Abstract
Simian foamy viruses (SFV) are complex retroviruses that are ubiquitous in nonhuman primates (NHP) and are zoonotically transmitted to humans, presumably through NHP saliva, by licking, biting, and other behaviors. We have studied SFV in free-ranging rhesus macaques in Bangladesh. It has been previously shown that SFV in immunocompetent animals replicates to detectable levels only in superficial epithelial cells of the oral mucosa, although latent proviruses are found in most, if not all, tissues. In this study, we compare DNA sequences from latent SFV proviruses found in blood cells of 30 Bangladesh rhesus macaques to RNA sequences of transcriptionally active SFV from buccal swabs obtained from the same animals. Viral strains, defined by differences in SFV gag sequences, from buccal mucosal specimens overlapped with those from blood samples in 90% of animals. Thus, latent proviruses in peripheral blood mononuclear cells (PBMC) are, to a great extent, representative of viruses likely to be transmitted to other hosts. The level of SFV RNA in buccal swabs varied greatly between macaques, with increasing amounts of viral RNA in older animals. Evidence of APOBEC3-induced mutations was found in gag sequences derived from the blood and oral mucosa.
INTRODUCTION
Foamy viruses (FV) are retroviruses with many properties distinct from those of orthoretroviruses. Notably, FV undergo reverse transcription during viral assembly and/or budding, leading to production of infectious DNA-containing viruses (1). In immunocompetent animals, FV replication is limited to superficial epithelial cells of the oral mucosa (permissive cells) (2, 3, 4). However, latent proviruses are present in many, if not all, tissues, including peripheral blood (3). Simian foamy viruses (SFV) are ubiquitous in adult nonhuman primates (NHP), including rhesus macaques (Macaca mulatta) in South Asia (5, 6). Transmission among NHP appears to occur via subcutaneous introduction of saliva through biting or other behaviors (7, 8). SFV can be zoonotically transmitted to humans in close contact with NHP, presumably through bites and scratches. Humans who are infected by SFV, such as veterinarians, lab workers, or bush meat hunters in Africa, have reported occupational contacts with NHP (9, 10). In Asia, where free-ranging urban NHP and temple monkeys are common, exposure to NHP occurs frequently in the course of daily activities and can also lead to SFV infection (11, 12). Although much remains to be learned about SFV transmission, transfusion studies in monkeys (13, 14) have generated the following picture. After introduction of SFV into a naive host, the first signs of infection are antibody production and presence of proviral DNA in peripheral blood mononuclear cells (PBMC). It takes several months for viral transcription in the oral mucosa to be detectable. Thus, initial infection does not involve replication in permissive tissue of the new host. After acquisition of SFV via saliva, PBMC become latently infected. Latently infected PBMC migrate throughout the animal, and when an infected PBMC migrates to oral mucosal tissue, cell-cell interactions or signal transductions lead to production of infectious FV. Subsequently, uninfected PBMC migrate to the oral mucosa and become latently infected. FV infection in natural hosts appears to persist for life (15).
As reported elsewhere (16), we have characterized latent proviral DNA in PBMCs of a large number of free-ranging rhesus macaques in Bangladesh. In these animals, we found a number of different strains of SFV as defined by gag sequence clustering. Among the urban animals, these strains largely mapped to different geographic areas (16). A map of Bangladesh showing the areas under study is found in reference 16. We are interested in determining which SFV strains are zoonotically transmitted and whether efficiency of transcription as measured by RNA levels in oral mucosal tissues influences transmission to macaques and humans. In our previous work (16), we focused on the sequences of SFV genomes in PBMC, with the assumption that these are representative of actively transcribed viruses, according to the above model. However, there are no published data comparing transcribed and latent viruses in individuals of any host species.
In the current study, we compare the gag sequences of actively transcribing viruses in permissive cells of the oral mucosa to latent proviruses in the blood of individual free-ranging macaques. The level of viral RNA in buccal swab samples was quantified for a subset of the animals. We found that latent proviruses are representative of the actively transcribing viruses found in the oral mucosa. Further, individual macaques differed in SFV RNA levels in buccal swab samples, with age at the time of sampling being an important factor. Some macaques had proviral DNA, but we could not detect SFV RNA in the buccal swabs. This was particularly true for younger animals.
Hosts are known to synthesize proteins that can restrict retroviruses. Much work has been done on the APOBEC cytidine deaminases, reviewed in reference 17. A number of groups have shown that APOBEC3G deaminates cytidine in the HIV genome and that HIV encodes a protein (Vif) to specifically counter APOBEC3G (18). Recently, it has been shown that macaques synthesize APOBEC3F and APOBEC3DE, but little APOBEC3G in PBMC (19). In this report, we present evidence that some SFV in naturally infected rhesus macaques exhibit hypermutations characteristic of APOBEC3 activity. This is true for both transcriptionally active viruses and latent proviruses.
MATERIALS AND METHODS
Sample collection.
The present study includes samples from 61 free-ranging rhesus macaques (Macaca mulatta) in Bangladesh, from 5 geographically distinct sites. The sites are described in reference 16. Capture and blood sample collection methods have been previously described (16). Oral mucosal samples were collected as buccal swabs from the anesthetized animals with a Whatman Sterile Omni Swab (Whatman, USA), a single-use swab made of absorbent material specifically designed for the collection of buccal cells. To ensure that the swabbing technique was consistent between animals, only two researchers (L.J.-E. and G.A.E.) were responsible for oral swab collections from the study animals. Researchers would swab the inside of one of the animal's cheeks 10 times using an up and down motion and then draw the swab across the surface of the animal's tongue and repeat the swabbing on the other cheek. The head of the swab was then ejected into a 1.8-ml Sarsdtedt vial containing 1 ml of RLT buffer (Qiagen, USA) and stored on cold packs until transfer to −80°C. All animal handling and sampling procedures have been reviewed and approved by the University of Washington's Institutional Animal Care and Use Committee (4233-01).
Isolation and cloning of gag DNA from whole blood.
Total genomic DNA was extracted from the whole blood of macaques using the QIAamp DNA blood minikit (Qiagen, USA) according to the manufacturer's protocol. The DNA extracted from 10 μl of whole blood was used for PCR amplification of a gag fragment as described in our previous paper (16). First-round primers G1 (+), 5′AGGATGGTGGGGACCAGCTA, and G2 (−), 5′ CAGGAAGAGGTACTCGGGG, were used under these conditions: denaturation at 95°C for 3 min, followed by two cycles of 95°C for 30 s, 38°C for 1 min, and 72°C for 1 min, followed by 25 cycles of 95°C for 30 s, 52°C for 1 min, and 72°C for 1 min. Water was used as a negative control. A 2-μl volume of the first-round PCR product was used as the template for the second-round PCR. Second-round primers were G3 (+), 5′CAACCTAGATGGAGAGCTGAAGG, and G4 (−), 5′GGGGACAAACTACGGCTGGG. The reaction mixtures were denatured at 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 52°C for 1 min, and 72°C for 1 min. The expected size of the gag PCR product is 1,235 bp after a two-round PCR. The purified fragments were ligated into the TOPO-TA cloning vector (Invitrogen, USA), and Escherichia coli TOP10 competent cells were transformed (Invitrogen, USA). Multiple colonies were individually selected from each DNA sample, and plasmid DNA was purified and sequenced. An average of 6 clones were analyzed per macaque. Sequencing was carried out, using M13 forward and reverse primers as well as internal forward and reverse primers, by GeneWiz Inc. (Seattle, WA, USA) or the sequencing facilities at the University of Washington.
Isolation of RNA from buccal swabs.
