Abstract
Parvoviruses are rapidly evolving viruses that infect a wide range of hosts, including vertebrates and invertebrates. Extensive methylation of the parvovirus genome has been recently demonstrated. A global pattern of methylation of CpG dinucleotides is seen in vertebrate genomes, compared to “fractional” methylation patterns in invertebrate genomes. It remains unknown if the loss of CpG dinucleotides occurs in all viruses of a given DNA virus family that infect host species spanning across vertebrates and invertebrates. We investigated the link between the extent of CpG dinucleotide depletion among autonomous parvoviruses and the evolutionary lineage of the infected host. We demonstrate major differences in the relative abundance of CpG dinucleotides among autonomous parvoviruses which share similar genome organization and common ancestry, depending on the infected host species. Parvoviruses infecting vertebrate hosts had significantly lower relative abundance of CpG dinucleotides than parvoviruses infecting invertebrate hosts. The strong correlation of CpG dinucleotide depletion with the gain in TpG/CpA dinucleotides and the loss of TpA dinucleotides among parvoviruses suggests a major role for CpG methylation in the evolution of parvoviruses. Our data present evidence that links the relative abundance of CpG dinucleotides in parvoviruses to the methylation capabilities of the infected host. In sum, our findings support a novel perspective of host-driven evolution among autonomous parvoviruses.
INTRODUCTION
The relative abundance of dinucleotides, particularly the CpG dinucleotide, has received much attention recently. The CpG dinucleotide content is normally expressed as a ratio of the actual (observed) number of CpG dinucleotides and the expected number of CpG dinucleotides (O/E ratio). The relative abundance of CpG dinucleotides varies greatly across viruses (1, 2). Depletion of CpG dinucleotides has been reported to occur in all small DNA viruses (<30 kb) infecting humans (2), while most large DNA viruses (>30 kb) infecting humans show little or no CpG dinucleotide depletion. Several possible reasons have been suggested to play a role in the depletion of CpG dinucleotides, including (i) lower transcription rate for CpG-containing codons (3), (ii) stimulation of Toll-like receptor 9-mediated innate immune response by unmethylated CpGs (4), and (iii) spontaneous deamination of methylated cytosines in CpG dinucleotides (5). Deamination of unmethylated cytosines results in C-to-U transitions which are amenable to mismatch repair mechanisms, whereas the deamination of 5-methylcytosine leads to C-to-T transitions that are often irreversible, resulting in high mutation rates in methylated CpGs and a depletion of CpG dinucleotides (6, 7).
In most vertebrate genomes, the O/E ratio for CpG dinucleotides is <0.35, suggesting that vertebrate genomes have lost about two-thirds of CpG dinucleotides (5, 8). In contrast, invertebrate genomes show minor or no depletion of CpG dinucleotides (5). Early investigations on insects showed that insect DNA lacked DNA methylation, suggesting the lack of DNA methylation apparatus in insects (9, 10). Subsequent studies unambiguously revealed the presence of several DNA methyltransferases in invertebrates (11). However, there are major differences between the methylation patterns of invertebrates and vertebrates. Vertebrate genomes show high levels of methylation (global methylation) of CpG dinucleotides (up to 90%) (12, 13) compared to the very low levels of CpG methylation seen among invertebrate genomes (13, 14).
Among viral genomes, methylation of CpG dinucleotides has been extensively studied among double-stranded DNA (dsDNA) viruses, including herpesviruses, papillomaviruses, and hepatitis B virus (15, 16, 17). In a recent report, Bonvicini et al. demonstrated for the first time CpG methylation of a single-stranded DNA (ssDNA) virus (18). They showed extensive CpG methylation of parvovirus B19 both in clinical samples and in cell culture models (18).
Studies demonstrating the loss of CpG dinucleotides in small DNA viruses (<30 kb) have focused only on viruses infecting humans or higher-order mammals. It remains unknown if the loss of CpG dinucleotide occurs in all viruses of a given DNA virus family that infect host species spanning across vertebrates and invertebrates. We hypothesized that within a given family of viruses, the genomes of those infecting invertebrates will show minimal or no CpG depletion, in keeping with the near-normal levels of CpG dinucleotides in invertebrates, while that of viruses of the same family infecting vertebrate hosts will have marked depletion of CpG dinucleotides. To test this hypothesis, we chose to investigate the CpG dinucleotide content of parvoviruses, which are single-stranded DNA viruses infecting a wide range of hosts, spanning from insects to primates, including humans. We believe that parvoviruses are an appropriate choice to test our hypothesis for the following reasons: (i) parvoviruses do not encode their own polymerase and their replication is dependent on the cellular replication machinery (19, 20), (ii) parvoviruses are known for their rapid adaption to the host (21), (iii) recent studies on parvovirus B19 confirm that parvovirus DNA is extensively methylated in humans (18), (iv) single-stranded methylated DNA is more rapidly deaminated than double-stranded methylated DNA (22), and (v) parvoviruses spanning across different subfamilies share common ancestral origins (23). Therefore, we believe that parvoviruses are suitable to study host-driven evolution of viruses.
The family Parvoviridae is comprised of small nonenveloped DNA viruses with ∼5-kb linear, single-stranded DNA genomes encapsidated within an icosahedral protein coat. The family Parvoviridae is divided into two subfamilies: Parvovirinae, infecting vertebrates, and Densovirinae, infecting invertebrates. Based on the phylogenetic analysis, the subfamily Parvovirinae is further classified into five genera, namely, Parvovirus, Amdovirus, Bocavirus, Dependovirus, and Erythrovirus. The subfamily Densovirinae is further classified into four genera, namely, Brevidensovirus, Densovirus, Iteravirus, and Pefudensovirus. The members of the genus Dependovirus require a helper virus for their replication (23).
In this study, we investigate the link between the evolutionary lineage of the infected host and the extent of CpG dinucleotide depletion in the infecting parvovirus genome. We believe that this study will shed light on the role of host methylation capabilities in driving parvovirus evolution.
MATERIALS AND METHODS
Parvovirus genomes analyzed.
