Abstract
Human endogenous retroviruses (HERVs) of the HERV-W group comprise hundreds of loci in the human genome. Deregulated HERV-W expression and HERV-W locus ERVWE1-encoded Syncytin-1 protein have been implicated in the pathogenesis of multiple sclerosis (MS). However, the actual transcription of HERV-W loci in the MS context has not been comprehensively analyzed. We investigated transcription of HERV-W in MS brain lesions and white matter brain tissue from healthy controls by employing next-generation amplicon sequencing of HERV-W env-specific reverse transcriptase (RT) PCR products, thus revealing transcribed HERV-W loci and the relative transcript levels of those loci. We identified more than 100 HERV-W loci that were transcribed in the human brain, with a limited number of loci being predominantly transcribed. Importantly, relative transcript levels of HERV-W loci were very similar between MS and healthy brain tissue samples, refuting deregulated transcription of HERV-W env in MS brain lesions, including the high-level-transcribed ERVWE1 locus encoding Syncytin-1. Quantitative RT-PCR likewise did not reveal differences in MS regarding HERV-W env general transcript or ERVWE1- and ERVWE2-specific transcript levels. However, we obtained evidence for interindividual differences in HERV-W transcript levels. Reporter gene assays indicated promoter activity of many HERV-W long terminal repeats (LTRs), including structurally incomplete LTRs. Our comprehensive analysis of HERV-W transcription in the human brain thus provides important information on the biology of HERV-W in MS lesions and normal human brain, implications for study design, and mechanisms by which HERV-W may (or may not) be involved in MS.
INTRODUCTION
Human endogenous retroviruses (HERVs) are remnants of exogenous retroviruses that formed proviruses in the genomes of germ cells, thus becoming inheritable throughout the evolution of species. Quite a number of phylogenetically different HERV groups are evidence of germ line infections by various exogenous retroviruses. Reinfection and intracellular amplifications could add up to sometimes thousands of loci per HERV group. About 8% of the human genome mass, distributed in about 700,000 loci, is due to the activity of retroviral sequences. Most HERV groups are evolutionarily old and no longer encode retroviral proteins, yet HERV transcripts can be detected in many cell and tissue types (for reviews, see references 1 to 4).
The human endogenous retrovirus group HERV-W comprises about 650 loci. Remarkably, about 180 loci of the HERV-W group are not due to a typical retroviral process for provirus formation but were formed by L1 retrotransposition machinery. Those loci lack the 5′ long terminal repeat (LTR) U3 region and the 3′ LTR U5 region or they display larger 5′ truncations (5–7). The HERV-W group was originally discovered as cDNA sequences generated from particle-associated RNA isolated from plasma or supernatants of cultured cells from patients with multiple sclerosis (MS). The identified sequences were therefore named “MS-associated retrovirus” (MSRV) (8–10).
Most HERV-W loci are coding deficient due to their evolutionary age (6, 11). However, the ERVWE1 locus in chromosome (chr.) 7q21.2, also named syncytin-1 or ERVW-1 (see below), is a HERV-W provirus converted into a cellular gene. The corresponding protein product, termed Syncytin-1, is involved in placenta development, participating in the fusion of cytotrophoblasts forming the syncytiotrophoblast layer (12).
Another HERV-W locus in chromosome Xq22.3, named ERVWE2, or ERVW-2, encodes an incomplete HERV-W envelope protein (N-Trenv) that can be expressed ex vivo and may also be expressed in vivo. However, the function of the protein is currently not clear (13).
MS is a chronic inflammatory demyelinating disease of the central nervous system of unknown etiology. Evidence suggests that genetic and environmental factors contribute to development of the disease (14). Detection of retroviral sequences in patients with MS stimulated research on the association of those sequences with MS and a potential pathogenic role. MSRV was proposed to be an exogenous replication-competent member of the HERV-W group that might be involved in MS (15–19). However, the existence of MSRV as a replication-competent exogenous retrovirus seems to have little support, as closer inspection of the reported MSRV sequences strongly suggests that they either originated from existing genomic HERV-W loci or were generated in vitro by reverse transcriptase (RT) switching templates, i.e., RNA transcripts from different HERV-W loci (20), during the generation of cDNA.
Nevertheless, a number of studies suggested a possible role of HERV-W in MS. For instance, monoclonal anti-HERV-W Env antibody 6A2B2 detected a protein expressed in acute demyelinating MS brain lesions (13, 16, 21, 22). Expression of Syncytin-1 in astrocytes was reported to result in oligodendrocyte cytotoxicity in vitro, and stereotactic implantation of Syncytin-1 into the corpus callosum caused oligodendrocyte loss and demyelination in a mouse model in vivo (22). Similarly, transgenic mice expressing Syncytin-1 under a glial fibrillary acidic protein promoter were reported to develop neuroinflammation and had reduced myelin levels in the corpus callosum (21). The surface domain of another HERV-W Env protein (GenBank accession number AAK18189.1) (13) was reported to exert proinflammatory effects by activating CD14/Toll-like receptor 4 (23). Recently, the same HERV-W Env protein was also reported to inhibit oligodendroglial precursor differentiation, possibly contributing to remyelination failure (24).
Several studies have investigated the transcription of HERV-W. Employing microarray strategies, HERV-W was found to be transcribed in testicular cancer and the placenta (25, 26) and in different human brain regions (27). High-resolution melting-temperature analysis indicated nonrandom patterns of HERV-W transcription and also provided evidence for variable transcript levels between individuals (28). A number of RT-PCR-based studies of HERV-W transcription were also performed in the MS context but have produced sometimes conflicting findings. HERV-W env RNA was detected at higher levels in autopsied brain tissue from patients with MS than in controls (16, 22, 29, 30). Upregulated HERV-W env transcript levels were likewise detected in peripheral blood mononuclear cells (PBMC) (16). However, another work did not detect such upregulation in PBMC (29). Potential technical flaws in two publications that reported upregulated HERV-W env transcription in patients with MS (29, 30) have been discussed recently (31).
Several HERV-W loci have been identified as being transcribed directly by assignment of HERV-W cDNA sequences to genomic HERV-W loci (20, 32, 33). A study by Laufer et al. (20) mapped HERV-W env cDNA sequences to specific genomic HERV-W env elements and thus identified seven different HERV-W loci, among them the ERVWE1/ERVW-1/syncytin-1 locus, as being transcribed in PBMC. Notably, no differences in relative transcript levels of specific HERV-W env loci were observed in PBMC from patients with MS and healthy controls in that study. Several HERV-W loci were also identified by a microarray-based strategy as being transcribed indirectly (26). In accordance with an initiative by the Human Gene Nomenclature Committee (HGNC) for assigning unique designations to transcribed HERV loci, several transcribed HERV-W loci were named ERVW-1 to ERVW-6 (34).
As for a possible role of HERV-W in MS, identification of the HERV-W loci actually transcribed in MS brain lesions seems of great relevance, since only the transcribed HERV-W loci, among them loci with potential protein-coding competence, such as ERVWE1/ERVW-1/syncytin-1 and ERVWE2/ERVW-2 in Xq22.3, can be of biological relevance in regard to gene products. We therefore comprehensively identified transcribed HERV-W loci in autopsied brain lesions from patients with MS and white matter brain tissue samples from healthy controls employing high-throughput amplicon sequencing. We also quantified general and locus-specific HERV-W env transcript levels in those brain samples and assayed the promoter activity of HERV-W LTRs. Our study thus provides important information on the biology of HERV-W, especially in the context of MS.
MATERIALS AND METHODS
Brain tissue samples and cell lines.
Postmortem brain tissues from patients with MS and controls were obtained from The Netherlands Brain Bank (NBB), Netherlands Institute for Neuroscience, Amsterdam, Netherlands. All material was collected from donors for or from whom written informed consent for a brain autopsy and the use of the material and clinical information for research purposes had been obtained by the NBB. Specifically, 7 neuropathologically confirmed white matter MS brain lesions from 6 patients with MS and 7 white matter brain tissue samples from 7 controls were obtained from the NBB. The controls had no history of neurological disease during life and had died of nonneurological conditions, e.g., cardiac infarction, heart failure, postoperative retroperitoneal bleeding, multiorgan failure, or pulmonary infection. Thorough neuropathological examination revealed no significant cerebral abnormalities in any of the controls. Further information on the patients and controls is given in Table 1. The tissue samples were stored at −80°C. JEG-3 cells were cultivated in Ham's F-12 medium (PAA Laboratories GmbH, Pasching, Austria) supplemented with 10% (vol/vol) fetal calf serum (FCS), 10,000 U/ml penicillin, 10 mg/ml streptomycin at 37°C in a humidified 5% (vol/vol) CO2 atmosphere.
Table 1.
Brain tissue sample details
| Samplea | NBB no.b | Autopsy no.b | Sexc | Age (yr)d | PMD (h:min)e |
|---|---|---|---|---|---|
| MS1 | 2001-130 | S01/298 | F | 53 | 10:45 |
| MS2 | 2001-135 | S01/316 | M | 43 | 08:30 |
| MS3 | 2002-055 | S02/156 | F | 48 | 04:50 |
| MS4 | 2006-045 | S06/139 | M | 56 | 08:00 |
| MS5 | 2006-045 | S06/139 | M | 56 | 08:00 |
| MS6 | 2007-010 | S07/051 | M | 47 | 07:15 |
| MS7 | 2009-067 | S09/219 | M | 44 | 12:00 |
| H1 | 1991-124 | 91/225.4 | M | 38 | 7:00 |
| H2 | 1996-057 | S96/163 | F | 69 | 08:30 |
| H3 | 2010-070 | S10/196 | F | 60 | 07:30 |
| H4 | 1991-125 | *91/230 | M | 61 | 05:40 |
| H5 | 1997-043 | S97/133 | M | 68 | 10:10 |
| H6 | 1998-127 | S98/235 | M | 56 | 05:25 |
| H7 | 2009-003 | S09/007 | M | 62 | 07:20 |
Samples MS1 to MS7 were from patients with MS, and the respective tissue samples were from MS brain lesions. Samples H1 to H7 are white matter tissue from controls with no history of neurological disease during life.
Netherlands Brain Bank-specific information. Note that samples MS4 and MS5 were obtained during the same autopsy.
Note that the same numbers of samples from male (M) and female (F) donors were investigated in both groups of samples.
Ages for the control group were slightly higher than those for the group of patients with MS.
Postmortem delays (PMD) were similar for both groups (see Materials and Methods for further details).
RNA isolation and RT-PCR amplification of HERV-W.
RNA was isolated from brain samples using the RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The brain samples (∼0.25 cm3 each) were cut into small pieces while still frozen and homogenized in 1.5 ml TRIzol (Invitrogen, Life Technologies, Carlsbad, CA, USA). The homogenate was applied to a QIAshredder column (Qiagen, Hilden, Germany), centrifuged at 15,000 × g for 2 min, transferred into a 15-ml Falcon tube, and incubated at 30°C for 5 min. After addition of 400 μl chloroform each, the samples were vortexed for 40 s, incubated in a 30°C water bath for 3 min, and centrifuged at 8,600 × g and 4°C for 20 min. The upper phase was mixed with 1 volume of 70% (vol/vol) ethanol and applied to an RNeasy Mini spin column. Rigorous DNase treatment of isolated RNA and generation of cDNA were performed as previously described (35).