Frozen buccal swab samples in 500 μl of RLT buffer (Qiagen, USA) were thawed at 37°C for 15 min with 40 U of the RNaseOUT RNase inhibitor (Invitrogen, USA) and 1 U of RNase-free DNase I (Promega, USA) immediately prior to RNA extraction. Total RNA was extracted according to the standard protocol using the RNeasy minikit (Qiagen, USA). RNA was treated again with 40 U of RNaseOUT and 1 U of RNase-free DNase I at 37°C for 1 h. Samples were then incubated at 65°C for 15 min to inactivate enzymes. RNA was stored at −20°C for subsequent use. The final volume of RNA from each buccal swab sample was 50 μl.
RT-PCR of buccal swab RNA.
Reverse transcription-PCR (RT-PCR) was performed to clone partial gag sequences (1,235 bp) from buccal swab RNA. First-strand cDNA was synthesized using the Thermoscript RT-PCR system (Invitrogen, USA) or the Maxima H Minus First Strand cDNA synthesis kit (Thermo Scientific, USA) according to the manufacturers' protocols using the oligo(dT)18 primer. The RT reactions were inactivated at 85°C for 5 min. The resulting cDNAs were used as the template for two-round PCR as described above for whole-blood FV DNA amplification. PCR products were cloned and sequenced as described above for whole-blood genomic DNA. The gag sequences from 61 macaque buccal swabs from an average of 6 clones were analyzed.
Quantitation of FV RNA.
Quantitative RT-PCR (qRT-PCR) was performed using 10 μl of each RNA sample, using the TaqMan One-Step RT-PCR Master Mix Reagent kit and analyzed using the ABI 7900 Real Time PCR system (Applied Biosystems, USA) according to the manufacturer's instructions. Primers and probes were designed to detect an 80-nucleotide highly conserved region near the C terminus of the gag gene as described previously (3). The primers used were gag1759F, 5′ACGAACATCTGGTGCGGG, and gag1835R, 5′CTGCGTTTCCACCAGCTGA. The probe was gag1789 6-carboxyfluorescein (FAM)-AGGAAGAGGGAACCAAAACCGAAACCA-6 carboxyltetramethylrhodamine (TAMRA). The quantitative RT-PCR (qRT-PCR) conditions were as follows: 48°C for 30 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Nonamplification (reaction with no enzyme mix) and nontemplate controls (reaction with water as the template) were included to rule out the possibility of plasmid contamination.
To generate standards for the qRT-PCR, the gag PCR target sequence was subcloned from a TOPO-TA vector (Life Technologies) into the pNEB vector (New England BioLabs, USA) containing a T7 promoter. In vitro transcription was performed using the Riboprobe in Vitro Transcription System (Promega) according to standard protocols. The amount of RNA was determined by spectrophotometry. Tenfold dilutions (ranging from 107 to 102 copies) of the viral RNA were used to generate a standard curve for each assay. RT-PCR for 18S rRNA was performed using a commercially available primer-probe set (Applied Biosystems, USA). An 18S standard curve was generated by adding 10-fold dilutions of Telo-RF cell total RNA (ranging from 105 to 10 cell equivalents) to the reaction mixtures. Telo-RF cells are macaque fibroblast cells (described in reference 20). Buccal swab FV RNA and 18S rRNA were tested in duplicate.
Quantitation of FV DNA from blood.
DNA was isolated as above, and qPCR was performed using iTaq Universal Probes Supermix (Bio-Rad, USA) and analyzed using the ABI 7900 Real Time PCR system (Applied Biosystems, USA). The same primers and probes used for qRT-PCR were used here to detect gag. The qPCR conditions were as follows: 95°C for 3 min and 40 cycles of 95°C for 15 s and 60°C for 1 min.
The same plasmid DNA containing the gag PCR target sequence used above was used for the qPCR to generate standards. Tenfold serial dilutions of the plasmid DNA (ranging from 106 to 101 copies) were used to generate a standard curve in every individual assay. We were able to detect one copy of the gag plasmid in our qPCR assay. Each assay also included a nontemplate control and an SFV-negative control (Telo-RF genomic DNA). A c-myc cellular DNA qPCR was done for each sample in each assay to normalize for cell equivalent. The primers for c-myc were as follows: c-mycF1, 5′ GCCCCTCAACGTTAGCTTCA; c-mycR1, CGCATGAGAAATACGGCTGCA; probe, FAM-CAACAGGAACTATGACCTCGACTACGACTCG-TAMRA. All assays included a c-myc standard curve generated using 10-fold dilutions of Telo-RF DNA (ranging from 105 to 101 cell equivalents). All samples tested were in duplicate.
Bioinformatics analysis: hypermutation detection.
Paired blood and buccal swab sample FV sequences were aligned using MUSCLE v3.8.31 (21) together with additional proviral blood sequences as previously described (16).
We noted sequences that had G to A mutations, possibly indicative of APOBEC3 activity, and analyzed these sequences using methodologies described in another work (F. A. Matsen, C. T. Small, K. Soliven, G. A. Engel, M. M. Feeroz, X. Wang, K. L. Craig, M. K. Hasan, M. Emerman, M. L. Linial, L. Jones-Engel), submitted for publication. This analysis identified sequences, and sites within these sequences, suspected of hypermutation. Sites suspected of hypermutation were removed from the sequence alignment, producing a putatively hypermutation-free sequence alignment. This alignment was used for the remainder of the analyses in this study, in order to avoid conflation of hypermutation and genuine phylogenetic signal.
Bioinformatics analysis: strain classification.
As in Feeroz et al. (16), representative sequences from the parental strains (dhamrai, dokhola, charmaguria, and karamjal) were used as genotype representatives for cBrother v2.0 (22) analyses. From each cluster in the strain analysis, a representative sequence was evaluated for recombinant relationships to the parental strains. Two independent cBrother runs of 1.1 million generations were run for each cluster's representative sequence, with the initial 10% discarded as burn-in and sampling every 1,000 generations. Convergence was assessed using the Gelman-Rubin diagnostic included with the cBrother distribution. Before being run through this analysis, the same parental strain genotype representatives used in reference 16 were aligned with the hypermutation-free strain representatives from these strain clusters using MUSCLE's profile alignment functionality. Portions of evaluated sequences that were found to be descendant from a particular parental strain at a posterior probability of 90% or higher for at least 200 contiguous base pairs were identified as bearing a recombinant relationship to that parental strain.
Phylogenetic analysis of intrahost diversity.
Phylogenetic trees were constructed from sequences obtained from several of the animals. For each of these animals, the corresponding sequences were selected from the global hypermutation-free sequence alignment. Trees were constructed from these selections using PhyML 3.0 (23) using an HKY85 (24) model and rendered using Archeopteryx (25).
Statistical analysis.
Buccal swab SFV RNA copy numbers were compared using a Wilcoxon signed-rank test. Samples for which neither 18S rRNA nor SFV copy number could be established were left out of the analysis. Samples for which 18S rRNA but no SFV RNA was found were treated as zero copy number per cell equivalent but excluded from the computation of medians. The Wilcoxon test was evaluated using the R (26) function “wilcox.test.”