A total of 259 full-length sequences (all the full-length sequences available for autonomous parvoviruses) were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/); this includes 35 sequences of parvoviruses infecting invertebrates (densoviruses; n = 35), parvoviruses infecting vertebrates (n = 56), human bocaviruses (n = 144), gorilla bocaviruses (n = 2), and animal bocaviruses (includes bovine, canine, and feline bocaviruses; n = 22) (all the accession numbers are provided in Table 1). The keywords used to search the full-length genomes in the NCBI GenBank sequence database were “parvovirus complete genome” and “bocavirus complete genome.” The majority of the full-length sequences retrieved did not contain the terminal palindromic sequences, while 50 sequences contained the terminal palindromic sequences. The terminal palindromic regions were removed from the full-length sequences using palindrome prediction software (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::palindrome). A total of 259 sequences (includes 209 sequences used as available from GenBank and 50 sequences from GenBank in which the terminal palindromic sequences were trimmed) were used for analysis. Analysis using all the sequences (n = 259) as available from GenBank (without trimming the terminal palindromes) caused no significant changes to any of the findings reported in this study (data not shown). The genus Dependovirus was not included in this study, as members are not capable of autonomous replication and are dependent on a helper adenovirus or herpesvirus for active replication (19, 23). In addition, the mode of cellular entry, infection, and replication of dependoviruses is different from that of autonomous parvoviruses (19). Hence, our study was restricted to autonomous parvoviruses. Despite their ability to replicate autonomously, goose and Muscovy duck parvoviruses were excluded from the study, as they are genetically related to dependoviruses (24, 25). With a few exceptions, parvoviruses are known to cause acute infections. Human parvovirus B19 (26, 27), canine parvovirus (28, 29), and Aleutian mink disease virus (30, 31) are known to cause chronic/persistent infection. Extended virus-host interaction during chronic/persistent infection will have a bearing on virus evolution and may potentially influence the results of this study; hence, human parvovirus B19, canine parvovirus, and Aleutian mink disease virus were excluded from this study. The inclusion of human parvovirus B19, canine parvovirus, and Aleutian mink disease virus caused no significant changes to findings reported in this study (data not shown).
Table 1.
Accession numbers of autonomous parvovirus sequences analyzeda
n = 259 sequences. Accession numbers are grouped based on the infected host.
Calculation of dinucleotide frequencies.
The observed and expected frequency of dinucleotides was calculated using a program written in PHP (hypertext preprocessor). The observed/expected ratios for the dinucleotide XpY [(O/E)XpY] were calculated as the observed frequency of the dinucleotide f(XY) relative to the product of the frequency of the individual nucleotides f(X) and f(Y) weighted by the length of the genome G. In other words, (O/E)XpY = [f(XY)/f(X)f(Y)] × G.
The loss of CpG dinucleotides was calculated by using the following formula: Loss of CpG dinucleotides = 1 − (O/E)CpG.
The average gain in TpG and CpA dinucleotides was calculated using the following formula: Average gain in TpG and CpA dinucleotide frequencies = {[(O/E)TpG − 1] + [(O/E)CpA − 1]}/2.
The deviation of the actual O/E ratio from 1 (i.e., if the observed frequency equals the expected frequency) for each dinucleotide (XpY) was calculated using the following formula: Deviation of O/E ratio for dinucleotide (XpY) = 1 − (O/E)XpY.
Statistical analysis.
Data were analyzed using Student's t test, Pearson's correlation coefficient (r2), and analysis of variance (ANOVA) as appropriate. Box plots and scatter plots were created using Matlab. The minimum value, lower quartile (Q1), median, upper quartile (Q3), and maximum value were used for constructing box plots. On each box, the central horizontal line marks the median, the edges represent first and third quartiles, and the interquartile range (IQR) within the box includes the central 50% of the data. Scatter plots were used to analyze the correlation between 2 parameters. Results were considered statistically significant at a P value of <0.05.
RESULTS
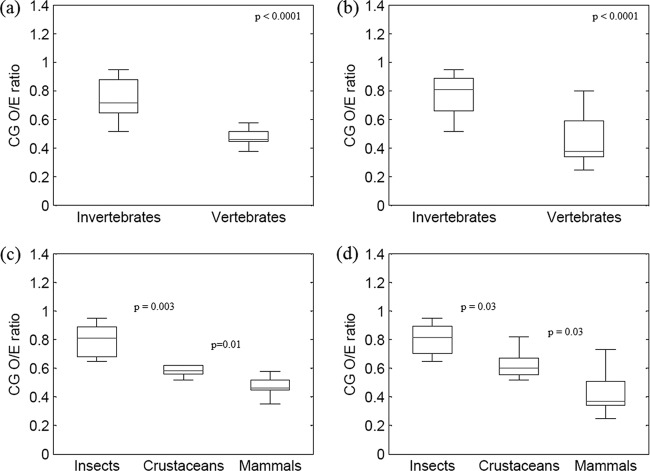
Autonomous parvoviruses infecting invertebrates had a significantly higher CpG O/E ratio than those infecting vertebrates (mean ± standard deviation [SD], 0.76 ± 0.19 compared to 0.50 ± 0.15; P < 0.0001) (Fig. 1a). The distribution of CpG dinucleotide O/E ratios for parvoviruses infecting different classes of host species is shown in a box plot (Fig. 1c). Among parvoviruses infecting invertebrates, those infecting insects had significantly higher CpG O/E ratios than those infecting crustaceans (mean ± SD, 0.82 ± 0.18 compared to 0.62 ± 0.11; P = 0.003). Among all autonomous parvoviruses studied, the lowest CpG O/E ratios were observed in parvoviruses infecting mammals (mean ± SD, 0.50 ± 0.15). The CpG O/E ratios observed in parvoviruses infecting mammals were significantly lower than those observed in parvoviruses infecting insects (mean ± SD, 0.50 ± 0.15 compared to 0.82 ± 0.18; P < 0.0001) and crustaceans (mean ± SD, 0.50 ± 0.15 compared to 0.62 ± 0.11; P = 0.01).
Fig 1.