To amplify as many different HERV-W loci as possible in one PCR, primers to be used in HERV-W env cDNA-specific PCRs were optimized. Since many HERV-W loci lack parts or all of the 5′ env region, two amplicons of ∼280 bp and ∼330 bp, designated 5′ env and 3′ env, respectively, were selected, representing the env 5′ region, ranging from nucleotides (nt) 7548/7568 to 7821, and the 3′ region, from nt 8349 to 8587, respectively, in the HERV17 consensus sequence included in Repbase (36). In the case of 5′ env, combinations of two partially overlapping forward primers and two reverse primers, reflecting nucleotide differences in the primer binding regions of the various HERV-W target sequences, were used. Primer variants were combined at ratios correlating roughly with the number of potential targets for each variant. Forward primers 17_7548_for (5′-GAACAATGGAACAACTTCAGCAC-3′) and 17_7568_for (5′-GCACAGAAATAAACACCACTTCC-3′) and reverse primers 17_7821_rev (5′-CACTAAGAATGAGAGGAAGCAC-3′) and 17_7821_rev2 (5′-CACTAAGAATGACAGGAAGCAC-3′) were each combined in a 1:1 ratio. In the case of 3′env, combinations of 4 forward and 7 reverse primers were used. Forward primers 17_8349_for1 (5′-CCTCCTTGTTAAGTTTGTCTC-3′), 17_8349_for2 (5′-CCTCCTTGTTAACTTTGTCTC-3′), 17_8349_for3 (5′-CCTCCTTGTTAAGTTTGTCTT-3′), and 17_8349_for4 (5′-CCTCCTTATTAAATTGGTCTC-3′) were combined in a 27:1:1:1 ratio. Reverse primers 17_8587_rev1 (5′-AACCCAAGTGCTGTTGGGGA-3′), 17_8587_rev2 (5′-AACCTAAGTGCTGTTGGGGA-3′), 17_8587_rev3 (5′-AACCCAAGTGCTGCTGGGGA-3′), 17_8587_rev4 (5′-AACCCAACTGCTGTTGGGGA-3′), 17_8587_rev5 (5′-AACCCAAGTGCTTTTGGGGA-3′), 17_8587_rev6 (5′-AACCAAAGTGCTGTGGGGGA-3′), and 17_8587_rev7 (5′-AACTCAAGTGCTGTTGGGGT-3′) were combined in a 54:1:1:1:1:1:1 ratio. The PCR mixture contained forward and reverse primer mixtures at 0.4 μM each, 2.5 U Hot FirePol DNA Polymerase (Solis Biodyne, Tartu, Estonia), 2.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP), 1× PCR buffer B1 (as provided by the manufacturer), and 1 μl of cDNA in a 30-μl reaction mixture. A water control was included, as well as a negative control for each sample, containing 1 μl of the respective RT-lacking reaction mixture as the template (see reference 35). The cycling conditions were as follows: initial denaturation for 15 min at 95°C; 30 to 35 cycles of 1 min at 95°C, 1 min at 53°C, and 45 s at 72°C; and final elongation for 10 min at 72°C. The low-stringency annealing temperature was expected to contribute to amplification of HERV-W loci with imperfectly matching primer regions. The RT-PCR products were separated by agarose gel electrophoresis and purified using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The RT-PCR products were subsequently sequenced using 454/FLX technology (454 Life Sciences, Branford, CT, USA). Samples MS1 to MS3 and H1 to H3 and samples MS4 to MS7 and H4 to H7 were analyzed in two different sequencing runs. For initial RT-PCR amplification, primers with appropriate adapter and key sequences added to the 5′ ends were used. In a first run, adapter sequences 5′-CGTATCGCCTCCCTCGCGCCATCAG-3′ and 5′-CTATGCGCCTTGCCAGCCCGCTCAG-3′ were used for the forward and reverse primers, respectively. For the second run, adapter sequences 5′-CCATCTCATCCCTGCGTGTCTCCGACGACT-3′ and 5′-CCTATCCCCTGTGTGCCTTGGCAGTCGACT-3′ were used. The key sequences are underlined. 454/FLX sequencing was performed at the Department of Epigenetics, Saarland University (Saarbrücken, Germany), using a Roche GS FLX Titanium sequencer. RT-PCR products from samples MS4 to MS7 and H4 to H7 were also sequenced, using a MiSeq sequencer (Illumina, San Diego, CA, USA). For initial RT-PCR amplification, primers without additional adapter sequences were used. MiSeq library preparation and sequencing was performed by Seq-IT GmbH (Kaiserslautern, Germany).
PCR amplification from genomic DNA.
PCR primers for 5′ env and 3′ env amplicons, with or without FLX adaptors, were verified on genomic DNA. The PCR mixture contained forward and reverse primer mixtures at 0.5 μM each, 1 U recombinant Taq DNA polymerase (Invitrogen, Life Technologies, Carlsbad, CA, USA), 1.5 mM MgCl2, 0.2 mM each dNTP, 1× PCR buffer, and 1 μl of cDNA in a 20-μl reaction mixture. A water control was included. The cycling conditions were as follows: initial denaturation for 3 min at 95°C; 40 cycles of 50 s at 95°C, 50 s at 53°C, and 1 min at 72°C; and final elongation for 10 min at 72°C. The PCR products were cloned into the pGEM-T Easy vector (Promega, Fitchburg, WI, USA), ligations were transformed into chemocompetent Escherichia coli DH5α cells, insert-containing clones were identified by colony PCR using vector-specific M13 primers, and plasmid DNA of positive clones was isolated in a 96-well format using the Agencourt CosMCPrep Kit (Beckman Coulter Genomics, Danvers, MA, USA). The sequences of cloned PCR products were generated by Seq-IT GmbH (Kaiserslautern, Germany) using the vector-specific T7 primer and an Applied Biosystems 3730 DNA Analyzer.
Assignment of cDNA sequences to genomic HERV-W loci.
For each sample and amplicon, the generated 454/FLX sequences were multiply aligned using MAFFT (37), and poor-quality reads, as well as short sequences and primer dimers, were excluded from further analysis. PCR primer sequences were removed from cDNA sequences using Geneious software (Biomatters Ltd., Auckland, New Zealand). In the case of MiSeq, short sequences were filtered, and primer portions were removed using various tools provided by the Galaxy public server (38–40). Trimmed cDNA sequences were assigned to genomic HERV-W loci by employing a local BLAT installation (41) and the human NCBI36/hg18 reference genome sequence (42). A cDNA sequence assignment was defined as unambiguous if there was only one best match with less than three mismatches to the corresponding genomic sequence and a second-best match displaying at least one more mismatch. Sequences with more than three mismatches were excluded from analysis, allowing up to ∼1% sequence difference due to RT-PCR and sequencing errors and interindividual differences. Relative cDNA frequencies were calculated for each HERV-W locus and sample based on the number of cDNA sequences assigned to a locus in a given sample relative to the total number of cDNA sequences assigned to loci in that sample. Potential differences in observed cDNA frequencies per locus were statistically tested by Wilcoxon-Mann-Whitney tests by calculating adjusted probability values.
qRT-PCR.
Real-time PCR was used to quantify HERV-W transcription. The quantitative RT-PCR (qRT-PCR) system, master mixture, and cycling conditions were as previously described (35). For HERV-W group-specific amplifications, the above-described 5′ env PCR primer mixtures were used. For ERVWE-1/ERVW-1/syncytin-1-specific amplification primers syncytin-1_for (5′-TTCACTGCCCACACCCAT-3′) (20) and syncytin-1_rev (5′-CCCCATCAGACATACCAGTT-3′) (43) were used to generate a 169-bp product. For ERVWE2/ERVW-2-specific amplification, primers Xq22.3_for (5′-GCTGCTGTACAACCAGTAGCTC-3′) and Xq22.3_rev (5′-TTCTCTTGCCTGACCTTGAAT-3′) (13) were used to generate a 305-bp product. The specificity of primer pairs was verified by cloning of PCR products amplified from genomic DNA and sequencing of a number of randomly selected clones (J. Mayer, G. Laufer, and K. Ruprecht, unpublished results). Normalization of HERV-W, ERVWE-1/ERVW-1/syncytin-1, and ERVWE2/ERVW-2 transcript levels and further analysis of measured cDNA levels were done using StepOne Software v2.2.2 (Applied Biosystems, Life Technologies) and as previously described (35). Potential differences in observed relative transcript levels were statistically tested by Wilcoxon-Mann-Whitney (WMW) tests and by calculating adjusted probability values.
Reporter gene assays of HERV-W LTRs.
5′ and 3′ LTRs of selected transcribed HERV-W elements were assayed for promoter activity by employing a luciferase-based reporter system. Selected HERV-W LTRs were amplified from genomic DNA, yielding PCR products between 203 bp and 919 bp in length for the various loci (primer sequences are available on request). The PCR mixture contained forward and reverse primers at 0.5 μM each, 2.5 U Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), 1.5 mM MgCl2, 0.2 mM each dNTP, 1× PCR buffer, and 50 ng DNA in a 50-μl reaction mixture. A water control was included. The cycling conditions were as follows: initial denaturation for 5 min at 95°C; 40 cycles of 50 s at 95°C, 50 s at 54°C, and 45 s at 72°C; and final elongation for 10 min at 72°C. The PCR products were cloned into the pGEM-T Easy vector. The cloned HERV-W LTR-harboring PCR products were released by EcoRI or BamHI restriction digestion and cloned into pGLuc-Basic (NEB, Ipswich, MA, USA). The empty pGLuc-Basic vector served as a negative control, and pCMV-GLuc, harboring a strong cytomegalovirus (CMV) promoter, served as the positive control. JEG-3 cells were seeded into 12-well plates at 6 × 104 cells per well and cultured at 37°C in a humidified 5% (vol/vol) CO2 atmosphere for 24 h. The cells were cotransfected with 1 μg of a pGLuc-LTR construct, or control vector, and 0.2 μg of pCMVβ vector (Clontech Laboratories, Mountain View, CA, USA) each for normalization purposes, using TurboFect Transfection Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Luciferase and β-galactosidase assays were performed 48 h after transfection using the BioLux Gaussia Luciferase Assay Kit (NEB, Ipswich, MA, USA) and the β-Galactosidase Enzyme Assay System with Reporter Lysis Buffer (Promega, Fitchburg, WI, USA) according to the manufacturers' instructions. Briefly, the medium was removed and the cells were washed with 1 ml of 1× DPBS (Gibco, Life Technologies, Carlsbad, CA, USA) per well. The cells were lysed in 300 μl 1× Luciferase Cell Lysis Buffer (NEB, Ipswich, MA, USA) per well, and the lysates were transferred into 1.5-ml Eppendorf tubes, followed by vortexing of the lysates for 15 s each and pelleting of cellular debris for 1 min at 14,000 rpm in a tabletop centrifuge. Actual luciferase and β-galactosidase measurements were performed using 5 μl and 100 μl, respectively, of cell lysate each, a Berthold Technologies Lumat LB 9507 luminometer, and a NanoDrop ND-2000 (peqlab, Erlangen, Germany) photometer. All plasmid constructs were measured in triplicate, and the experiment was repeated three times.
Nucleotide sequence accession numbers.
The cDNA sequences for the HERV-W loci discussed in this paper have been registered with the EMBL database under accession numbers HG421036 to HG421067.
RESULTS
An optimized HERV-W amplification strategy.
Transcription of HERV-W, and of specific HERV-W loci in particular, appears to be an unresolved issue, especially in the context of MS. We therefore aimed to detect transcribed HERV-W loci comprehensively. The RT-PCR primers employed for HERV-W cDNA generation were optimized for amplification of as many HERV-W loci as possible in a straightforward RT-PCR experiment. HERV-W loci were often formed by L1-mediated retrotransposition, and therefore, they often lack 5′ proviral regions of variable length and the U5 region of the 3′ LTR (6, 7). For an optimized amplification strategy, we compiled HERV-W sequences from the hg18 human reference genome sequence and generated a multiple alignment of 176 genomic loci harboring, in particular, HERV-W env sequence portions, as that proviral 3′ region is most often present in HERV-W loci. A previous study by Laufer et al. (20) had investigated HERV-W transcription in PBMC of MS patients and healthy controls by employing an ∼690-bp amplicon located in the 5′ region of HERV-W env. That amplicon region was not entirely present in several HERV-W loci. We therefore designed a shorter PCR amplicon, approximately 280 bp in length, overlapping the Laufer et al. (20) amplicon's 3′ region but amplifying a greater number of HERV-W loci. However, many HERV-W loci display a much longer 5′ truncation and therefore lack that amplicon region within env (see Fig. S1 in the supplemental material). We therefore designed a second PCR amplicon approximately 330 bp in length and located further downstream in the HERV-W env 3′ region. To further ensure amplification of as many HERV-W loci as possible, we used mixtures of several forward and reverse primers representing major sequence variants of HERV-W loci within primer binding regions (35). Assuming amplification of HERV-W loci with up to two mismatches in the primer binding region (not located at the primer's 3′ end), our primer design should, in principle, be able to amplify at least 23 HERV-W loci for the 5′ env amplicon and 147 HERV-W loci for the 3′ env amplicon, with an overlap of 20 HERV-W loci amplifiable by both amplicons. To verify unbiased PCR amplification of HERV-W loci, 5′ env and 3′env amplicons were amplified from genomic DNA, the PCR products were cloned, and randomly selected clones were sequenced. Primers with and without FLX adaptors (see below) were used in the control experiment to further exclude adaptor interference during PCRs. A total of 136 5′ env amplicon-derived and 149 3′ env amplicon-derived sequences could be assigned unambiguously to genomic HERV-W loci. Twenty-two and 83 different HERV-W loci were identified for the 5′ env and 3′ env amplicons, respectively. The relative cloning frequencies of the amplified loci did not indicate biased amplification of specific HERV-W loci, arguing against preferential amplification of some HERV-W loci.