The significance of observed congruence between strain classifications was analyzed using a simulation method. In each of the simulations performed, blood strain assignments were held fixed, while the buccal assignments were shuffled between animals. If an animal had more than one strain in its buccal sample, those strains were reassigned together as a unit in this shuffling. The congruency score for a set of animals was defined as the sum total of the number of strains shared between blood and buccal assignments for each animal. The simulations represent the distribution of strain assignments in the absence of correlation between strains found in buccal and blood samples from an animal. Thus, if the actual observed strain classifications for a given set of animals had a significantly higher congruency score than the score of the simulations, we can conclude that the strain level classifications in blood sequences are a good reflection of what is found to be transcriptionally active in the oral mucosa. Indeed, the fraction of simulations that have a higher score than the actual observed classifications can be taken as the P value of the hypothesis that strains observed in buccal samples are correlated with the strains observed in blood. Two such simulation experiments were performed, with one million simulations each, one for the entire set of animals and one for the subset of animals from locations other than Charmaguria and Dhamrai. This subset was chosen to exclude the possibility of the perfect strain congruence observed in Charmaguria and Dhamrai inflating the significance of these results. The P values reported from these experiments are multiplied by two for Bonferroni's correction.
Nucleotide sequence accession numbers.
The sequences newly determined in this study have been deposited in GenBank under accession numbers KF684969 through KF685045 and KF685335 through KF685646.
RESULTS
The overall goal of the work described here is to determine whether SFV proviral DNA sequences in blood of free-ranging macaques are indicative of viruses producing RNA in buccal swabs. In order to be sure that buccal swab sequences are from transcribed RNA and not latent proviruses, we attempted to quantitate the SFV RNA levels in oral samples. In samples in which RNA levels were detectable and quantitated, we could be sure that oral sequences arose from RNA and not the much rarer proviral DNAs. Although we could obtain clones from blood and buccal swabs of 61 animals and we could obtain quantitative RNA levels in only 43 animals, in many cases the animals in these two groups did not completely overlap. Only 30 animals yielded both quantitative RNA levels and paired buccal and blood sequences.
Comparison of SFV sequences of latent proviruses in PBMC and transcriptionally active viruses in buccal swabs.
In this study, we have compared SFV gag sequences in blood and buccal swabs obtained from the same animals. Buccal swab sequences are primarily from SFV that are transcriptionally active and presumably reflect replicating viruses that can be transmitted through saliva. For each macaque from which we were able to obtain SFV sequences from blood cells, we attempted to sequence the same 1,125-nucleotide portion of gag from buccal swab RNA. In order to confirm that buccal sequences are from RNA, we measured the level of SFV RNA in the oral mucosal tissues.
Paired blood and buccal partial gag clones from a total of 61 animals from five different regions were sequenced (Table 1). However, buccal swab SFV RNA was only confirmed by qRT-PCR for 30 of these macaques. Unfortunately, there was not enough RNA to determine FV copy number for the remaining 31 samples. It is likely that the SFV buccal swab-derived sequences are from RNA in all cases because we treat the RNA preps with DNase before amplification and cloning. However, since we cannot definitively rule out amplification of latent proviral DNA from contaminating blood cells, we have limited our conclusions to the results from the 30 animals with known SFV RNA levels in buccal swabs. Using quantitative PCR and RT-PCR, we determined that in these animals there are at least 5,000 copies of SFV RNA per 104 cells compared to 8 copies of latent proviral DNA per 104 cells (data not shown), so it is highly likely that buccal swab clones obtained are from SFV RNA. For both blood and buccal samples, we sequenced an average of 6 clones, with a range of 2 to 15 clones per sample. The relatively small number of clones sequenced per sample could preclude identification of all strains that were present.
Table 1.
Comparison of strains in PBMC and buccal swab samples from five sites
| Location and ID of SFV+ macaques | Straina (no. of clones) in: |
FV RNA | |
|---|---|---|---|
| PBMCs | Buccal swabs | ||
| Charmaguria (n = 10) and Dhamrai (n = 18) | |||
| MBG54, 61, 67 | CM (5, 6, 5) | CM (3, 3, 4) | Yes |
| MBG184, 186, 188, 226, 228, 232 | DM (6, 6, 6, 8, 6, 4) | DM (5, 9, 7, 5, 6, 6) | Yes |
| MBG6,175,177,180,182,183,185,187, 229–331, 233 | DM | DM | No |
| MBG55, 57, 58, 59, 60, 65, 68 | CM | CM | No |
| Bormi (n = 13) | |||
| MBG202, 208, 214 | BM1 (6, 6, 7) | BM1 (6, 4, 3) | Yes |
| MBG204, 205 | BM2 (6, 7) | BM2 (3, 4) | Yes |
| MBG206 | BM1 (8) | BM1 (6), BM2 (1) | Yes |
| MBG212 | BM1 (5) | DK (6) | Yes |
| MBG234 | BM1 (3), BM2 (2), NCS (1) | BM2 (6) | Yes |
| MBG237 | BM2 (4) | BM1 (5) | Yes |
| MBG239 | BM1 (4) | BM1 (1), DK(1), BM2 (4) | Yes |
| MBG85 | BM1 (4), BM2 (3) | BM1 (2) | Yes |
| MBG238 | BM1 (2), DK (1), BM2 (1), NCS (2) | BM1 | Yes |
| MBG236 | DM, DK | DM, DK | No |
| Narayanganj (n = 10) | |||
| MBG269 | DM (6) | DM (5), NCS (1) | Yes |
| MBG270, 271 | DM (6, 6) | DM (6, 6) | Yes |
| MBG109 | DM | NCS | No |
| MBG189, 196, 197 | DM | DM | No |
| MBG193 | NCS | DM | No |
| MBG108 | BM2, NCS | DM | No |
| MBG192 | DM, NCS | DM | No |
| Dokhola (n = 10) | |||
| MBG164 | DM (4), BM2 (1), NCS (1) | DM (3) | Yes |
| MBG241 | DK (5), NCS (1) | DK (6) | Yes |
| MBG243 | DK (1), DM (5) | DK (3) | Yes |
| MBG244 | DK (4), BM2 (1), NCS (1) | DK (2), DM (4) | Yes |
| MBG246 | DK (5), BM2 (1) | DK (5), BM2 (1) | Yes |
| MBG247 | DK (5) | DM (4) | Yes |
| MBG157 | DK | DM, DK | No |
| MBG159 | DK, DM | DM | No |
| MBG240 | DK | DK | No |
| MBG242 | DM, DK | DK | No |
Strain abbreviations: CM, charmaguria; DM, dhamrai; DK, dokhola; BM1 and BM2, bormi1 and bormi2, respectively; NCS, noncore strains.
As previously reported (16), we found multiple strains of SFV in macaque blood samples. For the most part, the SFV strains cosegregated with the macaque populations and were named according to the geographical region in which they were predominantly found. A detailed phylogenetic analysis of these strains is reported in Feeroz et al. (16). The core strains, based on the gag sequences, are representative of the viruses that tightly cluster in each region and are named charmaguria (CM), dhamrai (DM), bormi1 (BM1), bormi2 (BM2), and dokhola (DK). In this study, at least one-third of the macaques were infected with more than one strain, and two animals had evidence of infection with 4 strains (Table 1). Of the 30 macaques with confirmed SFV RNA, 17 (57%) had perfect strain congruence in clones derived from buccal and blood samples. In 10 of the 30 macaques (33%), there was some but not perfect congruence between strains observed in buccal and blood samples (at least one strain in common). Only 3 animals (10%) had buccal and blood samples in which there was no overlap in strains. Of the 31 animals for which we could not confirm the presence of SFV buccal swab RNA, 24 had perfect congruence between buccal and blood strains.
The degree of congruence between strains observed in blood and buccal samples varied between sampling locations. In Charmaguria and Dhamrai, each of which exhibits a distinct local strain (Table 1), perfect strain congruence was observed in all animals. In the remaining populations (Bormi, Narayanganj, and Dokhola), a greater variety of viral strains and, in some cases, recombinant viruses annotated as noncore strain (NCS) were observed. There was less congruence between strains identified in buccal and blood samples in Bormi, Narayanganj, and Dokhola (Table 1).
