbogen RA, Galloway DA. 2009. Papillomavirus E6 proteins. Virology 384:324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talis AL, Huibregtse JM, Howley PM. 1998. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J. Biol. Chem. 273:6439–6445 [DOI] [PubMed] [Google Scholar]
- 14.Nakagawa S, Huibregtse JM. 2000. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 20:8244–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley ML, Keiger KE, Lee CJ, Huibregtse JM. 2005. The global transcriptional effects of the human papillomavirus E6 protein in cervical carcinoma cell lines are mediated by the E6AP ubiquitin ligase. J. Virol. 79:3737–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Yuan H, Fu B, Disbrow GL, Apolinario T, Tomaic V, Kelley ML, Baker CC, Huibregtse J, Schlegel R. 2005. The E6AP ubiquitin ligase is required for transactivation of the hTERT promoter by the human papillomavirus E6 oncoprotein. J. Biol. Chem. 280:10807–10816 [DOI] [PubMed] [Google Scholar]
- 17.Handa K, Yugawa T, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. 2007. E6AP-dependent degradation of DLG4/PSD95 by high-risk human papillomavirus type 18 E6 protein. J. Virol. 81:1379–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shai A, Pitot HC, Lambert PF. 2010. E6-associated protein is required for human papillomavirus type 16 E6 to cause cervical cancer in mice. Cancer Res. 70:5064–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuballa P, Matentzoglu K, Scheffner M. 2007. The role of the ubiquitin ligase E6-AP in human papillomavirus E6-mediated degradation of PDZ domain-containing proteins. J. Biol. Chem. 282:65–71 [DOI] [PubMed] [Google Scholar]
- 20.Katzenellenbogen RA, Egelkrout EM, Vliet-Gregg P, Gewin LC, Gafken PR, Galloway DA. 2007. NFX1-123 and poly(A) binding proteins synergistically augment activation of telomerase in human papillomavirus type 16E6 expressing cells. J. Virol. 81:3786–3796. 10.1128/JVI.02007-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katzenellenbogen RA, Vliet-Gregg P, Xu M, Galloway DA. 2009. NFX1-123 increases hTERT expression and telomerase activity posttranscriptionally in human papillomavirus type 16 E6 keratinocytes. J. Virol. 83:6446–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gewin L, Myers H, Kiyono T, Galloway DA. 2004. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 18:2269–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu M, Luo W, Elzi DJ, Grandori C, Galloway DA. 2008. NFX1 interacts with mSin3A/histone deacetylase to repress hTERT transcription in keratinocytes. Mol. Cell. Biol. 28:4819–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branca M, Giorgi C, Ciotti M, Santini D, Di Bonito L, Costa S, Benedetto A, Bonifacio D, Di Bonito P, Paba P, Accardi L, Mariani L, Ruutu M, Syrjanen S, Favalli C, Syrjanen K. 2006. Upregulation of telomerase (hTERT) is related to the grade of cervical intraepithelial neoplasia, but is not an independent predictor of high-risk human papillomavirus, virus persistence, or disease outcome in cervical cancer. Diagn. Cytopathol. 34:739–748 [DOI] [PubMed] [Google Scholar]
- 25.Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84–88 [DOI] [PubMed] [Google Scholar]
- 26.Lambertini C, Pantano S, Dotto GP. 2010. Differential control of Notch1 gene transcription by Klf4 and Sp3 transcription factors in normal versus cancer-derived keratinocytes. PLoS One 5:e10369. 10.1371/journal.pone.0010369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yugawa T, Narisawa-Saito M, Yoshimatsu Y, Haga K, Ohno S, Egawa N, Fujita M, Kiyono T. 2010. DeltaNp63alpha repression of the Notch1 gene supports the proliferative capacity of normal human keratinocytes and cervical cancer cells. Cancer Res. 70:4034–4044 [DOI] [PubMed] [Google Scholar]
- 28.Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. 2007. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol. Cell. Biol. 27:3732–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talora C, Sgroi DC, Crum CP, Dotto GP. 2002. Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev. 16:2252–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rangarajan A, Syal R, Selvarajah S, Chakrabarti O, Sarin A, Krishna S. 2001. Activated Notch1 signaling cooperates with papillomavirus oncogenes in transformation and generates resistance to apoptosis on matrix withdrawal through PKB/Akt. Virology 286:23–30 [DOI] [PubMed] [Google Scholar]
- 31.Lathion S, Schaper J, Beard P, Raj K. 2003. Notch1 can contribute to viral-induced transformation of primary human keratinocytes. Cancer Res. 63:8687–8694 [PubMed] [Google Scholar]