Identification of transcribed HERV-W loci in MS and control brain samples using next generation sequencing.
RT-PCR products were generated from total RNA isolated from postmortem samples of 7 MS lesions from 6 patients with MS and white matter brain tissue samples from 7 healthy controls (Table 1). The absence of genomic DNA was verified subsequent to rigorous DNase treatment of the RNA. RT-PCR from total RNA also included controls for DNA contamination during PCR. We then subjected the PCR products to next-generation amplicon sequencing using FLX/454 technology. In total, nearly 62,000 454/FLX sequence reads were generated from the 14 samples investigated (see below for the Illumina/MiSeq data set). We further curated the sequence data set by filtering for artifactual short sequencing reads and removal of primer sequences. For the 454/FLX data set, ∼56,500 sequence reads were assigned to genomic HERV-W loci located in the human reference genome sequence (hg18/NCBI Build 36.1) using local BLAT (41). HERV-W locus sequences differ by ∼5% from each other, making assignment of cloned HERV-W sequences to specific HERV-W loci reasonably reliable. For the 5′ env amplicon, an average of 74% (maximum, 79%; minimum; 66%) of sequences amplified from the 14 brain tissue samples displayed zero mismatches to the best BLAT match, 18% (maximum, 24%; minimum, 14%) of the sequences displayed 1 mismatch, 6% (maximum, 11%; minimum, 3%) of the sequences displayed 2 mismatches, and 1% (maximum, 2%; minimum, 1%) of the sequences displayed 3 mismatches. The numbers for the 3′ env amplicon were very similar: 69% (maximum, 84%; minimum, 60%) of the sequences displayed zero mismatches to the best BLAT match, 23% (maximum, 30%; minimum, 12%) of the sequences displayed 1 mismatch, 6% (maximum, 10%; minimum, 2%) of the sequences displayed 2 mismatches, and 2% (maximum, 4%; minimum, 1%) of the sequences displayed 3 mismatches. Between 0% and 4% of the sequences displayed 4 or more mismatches to the best BLAT match. Nevertheless, we applied a rather stringent mapping strategy. Sequence reads with greater than 3 mismatches to the best BLAT match were excluded from analysis, allowing only up to ∼1% sequence differences due to PCR and sequencing errors and presumably sometimes single-nucleotide or other polymorphisms present in HERV-W loci. In total, ∼92% of all generated sequences could be unambiguously mapped to a specific HERV-W locus (Table 2; see Table S1 in the supplemental material). We interpret the remaining 8% of unassigned sequences as reads with unrecognized sequencing artifacts or potential recombinants of transcripts from different HERV-W loci generated during RT-PCR (20, 44).
Table 2.
Transcribed HERV-W loci and relative cDNA sequence frequencies of specific HERV-W loci in MS and control brain tissue samplesa
| Chrb | Positionb |
Bandc | HGNC: ERVWc | Strande | 5′ LTRe | 3′ LTRe | Pr. ps.g | Relative frequency (%) of cDNA sequences assignable unambiguously to the respective HERV-W locush |
No. of reads in Caltech RNA-Seq data set for cell linei: |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | MS1 | MS2 | MS3 | MS4 | MS5 | MS6 | MS7 | H1 | H2 | H3 | H4 | H5 | H6 | H7 | Mean | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||||||
| 1 | 42182714 | 42188567 | 1p34.2 | + | − | Δ325–780 | + | 1.36 | 1.72 | 0.89 | 0.05 | 0.16 | 1.13 | 0.13 | 0.89 | 0.12 | 0.72 | 1 | 5 | 2 | 1 | 3 | 1 | |||||||
| 1 | 55149269 | 55157786 | 1p32.3B | − | + | + | 0.19 | 0.42 | 0.14 | 1.54 | 0.02 | 0.05 | 0.12 | 0.35 | 5 | 10 | 1 | 3 | 4 | 7 | ||||||||||
| 1 | 210095762 | 210098886 | 1q32.3 | − | − | Δ325–780 | + | 0.32 | 0.07 | 0.10 | 0.14 | 0.06 | 0.05 | 0.12 | 0.12 | 2 | 2 | 5 | ||||||||||||
| 2 | 136675926 | 136677399 | 2q22.1 | − | − | Δ326–780 | + | 0.05 | 0.05 | 6 | 1 | 2 | ||||||||||||||||||
| 2 | 175898116 | 175899503 | 2q31.1B | + | − | Δ325–780 | + | 0.03 | 0.03 | 1 | 1 | |||||||||||||||||||
| 3 | 27017519 | 27022905 | 3p24.1 | + | Δ1–261 | − | 0.46 | 0.23 | 0.22 | 0.32 | 0.05 | 0.14 | 0.24 | 2 | 1 | 1 | 1 | 4 | ||||||||||||
| 3 | 97868008 | 97876978 | 3q11.2 | − | Δ774–780 | Δ774–780 | 0.19 | 0.05 | 0.05 | 0.03 | 0.04 | 0.17 | 0.09 | 3 | 2 | 1 | 2 | 1 | 3 | |||||||||||
| 3 | 143021321 | 143023045 | 3q23A | 3 | + | − | Δ327–780 | + | 0.02 | 0.19 | 0.20 | 0.14 | 0.06 | 0.12 | 1 | 2 | ||||||||||||||
| 4 | 74010156 | 74017767 | 4q13.3 | − | + | + | 0.17 | 0.23 | 0.02 | 0.15 | 0.14 | 3 | 1 | 3 | 2 | 2 | 5 | 1 | ||||||||||||
| 4 | 139762390 | 139767803 | 4q31.1 | − | Δ140–780 | + | 0.61 | 0.65 | 1.01 | 0.35 | 0.97 | 1.21 | 0.74 | 0.14 | 0.71 | 1 | 1 | 3 | 1 | |||||||||||
| 4 | 165796233 | 165798754 | 4q32.3 | + | − | Δ327–780 | + | 0.58 | 0.19 | 0.19 | 2.16 | 2.91 | 0.04 | 0.13 | 1.68 | 0.99 | 1 | 2 | 1 | 1 | ||||||||||
| 5 | 44146946 | 44150936 | 5p12 | + | + | + | 0.02 | 0.03 | 0.03 | 1 | 1 | 2 | 2 | 1 | ||||||||||||||||
| 5 | 56851606 | 56854473 | 5q11.2d | − | − | Δ325–780 | + | 0.80 | 1.35 | 2.35 | 0.43 | 0.49 | 0.25 | 0.06 | 0.05 | 0.14 | 0.17 | 0.61 | 1 | 5 | 1 | 10 | 1 | 1 | ||||||
| 5 | 107937705 | 107942182 | 5q21.3 | − | − | + | 0.10 | 0.09 | 0.34 | 0.18 | 2 | 1 | 3 | 2 | ||||||||||||||||
| 6 | 106782704 | 106790382 | 6q21Ad | 17 | + | Δ257–780 | Δ327–780 | + | 13.15 | 10.34 | 13.66 | 12.52 | 8.75 | 11.15 | 33.27 | 14.07 | 9.84 | 8.57 | 8.06 | 11.79 | 9.03 | 6.77 | 12.21 | 3 | 11 | 3 | 8 | 2 | 3 | 13 |
| 7 | 91935248 | 91945442 | 7q21.2d | 1 | − | + | Δ707–780 | 38.44 | 44.20 | 35.98 | 31.72 | 40.90 | 38.74 | 24.72 | 38.55 | 46.95 | 52.31 | 58.42 | 56.6 | 59.66 | 54.82 | 44.43 | 11 | 3 | 11 | 10 | 5 | 8 | 7 | |
| 9 | 113138513 | 113140280 | 9q31.3 | + | - | Δ325–780 | + | 0.42 | 0.60 | 0.95 | 1.21 | 0.39 | 1.08 | 0.42 | 0.43 | 0.24 | 1.47 | 0.17 | 0.67 | |||||||||||
| 11 | 9325928 | 9327740 | 11p15.4 | − | − | Δ325–780 | + | 0.14 | 0.02 | 0.08 | 1 | 4 | 1 | 2 | 2 | |||||||||||||||
| 12 | 49582525 | 49593417 | 12q13.13 | 24 | − | +f | + | 0.22 | 1.39 | 1.03 | 0.60 | 0.91 | 2.16 | 2.12 | 1.14 | 0.95 | 1.29 | 1.18 | 2.73 | 0.85 | 1.27 | 2 | 5 | 2 | 4 | 4 | 5 | |||
| 14 | 44558438 | 44562647 | 14q21.3d | 26 | − | − | Δ327–780 | + | 20.57 | 24.91 | 21.25 | 29.23 | 26.11 | 18.07 | 16.85 | 24.89 | 26.21 | 22.19 | 26.19 | 20.75 | 20.38 | 17.94 | 22.54 | 6 | 1 | 2 | 2 | 3 | ||
| 15 | 53384371 | 53391866 | 15q21.3d | 4 | − | Δ257–780 | Δ327–780 | + | 9.14 | 4.64 | 9.08 | 15.06 | 9.66 | 12.99 | 15.62 | 7.5 | 4.63 | 2.68 | 0.58 | 3.3 | 0.84 | 2.2 | 6.99 | 16 | 17 | 7 | 5 | 10 | 14 | 15 |
| 17 | 32764002 | 32767735 | 17q12Bd | 28 | + | − | Δ327–780 | + | 1.19 | 2.74 | 3.31 | 0.55 | 0.10 | 0.43 | 1.15 | 1.88 | 1.65 | 2.1 | 1.58 | 0.59 | 1.44 | 1 | 1 | 1 | 1 | 2 | ||||
| 20 | 53398912 | 53402997 | 20q13.2 | + | − | Δ327–780 | + | 0.05 | 0.05 | 3 | 9 | 2 | 3 | 1 | ||||||||||||||||
| X | 7627209 | 7632109 | Xp22.31 | − | Δ1–255 | Δ325–780 | + | 0.29 | 0.05 | 0.05 | 0.12 | 0.13 | 3 | 2 | 2 | 1 | 2 | |||||||||||||
| X | 106182016 | 106184757 | Xq22.3Bd | 2 | − | − | Δ327–780 | + | 12.18 | 6.26 | 9.73 | 8.58 | 12.07 | 13.53 | 3.19 | 7.84 | 6.85 | 8.94 | 3.02 | 5.07 | 4.2 | 16.92 | 8.46 | 2 | 8 | 2 | 43 | 8 | 9 | 2 |
| Total no, of readsj | 4,123 | 2,156 | 4,174 | 2,005 | 1,988 | 924 | 3,637 | 5,572 | 1,576 | 1,902 | 695 | 848 | 476 | 591 | 68 | 86 | 40 | 102 | 50 | 67 | 72 | |||||||||
| Total no. of locik | 20 | 18 | 18 | 14 | 12 | 10 | 13 | 17 | 14 | 14 | 11 | 12 | 8 | 9 | 18 | 19 | 13 | 20 | 15 | 19 | 19 | |||||||||
The numbers presented are based on the 5′ env amplicon-454/FLX data set; loci identified by the 3′ env data set were not included because relative cDNA frequencies would be misleading, as many more loci were identified by the 3′ env amplicon (see the supplemental material).