The extent of CpG depletion is closely linked to host methylation capabilities. (a) Box plot showing CpG O/E ratios in parvoviruses infecting different classes of host species; all autonomous parvovirus sequences studied were analyzed (n = 259). Parvoviruses infecting invertebrates have significantly higher CpG O/E ratios than parvoviruses infecting vertebrates (P < 0.0001). (b) Box plot showing the CpG O/E ratios in parvoviruses infecting different classes of host species in a subset of sequences (one sequence each for every autonomous parvovirus; n = 56) (P < 0.0001). (c) CpG depletion is more pronounced among parvoviruses infecting mammals than among those infecting crustaceans and insects when analyzing all the sequences studied (n = 259) (mammals compared to crustaceans, mean ± SD, 0.50 ± 0.15 compared to 0.62 ± 0.11, P = 0.01; mammals compared to insects, mean ± SD, 0.50 ± 0.15 compared to 0.82 ± 0.18, P < 0.0001). (d) CpG depletion is more pronounced among parvoviruses infecting mammals compared to those infecting crustaceans and insects in the subset of sequences analyzed (n = 56) (mammals compared to crustaceans, mean ± SD, 0.44 ± 0.17 compared to 0.63 ± 0.12, P = 0.03; mammals compared to insects, mean ± SD, 0.44 ± 0.17 compared to 0.84 ± 0.19, P < 0.0001).
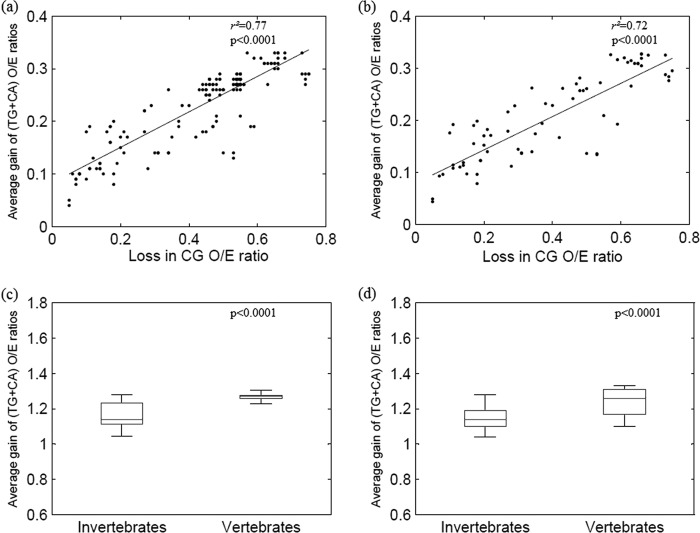
The loss of CpG dinucleotides by deamination of methylated cytosines within CpG dinucleotides results in a gain of TpG and CpA dinucleotides (5). Interestingly, we found a strong correlation between the loss of CpG dinucleotides and the average gain in TpG and CpA dinucleotides in parvoviruses (r2 = 0.77; P < 0.0001), as shown in Fig. 2a. The average gain in TpG and CpA dinucleotides in parvoviruses infecting invertebrates was significantly higher than those infecting vertebrates (1.25 ± 0.05 compared to 1.10 ± 0.03; P < 0.0001) (Fig. 2c).
Fig 2.
Loss of the CpG dinucleotide is linked to deamination of 5-methylcytosine. (a) Scatter plot demonstrating a positive correlation between the loss of CpG O/E ratios (x axis) and the average gain in TpG and CpA O/E ratios (y axis); all autonomous parvovirus sequences studied were analyzed (n = 259). (b) Scatter plot demonstrating a positive correlation between loss of CpG O/E ratios (x axis) and average gain in TpG and CpA O/E ratios (y axis); analysis done using a subset of sequences (one sequence each for every autonomous parvovirus; n = 56). (c) Box plot comparing the average gain in TpG and CpA ratios between parvovirus infecting vertebrates and invertebrates when analyzing all the sequences studied (n = 259). The average gain in TpG and CpA O/E ratios is higher among parvoviruses infecting vertebrates than among those infecting invertebrates (1.25 ± 0.05 compared to 1.10 ± 0.03; P < 0.0001). (d) Box plot comparing the average gain in TpG and CpA ratios between parvovirus infecting vertebrates and invertebrates; analysis done using a subset of sequences (n = 56) (1.24 ± 0.08 compared to 1.14 ± 0.09; P < 0.0001).
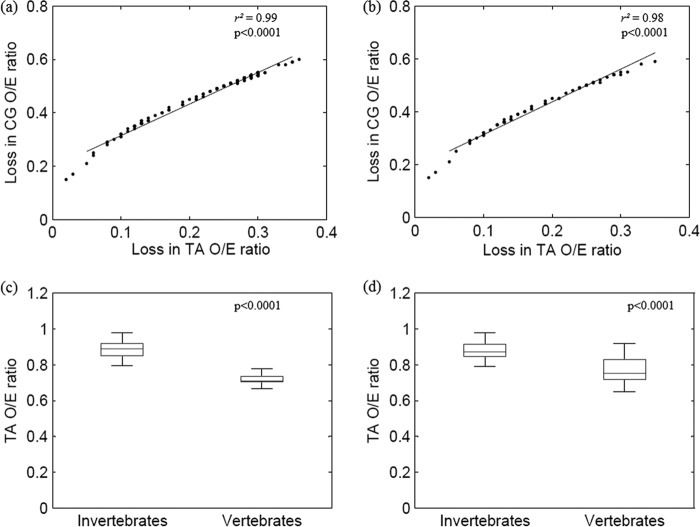
A strong correlation between the loss of CpG dinucleotides and the loss of TpA dinucleotides among parvoviruses is shown in Fig. 3a (r2 = 0.99; P < 0.0001). The depletion of TpA dinucleotides among parvoviruses infecting vertebrates was much higher than that observed among parvoviruses infecting invertebrates (0.89 ± 0.049 compared to 0.73 ± 0.049; P < 0.0001) (Fig. 3c).
Fig 3.
Depletion of CpG dinucleotides is linked to the loss of TpA dinucleotides. (a) Scatter plot demonstrating a strong correlation between the loss of CpG dinucleotides (y axis) and the loss of TpA dinucleotides (x axis); all autonomous parvovirus sequences studied were analyzed (n = 259). (b) Scatter plot demonstrating a strong correlation between the loss of CpG dinucleotides (y axis) and the loss of TpA dinucleotides (x axis) in a subset of sequences (one sequence each for every autonomous parvovirus; n = 56). (c) Box plot comparing the TpA O/E ratios between parvoviruses infecting invertebrates and vertebrates; all autonomous parvovirus sequences studied were analyzed (n = 259). The TpA O/E ratios are higher among parvoviruses infecting invertebrates than among those infecting vertebrates (0.89 ± 0.049 compared to 0.73 ± 0.049; P < 0.0001). (d) Box plot comparing the TpA O/E ratios between parvovirus infecting invertebrates and vertebrates in a subset of sequences analyzed (n = 56) (0.88 ± 0.05 compared to 0.77 ± 0.07; P < 0.0001).