- 32.Bajaj J, Maliekal TT, Vivien E, Pattabiraman C, Srivastava S, Krishnamurthy H, Giri V, Subramanyam D, Krishna S. 2011. Notch signaling in CD66+ cells drives the progression of human cervical cancers. Cancer Res. 71:4888–4897 [DOI] [PubMed] [Google Scholar]
- 33.Weijzen S, Zlobin A, Braid M, Miele L, Kast WM. 2003. HPV16 E6 and E7 oncoproteins regulate Notch-1 expression and cooperate to induce transformation. J. Cell Physiol. 194:356–362 [DOI] [PubMed] [Google Scholar]
- 34.Henken FE, De-Castro AJ, Rosl F, Bosch L, Meijer CJ, Snijders PJ, Steenbergen RD. 2012. The functional role of Notch signaling in HPV-mediated transformation is dose-dependent and linked to AP-1 alterations. Cell Oncol. 35:77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. 1995. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc. Natl. Acad. Sci. U. S. A. 92:6414–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M. 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartz SR, Vodicka MA. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337–342 [DOI] [PubMed] [Google Scholar]
- 38.Borggrefe T, Oswald F. 2009. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol. Life Sci. 66:1631–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozlov G, Menade M, Rosenauer A, Nguyen L, Gehring K. 2010. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J. Mol. Biol. 397:397–407 [DOI] [PubMed] [Google Scholar]
- 40.Grishin NV. 1998. The R3H motif: a domain that binds single-stranded nucleic acids. Trends Biochem. Sci. 23:329–330 [DOI] [PubMed] [Google Scholar]
- 41.Lisso J, Altmann T, Mussig C. 2006. The AtNFXL1 gene encodes a NF-X1 type zinc finger protein required for growth under salt stress. FEBS Lett. 580:4851–4856 [DOI] [PubMed] [Google Scholar]
- 42.Stroumbakis ND, Li Z, Tolias PP. 1996. A homolog of human transcription factor NF-X1 encoded by the Drosophila shuttle craft gene is required in the embryonic central nervous system. Mol. Cell. Biol. 16:192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song Z, Krishna S, Thanos D, Strominger JL, Ono SJ. 1994. A novel cysteine-rich sequence-specific DNA-binding protein interacts with the conserved X-box motif of the human major histocompatibility complex class II genes via a repeated Cys-His domain and functions as a transcriptional repressor. J. Exp. Med. 180:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasyukova EG, Roshina NV, Mackay TF. 2004. Shuttle craft: a candidate quantitative trait gene for Drosophila lifespan. Aging Cell 3:297–307 [DOI] [PubMed] [Google Scholar]
- 45.Kunz J, Loeschmann A, Deuter-Reinhard M, Hall MN. 2000. FAP1, a homologue of human transcription factor NF-X1, competes with rapamycin for binding to FKBP12 in yeast. Mol. Microbiol. 37:1480–1493 [DOI] [PubMed] [Google Scholar]
- 46.Arlotta P, Miyazaki D, Copeland NG, Gilbert DJ, Jenkins NA, Ono SJ. 2002. Murine NFX.1: isolation and characterization of its messenger RNA, mapping of its chromosomal location and assessment of its developmental expression. Immunology 106:173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mussig C, Schroder F, Usadel B, Lisso J. 2010. Structure and putative function of NFX1-like proteins in plants. Plant Biol. 12:381–394 [DOI] [PubMed] [Google Scholar]
- 48.Tolias PP, Stroumbakis ND. 1998. The Drosophila zygotic lethal gene shuttle craft is required maternally for proper embryonic development. Dev. Genes Evol. 208:274–282 [DOI] [PubMed] [Google Scholar]
- 49.Asano T, Masuda D, Yasuda M, Nakashita H, Kudo T, Kimura M, Yamaguchi K, Nishiuchi T. 2008. AtNFXL1, an Arabidopsis homologue of the human transcription factor NF-X1, functions as a negative regulator of the trichothecene phytotoxin-induced defense response. Plant J. 53:450–464 [DOI] [PubMed] [Google Scholar]
- 50.Tan MJ, White EA, Sowa ME, Harper JW, Aster JC, Howley PM. 2012. Cutaneous beta-human papillomavirus E6 proteins bind Mastermind-like coactivators and repress Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 109:E1473–E1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brimer N, Lyons C, Wallberg AE, Vande Pol SB. 2012. Cutaneous papillomavirus E6 oncoproteins associate with MAML1 to repress transactivation and NOTCH signaling. Oncogene 31:4639–4646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyers JM, Spangle JM, Munger K. 2013. The HPV8 E6 protein interferes with NOTCH activation during keratinocyte differentiation. J. Virol. 87:4762–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White EA, Kramer RE, Tan MJ, Hayes SD, Harper JW, Howley PM. 2012. Comprehensive analysis of host cellular interactions with human papillomavirus E6 proteins identifies new E6 binding partners and reflects viral diversity. J. Virol. 86:13174–13186 [DOI] [PMC free article] [PubMed] [Google Scholar]