Chromosomal locations of transcribed HERV-W loci are given with respect to the NCBI36/hg18 human reference genome sequence as provided by UCSC Genome Browser (42).
Chromosomal-band and HGNC-approved designations of HERV-W loci are listed. Loci with chromosomal locations shown in italics have been identified in both the 5′ env and 3′ env data sets. Uppercase letters appended to chromosomal bands indicate more than one HERV-W locus in that chromosome band.
Locus previously identified as transcribed in PBMC from patients with MS and healthy controls (20).
Orientations of HERV-W loci in the respective chromosomes (Strand), as well as the structures of the 5′ and 3′ LTRs, with pluses and minuses meaning that an LTR is present or lacking, respectively, in its full length and numbers indicating the deleted regions with respect to the HERV-W LTR17 reference sequence as provided by Repbase (36).
A MER11A element is inserted into the LTR.
HERV-W loci as processed pseudogenes (Pr. ps.) generated by L1-mediated retrotransposition (+).
Note that the mean percentages for some loci display a somewhat wider range for some brain tissue samples, which may indicate interindividual transcription level differences. (However, also compare the overall close correspondence of 454/FLX and Illumina results shown in Fig. 3). The percentages are rounded; for instance, 0.02% of 4,123 reads means that 1 read was assigned to that locus in sample MS1. Mean number of transcribed HERV-W loci per sample: 15 for MS samples and 12 for control samples.
Total number of RNA-Seq reads from 7 different cell lines mapped to the respective HERV-W loci in data sets generated by ENCODE (see the text). The numbers represent cell lines as follows: 1, GM12878; 2, 1 H1 hESC; 3, HUVEC; 4, HeLa-S3; 5, Hep G2; 6, K562; 7,NHEK.
Total number of assigned FLX sequence reads per sample.
Total number of HERV-W loci detected as transcribed per sample.
Transcribed HERV-W loci based on the 454/FLX data set.
Based on the 454/FLX data set, we found evidence for transcription of, in total, approximately 140 HERV-W loci in the brain tissue samples investigated. Twenty-five different HERV-W loci were identified as transcribed by the 5′ env amplicon (Fig. 1 and Table 2) and 137 different HERV-W loci by the 3′ env amplicon (Fig. 2). Fourteen loci were detected by both amplicons. The overall number of transcribed loci did not vary strikingly between MS and control samples. The 5′ env amplicon detected an average of 15 different loci as transcribed in MS samples and 12 in healthy-control samples. The 3′ env amplicon detected an average of 68 active loci in MS samples and 60 in healthy-control samples. In accordance with an initiative by the HGNC (34), we assigned official designations to an additional 23 HERV-W loci transcribed at higher relative levels, officially named ERVW-7 to ERVW-29, that will be given in addition to chromosomal locations in this paper.
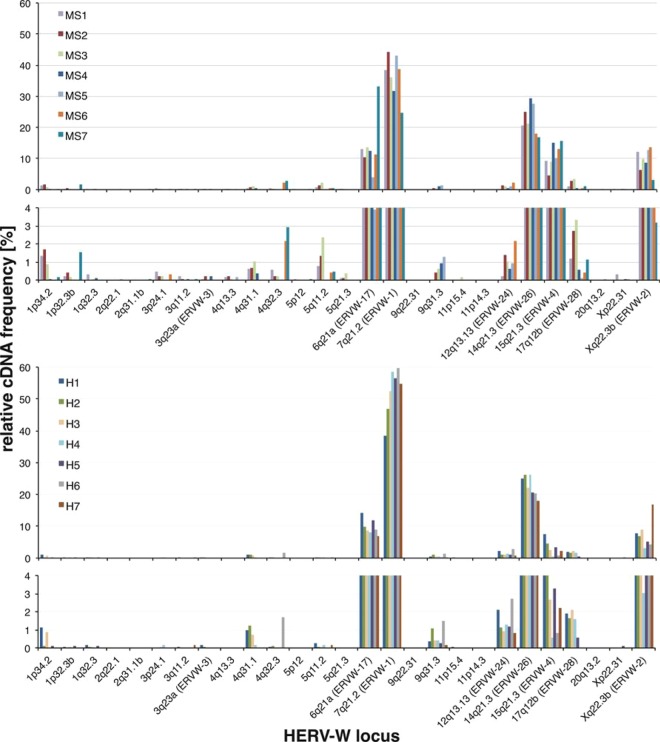
Fig 1.
Relative transcript levels of HERV-W loci in MS and healthy brain tissue samples. Shown are relative transcript levels of specific HERV-W loci in the various MS-derived and healthy brain tissue samples based on total numbers of cDNA sequences assignable to a locus relative to the total number of assignable cDNA sequences per sample. The numbers presented are from the 5′ env 454/FLX data set. The results from MS-derived (MS1 to MS7) and healthy-control (H1 to H7) brain tissue samples are presented in separate graphs that are further divided into sections depicting HERV-W loci with higher (top) and lower (<4%; bottom) relative cDNA sequence frequencies and thus transcript levels. HERV-W loci are designated according to their locations in chromosomal bands and HGNC-approved locus designations (Table 2; see the text and supplemental material).
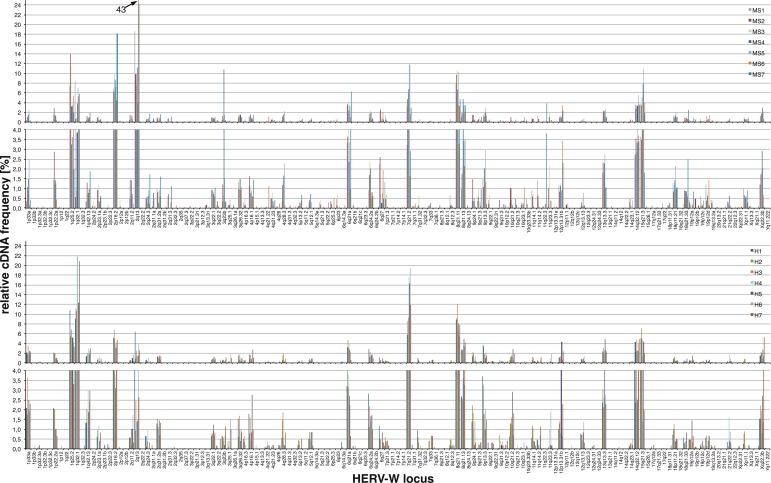
Fig 2.
Relative transcript levels of HERV-W loci in MS and healthy brain tissue samples based on the 3′ env amplicon data set. Depicted are relative transcript levels of specific HERV-W loci in MS lesion-derived and healthy brain tissue samples, basically as shown in Fig. 1 and the supplemental material.
Relative cDNA sequence frequencies, which roughly reflect the relative transcription levels of corresponding HERV-W loci, were calculated based on cDNA sequences assigned to a particular locus in a particular sample relative to the total number of locus-assignable cDNA sequence reads in that sample. Relative cDNA sequence frequencies, and thus relative transcript levels, of specific HERV-W loci differed significantly. Most HERV-W loci, for instance, loci located in chr. 2q22.1, 11p15.4, and 20q13.2, seem to be transcribed at low or very low levels, with few or very few cDNA sequences (sometimes only one) assigned to a locus in only a few of the samples. In contrast, the majority of HERV-W transcripts present in the various tissue samples always appeared to be derived from a few HERV-W loci displaying significantly higher relative cDNA sequence frequencies. More specifically, for the 5′ env amplicon and the 454/FLX data set, ∼96% of assignable cDNA sequences were derived from only 7 different HERV-W loci. For the 3′ env amplicon, ∼83% of cDNA sequences were derived from 25 HERV-W loci. For both 5′ env and 3′ env, all the remaining loci contributed, in total, less than 4% and 17%, respectively, of the cDNA sequence, and each locus less than 1%.
As for transcription levels of specific HERV-W loci, in regard to the 5′ env amplicon, the ERVWE1/ERVW-1/syncytin-1 locus appears to be transcribed at the highest level in all the brain tissue samples investigated, followed by HERV-W loci located in chr. 14q21.3/ERVW-26, 6q21A/ERVW-17, 15q21.3/ERVW-4, and the ERVWE2/ERVW-2 locus in chr. Xq22.3B, each of which displays an intermediate transcript level. Note that chromosomal-band designations with a letter appended indicate two or more HERV-W loci in that band (Table 2).
Overall similar findings were obtained for the 3′ env amplicon, though relative cDNA sequence frequencies differed somewhat, very likely due to the much greater number of detectable HERV-W loci (see above), also resulting in overall less pronounced differences in relative cDNA frequencies. A HERV-W locus in chr. 2q13/ERVW-13 displayed the highest relative frequency (∼9.8%), followed by loci in chr. 8q21.11/ERVW-20, 1q25.2/ERVW-9, 2p16.2/ERVW-12, 7q21.2/ERVWE1/ERVW-1/syncytin-1, 1q32.1/ERVW-10, and 15q21.3/ERVW-4 (∼9%, 8.2%, 7.6%, 7%, 6.1%, and 5%, respectively). Approximately 18 additional loci displayed relative frequencies between 3.6% and 1%, among them the ERVWE2/ERVW-2 locus in chr. Xq22.3B (1.8%). Another ∼112 loci displayed relative frequencies of <1% each.
Several of the more highly transcribed loci have previously been identified as transcribed in PBMC of patients with MS (20), specifically, HERV-W loci located in chr. 6q21A/ERVW-17, 14q21.3/ERVW-26, and 15q21.3/ERVW-4, and ERVWE1/ERVW-1/syncytin-1 located in 7q21.2 and ERVWE2/ERVW-2 in chr. Xq22.3B.
We next examined potentially different HERV-W transcription patterns between MS and control brain tissue samples. Very few HERV-W loci were identified exclusively in MS or control samples. For instance, HERV-W loci located in chr. 2q22.1, 2q31.1B, and 5p12 were found to be transcribed exclusively in some of the MS-derived samples, and a HERV-W locus in chr. 9q22.31 was found to be transcribed exclusively in one control sample. However, corresponding relative cDNA sequence frequencies, and thus presumably the transcription levels of those loci, appear to be very low; each of the samples showed a somewhat different transcription pattern of low-level-transcribed loci, and there was no strikingly different transcription pattern distinguishing MS samples from control samples. Of further note, some of the low-level-transcribed HERV-W loci displayed slightly higher relative transcript levels in a few samples. For instance, a locus in chr. 5q11.2 displayed relative cDNA sequence frequencies between 0.8% and 2.4% in three MS lesions, while relative transcript levels were generally lower in the healthy control samples. We currently interpret such seemingly sample- and locus-specific minor differences in our data set as stochastic phenomena very likely due to low cDNA sequence frequencies of corresponding loci.
More importantly in the context of the potential involvement of HERV-W sequences in MS, in regard to the HERV-W loci displaying higher relative transcription levels, our analysis demonstrated overall very similar relative transcript levels between MS and control brain tissue samples (Fig. 1 and Table 2; see Tables S1 and S2 in the supplemental material).
There were a few possible exceptions to overall very similar HERV-W locus transcription patterns. A HERV-W locus in chr. 15q21.3/ERVW-4 displayed seemingly higher relative cDNA frequencies in MS samples than in healthy tissue controls in the 454/FLX data sets generated for the 5′ env amplicon, with mean cDNA frequencies of 10.88% and 3.1% in MS and healthy-control samples, respectively. Those differences were statistically significant for the 5′ env data set (WMW test; adjusted P = 0.0082). However, there was no such significantly different cDNA frequency for the respective 3′ env data sets (WMW test; adjusted P = 1). Seemingly lower relative cDNA frequencies in MS than in healthy control samples for the ERVWE1/ERVW-1/syncytin-1 locus (mean values, 36.39% and 52.47%, respectively) were likewise significant for the 5′ env 454/FLX data set (WMW test; adjusted P = 0.014) but not statistically corroborated by the 3′ env data set (mean MS, 5.25%; healthy controls, 12.72%; WMW test; adjusted P = 0.069). Finally, for the 3′ env data set, cDNA frequencies of a HERV-W locus in chr. 1q32.1/ERVW-10 were significantly higher in healthy controls than in MS samples (14.02% versus 4.55%; WMW test; adjusted P = 0.015), and the cDNA frequencies of a locus in chr. 2q13/ERVW-13 were significantly lower in healthy controls than in MS samples (16.97% versus 2.47%; WMW test; adjusted P = 0.015). Furthermore, a HERV-W locus in chr. 6q21A/ERVW-17 displayed a considerably higher relative cDNA frequency in one MS-derived sample (MS7; 33%; seen in contrast to a mean frequency of ∼11.6% in the other MS-derived samples and 9.7% in the healthy-control samples in the 454/FLX 5′ env data set [Fig. 1]). Similar numbers were found for the Illumina/MiSeq data set (see below), specifically, 24% in MS7 versus 11.5% in the other MS-derived samples and 12.75% in the healthy-control samples.