The GC% of the parvovirus sequences used in this study ranged from 35% to 55%. There was no correlation between GC% and depletion of the CpG dinucleotide among the parvoviruses studied (Fig. 4a) (r2 = 0.06; P = 0.38).
Fig 4.
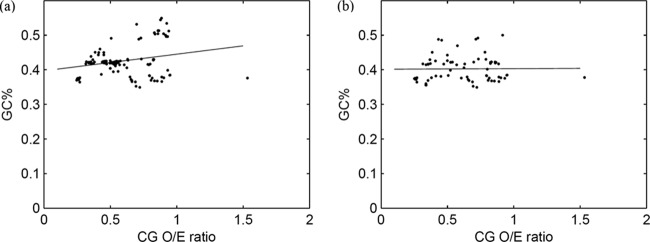
CpG depletion is not influenced by GC content. (a) Scatter plot demonstrating the lack of correlation between GC% (y axis) and CpG dinucleotide O/E ratio (x axis); all autonomous parvovirus sequences studied were analyzed (n = 259). (b) Scatter plot demonstrating the lack of correlation between GC% (y axis) and CpG dinucleotide O/E ratio (x axis); analysis done using a subset of sequences (one sequence each for every autonomous parvovirus; n = 56).
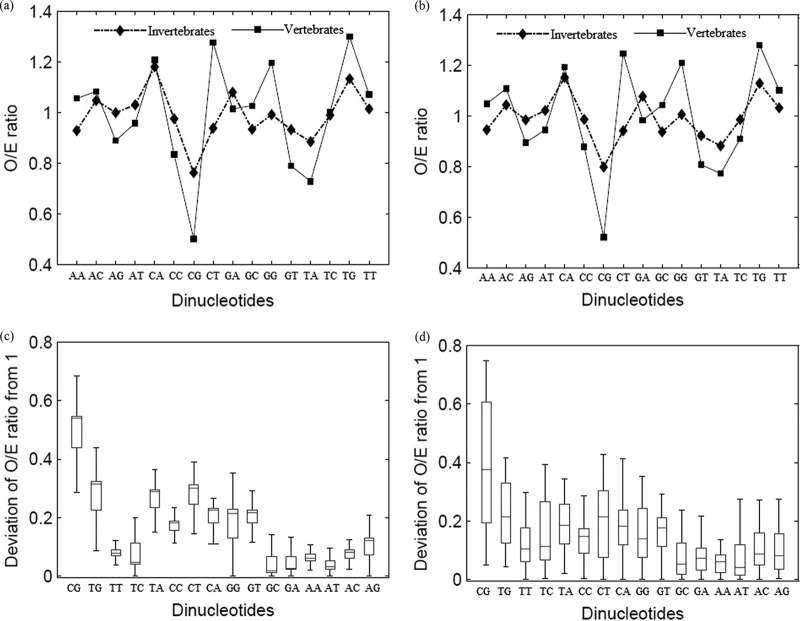
The O/E ratios for all dinucleotides are compared between parvoviruses infecting invertebrates and those infecting vertebrates in Fig. 5a. This figure demonstrates that among all dinucleotides, the CpG dinucleotide is the most depleted dinucleotide in both parvoviruses infecting invertebrates and vertebrates (depletion of CpG dinucleotide compared to depletion of any other dinucleotide, P < 0.0001). The deviation of the O/E ratio for each dinucleotide from 1 (the O/E ratio is 1 if the observed frequency equals the expected frequency) is shown in Fig. 5c. This figure demonstrates that among all dinucleotide O/E ratios, the O/E ratio for CpG dinucleotide is most deviant from 1 (i.e., if the observed frequency equals the expected frequency), suggesting that the loss of CpG dinucleotides is more pronounced than the loss or gain in any other dinucleotide among autonomous parvoviruses (deviation of CpG dinucleotide O/E ratio from 1 compared to deviation of any other dinucleotide O/E ratio from 1, P < 0.0001).
Fig 5.
Role of CpG depletion in parvovirus evolution. (a) Comparison of all dinucleotide O/E ratios between parvoviruses infecting invertebrates and those infecting vertebrates; all autonomous parvovirus sequences studied were analyzed (n = 259). CpG dinucleotides are the most depleted dinucleotides among parvoviruses infecting invertebrates and vertebrates (depletion of CpG dinucleotide compared to depletion of any other dinucleotide, P < 0.0001). (b) Comparison of all dinucleotide O/E ratios between parvoviruses infecting invertebrates and those infecting vertebrates in a subset of sequences (one sequence each for every autonomous parvovirus; n = 56). (c) Box plot comparing the deviation of the O/E ratio for each dinucleotide from 1 (the O/E ratio is 1 if the observed frequency equals the expected frequency); analysis done using all autonomous parvoviruses studied (n = 259). CpG dinucleotide O/E ratios are the most deviant among all dinucleotide ratios in autonomous parvoviruses, suggesting that CpG dinucleotide loss is more pronounced than the loss or gain of any other dinucleotide (deviation of CpG dinucleotide from 1 compared to deviation of any other dinucleotide from 1, P < 0.0001). (d) Box plot comparing the deviation of the O/E ratio for each dinucleotide from 1 (the O/E ratio is 1 if the observed frequency equals the expected frequency) in a subset of sequences (n = 56).
Bocavirus sequences are overrepresented among the sequences studied (168/259 sequences studied were bocavirus sequences). To ascertain if the overrepresentation of bocavirus sequences influenced any of the major findings reported in this study, we analyzed data using one sequence each for every autonomous parvovirus included in the study; this included autonomous parvoviruses infecting insects (n = 20), crustaceans (n = 5), and mammals (n = 31; including bocaviruses). The results of this analysis were in keeping with results obtained by analyzing all available sequences (Fig. 1b and d, 2b and d, 3b and d, 4b, and 5b and d), suggesting that the overrepresentation of bocavirus sequences did not affect any of the findings reported in this study.