Previously, we identified two viral strains from Bormi macaques, referred to here and in our previous work (16) as bormi1 (BM1) and bormi2 (BM2). Both viruses were also found in buccal swabs (Table 1). Three macaques had only BM1 in both tissues, and two had only BM2 in both tissues. None of the 12 Bormi animals with quantifiable SFV RNA had both strains in both tissues. The remaining animals had a combination of one or both strains in the blood and buccal swab samples. Only two animals had complete discordance between blood and buccal strains.
The DM viral strain was dominant in latent proviruses in Narayanganj macaques (Table 1). BM2, DK, and NCS, some of which were found to be recombinant with the core strains, were also present. We have performed controls to rule out PCR-generated recombination as the source of these strains, indicating that these recombinants arose naturally in macaques (16). We have confirmed the presence of SFV RNA in 3 buccal swabs from Narayanganj macaques. Two of these animals had congruent blood and buccal swab sequences. These three animals all had DM in both blood and buccal swabs, but one also had additional NCS viral sequences. The NCS sequence in the blood is different from the NCS sequence in the buccal swab. It is worth noting that many Narayanganj animals had an NCS (Table 1).
In Dokhola animals, the situation is complex (Table 1). No two animals had the same blood/buccal viral strain profile, and perfect congruence between blood and buccal SFV strains was observed in only one animal. However, most animals did have the DK strain in both blood and buccal samples. The DM strain was also often observed in buccal samples.
Congruence in strain classification between buccal and blood sequences was significantly greater than would be expected by chance (P < 1 × 10−6), as demonstrated through simulations in which buccal strain assignments were shuffled among the animals. These shuffled assignments were found to be significantly lower in congruence than what was observed in the actual data. When Dhamrai and Charmaguria animals were removed from this analysis, significance was maintained, indicating that the high level of perfect strain congruence observed in Dhamrai and Charmaguria animals was not the only driver of significance (P = 3 × 10−6).
Conservation of the Gag Late domain in SFV strains.
Each gag sequence was translated. The portion of gag chosen for amplification contains very few amino acids that are highly conserved in SFV from different species (reviewed in reference 27). This allows us to detect amino acid changes that might not interfere with viral replication and thus maximizes our ability to detect strain variation. The only known functional motif in this sequenced region of gag is PSAP. PSAP is the late domain, necessary for completion of nucleocapsid assembly and interaction with envelope glycoproteins in tissue culture cells (28, 29). We examined the predicted amino acid sequences from animals for which we had evidence of buccal swab SFV RNA. In 27 of these 30 animals, PSAP was conserved in all blood and buccal swab SFV clones. We saw PSAP mutated in the buccal swab sequences of only 2 animals. One animal had 1 of 6 clones with the sequence PGAP, and another animal had 1 of 5 clones with the sequence PNAP. These changes could be due to PCR-generated mutations. The third animal had 3 of 5 clones with the PSAP domain mutated to SSAP in the blood. The overall conservation of the PSAP motif in transcriptionally active virus is consistent with the importance of this domain in viral transmission and replication.
Levels of proviral SFV DNA in blood samples and SFV RNA in buccal swabs.
We quantitated the level of proviral DNA in PBMC from 6 of the rhesus macaques (2 from Bormi, 2 from Dhamrai, 1 from Narayanganj, and 1 from Dhokola). The qPCR used can detect as little as one copy of gag plasmid DNA (data not shown). No PCR result was detected using uninfected Telo-RF cell DNA. Thus, this assay is both sensitive and specific. There was a wide spread in the cellular copy number, ranging from 7.6 × 10−5 to 1.1 × 10−2 copies per cell. Four of the animals had values around 8 × 10−4, with a median SFV proviral copy number of 8.3 × 10−4 copies per cell (data not shown). We looked for RNA in blood samples. RT-PCR after DNase treatment failed to detect any SFV RNA (data not shown). We next examined SFV RNA levels in oral mucosal samples by quantitative RT-PCR (qRT-PCR). Our qRT-PCR assay employs primers and a probe to a small conserved region of gag and detects both genomic RNA and gag mRNA. The lower limit of detection of FV RNA was found to be about 100 RNA copies of FV per 10 μl of extracted RNA.
The 18S rRNA levels were not uniform in the buccal swab samples, although the same sampling protocol was used for each animal. Intrinsic differences in oral anatomy, recent food intake, and hydration could influence the composition of the samples. Alternatively, it is possible that in buccal swab-derived cells, SFV replication can lead to increased cell death and thereby decrease the level of 18S rRNA. Since there were large differences in 18S rRNA levels between samples (4 orders of magnitude), we are reporting the SFV RNA copy number per 10 μl of RNA extracted rather than normalizing to 18S rRNA, which would have yielded a copy number per cell equivalent. We report SFV RNA levels for every buccal swab sample for which we have evidence for either 18S rRNA or SFV RNA (Table 2). We determined FV copy number for 58 animals. These included the 30 samples for which we have PBMC and buccal swab sequence pairing as mentioned above. It is worth noting that the remaining 28 samples for which we determined FV copy numbers are different from the initial 61 animals for which we performed sequence comparison. Of the 58 samples, 23 were from Bormi and 35 from the other four sites. SFV RNA was not detected in 3 samples from Bormi and 12 samples from the other 4 sites, and these negative samples were not included in determining the median FV RNA levels. At each site, the SFV RNA levels varied greatly between individual animals (Fig. 1), ranging from 7 × 102 to 1.7 × 106 per 10 μl of RNA extracted. Such variation has been previously reported for laboratory macaques (3).
Table 2.
Summary of SFV RNA copy number analyses from MBG buccal swabs
| Location | No. of samples: |
Copy no.b |
|||
|---|---|---|---|---|---|
| Tested | Below detection levela | With copy numbers | Mean | Median | |
| Bormi | 23 | 3 | 20 | 1.85E+05 | 8.14E+04 |
| Dharmai | 9 | 3 | 6 | 2.30E+04 | 1.58E+04 |
| Narayanganj | 6 | 1 | 5 | 1.02E+05 | 8.32E+04 |
| Charmaguria | 10 | 5 | 5 | 3.21E+04 | 9.28E+03 |
| Dokhola | 10 | 3 | 7 | 3.68E+05 | 1.54E+05 |
| Total | 58 | 15 | 43 | 1.65E+05 | 6.64E+04 |
Samples with <100 FV RNA copies, the lower limit of detection.
Buccal swab RNA samples were in 50 μl, and 10 μl was tested for FV copy number.
Fig 1.
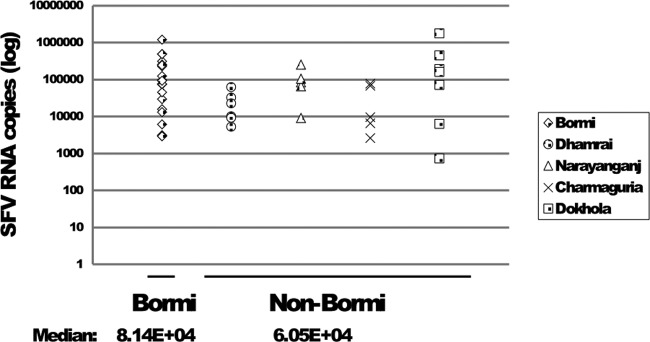
SFV RNA copy numbers from buccal swabs grouped according to location. FV RNA was quantified as discussed in Materials and Methods. The median values for Bormi and non-Bormi samples were calculated and are shown below the x axis. Samples with FV RNA below the level of detection are not shown.