Identification of transcribed HERV-W loci based on the Illumina/MiSeq data set.
Besides amplicon sequencing employing 454/FLX technology, we also subjected RT-PCR products generated from four MS-derived brain tissue samples (MS4 to MS7) and four normal brain tissue samples (C4 to C7) to amplicon sequencing using Illumina/MiSeq sequencing technology. In total, >8.5 million cDNA sequence reads were generated from the eight samples. Similar to the 454/FLX strategy, short (<185-bp) sequence reads and reads with more than one best match or more than 3 mismatches to their best BLAT match were excluded, leaving ∼6.2 million sequence reads for subsequent analysis (see Table S2 in the supplemental material).
The results for the Illumina/MiSeq data set were overall very similar to the results from the 454/FLX experiment (Fig. 3). A total of 183 different HERV-W loci were identified as transcribed in the 8 tissue samples investigated. Twenty-seven different HERV-W loci were identified by the 5′ env amplicon and 176 HERV-W loci by the 3′ env amplicon, with an overlap of 20 loci detected by both amplicons (see the supplemental material). For the 3′ env data set, an average of 124 different HERV-W loci were identified in every MS tissue and 117 in every control brain tissue sample. The overall higher number of HERV-W loci identified as transcribed is very likely due to the much higher number of generated and analyzed cDNA sequence reads compared to the 454/FLX approach.
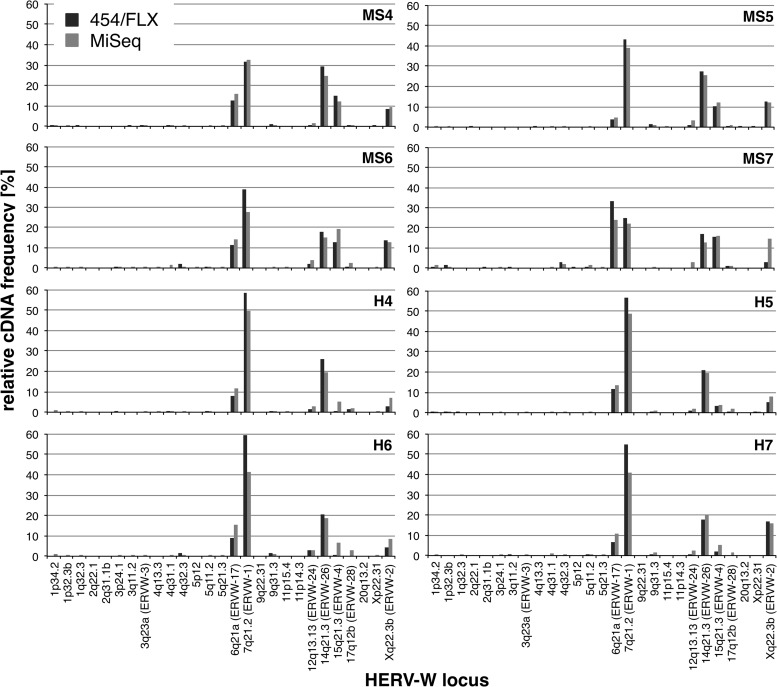
Fig 3.
No major differences were found in relative transcript levels of HERV-W loci for data sets generated by 454/FLX or Illumina/MiSeq amplicon-sequencing technology. Shown are the relative transcript levels of HERV-W loci obtained for the 5′ env data set generated by 454/FLX and Illumina/MiSeq amplicon sequencing deduced as described in the legend to Fig. 1. HERV-W loci with low relative cDNA frequencies are intentionally included to demonstrate results with little variation for those loci, as well. HERV-W loci are designated according to their locations in chromosomal bands and HGNC-approved locus designations (see the text). Detailed information on relative cDNA sequence frequencies in both data sets is provided in the supplemental material.
The relative frequencies of HERV-W loci were very similar to those obtained by the 454/FLX sequencing approach. Likewise, most HERV-W loci showed low or very low relative cDNA frequencies, and thus very low relative transcription levels, whereas the great majority of cDNA sequences were assigned to relatively few HERV-W loci that are thus transcribed at much higher relative levels (see above). HERV-W loci transcribed at higher levels matched those loci already identified by 454/FLX, including the ERVWE1/ERVW-1/syncytin-1 locus (Fig. 3). As for the 454/FLX results, the transcription patterns of HERV-W loci were overall very similar between MS-derived and control tissue samples. The Illumina/MiSeq-based analysis of transcription patterns of HERV-W loci therefore essentially replicated the findings of the 454/FLX-based analysis.
Structures and genomic locations of transcribed HERV loci.
We analyzed the structures and genomic locations of transcribed HERV-W loci. Most of the transcribed HERV-W loci are incomplete in that they represent processed pseudogenes generated by L1 retrotransposition machinery lacking the 5′ LTR U3 region or the entire 5′ LTR, or lacking proviral 5′ regions entirely, and the 3′ LTR U5 region. In contrast, about 35 transcribed HERV-W loci represent remnants of proviruses with both 5′ and 3′ LTRs. All the transcribed HERV-W loci, furthermore, harbor insertions and/or deletions of variable length, sometimes non-HERV-W repetitive elements, within the former retroviral genes and LTRs, owing to the evolutionary age of HERV-W loci (6). The ERVWE1/ERVW-1/syncytin-1 locus displaying a physiologically relevant open reading frame (ORF) is an exception to such highly defective states (Table 2; see Fig. S2 in the supplemental material).
We also analyzed the locations of transcribed HERV-W loci relative to cellular genes. Generally, the majority of transcribed HERV-W loci are not closely associated with cellular genes, that is, they are not located within gene introns and are several—up to many—kilobases distant from genes. However, a number of HERV-W loci are more closely associated with cellular genes (see the supplemental material). Some HERV-W loci are located within gene introns. For instance, the HERV-W locus in chr. 6q21A/ERVW-17, displaying a mean cDNA frequency of ∼12% for 5′ env in the 454/FLX data set, is located within an ∼46-kb intron of the ATG5 gene. Another locus in chr. 14q21.3/ERVW-26 identified as transcribed by the 5′ env 454/FLX data set, displaying a mean relative cDNA frequency of ∼23%, is located within a 16-kb intron of the FAM179B gene. Notably, an alternative splice variant of that gene, as indicated by an mRNA (GenBank accession number CR749557), harbors a 65-bp-long HERV-W portion. That HERV-W locus, therefore, provides splice donor and acceptor signals for the gene. Of further note, a low-level-transcribed HERV-W locus in chr. 22q22.2, identified by the 3′ env 454/FLX data set, comprises the first two exons, including the 5′ UTR and coding sequence starting in the first exon, of the IGSF5 gene. A transcribed HERV-W locus in chr. 1q25.2/ERVW-9 (3′ env; 454/FLX; ∼8% cDNA sequence frequency) is located within a 189-kb intron of the RASAL2 gene. A transcribed HERV-W locus in chr. 1q32.1/ERVW-10 (3′ env; 454/FLX; ∼6%) is located within an ∼28-kb intron of the LOC284581 gene, and another transcribed HERV-W locus in chr. 2p16.2/ERVW-12 (3′ env; 454/FLX; ∼7.6%) is located within an ∼14-kb intron of the ASB3 gene. Several low-level-transcribed HERV-W loci are also located within gene introns; for instance, a locus in chr. 1p34.2 (5′ env; 454/FLX; ∼0.72%; HIVEP3), a locus in chr. 17q12 (5′ env; 454/FLX; ∼1.6%; ACACA), and a locus in chr. 8q12.3 (3′ env; 454/FLX; ∼0.2%; CYP7B1).
Some other transcribed HERV-W loci are located relatively close to cellular genes; in particular, some are located downstream of genes with respect to the gene's direction of transcription. Examples are a HERV-W locus in chr. 19p12C (0.2%; 3′ env; 454/FLX) located just 100 bp downstream of the ZNF99 gene according to the corresponding RefSeq gene annotation, a locus in chr. 14q32.12/ERVW-27 (3.4%; 3′ env; 454/FLX) located just ∼300 bp downstream of the C14orf159 gene, and a locus in chr. 3q23A/ERVW-3 (0.07%; 3′ env; MiSeq data set) located ∼2.7 kb downstream of the GRK7 gene. As also discussed recently (13), the ERVWE2/ERVW-2 locus in chr. Xq22.3B (∼8.5%; 5′ env; 454/FLX) is located ∼7 kb downstream of the RBM41 gene.
A few other transcribed HERV-W loci are sandwiched between two genes located close together that are each transcribed toward the HERV-W locus. For instance, a HERV-W locus in chr. 18p11.21/ERVW-29 (∼1.1%; 3′ env; 454/FLX) is located ∼3.9 kb and 5.5 kb downstream of the LDLRAD4 and FAM210A genes, respectively. Another HERV-W locus in chr. 2q31.2B (0.004%; 3′ env; 454/FLX) is located ∼1 kb and 130 bp downstream of the DFNB59 and FKBP7 genes, respectively.
Other evidence for transcription of HERV-W loci.
We sought independent evidence of transcription of HERV-W loci. We examined Encyclopedia of DNA Elements (ENCODE) California Institute of Technology (Caltech) transcriptome-sequencing (RNA-Seq) data sets (45, 46) included in the University of California—Santa Cruz (UCSC) Genome Browser (47). Specifically, we analyzed whether the genomic coordinates of transcribed HERV-W loci overlapped locations of mapped single-pass 75-bp RNA-Seq reads generated from poly(A)+ RNA, using the UCSC Table Browser (48). We found evidence for transcription of a number of HERV-W loci in data sets derived from GM12878, 1 H1 hESC, HUVEC, HeLa-S3, Hep G2, K562, and NHEK cells (Table 2; see the supplemental material). The absolute number of RNA-Seq reads mapping within coordinates of transcribed HERV-W loci varied between 0 and 135 for the various cell lines. For instance, between 3 (1 H1 hESC) and 11 (GM12878 and HUVEC) RNA-Seq reads mapped within the ERVWE1/ERVW-1/syncytin-1 locus chromosome coordinates. Between 5 (HeLa-S3) and 17 (1 H1 hESC) RNA-Seq reads mapped to a HERV-W locus in chr. 15q21.3/ERVW-4. Forty-three HeLa-S3-derived RNA-Seq reads mapped to the ERVWE2/ERVW-2 locus in chr. Xq22.3B. Several other HERV-W loci were represented by fewer RNA-Seq reads, but still in several cell lines each. Overall, in all loci and cell lines combined, only 5 of the HERV-W loci we identified as transcribed in brain tissue were not represented by mapped RNA-Seq reads. We also note that HERV-W loci with higher transcription levels in brain tissue, as indicated by relative cDNA sequence frequencies in our analysis, were often also represented by a higher number of RNA-Seq reads in cell lines. Thus, RNA-Seq data sets generated by the ENCODE Consortium provide additional evidence for transcription of various HERV-W loci in various cell lines not originating from brain tissue.
Quantification of HERV-W transcription by qRT-PCR.