DISCUSSION
Host methylation capabilities may determine the extent of CpG depletion.
We investigated the depletion of CpG dinucleotides among autonomous parvoviruses that infect a wide range of host species, including insects, crustaceans, aves, and mammals. In our study, despite the similarities in genome organization, proteins encoded, and replication strategies, the CpG dinucleotide content of autonomous parvoviruses varied greatly depending on the infected host species. The depletion of CpG dinucleotides among parvoviruses infecting vertebrates was significantly higher than that among parvoviruses infecting invertebrates (0.50 ± 0.15 compared to 0.76 ± 0.19; P < 0.0001) (Fig. 1a).
Differences in DNA methylation capabilities between vertebrate and invertebrates are well documented. A global pattern of methylation is reported among vertebrate genomes compared to fractional methylation patterns among invertebrate genomes (12, 13), suggesting that vertebrates have superior methylation capabilities. The methylation of the cytosine of a CpG dinucleotide at the C5 position is catalyzed by enzymes known as DNA methyl transferases (DNMTs) (32). In vertebrate genomes, DNA methylation is recognized as a defense mechanism against intragenomic parasites (33), while studies on invertebrate genomes fail to support this role of DNA methylation among invertebrates (34). The near-normal levels of CpG dinucleotides among parvoviruses infecting invertebrates compared to major depletion of CpG dinucleotides among parvoviruses infecting vertebrates in our study may reflect the differences in DNA methylation capabilities between the two host lineages.
Interestingly, among all autonomous parvoviruses, minimal CpG depletion was seen in those infecting insects, the lowest lineage in evolution among hosts infected by parvoviruses, while the highest depletion of CpG dinucleotides was seen in parvoviruses infecting mammals, the highest lineage in evolution among hosts infected by parvoviruses (0.82 ± 0.18 compared to 0.50 ± 0.15; P < 0.0001) (Fig. 1c). Of note, studies investigating several vertebrate and invertebrate host genomes show that the lowest levels of DNA methylation occur in insect genomes, and the highest levels of DNA methylation are seen among mammalian genomes (12). Our study demonstrates for the first time a link between the evolutionary lineage of the infected host and the extent of CpG dinucleotide depletion in the infecting virus genome.
The presence of DNA methylation in the crustacean genomes and the number of DNMTs encoded by crustaceans are poorly studied. However, a limited number of studies investigating crustacean genomes show that crustaceans encode DNMT1, DNMT2, and DNMT3, while representative members of major insect lineages are believed to lack one or more of the DNMTs (35, 36). Parvoviruses infecting crustaceans infect primarily prawns of the genus Penaeus. In Penaeus spp., approximately 4.5% of CpGs are methylated (14). The average level of CpG methylation detected in Penaeus spp. is higher than that detected in most major insect lineages. Interestingly, in our study, among autonomous parvoviruses infecting invertebrates, significantly higher levels of CpG dinucleotide depletion were observed among those infecting crustaceans (higher-order invertebrates) compared to those infecting other insects (lower-order invertebrates) (0.62 ± 0.11 compared to 0.82 ± 0.18; P = 0.003) (Fig. 1c). This is in keeping with the detection of higher CpG methylation in Penaeus spp. than in most major insect lineages. This finding in our study further strengthens the link between host methylation capabilities and depletion of CpG dinucleotides in parvoviruses.
Loss of CpG dinucleotides in parvoviruses is linked to deamination of 5-methylcytosine.
The methylated cytosines in CpG dinucleotides are highly susceptible to spontaneous deamination (37) or enzyme-mediated deamination (38), resulting in thymine substitution causing T/G mismatch. This T/G mismatch is irreparable (6, 7), resulting in the formation of a CpA dinucleotide in the opposite strand. Therefore, the deamination of a methylated cytosine in a CpG dinucleotide leads to the loss of 2 CpG dinucleotides (one from each strand) and an increase in one TpG and one CpA dinucleotide (5). Other reasons proposed for the depletion of CpG dinucleotides, including avoidance of CpG dinucleotides for better transcription rates (3) or minimizing stimulation of undesired immune response (4), will result in random mutations at the CpG dinucleotides, leading to a loss of CpG dinucleotides but not necessarily an increase in the relative abundance of TpG and CpA dinucleotides. Hence, the correlation between the loss of CpG dinucleotides and the gain of TpG and CpA dinucleotides links the depletion of CpG dinucleotides to the deamination of methylated cytosines within the CpG dinucleotides (5).
In our study, we found a strong correlation between the depletion of CpG dinucleotide among autonomous parvoviruses and the gain in TpG and CpA dinucleotides (r2 = 0.77; P < 0.0001) (Fig. 2a). This suggests that methylation of the cytosine in CpG dinucleotides followed by deamination is the likely cause of CpG depletion in parvoviruses. It has been reported in literature that ssDNA serves as a better substrate for methyltransferases than dsDNA. Hence, the extent of methylation and subsequent deamination in ssDNA is higher than that in dsDNA (39); this is particularly relevant to the strong correlation between the depletion of CpG dinucleotide and the gain in TpG and CpA dinucleotides among autonomous parvoviruses that we found in our study.
The depletion of TpA dinucleotides is indirectly linked to loss of methylated cytosine in CpG dinucleotides by deamination. When methylated CpG dinucleotides are deaminated, TpG and CpA dinucleotides increase in number. This leads to an increase in number of T's and A's but not TpA. Hence, the loss of methylated CpG dinucleotides by deamination is linked to a decrease in the O/E ratio of TpA dinucleotides (40). In addition, the underrepresentation of TpA dinucleotide is more pronounced in DNA which is destined to become RNA. Two of the three stop codons, UAG and UAA, contain TpA dinucleotides, making TpA dinucleotide scarcity in the coding DNA sequence a prerequisite for mRNA stability (41). Second, cytoplasmic ribonucleases preferentially target UpA dinucleotides, and therefore coding DNA sequences are more likely to have TpA dinucleotide deficiencies than noncoding DNA sequences (41). Given that among parvoviruses most of the genome represents coding sequences, TpA deficiencies may occur in parvovirus genomes.