We examined whether macaques in different locations in this study had different SFV RNA levels. This is of interest because each macaque population differs in associated viral strain(s) (Table 1). When we looked at zoonotic infections in the different regions under study, there was a trend toward more infected humans in Bormi (12). We wondered whether Bormi monkeys in particular had higher levels of SFV RNA than monkeys in the other areas. The median FV RNA level in buccal swabs from Bormi macaques (8 × 104) was similar to that of the non-Bormi macaques (6 × 104) (Fig. 1). There is a trend toward higher levels of SFV RNA in Bormi macaques, but the difference is not significant (P = 0.068).
We detected only one of the two bormi strains in buccal swabs from 8 Bormi animals with quantifiable SFV RNA levels in buccal swabs. Additionally, three monkeys had both BM1 and BM2 in the buccal swab. Results of qRT-PCR showed no apparent difference in buccal swab viral RNA levels between samples from monkeys infected with either BM1 or BM2 or in the 3 animals with both strains (data not shown). Therefore, we have no evidence for differences in transcriptional activity between BM1 and BM2.
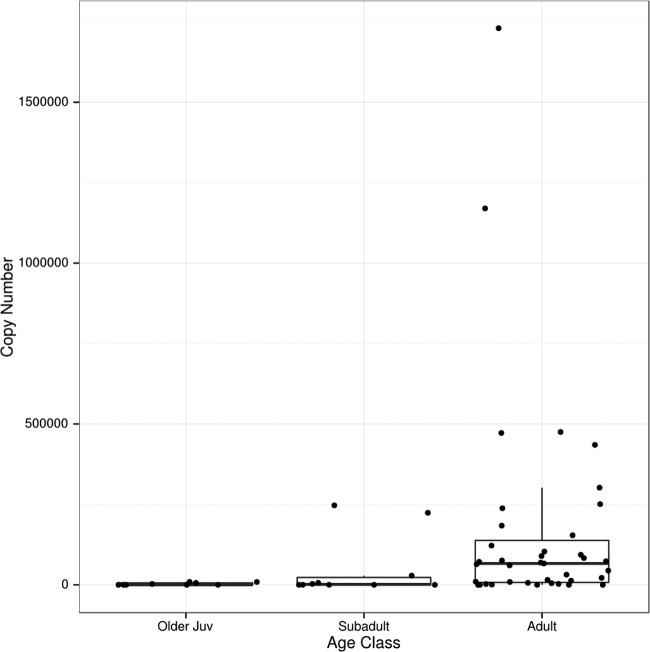
We found 16 free-ranging macaques that were PCR positive for proviruses in PBMC for which we could not detect FV RNA. Work from others has shown that it takes months to detect RNA in the oral mucosa after infection defined by antibody status and presence of DNA in blood (13, 14). Data from our study also show that RNA levels increase with age (Wilcoxon P = 4.3 × 10−4, comparing adult copy number to nonadult copy number), with very little or no SFV RNA detectable in animals less than 15 months of age (Fig. 2). From one macaque, we had buccal swab samples from 2 time points that yielded quantitative RNA levels. For this animal, we found 2.0 × 105 RNA copies at the first time point and 4.7 × 105 copies 3 years later. While the difference is not large, it is consistent with an increased level of SFV RNA with age. Ten of the 15 provirus-positive, FV RNA-negative animals are older juveniles or subadults and may have been sampled within months of initial infection. According to the FV infection model, we might expect very low levels of replicating virus early after infection. This leaves 4 adult females and 1 adult male that are PBMC SFV DNA positive and buccal swab SFV RNA negative. We do not know why this is the case.
Fig 2.
RNA levels in buccal swabs in different age groups. Ages were estimated by dental eruption patterns. Older juveniles (16 months to 3 years old), subadults (3 to 5 years old), and adults (older than 5 years). SFV RNA copy numbers are shown for 10 μl of extracted buccal swab RNA as described in Materials and Methods.
APOBEC3-generated hypermutation.
We examined the SFV sequences in blood and buccal swabs to determine whether there are changes that could be attributable to APOBEC3 enzymes. It is known that different APOBEC proteins exhibit specific dinucleotide specificities (30). We took advantage of this by considering whether the underlying probability of observing G-to-A mutation in a context associated with known APOBEC3 activity is statistically higher than the probability of observing such mutations in control contexts. A Bayesian method, as described in another work (Matsen et al., submitted), was used to make estimates, with uncertainty, of the ratio of the probability of a G-to-A mutation in an APOBEC3 context of interest to that in a control context, here called the relative probability ratio. From these estimates, we employed two metrics in the data analyses. The Q05 value represents a lower limit on the relative probability ratio (with 95% confidence). The maximum a posteriori (MAP) value represents the most likely relative probability ratio given the data. Higher Q05 values correspond to stronger evidence of genuine hypermutation; sequences with Q05 values greater than 1.0 were considered hypermutated. MAP values indicate the strength of hypermutation.
The hypermutation signal observed among buccal swab SFV RNA sequences from the 30 macaques with quantifiable RNA levels was generally low. In buccal swabs, only 5 of the 30 animals had sequences that we identified as hypermutation positive, and all were in a GG context. However, 3 of these animals exhibited only marginal evidence of hypermutation (Q05 values near 1). One of the remaining two animals had a sequence with a MAP value of 2.75, while the other animal had 4 positive sequences and MAP values as high as 5.15. None of these buccal swab sequences had stop codons when translated. In contrast, sequences obtained from blood samples showed evidence of hypermutation in GA, GG, GR (GG and GA), and GM (GA and GC) contexts. Ten of the 30 animals with buccal/blood sample pairings had blood sequences evaluated as hypermutated. The context in which sequences were most frequently found to be positive was GG. However, the strength of the hypermutation signal was highest in GA and GM contexts. MAP values from positive sequences ranged from 1.0 to 6.76, with a median value of 2.12 and a standard deviation of 1.5. Stop codons were observed in several of the more hypermutated blood sequences.
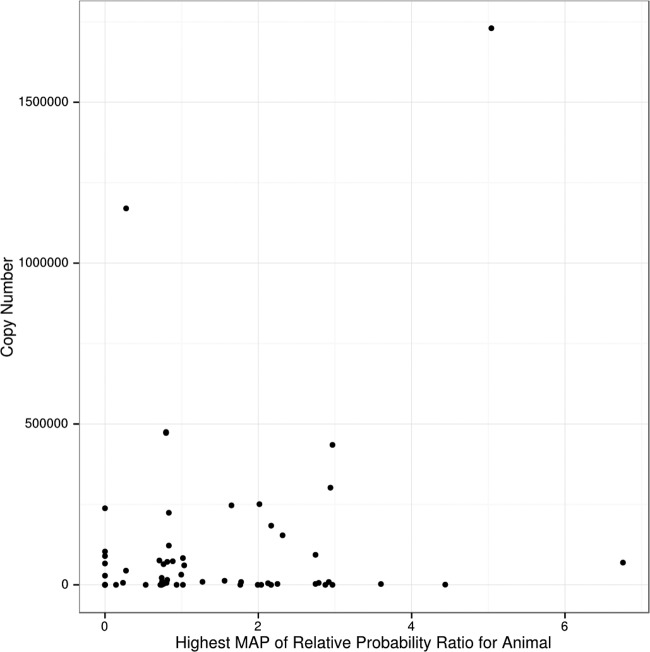
We examined whether animals with high levels of hypermutation in their latent proviral sequences have low levels of active replication in the oral mucosa. This would be consistent with the FV infection model wherein latent proviral sequences are representative of what is replicating in the oral mucosa, and hypermutated viruses are less likely to be replication competent. However, our data do not support this prediction. Indeed, there is no inverse correlation between hypermutation and levels of SFV RNA (Fig. 3).