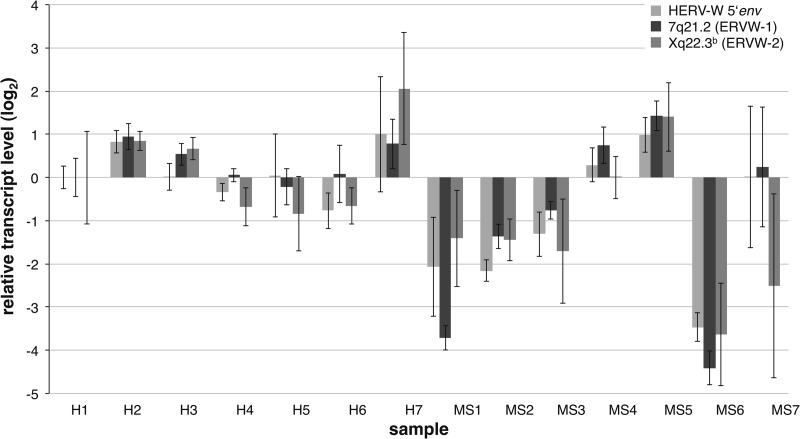
We were interested in quantifying HERV-W transcript levels in the various MS lesion and control brain tissue samples. We therefore determined for the investigated MS-derived and healthy brain tissue samples the relative HERV-W transcript levels based on the 5′ env amplicon and transcript levels of the ERVWE1/ERVW-1/syncytin-1 locus and the ERVWE2/ERVW-2 locus specifically, using semiquantitative real-time PCR. Specific detection of the ERVWE1/ERVW-1 and ERVWE2/ERVW-2 loci was done by employing previously established locus-specific primer pairs (13, 20, 43). HERV-W transcript levels were normalized to the transcript levels of G6PDH and RPII housekeeping genes. When transcript levels in healthy control tissue sample H1 were defined as a reference, most samples showed uniformly higher or lower transcript levels for all three amplicons than sample H1 (Fig. 4). However, HERV-W transcript levels varied significantly and uniformly between various individual samples, i.e., there appeared to be interindividual differences in HERV-W transcript levels. Mean transcript levels differed as much as ∼52-fold between samples. For instance, ERVWE1/ERVW-1/syncytin-1 transcript levels in MS-derived tissue sample MS6 were ∼5% those in sample H1, whereas transcript levels in MS-derived sample MS5 were ∼2,5-fold higher than in sample H1.
Fig 4.
Relative transcript levels of HERV-W and specific HERV-W loci measured by qRT-PCR. The relative levels of HERV-W transcripts were determined by semiquantitative RT-PCR for the 5′ env amplicon, which can detect transcripts/cDNA from a greater number of HERV-W loci. Transcript levels of the HERV-W loci ERVWE1/ERVW-1 and ERVWE2/ERVW-2 specifically were likewise determined using locus-specific primer sets (see the text). Relative transcript levels are given as log2-transformed fold changes, with the healthy brain tissue sample H1 set as the reference. The whiskers depict maximum and minimum changes observed in replicates of the experiment. Note that seemingly different transcript levels between healthy control and MS brain tissue sample entities are not statistically significant.
Comparing healthy brain tissue controls with MS-derived brain tissue samples, 5 MS samples displayed lower HERV-W transcript levels than were found for healthy-control samples for the 5′ env amplicon and the ERVWE1/ERVW-1/syncytin-1 and ERVWE2/ERVW-2 loci, or just the ERVWE2/ERVW-2 locus in the case of sample MS7. Three MS-derived samples displayed higher transcript levels than sample H1, yet those higher transcript levels were comparable to the transcript levels of some of the healthy-control samples (Fig. 4). However, when comparing transcript levels between the groups of MS and healthy-control brain tissue samples, there were no statistically significant differences in transcript levels (WMW tests; adjusted P > 0.22).
Taken together, our quantitation of HERV-W transcription indicated considerable interindividual variation regarding overall and locus-specific HERV-W transcript levels. Contrary to previous studies, but in accordance with our cDNA-sequencing-based description of HERV-W locus transcription patterns, we did not detect significantly upregulated transcription of the ERVWE1/ERVW-1/syncytin-1 locus or the ERVWE2/ERVW-2 locus, or generally upregulated HERV-W transcription, in MS brain lesions.
LTRs of various HERV-W loci display promoter activity.
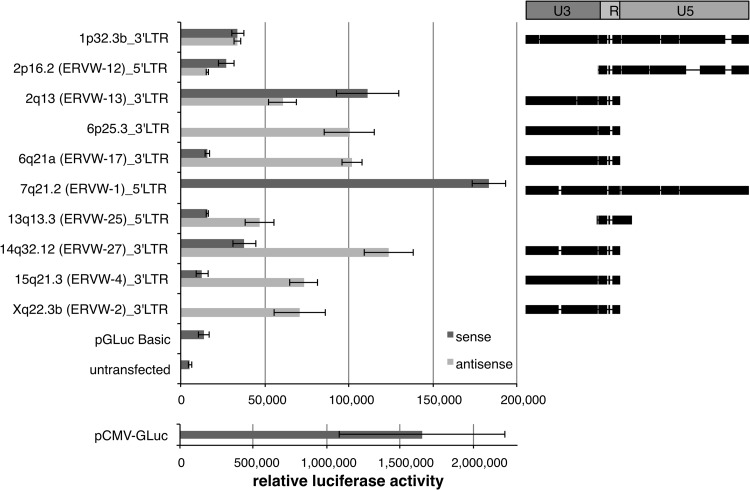
It is unclear how and where transcripts identified for various HERV-W loci were initiated. Either the HERV-W LTRs or unidentified flanking promoters initiated transcription. The HERV-W loci identified as transcribed in this study harbor both 5′ and 3′ LTRs at full length; some other loci lack the 5′ LTR U3 region, which is the promoter-housing LTR region, or they lack the 5′ LTR entirely, and some loci have only a 3′ LTR lacking U5. We therefore investigated how the various HERV-W loci identified as transcribed in our study may actually be transcribed in the genomic context, specifically, whether the respective (partial) HERV-W LTR sequences can initiate transcription. We assayed the promoter activities of various transcribed 5′ and 3′ LTRs, either full or partial length, sense and/or antisense, and including LTRs from high- and low-level-transcribed HERV-W loci. Corresponding HERV-W LTR sequences were amplified from normal genomic DNA and cloned into the promoterless pGLuc-Basic reporter gene vector. We examined the promoter activities of a total of 17 different HERV-W LTR constructs in luciferase reporter gene assays in the human choriocarcinoma cell line JEG-3, which was previously described as supporting HERV-W LTR promoter activity (49).
Both 5′ and 3′ LTRs of several of the tested HERV-W loci displayed significant promoter activity in the sense and/or antisense orientation in JEG-3 cells. Only 4 LTR constructs out of 17 did not display promoter activity significantly above the level of the promoterless pGLuc-Basic vector or untransfected negative controls. All other LTR constructs displayed promoter activity modestly or clearly higher than that of negative controls (Fig. 5). The intact ERVWE1/ERVW-1/syncytin-1 5′ LTR displayed the highest promoter activity in the sense orientation, about 10% that of the CMV-driven luciferase positive control. The 3′ LTRs of several loci displayed promoter activity in the antisense orientation. For instance, the 3′ LTRs of loci in chr. 6q21A/ERVW-17, 14q32.12/ERVW-27, and 15q21.3/ERVW-4 displayed higher antisense than sense promoter activity, or the LTR was inactive in the sense orientation. We note that all tested LTRs but the 3′ LTR of the locus in chr. 1p32.3 and the above-mentioned ERVWE1/ERVW-1/syncytin-1 5′ LTR are incomplete. For instance, the 3′ LTRs of transcribed HERV-W loci in chr. 2q13/ERVW-13, 6q21/ERVW-17, 6p25.3, 14q32.12/ERVW-17, 15q21.3/ERVW-4, and Xq22.3B/ERVWE2/ERVW-2 belong to loci generated by L1-mediated retrotransposition and therefore each lack the 3′ LTR U5 region. The 5′ LTR of the transcribed locus in chr. 2p16.2/ERVW-12 lacks the U3 region. The 5′ LTR of the locus in chr. 13q13.3/ERVW-25 consists only of a piece of U3 and the complete R region, yet it displays significant promoter activity, at least in the antisense orientation.
Fig 5.
HERV-W LTRs and remnants thereof often display promoter activity. Shown on the left is a representative result of normalized promoter activities of selected complete and incomplete HERV-W LTRs in the sense or antisense direction obtained from luciferase reporter gene assays in JEG-3 cells. LTR construct designations denote HGNC-approved locus names and the chromosomal position of a tested HERV-W 5′ or 3′ LTR, as well as the sense or antisense direction of the LTR within the luciferase reporter gene vector. A CMV-driven luciferase-expressing vector (pCMV-GLuc) served as the positive control and is presented separately to demonstrate ∼10-fold-higher promoter activity than the most active HERV-W LTR construct. A promoterless luciferase reporter vector (pGLuc-Basic) and untransfected JEG-3 cells served as negative controls. The error bars depict standard deviations observed for an experiment in triplicate. On the right are shown HERV-W LTRs or LTR portions, indicated by black bars, present within the various reporter gene constructs. HERV-W LTR sequences are depicted in comparison to the 780-bp-long HERV-W LTR17 reference sequence provided by Repbase (36). Note that some LTRs harbor insertions compared to the reference sequence. The HERV-W LTR U3 (harboring a TATA box toward the 3′ end), R (the boundaries of which define the start and endpoints of proviral transcription), and U5 regions are indicated at the top, further showing that several of the tested LTRs have been formed by L1-mediated retrotransposition and therefore lack certain LTR portions.
We conclude that a majority of HERV-W LTRs display promoter activity in the sense and/or antisense orientation, even if structurally incomplete. Therefore, most of the HERV-W loci identified as transcribed in this study may in fact be transcribed by remnants of their own 5′ and/or 3′ LTRs in the sense and/or antisense direction.
DISCUSSION
Transcription of HERV-W in the context of MS appeared to be an unresolved issue, as previous studies sometimes reported heterogeneous or even conflicting results (see the introduction), some of which might have been due to the repetitive nature of HERV-W requiring, for instance, meticulous design of PCR primer pairs. We previously described transcriptional activities of specific loci of various HERV groups under different biological and clinical conditions. We chose a direct approach in those studies by generating cDNA sequences from RNA transcripts from HERV loci and reassigning those cDNA sequences to specific HERV loci. Such a strategy not only identifies HERV loci actually transcribed, but also generates data on relative transcription levels of HERV loci (35, 44, 50). Using this strategy, we recently also investigated the transcription of HERV-W in PBMC and identified a total of seven loci as being transcribed. Notably, we did not detect significantly different transcription patterns when comparing HERV-W transcription in PBMC from patients with MS and from healthy controls (20).
We have now studied HERV-W transcription in brain lesions from patients with MS compared to white matter brain tissue from healthy controls and present a comprehensive picture of HERV-W locus transcription in those samples. We employed an optimized strategy for generating cDNA from as many HERV-W loci as possible in a straightforward RT-PCR experiment. We used second-generation amplicon sequencing to produce a great number of HERV-W cDNA sequence reads per tissue sample, on average ∼2,200 analyzed reads per sample for 454/FLX and ∼348,000 for Illumina/MiSeq, significantly reducing potentially skewed relative cDNA sequence frequencies when generating relatively small numbers of cDNA sequences and, furthermore, enabling comprehensive detection of HERV-W loci with lower relative transcript levels that are less well represented in the cDNA pool. Assignment of HERV-W cDNA sequences to specific HERV-W loci, furthermore, was straightforward, even for relatively short second-generation sequencing reads because of the evolutionary age of, and thus sequence differences between, the various HERV-W loci in the human genome. This was further demonstrated by the fact that two independent second-generation sequencing strategies, 454/FLX and Illumina/MiSeq, produced overall very similar results. We therefore suggest that the strategy employed here is suitable for generating a high-resolution picture of HERV-W transcription in a biological sample of interest. The strategy employed here may be equally suited to depicting transcription patterns of other high-copy-number HERV groups, e.g., HERV-H, HERV-L, and HERV-E. We also note that a high-throughput amplicon-sequencing approach for identifying transcribed HERV loci may also identify biologically relevant somatic mutations in transcribed HERV sequences. This appears to be especially important in light of the fact that assignment of cDNA sequences must rely on a reference genome sequence that probably lacks many allelic variants. Repeatedly detected sequence variants would thus be indicative of allelic variants rather than experimental artifacts. Raw sequence data would have to be thoroughly examined for such potential variants.