We demonstrate a strong correlation between depletion of CpG dinucleotides and the loss of TpA dinucleotides among parvoviruses (Fig. 5a) (r2 = 0.99; P < 0.0001). Our results clearly indicate that the loss of CpG dinucleotides in parvoviruses is linked to a gain in TpG and CpA dinucleotides (Fig. 2a) and also to a loss in TpA dinucleotides (Fig. 3a). This finding suggests that the loss of CpG dinucleotides in parvoviruses is linked to deamination of methylated cytosines within the CpG dinucleotide leading to the gain of TpG and CpA dinucleotides that in turn results in the loss of TpA dinucleotides. These results argue against random mutations leading to the loss of CpG dinucleotides in parvoviruses and also against inherent differences in CpG dinucleotide composition among parvoviruses. We report here for the first time a link between the evolutionary lineage of the infected host and TpA depletion in the infecting virus (Fig. 3c).
Apart from the proposed role of methylation capabilities of the infected host, differences in the extent of CpG methylation among autonomous parvoviruses infecting a given host could potentially influence the depletion of CpG dinucleotides. In addition, depletion of CpG containing codons by random mutations in the CpG dinucleotide may allow better transcription rates (3). It is also possible that random mutations occurring during virus replication leading to the loss of CpG dinucleotides may help the virus survive host defense mechanisms, and vertebrate parvovirus strains with these mutations may be positively selected. However, our findings demonstrate that the loss of CpGs in parvoviruses is primarily by mutation of CpG dinucleotides to TpG/CpA dinucleotides and not by random mutation of the CpG dinucleotide. Therefore, it is unlikely that the loss of CpG dinucleotides, either for avoidance of stimulation of TLR-9-mediated innate immune responses or for improved transcription rates, is a major mechanism driving the depletion of CpG dinucleotide among parvoviruses.
GC content does not influence CpG depletion.
It is well recognized that among vertebrate genomes the CpG dinucleotide O/E ratios positively correlate with the GC% of the genome (42, 43). Among DNA viruses that infect humans, minimal or no CpG depletion is seen in those with a high G+C content, with a few exceptions (2, 44). Among autonomous parvoviruses, there is no correlation between the GC% and the extent of CpG depletion (r2 = 0.06; P = 0.38) (Fig. 4a). This finding suggests the observed differences in CpG depletion among parvoviruses infecting hosts of different evolutionary lineages is not linked to differences in GC%. Based on this finding and the fact that parvoviruses have diverged from a single common ancestor (23, 45), it may not be speculative to argue that the differences in CpG depletion among parvoviruses is not due to the inherent differences in nucleotide composition of the parvovirus genomes but is more likely due to the host-driven component of virus evolution, which in this case is linked to host methylation capabilities.
Not all small DNA viruses are CpG depleted.
Based on analysis of viruses infecting humans, it is generally accepted that CpG dinucleotides are depleted in all small DNA viruses (<30 kb) but not in large DNA viruses (2, 44). To our knowledge, this is the first study to investigate CpG depletion among small DNA viruses infecting a wide variety of hosts. Our data show that small DNA viruses are not necessarily CpG depleted. In fact, depending on the infected host species, parvoviruses could have CpG O/E ratios comparable to those reported for large DNA viruses. Cytomegalovirus is a large DNA virus with a high G+C content and a CpG dinucleotide O/E ratio of 1.19, the highest reported CpG O/E ratio for any virus in the literature (44). Surprisingly, in our study, the CpG dinucleotide O/E ratio for the parvovirus infecting Planococcus citri is 1.53, making it the virus with the highest relative abundance of CpG dinucleotides. Planococcus citri, also known as citrus mealybug, is known for the unusual epigenetic mechanisms involved in embryonic development (46, 47). It has been demonstrated that in mealybugs, CpG methylation is used primarily to mark the parental origin of chromosomes and that CpG methylation does not play a role in gene silencing (48). In addition, cytosine methylation does not play a role either in heterochromatin formation or imprinting in mealybugs (45). Our finding of a high relative abundance of CpG dinucleotides (CpG O/E ratio of 1.53) in parvoviruses infecting Planococcus citri may reflect unusual host methylation capabilities.
Role of CpG depletion in parvovirus evolution.
Among parvoviruses infecting invertebrates and vertebrates, the CpG dinucleotide is the most depleted dinucleotide (Fig. 5a). In addition, CpG dinucleotide O/E ratios are the most deviant (maximum difference between observed frequency and expected frequency) among all dinucleotide ratios in autonomous parvoviruses (Fig. 5c; deviation of CpG dinucleotide O/E ratio from expected compared to deviation of any other dinucleotide O/E ratio from expected, P < 0.0001). This finding suggests that the loss of CpG dinucleotides is more pronounced than the loss or gain in any other dinucleotide among autonomous parvoviruses. Therefore, depletion of CpG dinucleotides is likely to play an important role in the evolution of parvoviruses.
Relative abundance of CpG dinucleotides in bocaviruses.
Parvoviruses infecting vertebrates showed marked CpG dinucleotide depletion (Fig. 1) with a few exceptions. Modest CpG dinucleotide depletion is observed among animal bocaviruses, including bovine parvovirus (mean CpG O/E = 0.70), minute virus of canines (mean CpG O/E = 0.79), feline bocaviruses (mean CpG O/E = 0.82), porcine bocaviruses (mean CpG O/E = 0.87), and canine bocavirus (mean CpG O/E = 0.93). The CpG dinucleotide depletion in bocaviruses infecting primates, including human bocaviruses (mean CpG O/E = 0.47) and gorilla bocavirus (mean CpG O/E = 0.54), was similar to that observed for other parvoviruses infecting vertebrates. Gorilla bocavirus and human bocaviruses are genetically closely related and share a common ancestry (49, 50). Recent studies demonstrate that feline bocaviruses (51), minute virus of canines (51), porcine bocaviruses (52), and canine bocavirus (53) are genetically distinct from other related bocaviruses, including bocaviruses infecting primates. In addition, differences in splicing (54), length of nonstructural proteins (54), and replication strategies (55) have been documented between bocaviruses infecting primates and bocaviruses infecting other animals. Differences in genome organization and the lack of evidence supporting common ancestry and between bocaviruses infecting primates and those infecting other animals could potentially explain the differences in the relative abundance of CpG dinucleotides between these viruses.