Fig 3.
Relationship of RNA levels in buccal swabs and intensity of APOBEC-induced mutations in latent proviruses in blood cells. Hypermutation strength is represented by the maximum a posteriori (MAP) value as described in Results. The higher the MAP number, the more hypermutation there is.
Sequence similarity between latent proviruses and transcriptionally active viruses.
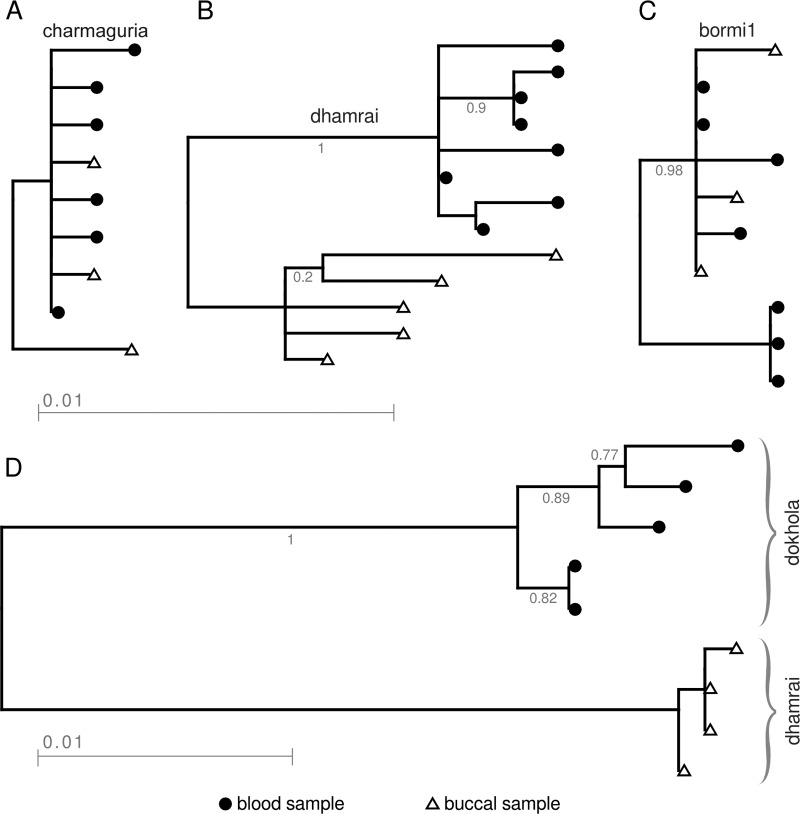
To better understand the relationship between latent proviruses in blood and transcriptionally active viruses in the buccal swabs, we performed phylogenetic analysis of the SFV clones from animals infected with only a single SFV strain. In the 15 animals examined, we observed 3 patterns. In some, the buccal and blood SFV clones clustered together. In others, there was separation between buccal and blood clones. Finally, in some animals there were intermediate patterns with some clones from buccal and blood clustering together and others separately. In all animals infected with CM (3 animals), we found interspersion of blood and buccal sequences (Fig. 4A). At the other extreme, we examined 7 macaques with DM, and 6 of them showed two clusters, one from blood and one from buccal swab (Fig. 4B). Although the blood and buccal sequences segregate into distinct clades, the overall sequence identity was 98.5% between buccal and blood viruses, so the branch lengths are very short. Of the 6 core strains studied, DM is the most diverse strain. Some animals showed intermediate clustering patterns. An example is shown for an animal with BM1 (Fig. 4C). Because deep sequencing was not performed, it is not possible to know how each of these trees based on about 10 clones reflects the complexity in the animal. For contrast, a phylogenetic tree was also constructed for an animal in which a different strain was observed in each compartment (Fig. 4D). As expected, the branch lengths are longer.
Fig 4.
Phylogenetic trees of viruses from 4 animals, representing the different levels of diversity observed within animals. Triangles correspond to sequences from buccal swab samples and circles to sequences from blood samples. Animals MBG61 (A), and MBG226 (B), and MBG214 (C) each had a single strain (charmaguria, dhamrai, and bormi1, respectively) in the two compartments. In animal MBG247 (D), dokhola is observed in blood samples and dhamrai in buccal swab samples.
We also analyzed the phylogenetics of individual strains in all monkey blood and buccal samples from animals in which the same virus was in blood and buccal samples. Figure 5 shows the phylogenetic tree for bormi1 for all animals that had bormi1 in both buccal and blood. We chose BM1 because the strain diversity is representative of all strains except DM, which is the most diverse. For the most part, the BM1 sequences in each animal clustered well, although in a few cases, they were a bit more diverse in the two compartments. Overall, the data support the findings for the individual animals, shown in Fig. 4A to C, indicating that viruses in individual animals tend to cluster more than viruses in blood samples or buccal swabs of different animals. A similar level of intrastrain diversity was seen in blood and in buccal samples. These data are consistent with the model that latent SFV in blood is the progenitor of transcriptionally active SFV in oral tissue, which in turn can lead to new proviral sequences in blood.
Fig 5.
Phylogenetic tree representing viral diversity within the bormi1 strain. Sequences are included only from animals that have both buccal swab (BS) and whole-blood (WB) sequences classified as bormi1. The tile grid adjacent to the tree indicates the animal and sample type from which the sequence was obtained. The rows of the tile grid are aligned with the tree tips to which they correspond; the columns of this grid indicate which animals the sequence is from.
DISCUSSION
Simian foamy viruses (SFV) are often cytopathic when studied in tissue culture cell lines such as fibroblasts and epithelium-derived cells in which they are able to replicate. In rhesus macaques in vivo, latent proviruses are found in most, if not all, tissues. Detectable transcription has been documented only in the oral mucosa in immunocompetent animals (2, 3), where the infected cells are terminally differentiating epithelial cells (4). It is not known whether SFV replication in vivo perturbs the cells in which it replicates, and there is no tissue culture system for superficial oral mucosal epithelial cells. However, since essentially all adult members of free-ranging host populations are infected, SFV should be considered part of the natural microbiome. It is not possible to know whether there are consequences of SFV replication in the normal host since there are not uninfected free-ranging adult animals for comparison.
All published studies including sequence analysis of SFV in vivo have been limited to latent proviral DNA in peripheral blood (5, 16, 31) or viral RNA in fecal samples (32, 33). The source of this fecal RNA, indicative of viral replication, is not known. There are no previous reports describing the sequences of transcriptionally active SFV isolated from oral mucosal samples from free-ranging animals, and it is the virus replicating in this compartment that will likely be transmitted to other animals and/or humans.
In this study, we report for the first time the sequences of transcriptionally active SFV in free-ranging macaques. We first quantitated levels of SFV RNA in buccal swab samples obtained from macaques in different areas of Bangladesh. We measured the total SFV RNA copy number but did not measure the contribution of different viral strains to the SFV RNA level. The variation in SFV levels was very large, up to 4 orders of magnitude between macaques. Although the same methodology was used to obtain the buccal swab samples from each monkey, the contribution made by cells and saliva to each sample could differ greatly. As discussed in Results, SFV RNA levels were higher in adults than in subadults and juveniles (Fig. 2). It is interesting that Bormi was the site with the largest number of cases of zoonotic transmission of SFV (12). However, we did not find a significant correlation with higher FV RNA levels from buccal samples from Bormi monkeys. Ultimately, to understand whether viral strains replicate to different levels, isolation of individual strains and infection of tissue culture cells will be required. During the current study, we were able to isolate infectious virus from several of the buccal swab samples that had been stored in tissue culture media. Thus far, we have isolated the DM and BM2 viruses.