We also stress that the strategy employed here for identifying transcribed HERV-W loci intentionally does not distinguish the direction of transcription, as HERV-W loci transcribed in antisense may be equally biologically relevant by, for instance, regulating expression of HERV-W loci or cellular genes by RNA interference (RNAi) (51, 52).
In the context of HERV-W and MS, an important result of our study is that the optimized and comprehensive strategy employed here for describing HERV-W transcription revealed overall very similar transcription patterns of HERV-W loci in MS and healthy brain tissues. Especially regarding the ERVWE1/ERVW-1/syncytin-1 locus, relative transcription levels were not clearly different between MS-derived and healthy tissue samples. Slightly lower transcript levels of the ERVWE1/ERVW-1/syncytin-1 locus in MS brain lesions were statistically significant only for the 5′ env data set and not for the 3′ env data set, and quantitative RT-PCR revealed no significant differences between the groups of patients with MS and controls with respect to ERVWE1/ERVW-1/syncytin-1 locus transcription. Likewise, a HERV-W locus in chr. 15q21.3/ERVW-4 appeared to be transcribed at somewhat higher relative levels in MS than in healthy brain tissue, yet this finding was statistically significant only for the 5′ env amplicon and not for the 3′ env amplicon. Furthermore, no significant differences were found for the ERVWE2/ERVW-2 locus in chr. Xq22.3B and various other higher-level-transcribed HERV-W loci. We therefore interpret differences in cDNA sequence frequencies seen for a few HERV-W loci that also were opposite for 5′ env and 3′ env amplicons to be very likely due to stochastic variations in the experiment rather than true differences in relative transcription levels.
Potential minor transcriptional deregulation of HERV-W loci in MS-derived brain tissue samples would have to be specifically investigated, though the biological significance of such potential minor deregulation is uncharted. While transcription of HERV-W loci may be deregulated for as yet unknown reasons, it is also conceivable that epigenetic differences in HERV-W locus-harboring genome regions may also result in slightly higher-level transcription of HERV-W loci in MS brain tissue cells. It is known that CpG methylation can strongly influence HERV-W transcription, as revealed by studies on the ERVWE1/ERVW-1/syncytin-1 locus (53–55). Since MS-derived brain tissue samples investigated in our study are from inflammatory plaque regions, it is also conceivable that cell types other than brain cells activated in the course of inflammatory processes contributed transcript from specific HERV-W loci.
Our findings are in contrast to previous studies that reported upregulated or strongly upregulated HERV-W (env) transcription in MS samples. Therefore, according to our results, if HERV-W is involved in the pathogenesis of MS, deregulated transcription of specific HERV-W loci, especially protein-encoding HERV-W loci, is unlikely to be the underlying mechanism. This may be an important finding for future studies addressing the role of HERV-W and HERV-W-encoded proteins in MS. On the other hand, the fact that both ERVWE1/ERVW-1 and ERVWE2/ERVW-2 are transcribed in normal brain tissue implies translation of the respective proteins in the brain, in line with immunohistochemical detection of a HERV-W Env protein in brain tissue by monoclonal antibody 6A2B2 (13). However, physiological functions of ERVWE1/ERVW-1-encoded Syncytin-1 and/or ERVWE2/ERVW-2-encoded N-Trenv in the brain remain enigmatic.
Furthermore, relative cDNA sequence frequencies demonstrated, on one hand, dominant transcription of a limited number of HERV-W loci and, on the other hand, transcription of a greater number of HERV-W loci at lower or much lower transcript levels. Notably, several of the HERV-W loci dominantly transcribed in the brain were also dominantly transcribed in PBMC based on cDNA cloning frequencies (20). Some HERV-W loci were identified in our study as being transcribed by only one cDNA sequence read in one tissue sample. While we rigorously removed DNA from RNA preparations and very carefully verified the absence of DNA contamination in RT-PCR experiments, it is nevertheless very difficult to remove all traces of DNA from an RNA preparation. One therefore cannot formally exclude the presence of residual DNA in RNA used for cDNA generation. HERV-W loci represented by very few or just one cDNA sequence in the entire sequence data set, therefore, may be due to residual traces of DNA, especially when generating hundreds of thousands of reads per sample. It is thus difficult to draw a line for false-positive detection of transcribed HERV-W loci. However, we note in this context that many regions of the human genome are apparently transcribed at low levels, with some regions represented in RNA-Seq data sets by very few reads (56, 57). Many HERV-W loci could likewise be transcribed at such low levels, perhaps because they are located within such low-level-transcribed genome regions. Also, some of the HERV-W loci with very low relative cDNA sequence frequencies identified here are located within gene introns, and it is possible that cDNA from such loci might have been generated from unspliced hnRNA. Nevertheless, a greater number of HERV-W loci were represented by higher relative cDNA sequence frequencies, and we are confident that those loci are indeed transcribed in brain tissue, at least at somewhat lower levels. Transcription of quite a number of HERV-W loci was further supported by RNA-Seq data sets generated by the ENCODE Consortium from various human cell lines in that many of the loci we identified were also identified as transcribed by variable numbers of RNA-Seq reads, sometimes in several of the cell lines, with RNA-Seq read numbers for some loci roughly correlating with relative cDNA sequence frequencies observed by us in brain tissues.
Altogether, when considering rather strict thresholds, we conclude that, in brain tissue, about 28 HERV-W loci are transcribed at higher relative levels and another approximately 120 HERV-W loci are transcribed at low relative levels. HERV-W loci ERVWE1/ERVW-1/syncytin-1 and ERVWE2/ERVW-2, which may be relevant in the context of MS, are among the higher-level-transcribed HERV-W loci, and their respective relative transcription levels do not significantly differ between MS and healthy brain tissue.
Similar transcription levels of HERV-W loci were further corroborated by our qRT-PCR results. When determining overall HERV-W transcript levels, or transcript levels specifically for the ERVWE1/ERVW-1/syncytin-1 and ERVWE2/ERVW-2 loci, with locus-specific PCR primers (20) relative to two different housekeeping gene transcript levels, we did not find statistically significant differences in the respective transcript levels between MS lesions and healthy tissue. Several of the MS lesion samples displayed HERV-W transcript levels lower than those in healthy controls, yet those differences were not statistically significant in regard to MS lesions and healthy-control samples, each as a group. Nevertheless, we detected sometimes pronounced differences in HERV-W transcript levels between tissue samples, overall and locus specific, pointing to interindividual differences in HERV-W transcript levels. Such interindividual variation in HERV transcript levels was also observed in previous microarray-based experiments (27, 50), and a strategy involving high-resolution melting temperatures likewise suggested interindividual differences in HERV-W transcript levels (28). Interindividual variation in cellular-gene transcription levels is a well-known phenomenon caused by genetic variation (58–60). Therefore, genetic differences may also influence HERV-W transcript levels in individuals. Our findings thus suggest a more detailed and specific investigation as to whether such overall interindividual variation in HERV-W transcript levels correlates with MS. It is furthermore conceivable that overall HERV-W transcript levels may differ between brain areas. This is supported by relative HERV-W transcript levels in samples MS4 and MS5, which were obtained from the same patient, yet the MS5 sample displayed slightly higher relative transcript levels. The above-mentioned high-resolution melting-temperature-based study also suggested differences in transcript levels between brain areas (28). However, specifically designed studies will be required to answer those questions. Last but not least, it is conceivable that previously observed MS-specific differences in HERV-W transcript levels were influenced by interindividual differences in HERV-W transcript levels.
We also found that remnants of HERV-W LTRs associated with different HERV-W loci often display promoter activity. Results from those reporter gene assays therefore indicate that, in principle, HERV-W loci may often be transcribed from a HERV-W locus' own promoter present within remnants of the HERV-W LTR. While we performed reporter gene assays in JEG-3 cells that support transcription of HERV-W LTRs (49), the actual activity of these and other HERV-W LTRs may differ in brain tissue cells. Nevertheless, HERV-W LTRs, in principle, likely can initiate transcription in many cases also in brain tissue cells, thus potentially explaining the transcription of many HERV-W loci. Our results also indicate that HERV-W LTRs retaining U3R mainly give rise to antisense transcripts, while those LTRs retaining RU5, and whole LTRs, display a preference for sense transcription. Results reported by Gimenez et al. (25) suggest that HERV-W LTR U3 and R regions can initiate transcription. Lee et al. (61) reported transcriptional activity for the HERV-W LTR R/U5 region. Notably, the HERV-W LTR's R region is usually present in at least one copy, and the U3 region is present in the 3′ LTR in HERV-W loci formed by L1-mediated retrotransposition. However, more specific studies of HERV-W LTRs are required to reveal the contributions of U3, R, and U5 to sense and antisense transcriptional activity of incomplete HERV-W LTRs. In any case, our results lend further support to HERV-W loci identified in our study as transcribed harboring LTR regions with potential promoter activity—besides HERV-W loci with full-length 5′ and 3′ LTRs.
Our finding of apparently many HERV-W LTRs displaying promoter activity also contributes to a better understanding of the effects HERV-W sequences may have on neighboring cellular genes. Many HERV-W LTRs, either HERV-W locus associated or solitary, or even incomplete because they belong to HERV-W processed pseudogene loci, likely still display promoter activity and thus may provide alternative transcripts to many cellular genes and may become deregulated in certain cell types and disease conditions.
Nevertheless, some HERV-W loci may also be transcribed because of close proximity to a cellular gene. For instance, read-through transcripts of the cellular genes ZNF99, C14orf159, RBM41, and GRK7 may cotranscribe, or contribute to the transcription of, HERV-W loci located up to a few kilobases downstream of those genes. This may also apply to loci sandwiched between two cellular genes. However, only a minority of HERV-W loci seem to be transcribed or cotranscribed indirectly due to the transcription of cellular genes.
Finally, we previously analyzed published MSRV sequences in the context of template switching of reverse transcriptase during cDNA generation in vitro. We argued that reported MSRV sequences were either derived from or could be explained as recombinants of HERV-W loci located in chr. 3p12.3, 3q23A, 3q26.32/ERVW-5, 5p12, 15q21.3/ERVW-4, 18q21.32, and Xq22.3B/ERVWE2/ERVW-2. However, only the loci in chr. 15q21.3/ERVW-4 and Xq22.3B/ERVWE2/ERVW-2 were identified as transcribed in PBMC (20). The data presented here demonstrate that HERV-W loci in chr. 3p12.3, 3q23A, 3q26.32/ERVW-5, 5p12, and 18q21.32 can also be transcribed, though some of them at lower relative levels in the brain, but nevertheless lending further support to the concept that those previously described MSRV sequences that are not directly assignable to genomic HERV-W loci were indeed generated by in vitro recombinations.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from Deutsche Forschungsgemeinschaft (grant no. Ma2298/8-1) to J.M.
We are greatly indebted to Jasmin Gries for help with FLX sequencing and Pavlo Lutsik and Nicole Souren for preprocessing of FLX sequence reads. We also thank Karen Rother and Sindy Suhr for kindly providing JEG-3 cells.
Footnotes
Published ahead of print 9 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02388-13.