Conclusion.
We demonstrate major differences in the relative abundance of CpG dinucleotide among viruses of the same family with similar genome organization and life cycle depending on the infected host species. Contrary to the general perception, all small DNA viruses are not necessarily CpG depleted. We present evidence that CpG dinucleotide depletion in parvoviruses is linked to the methylation capabilities of the infected host. The strong correlation of CpG dinucleotide depletion with a gain in TpG/CpA dinucleotides and a loss of TpA dinucleotides among parvoviruses suggests a major role for CpG methylation in the evolution of parvoviruses. Taken together, our data suggest a strong link between the evolutionary lineage of the infected host and the extent of CpG dinucleotide depletion in the infecting parvovirus genome. Our findings provide evidence for a novel perspective of host-driven evolution among parvoviruses.
Footnotes
Published ahead of print 9 October 2013
REFERENCES
- 1.McGeoch DJ, Crawford LV, Follett EA. 1970. The DNAs of three parvoviruses. J. Gen. Virol. 6:33–40 [DOI] [PubMed] [Google Scholar]
- 2.Hoelzer K, Shackelton LA, Parrish CR. 2008. Presence and role of cytosine methylation in DNA viruses of animals. Nucleic Acids Res. 36:2825–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medvedeva YA, Fridman MV, Oparina NJ, Malko DB, Ermakova EO, Kulakovskiy IV, Heinzel A, Makeev VJ. 2010. Intergenic, gene terminal, and intragenic CpG islands in the human genome. BMC Genomics 11:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chinnery HR, McLenachan S, Binz N, Sun Y, Forrester JV, Degli-Esposti MA, Pearlman E, McMenamin PG. 2012. TLR9 ligand CpG-ODN applied to the injured mouse cornea elicits retinal inflammation. Am. J. Pathol. 180:209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird AP. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8:1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274:775–780 [DOI] [PubMed] [Google Scholar]
- 7.Wiebauer K, Neddermann P, Hughes M, Jiricny J. 1993. The repair of 5-methylcytosine deamination damage. EXS 64:510–522 [DOI] [PubMed] [Google Scholar]
- 8.Schorderet DF, Gartler SM. 1992. Analysis of CpG suppression in methylated and nonmethylated species. Proc. Natl. Acad. Sci. U. S. A. 89:957–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rae PM, Steele RE. 1979. Absence of cytosine methylation at C-C-G-G and G-C-G-C sites in the rDNA coding regions and intervening sequences of Drosophila and the rDNA of other insects. Nucleic Acids Res. 6:2987–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchi NO, Vidal-Rioja L, Cleaver JE. 1986. Direct visualization of the sites of DNA methylation in human, and mosquito chromosomes. Chromosoma 94:362–366 [DOI] [PubMed] [Google Scholar]
- 11.Lyko F, Maleszka R. 2011. Insects as innovative models for functional studies of DNA methylation. Trends Genet. 27:127–131 [DOI] [PubMed] [Google Scholar]
- 12.Tweedie S, Charlton J, Clark V, Bird A. 1997. Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol. Cell. Biol. 17:1469–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrich B, Tweedie S. 2003. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 19:269–277 [DOI] [PubMed] [Google Scholar]
- 14.Regev A, Lamb MJ, Jablonka E. 1998. The role of DNA methylation in invertebrates: developmental regulation or genome defense? Mol. Biol. Evol. 15:880–891 [Google Scholar]
- 15.Fernandez AF, Rosales C, Lopez-Nieva P, Grana O, Ballestar E, Ropero S, Espada J, Melo SA, Lujambio A, Fraga MF, Pino I, Javierre B, Carmona FJ, Acquadro F, Steenbergen RD, Snijders PJ, Meijer CJ, Pinea 2009. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 19:438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vivekanandan P, Daniel HD, Kannangai R, Martinez-Murillo F, Torbenson M. 2010. Hepatitis B virus replication induces methylation of both host and viral DNA. J. Virol. 84:4321–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vivekanandan P, Thomas D, Torbenson M. 2009. Methylation regulates hepatitis B viral protein expression. J. Infect. Dis. 199:1286–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonvicini F, Manaresi E, Di Furio F, De Falco L, Gallinella G. 2012. Parvovirus b19 DNA CpG dinucleotide methylation and epigenetic regulation of viral expression. PLoS One 7:e33316. 10.1371/journal.pone.0033316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berns KI. 1990. Parvovirus replication. Microbiol. Rev. 54:316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotmore SF, Tattersall P. 1996. Parvovirus DNA replication, p 799–813 In De Pamphilis M. (ed), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21.Shackelton LA, Parrish CR, Truyen U, Holmes EC. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. U. S. A. 102:379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan K, Sterling JF, Roberts SA, Bhagwat AS, Resnick MA, Gordenin DA. 2012. Base damage within single-strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent. PLoS Genet. 8:e1003149. 10.1371/journal.pgen.1003149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotmore SF, Tattersall P. 2007. Parvoviral host range and cell entry mechanisms. Adv. Virus Res. 70:183–232 [DOI] [PubMed] [Google Scholar]
- 24.Zadori Z, Stefancsik R, Rauch T, Kisary J. 1995. Analysis of the complete nucleotide sequences of goose and Muscovy duck parvoviruses indicates common ancestral origin with adeno-associated virus 2. Virology 212:562–573 [DOI] [PubMed] [Google Scholar]
- 25.Brown KE, Green SW, Young NS. 1995. Goose parvovirus—an autonomous member of the dependovirus genus? Virology 210:283–291 [DOI] [PubMed] [Google Scholar]
- 26.Kurtzman GJ, Cohen B, Meyers P, Amunullah A, Young NS. 1988. Persistent B19 parvovirus infection as a cause of severe chronic anaemia in children with acute lymphocytic leukaemia. Lancet ii:1159–1162 [DOI] [PubMed] [Google Scholar]
- 27.Lowry SM, Brent LH, Menaldino S, Kerr JR. 2005. A case of persistent parvovirus B19 infection with bilateral cartilaginous and ligamentous damage to the wrists. Clin. Infect. Dis. 41:e42–e44 [DOI] [PubMed] [Google Scholar]