For our SFV genetic analyses, we have used a portion of the gag gene that is highly divergent in SFV isolated from different nonhuman primate species. Other groups examining SFV infection in animals have studied a portion of the pol gene, encoding integrase, which is highly conserved (34). Analysis of this nonconserved gag region has allowed us to identify a number of strains latent in blood cells in rhesus macaques. In the current study, we have compared SFV sequences obtained from buccal swabs and blood samples from the same animal. In 90% of the animals, there was at least some overlap in the strain classifications of blood and buccal samples. In more than one-half of the cases (57%), there was complete strain congruence between the two compartments. As we have examined only about 6 viral clones per tissue sample, we have a somewhat limited view of the relative contribution of viral strains and variants to overall transcription. In the future, deep sequencing could provide a more comprehensive view of the differences between latent proviruses and viruses that are capable of replication in permissive cells.
Here, we show that all the strains detected as latent proviruses in blood cells are capable of transcription in the oral mucosal tissues. RNA from the dhamrai (DM) strain is found in the buccal swabs of monkeys from many locations. It is not clear whether this is a result of human-assisted translocation of SFV-infected monkeys or the movement of animals and their viruses when forest cover facilitated natural migration.
In our previous studies on latent proviruses, we found that individual animals are frequently infected with more than one strain (16). In some animals, particularly those from Dokhola, we detected more than one transcriptionally active strain. Infection by multiple strains could allow for recombination and the generation of new variants (16, 35). In the case of lentiviruses, recombination between nonpathogenic primate viruses led to the emergence of strains that were more easily transmissible to humans (36). We have previously shown that some SFV strains currently infecting Bangladeshi macaques and humans are established recombinants, for example bormi2 (12, 16). In this study, we confirmed that these recombinant noncore strains are present in macaque buccal swabs.
We analyzed changes in the conserved PSAP late domain in gag genes cloned from individual macaques. We found latent proviruses with mutations that lead to predicted amino acid changes in the PSAP motif, such as SSAP. However, very few clones derived from RNA in buccal swabs showed mutations in PSAP. This is consistent with the requirement of the PSAP domain for viral replication in vivo, as previously shown in tissue culture cells (28, 29; reviewed in reference 27).
Hosts have developed innate immunity to counteract viral infections. Viruses in turn have evolved ways to counteract such host factors. Although the host has enzymes designed to inactivate genomes of pathogens, instances in which genetic modifications could benefit a virus could arise. One important group of enzymes that can modify viral genomes is the APOBEC3 family of cytidine deaminases. Using novel methods for detection of hypermutation (Matsen et al., submitted), we have shown that APOBEC3 enzymes are active in vivo in macaque hosts. In HIV, APOBEC3G has been shown to mutate G to A at GG dinucleotides in single-stranded DNA (ssDNA) (37). HIV-1 encodes a protein (Vif) that protects the virus from APOBEC3G hypermutation (18). SFV have been shown in vitro to be susceptible to APOBEC3G mutation (38, 39), but there was little published evidence of this in the natural host prior to this study. It is unclear whether the SFV accessory protein Bet can counter APOBEC3G activity (38, 40, 41), although evidence is increasing that Bet is important in protecting FV from restriction by APOBEC (42, 43). Recently, APOBEC3 proteins from rhesus macaque PBMC have been characterized (19). In macaques, different APOBEC3 activities have been shown to mutate G to A in GM (M being the degenerate IUPAC code for A or C). We have reported evidence of human APOBEC3G hypermutation in Bangladeshi humans who were zoonotically infected (Matsen et al., submitted). Since these particular mutation patterns are not observed in either buccal swab or blood samples from rhesus macaque SFV with the same intensity or frequency, it is most likely that the mutations arose after zoonotic infection of humans.
Overall, our work in natural monkey populations highlights the diversity of SFV strains. Many factors can contribute to the formation of viral strains. We found viral strains that cosegregate with monkey populations localized to different geographic areas. The genetic differences between strains could be the result of viral recombination or reverse transcriptase errors and also of host factors such as the APOBEC enzymes. Details are given here and in other publications from our group (12, 16; Matsen et al., submitted). Any of these factors could lead to the emergence of SFV with new biological properties, including pathogenic consequences for the natural hosts or zoonotically infected humans. Thus far, fortunately, no pathology associated with SFV has been found, but given the number of zoonotic cases and potential for genetic change, this virus merits continued surveillance.
ACKNOWLEDGMENTS
This research was greatly aided by the efforts of student assistants and faculty in the Wildlife Branch of the Department of Zoology at Jahangirnagar University, Bangladesh. We also thank the Bangladesh Forest Department for their constant support of our research program. We thank Dana Jackson at the Fred Hutchinson Cancer Research Center (FHCRC) for her technical assistance.
This research was supported by funding from NIH-NIAID grants R01 AI078229, R01AI078229-03S1, and R03 AI064865, NIH-NCI grant CA18282, NIH-NCRR grant P51 RR000166, and New Development Institutional Support from the FHCRC.
Footnotes
Published ahead of print 9 October 2013
REFERENCES
- 1.Yu SF, Sullivan MD, Linial ML. 1999. Evidence that the human foamy virus genome is DNA. J. Virol. 73:1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falcone V, Leupold J, Clotten J, Urbanyi E, Herchenröder O, Spatz W, Volk B, Bölm N, Toniolo A, Neumann-Haefelin D, Schweizer M. 1999. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 257:7–14 [DOI] [PubMed] [Google Scholar]
- 3.Murray SM, Picker LJ, Axthelm MK, Linial ML. 2006. Expanded tissue targets for foamy virus replication with simian immunodeficiency virus-induced immunosuppression. J. Virol. 80:663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray SM, Picker LJ, Axthelm MK, Hudkins K, Alpers CE, Linial ML. 2008. Replication in a superficial epithelial cell niche explains the lack of pathogenicity of primate foamy virus infections. J. Virol. 82:5981–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones-Engel L, Steinkraus KA, Murray SM, Engel GA, Grant R, Aggimarangsee N, Lee BPY-H, May C, Schillaci MA, Somgird C, Sutthipat T, Vojtech L, Zhao J-Y, Linial ML. 2007. Sensitive assays for simian foamy viruses reveal a high prevalence of infection in commensal, free-ranging, Asian monkeys. J. Virol. 81:7330–7337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falcone V, Schweizer M, Neumann-Haefelin D. 2003. Replication of primate foamy viruses in natural and experimental hosts, p 161–180 In Rethwilm A. (ed), Foamy viruses. Springer, Berlin, Germany: [DOI] [PubMed] [Google Scholar]
- 7.Blewett EL, Black DH, Lerche NW, White G, Eberle R. 2000. Simian foamy virus infections in a baboon breeding colony. Virology 278:183–193 [DOI] [PubMed] [Google Scholar]