REFERENCES
- 1.Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42:709–732 [DOI] [PubMed] [Google Scholar]
- 2.Mayer J, Meese E. 2005. Human endogenous retroviruses in the primate lineage and their influence on host genomes. Cytogenet. Genome Res. 110:448–456 [DOI] [PubMed] [Google Scholar]
- 3.Stoye JP. 2001. Endogenous retroviruses: still active after all these years? Curr. Biol. 11:R914–R916 [DOI] [PubMed] [Google Scholar]
- 4.Kurth R, Bannert N. 2010. Beneficial and detrimental effects of human endogenous retroviruses. Int. J. Cancer 126:306–314 [DOI] [PubMed] [Google Scholar]
- 5.Belshaw R, Katzourakis A, Paces J, Burt A, Tristem M. 2005. High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol. Biol. Evol. 22:814–817 [DOI] [PubMed] [Google Scholar]
- 6.Costas J. 2002. Characterization of the intragenomic spread of the human endogenous retrovirus family HERV-W. Mol. Biol. Evol. 19:526–533 [DOI] [PubMed] [Google Scholar]
- 7.Pavlícek A, Paces J, Elleder D, Hejnar J. 2002. Processed pseudogenes of human endogenous retroviruses generated by LINEs: their integration, stability, and distribution. Genome Res. 12:391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blond JL, Besème F, Duret L, Bouton O, Bedin F, Perron H, Mandrand B, Mallet F. 1999. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 73:1175–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komurian-Pradel F, Paranhos-Baccala G, Bedin F, Ounanian-Paraz A, Sodoyer M, Ott C, Rajoharison A, Garcia E, Mallet F, Mandrand B, Perron H. 1999. Molecular cloning and characterization of MSRV-related sequences associated with retrovirus-like particles. Virology 260:1–9 [DOI] [PubMed] [Google Scholar]
- 10.Perron H, Garson JA, Bedin F, Beseme F, Paranhos-Baccala G, Komurian-Pradel F, Mallet F, Tuke PW, Voisset C, Blond JL, Lalande B, Seigneurin JM, Mandrand B. 1997. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The Collaborative Research Group on Multiple Sclerosis. Proc. Natl. Acad. Sci. U. S. A. 94:7583–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voisset C, Blancher A, Perron H, Mandrand B, Mallet F, Paranhos-Baccalà G. 1999. Phylogeny of a novel family of human endogenous retrovirus sequences, HERV-W, in humans and other primates. AIDS Res. Hum. Retroviruses 15:1529–1533 [DOI] [PubMed] [Google Scholar]
- 12.Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roebke C, Wahl S, Laufer G, Stadelmann C, Sauter M, Mueller-Lantzsch N, Mayer J, Ruprecht K. 2010. An N-terminally truncated envelope protein encoded by a human endogenous retrovirus W locus on chromosome Xq22.3. Retrovirology 7:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Compston A, Coles A. 2008. Multiple sclerosis. Lancet 372:1502–1517 [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Montojo M, Dominguez-Mozo M, Arias-Leal A, Garcia-Martinez A, De las Heras V, Casanova I, Faucard R, Gehin N, Madeira A, Arroyo R, Curtin F, Alvarez-Lafuente R, Perron H. 2013. The DNA copy number of human endogenous retrovirus-W (MSRV-type) is increased in multiple sclerosis patients and is influenced by gender and disease severity. PLoS One 8:e53623. 10.1371/journal.pone.0053623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mameli G, Astone V, Arru G, Marconi S, Lovato L, Serra C, Sotgiu S, Bonetti B, Dolei A. 2007. Brains and peripheral blood mononuclear cells of multiple sclerosis (MS) patients hyperexpress MS-associated retrovirus/HERV-W endogenous retrovirus, but not Human herpesvirus 6. J. Gen. Virol. 88:264–274 [DOI] [PubMed] [Google Scholar]
- 17.Mameli G, Poddighe L, Astone V, Delogu G, Arru G, Sotgiu S, Serra C, Dolei A. 2009. Novel reliable real-time PCR for differential detection of MSRVenv and syncytin-1 in RNA and DNA from patients with multiple sclerosis. J. Virol. Methods 161:98–106 [DOI] [PubMed] [Google Scholar]
- 18.Dolei A, Perron H. 2009. The multiple sclerosis-associated retrovirus and its HERV-W endogenous family: a biological interface between virology, genetics, and immunology in human physiology and disease. J. Neurovirol. 15:4–13 [DOI] [PubMed] [Google Scholar]
- 19.Perron H, Perin JP, Rieger F, Alliel PM. 2000. Particle-associated retroviral RNA and tandem RGH/HERV-W copies on human chromosome 7q: possible components of a ‘chain-reaction' triggered by infectious agents in multiple sclerosis? J. Neurovirol. 6(Suppl 2):S67–S75 [PubMed] [Google Scholar]
- 20.Laufer G, Mayer J, Mueller BF, Mueller-Lantzsch N, Ruprecht K. 2009. Analysis of transcribed human endogenous retrovirus W env loci clarifies the origin of multiple sclerosis-associated retrovirus env sequences. Retrovirology 6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antony JM, Ellestad KK, Hammond R, Imaizumi K, Mallet F, Warren KG, Power C. 2007. The human endogenous retrovirus envelope glycoprotein, syncytin-1, regulates neuroinflammation and its receptor expression in multiple sclerosis: a role for endoplasmic reticulum chaperones in astrocytes. J. Immunol. 179:1210–1224 [DOI] [PubMed] [Google Scholar]
- 22.Antony JM, van Marle G, Opii W, Butterfield DA, Mallet F, Yong VW, Wallace JL, Deacon RM, Warren K, Power C. 2004. Human endogenous retrovirus glycoprotein-mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nat. Neurosci. 7:1088–1095 [DOI] [PubMed] [Google Scholar]
- 23.Rolland A, Jouvin-Marche E, Viret C, Faure M, Perron H, Marche PN. 2006. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J. Immunol. 176:7636–7644 [DOI] [PubMed] [Google Scholar]
- 24.Kremer D, Schichel T, Forster M, Tzekova N, Bernard C, van der Valk P, van Horssen J, Hartung HP, Perron H, Kury P. 2013. HERV-W envelope protein inhibits oligodendroglial precursor cell differentiation. Ann. Neurol. [Epub ahead of print.] 10.1002/ana.23970 [DOI] [PubMed] [Google Scholar]
- 25.Gimenez J, Montgiraud C, Pichon JP, Bonnaud B, Arsac M, Ruel K, Bouton O, Mallet F. 2010. Custom human endogenous retroviruses dedicated microarray identifies self-induced HERV-W family elements reactivated in testicular cancer upon methylation control. Nucleic Acids Res. 38:2229–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perot P, Mugnier N, Montgiraud C, Gimenez J, Jaillard M, Bonnaud B, Mallet F. 2012. Microarray-based sketches of the HERV transcriptome landscape. PLoS One 7:e40194. 10.1371/journal.pone.0040194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank O, Giehl M, Zheng C, Hehlmann R, Leib-Mosch C, Seifarth W. 2005. Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders. J. Virol. 79:10890–10901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nellåker C, Li F, Uhrzander F, Tyrcha J, Karlsson H. 2009. Expression profiling of repetitive elements by melting temperature analysis: variation in HERV-W gag expression across human individuals and tissues. BMC Genomics 10:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antony JM, Izad M, Bar-Or A, Warren KG, Vodjgani M, Mallet F, Power C. 2006. Quantitative analysis of human endogenous retrovirus-W env in neuroinflammatory diseases. AIDS Res. Hum. Retroviruses 22:1253–1259 [DOI] [PubMed] [Google Scholar]
- 30.Antony JM, Zhu Y, Izad M, Warren KG, Vodjgani M, Mallet F, Power C. 2007. Comparative expression of human endogenous retrovirus-W genes in multiple sclerosis. AIDS Res. Hum. Retroviruses 23:1251–1256 [DOI] [PubMed] [Google Scholar]
- 31.Garson JA, Huggett JF, Bustin SA, Pfaffl MW, Benes V, Vandesompele J, Shipley GL. 2009. Unreliable real-time PCR analysis of human endogenous retrovirus-W (HERV-W) RNA expression and DNA copy number in multiple sclerosis. AIDS Res. Hum. Retroviruses 25:377–378 (Author's reply, 25:379–381.) [DOI] [PubMed] [Google Scholar]
- 32.Nellåker C, Yao Y, Jones-Brando L, Mallet F, Yolken RH, Karlsson H. 2006. Transactivation of elements in the human endogenous retrovirus W family by viral infection. Retrovirology 3:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao Y, Schroder J, Nellaker C, Bottmer C, Bachmann S, Yolken RH, Karlsson H. 2008. Elevated levels of human endogenous retrovirus-W transcripts in blood cells from patients with first episode schizophrenia. Genes Brain Behav. 7:103–112 [DOI] [PubMed] [Google Scholar]
- 34.Mayer J, Blomberg J, Seal RL. 2011. A revised nomenclature for transcribed human endogenous retroviral loci. Mob. DNA 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. 2013. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol. Evol. 5:307–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110:462–467 [DOI] [PubMed] [Google Scholar]
- 37.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19:Unit 19.10.11-21. 10.1002/0471142727.mb1910s89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15:1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Parseval N, Lazar V, Casella J-F, Benit L, Heidmann T. 2003. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 77:10414–10422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flockerzi A, Maydt J, Frank O, Ruggieri A, Maldener E, Seifarth W, Medstrand P, Lengauer T, Meyerhans A, Leib-Mosch C, Meese E, Mayer J. 2007. Expression pattern analysis of transcribed HERV sequences is complicated by ex vivo recombination. Retrovirology 4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Trapnell C, Pop M, Salzberg S. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- 47.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, Raney BJ, Pohl A, Malladi VS, Li CH, Lee BT, Learned K, Kirkup V, Hsu F, Heitner S, Harte RA, Haeussler M, Guruvadoo L, Goldman M, Giardine BM, Fujita PA, Dreszer TR, Diekhans M, Cline MS, Clawson H, Barber GP, Haussler D, Kent WJ. 2013. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 41:D64–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32:D493–D496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prudhomme S, Oriol G, Mallet F. 2004. A retroviral promoter and a cellular enhancer define a bipartite element which controls env ERVWE1 placental expression. J. Virol. 78:12157–12168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frank O, Verbeke C, Schwarz N, Mayer J, Fabarius A, Hehlmann R, Leib-Mösch C, Seifarth W. 2008. Variable transcriptional activity of endogenous retroviruses in human breast cancer. J. Virol. 82:1808–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gogvadze E, Stukacheva E, Buzdin A, Sverdlov E. 2009. Human-specific modulation of transcriptional activity provided by endogenous retroviral insertions. J. Virol. 83:6098–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gosenca D, Gabriel U, Steidler A, Mayer J, Diem O, Erben P, Fabarius A, Leib-Mosch C, Hofmann WK, Seifarth W. 2012. HERV-E-mediated modulation of PLA2G4A transcription in urothelial carcinoma. PLoS One 7:e49341. 10.1371/journal.pone.0049341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gimenez J, Montgiraud C, Oriol G, Pichon JP, Ruel K, Tsatsaris V, Gerbaud P, Frendo JL, Evain-Brion D, Mallet F. 2009. Comparative methylation of ERVWE1/syncytin-1 and other human endogenous retrovirus LTRs in placenta tissues. DNA Res. 16:195–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matousková M, Blazková J, Pajer P, Pavlícek A, Hejnar J. 2006. CpG methylation suppresses transcriptional activity of human syncytin-1 in non-placental tissues. Exp. Cell Res. 312:1011–1020 [DOI] [PubMed] [Google Scholar]
- 55.Trejbalova K, Blazkova J, Matouskova M, Kucerova D, Pecnova L, Vernerova Z, Heracek J, Hirsch I, Hejnar J. 2011. Epigenetic regulation of transcription and splicing of syncytins, fusogenic glycoproteins of retroviral origin. Nucleic Acids Res. 39:8728–8739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hangauer MJ, Vaughn IW, McManus MT. 2013. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 9:e1003569. 10.1371/journal.pgen.1003569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 8:e1000371. 10.1371/journal.pbio.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. 2003. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 33:422–425 [DOI] [PubMed] [Google Scholar]
- 59.Cheung VG, Nayak RR, Wang IX, Elwyn S, Cousins SM, Morley M, Spielman RS. 2010. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol. 8:pii: e1000480. 10.1371/journal.pbio.1000480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheung VG, Spielman RS. 2009. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat. Rev. Genet. 10:595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee WJ, Kwun HJ, Kim HS, Jang KL. 2003. Activation of the human endogenous retrovirus W long terminal repeat by herpes simplex virus type 1 immediate early protein 1. Mol. Cells 15:75–80 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.