- 28.Lenghaus C, Studdert MJ. 1984. Acute and chronic viral myocarditis. Acute diffuse nonsuppurative myocarditis and residual myocardial scarring following infection with canine parvovirus. Am. J. Pathol. 115:316–319 [PMC free article] [PubMed] [Google Scholar]
- 29.Lenghaus C, Studdert MJ, Finnie JW. 1980. Acute and chronic canine parvovirus myocarditis following intrauterine inoculation. Aust. Vet. J. 56:465–468 [DOI] [PubMed] [Google Scholar]
- 30.Storgaard T, Christensen J, Aasted B, Alexandersen S. 1993. cis-acting sequences in the Aleutian mink disease parvovirus late promoter important for transcription: comparison to the canine parvovirus and minute virus of mice. J. Virol. 67:1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storgaard T, Oleksiewicz M, Bloom ME, Ching B, Alexandersen S. 1997. Two parvoviruses that cause different diseases in mink have different transcription patterns: transcription analysis of mink enteritis virus and Aleutian mink disease parvovirus in the same cell line. J. Virol. 71:4990–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goll MG, Bestor TH. 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74:481–514 [DOI] [PubMed] [Google Scholar]
- 33.Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335–340 [DOI] [PubMed] [Google Scholar]
- 34.Field LM, Lyko F, Mandrioli M, Prantera G. 2004. DNA methylation in insects. Insect Mol. Biol. 13:109–115 [DOI] [PubMed] [Google Scholar]
- 35.Glastad KM, Hunt BG, Yi SV, Goodisman MA. 2011. DNA methylation in insects: on the brink of the epigenomic era. Insect Mol. Biol. 20:553–565 [DOI] [PubMed] [Google Scholar]
- 36.Albalat R. 2008. Evolution of DNA-methylation machinery: DNA methyltransferases and methyl-DNA binding proteins in the amphioxus Branchiostoma floridae. Dev. Genes Evol. 218:691–701 [DOI] [PubMed] [Google Scholar]
- 37.Kow YW. 2002. Repair of deaminated bases in DNA. Free Radic. Biol. Med. 33:886–893 [DOI] [PubMed] [Google Scholar]
- 38.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. 2004. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 279:52353–52360 [DOI] [PubMed] [Google Scholar]
- 39.Christman JK, Sheikhnejad G, Marasco CJ, Sufrin JR. 1995. 5-Methyl-2′-deoxycytidine in single-stranded DNA can act in cis to signal de novo DNA methylation. Proc. Natl. Acad. Sci. U. S. A. 92:7347–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duret L, Galtier N. 2000. The covariation between TpA deficiency, CpG deficiency, and G+C content of human isochores is due to a mathematical artifact. Mol. Biol. Evol. 17:1620–1625 [DOI] [PubMed] [Google Scholar]
- 41.Beutler E, Gelbart T, Han JH, Koziol JA, Beutler B. 1989. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage. Proc. Natl. Acad. Sci. U. S. A. 86:192–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernardi G, Olofsson B, Filipski J, Zerial M, Salinas J, Cuny G, Meunier-Rotival M, Rodier F. 1985. The mosaic genome of warm-blooded vertebrates. Science 228:953–958 [DOI] [PubMed] [Google Scholar]
- 43.Jabbari K, Bernardi G. 1998. CpG doublets, CpG islands and Alu repeats in long human DNA sequences from different isochore families. Gene 224:123–127 [DOI] [PubMed] [Google Scholar]
- 44.Karlin S, Doerfler W, Cardon LR. 1994. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 68:2889–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bando H, Kusuda J, Gojobori T, Maruyama T, Kawase S. 1987. Organization and nucleotide sequence of a densovirus genome imply a host-dependent evolution of the parvoviruses. J. Virol. 61:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buglia G, Predazzi V, Ferraro M. 1999. Cytosine methylation is not involved in the heterochromatization of the paternal genome of mealybug Planococcus citri. Chromosome Res. 7:71–73 [DOI] [PubMed] [Google Scholar]
- 47.Bongiorni S, Cintio O, Prantera G. 1999. The relationship between DNA methylation and chromosome imprinting in the coccid Planococcus citri. Genetics 151:1471–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bongiorni S, Prantera G. 2003. Imprinted facultative heterochromatization in mealybugs. Genetica 117:271–279 [DOI] [PubMed] [Google Scholar]
- 49.Kapoor A, Mehta N, Esper F, Poljsak-Prijatelj M, Quan PL, Qaisar N, Delwart E, Lipkin WI. 2010. Identification and characterization of a new bocavirus species in gorillas. PLoS One 5:e11948. 10.1371/journal.pone.0011948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babkin IV, Tyumentsev AI, Tikunov AY, Kurilshikov AM, Ryabchikova EI, Zhirakovskaya EV, Netesov SV, Tikunova NV. 2013. Evolutionary time-scale of primate bocaviruses. Infect. Genet. Evol. 14:265–274 [DOI] [PubMed] [Google Scholar]
- 51.Lau SK, Woo PC, Yeung HC, Teng JL, Wu Y, Bai R, Fan RY, Chan KH, Yuen KY. 2012. Identification and characterization of bocaviruses in cats and dogs reveals a novel feline bocavirus and a novel genetic group of canine bocavirus. J. Gen. Virol. 93:1573–1582 [DOI] [PubMed] [Google Scholar]
- 52.Yang WZ, Yu JM, Li JS, Cheng WX, Huang CP, Duan ZJ. 2012. Genome characterization of a novel porcine bocavirus. Arch. Virol. 157:2125–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kapoor A, Mehta N, Dubovi EJ, Simmonds P, Govindasamy L, Medina JL, Street C, Shields S, Lipkin WI. 2012. Characterization of novel canine bocaviruses and their association with respiratory disease. J. Gen. Virol. 93:341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kapoor A, Simmonds P, Slikas E, Li L, Bodhidatta L, Sethabutr O, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besser J, Bartkus J, Delwart E. 2010. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 201:1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapoor A, Hornig M, Asokan A, Williams B, Henriquez JA, Lipkin WI. 2011. Bocavirus episome in infected human tissue contains nonidentical termini. PLoS One 6:e21362. 10.1371/journal.pone.0021362 [DOI] [PMC free article] [PubMed] [Google Scholar]