- 8.Calattini S, Wanert F, Thierry B, Schmitt C, Bassot S, Saib A, Herrenschmidt N, Gessain A. 2006. Modes of transmission and genetic diversity of foamy viruses in a Macaca tonkeana colony. Retrovirology 3:23. 10.1186/1742-4690-3-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boneva RS, Switzer WM, Spira TJ, Bhullar VB, Shanmugam V, Cong Me Lam L, Heneine W, Folks TM, Chapman LE. 2007. Clinical and virological characterization of persistent human infection with simian foamy viruses. AIDS Res. Hum. Retroviruses 23:1330–1337 [DOI] [PubMed] [Google Scholar]
- 10.Betsem E, Rua R, Tortevoye P, Froment A, Gessain A. 2011. Frequent and recent human acquisition of simian foamy viruses through apes' bites in central Africa. PLoS Pathog. 7(10):e1002306. 10.1371/journal.ppat.1002306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones-Engel L, May CC, Engel GA, Steinkraus KA, Schillaci MA, Fuentes A, Rompis A, Chalise MK, Aggimarangsee N, Feeroz MM, Grant R, Allan JS, Putra A, Wandia N, Watanabe R, Kuller L, Thongsawat S, Chaiwarith R, Kyes RC, Linial ML. 2008. Diverse contexts of zoonotic transmission of simian foamy viruses in Asia. Emerg. Infect. Dis. 14:1200–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engel GA, Small CT, Soliven K, Feeroz MM, Wang X, Hasan K, Gunwha O, Alam S, Craig K, Jackson D, Matsen FA, IV, Linial ML, Jones-Engel L. 2013. Zoonotic simian foamy virus in Bangladesh reflects diverse patterns of transmission and co-infections among humans. Emerg. Microbes Infect. 2:e58. 10.1038/emi.2013.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan AS, Kumar D. 2006. Simian foamy virus infection by whole-blood transfer in rhesus macaques: potential for transfusion transmission in humans. Transfusion 46:1352–1359 [DOI] [PubMed] [Google Scholar]
- 14.Brooks JI, Merks HW, Fournier J, Boneva RS, Sandstrom PA. 2007. Characterization of blood-borne transmission of simian foamy virus. Transfusion 47:162–170 [DOI] [PubMed] [Google Scholar]
- 15.Hooks JJ, Burns W, Hayashi K, Geis S, Notkins AL. 1976. Viral spread in the presence of neutralizing antibody: mechanisms of persistence in foamy virus infection. Infect. Immun. 14:1172–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feeroz M, Soliven K, Small C, Engel G, Pacheco M, Yee J, Wang X, Hasan K, Oh G, Levine K, Alam S, Craig K, Jackson D, Lee E-G, Barry P, Lerche N, Escalante A, Matsen F, Linial M, Jones-Engel L. 2013. Population dynamics of rhesus macaques and associated foamy virus in Bangladesh. Emerg. Microbes Infect. 2:e29. 10.1038/emi.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cullen BR. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80:1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398–1403 [DOI] [PubMed] [Google Scholar]
- 19.Zhang A, Bogerd H, Villinger F, Das Gupta J, Dong B, Klein EA, Hackett J, Jr, Schochetman G, Cullen BR, Silverman RH. 2011. In vivo hypermutation of xenotropic murine leukemia virus-related virus DNA in peripheral blood mononuclear cells of rhesus macaque by APOBEC3 proteins. Virology 421:28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchoff V, Wong S, St JS, Pari GS. 2002. Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch. Virol. 147:321–333 [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang F, Ding J, Minin VN, Suchard MA, Dorman KS. 2007. cBrother: relaxing parental tree assumptions for Bayesian recombination detection. Bioinformatics 23:507–508 [DOI] [PubMed] [Google Scholar]
- 23.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 24.Hasegawa M, Kishino H, Yano TA. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160–174 [DOI] [PubMed] [Google Scholar]
- 25.Han MV, Zmasek CM. 2009. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinform. 10:356. 10.1186/1471-2105-10-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Core Team R 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 27.Mullers E. 2013. The foamy virus Gag proteins: what makes them different? Viruses 5:1023–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stange A, Mannigel I, Peters K, Heinkelein M, Stanke N, Cartellieri M, Gottlinger H, Rethwilm A, Zentgraf H, Lindemann D. 2005. Characterization of prototype foamy virus gag late assembly domain motifs and their role in particle egress and infectivity. J. Virol. 79:5466–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patton GS, Morris SA, Chung W, Bieniasz PD, McClure MO. 2005. Identification of domains in gag important for prototypic foamy virus egress. J. Virol. 79:6392–6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beale RCL, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585–596 [DOI] [PubMed] [Google Scholar]
- 31.Calattini S, Nerrienet E, Mauclere P, Georges-Courbot MC, Saib A, Gessain A. 2004. Natural simian foamy virus infection in wild-caught gorillas, mandrills and drills from Cameroon and Gabon. J. Gen. Virol. 85:3313–3317 [DOI] [PubMed] [Google Scholar]
- 32.Liu W, Worobey M, Li Y, Keele BF, Bibollet-Ruche F, Guo Y, Goepfert PA, Santiago ML, Ndjango JB, Neel C, Clifford SL, Sanz C, Kamenya S, Wilson ML, Pusey AE, Gross-Camp N, Boesch C, Smith V, Zamma K, Huffman MA, Mitani JC, Watts DP, Peeters M, Shaw GM, Switzer WM, Sharp PM, Hahn BH. 2008. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 4:e1000097. 10.1371/journal.ppat.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blasse A, Calvignac-Spencer S, Merkel K, Goffe AS, Boesch C, Mundry R, Leendertz FH. 2013. Mother-offspring transmission and age-dependent accumulation of simian foamy virus in wild chimpanzees. J. Virol. 87:5193–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hussain AI, Shanmugam V, Bhullar VB, Beer BE, Vallet D, Gautier-Hion A, Wolfe ND, Karesh WB, Kilbourn AM, Tooze Z, Heneine W, Switzer WM. 2003. Screening for simian foamy virus infection by using a combined antigen Western blot assay: evidence for a wide distribution among Old World primates and identification of four new divergent viruses. Virology 309:248–257 [DOI] [PubMed] [Google Scholar]
- 35.Galvin TA, Ahmed IA, Shahabuddin M, Bryan T, Khan AS. 2013. Identification of recombination in the envelope gene of simian foamy virus serotype 2 isolated from Macaca cyclopis. J. Virol. 87:8792–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713–1716 [DOI] [PubMed] [Google Scholar]
- 37.Chaipan C, Smith JL, Hu WS, Pathak VK. 2013. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J. Virol. 87:444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delebecque F, Suspene R, Calattini S, Casartelli N, Saib A, Froment A, Wain-Hobson S, Gessain A, Vartanian JP, Schwartz O. 2006. Restriction of foamy viruses by APOBEC cytidine deaminases. J. Virol. 80:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gartner K, Wiktorowicz T, Park J, Mergia A, Rethwilm A, Scheller C. 2009. Accuracy estimation of foamy virus genome copying. Retrovirology 6:32. 10.1186/1742-4690-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkovic M, Schmidt S, Marino D, Russell RA, Stauch B, Hofmann H, Kopietz F, Kloke BP, Zielonka J, Strover H, Hermle J, Lindemann D, Pathak VK, Schneider G, Lochelt M, Cichutek K, Munk C. 2009. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein Bet. J. Biol. Chem. 284:5819–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell RA, Wiegand HL, Moore MD, Schafer A, McClure MO, Cullen BR. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724–8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaguva Vasudevan AA, Perkovic M, Bulliard Y, Cichutek K, Trono D, Haussinger D, Munk C. 2013. Prototype foamy virus Bet impairs the dimerization and cytosolic solubility of human APOBEC3G. J. Virol. 87:9030–9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukic D, Hotz-Wagenblatt A, Lei J, Rathe A-M, Muhle M, Denner J, Munk C, Lochelt M. 2013. Identification of the feline foamy virus Bet domain essential for APOBEC3 counteraction. Retrovirology 10:76. 10.1186/1742-4690-10-76 [DOI] [PMC free article] [PubMed] [Google Scholar]