Abstract
Hepatitis B virus (HBV) causes chronic hepatitis in hundreds of millions of people worldwide, which can eventually lead to hepatocellular carcinoma (HCC). The molecular mechanisms underlying HBV persistence are not well understood. In this study, we found that HBV inhibited the chemotherapy drug etoposide-induced apoptosis of hepatoma cells. Further analysis revealed that HBV mRNAs possess a microRNA 15a/16 (miR-15a/16)-complementary site (HBV nucleotides [nt] 1362 to 1383) that acts as a sponge to bind and sequester endogenous miR-15a/16. Consequently, Bcl-2, known as the target of miR-15a/16, was upregulated in HBV-infected cells. The data from HBV-transgenic mice further confirmed that HBV transcripts cause the reduction of miR-15a/16 and increase of Bcl-2. More importantly, we examined the levels of HBV transcripts and miR-15a/16 in HBV-infected HCC from patients and found that the amount of HBV mRNA and the level of miR-15a/16 were negatively correlated. Consistently, the level of Bcl-2 mRNA was upregulated in HBV-infected patients. In conclusion, we identified a novel HBV mRNA–miR-15a/16–Bcl-2 regulatory pathway that is involved in inhibiting etoposide-induced apoptosis of hepatoma cells, which may contribute to facilitating chronic HBV infection and hepatoma development.
INTRODUCTION
There are approximately 350 million chronic hepatitis B virus (HBV) carriers worldwide, and chronic HBV infection is the major etiological factor in hepatocellular carcinoma (HCC) (1, 2). The relative risk for the development of HCC in chronic hepatitis B (CHB) patients is estimated to be 25 to 37 times higher than that in those without infection (1, 3, 4).
HBV is an enveloped, partially double-stranded DNA virus with a genome size of 3.2 kb. The HBV genome contains four overlapping open reading frames (ORFs). The RNA transcripts are polyadenylated and capped and are named the pre-C/C or pregenomic RNA (pgRNA) and the pre-S, S, and X mRNAs. These mRNAs encode several viral proteins, including the polymerase, core, HBe, pre-S1, S2, S, and X proteins (5). HBV has been reported to play an important role in regulating apoptosis. For example, HBV core protein inhibits TRAIL-induced apoptosis of hepatocytes by blocking DR5 expression (6). HBx can bind to the C terminus of p53 and inhibit p53-mediated cellular processes, including transcriptional transactivation and apoptosis (7). But the HBx protein was also found to sensitize cells to apoptotic killing by tumor necrosis factor alpha (8) and to inhibit Fas-mediated apoptosis associated with upregulation of the SAPK/JNK pathway in Chang cells (9).
MicroRNAs (miRNAs) are single-stranded noncoding RNAs which negatively regulate gene expression at the posttranscriptional level, primarily through base pairing to the 3′-untranslated region (UTR) of target mRNA (10). Growing evidence indicates that microRNAs control basic cell functions, ranging from proliferation to apoptosis, by direct targeting (11, 12). For instance, miR-101 exerts a proapoptotic function by targeting Mcl-1 (13), and miR-29c inhibits cell proliferation and induces apoptosis by targeting TNFAIP3 (14).
miR-15a and miR-16-1 are transcribed as a cluster (miR-15a/16) that resides in the 13q14 chromosomal region (15). miR-15a/16 can downregulate Bcl-2 expression, and correspondingly, miR-15a/16 is often deleted or downregulated in tumor cells (16). Bcl-2 is an important target of the miR-15a/16 cluster and is known to act as an antiapoptotic protein by inhibiting caspase activity by preventing the release of cytochrome c from the mitochondria and/or binding to the apoptosis-activating factor (Apaf-1) (17–20).
It has been reported that highly abundant viral transcripts can downregulate cellular microRNAs, which is very important for efficient virus replication. For instance, murine cytomegalovirus (MCMV) m169 transcript-mediated degradation of miR-27a/b is important for MCMV replication (21), and HBV mRNAs can sequester endogenous miR-122 to facilitate HBV replication (22). It has also been reported that the HBx protein can downregulate the expression of the miR-16 family in hepatoma cells (23). Based on the observations that miR-15a/16 is decreased in HBV-infected cells (24) and that HBV inhibits apoptosis of hepatoma cells, the objective of this study was to explore the mechanism of how HBV downregulates miR-15a/16 and affects the apoptosis of hepatoma cells. Our results demonstrate that HBV mRNAs possess a miR-15a/16-complementary site that acts as a sponge to bind and sequester endogenous miR-15a/16. Consistently, Bcl-2, the target of miR-15a/16, was increased significantly in HBV-transfected cells. Furthermore, we found that the miR-15a/16 cluster was downregulated, while Bcl-2 was upregulated, in HBV-infected HCC from patients. Our results reveal a novel mechanism by which HBV inhibits apoptosis through decreasing miR-15a/16 by its own transcripts during chronic HBV infection.
MATERIALS AND METHODS
Patients and human specimens.
HCC liver tissues from 40 patients were collected at the 302 Hospital of PLA, Beijing, China. The patients were hospitalized during June 2012 to July 2013. The clinical characteristics of enrolled subjects are listed in Table 1. Written informed consent was provided by all study participants. Patient samples were assigned arbitrary identifications based on the order of enrollment in our study. The study protocol was approved by the ethics committee of the 302 Hospital of PLA.
Table 1.
Clinical characteristics of studied HCC subjects
| Parameter | Value |
|---|---|
| No. of cases | 40 |
| Age (yr) (mean [range]) | 56 (27–71) |
| No. of males/no. of females | 22/18 |
| Alanine aminotransferase level (U/liter) (mean [range]) | 69 (11–187) |
| HBV load (no. of DNA copies/ml serum) | 0–2.88 × 107 |
Mice.
HBV-transgenic BALB/c mice (female, 6 to 8 weeks old) were purchased from the Transgenic Engineering Lab, Infectious Disease Center (Guangzhou, China). They were generated with a viral DNA construct, pHBV1.3, containing 1.3 copies of the HBV genome (D genotype). All transgenic mice were positive for serum HBV surface antigen (HBsAg), virus DNA, and HBV core proteins (HBcAg) in hepatocytes in the liver (25). Animals received humane care, and the study of mice was permitted by the Research Ethics Committee of the Institute of Microbiology.
Plasmid constructs.
The HBV genome (nucleotides [nt] 1300 to 1500) was amplified by PCR using the following primers: sense, 5′-CGGAATTCGCAACCGGTCTGGAGCGAAACTT-3′; and antisense, 5′-GCTCTAGACGAAGAAGGGGACGATAGAGGCC-3′. The PCR product was cloned into the dual-luciferase reporter pmirGLO vector (Promega) at the EcoRI and XbaI sites, and the resultant clone was named UTR-HBV-wt. UTR-HBV-mut, with mutations in the seed region of the miR-15a/16-binding site in the HBV genome, was generated by site-directed mutagenesis. A full-length HBV genome mutant (pHBV1.3-mut) with mutations in the miR-15a/16-binding site was also generated by site-directed mutagenesis.
Reagents and antibodies.
Chemically synthesized miR-15a and miR-16 inhibitors (CACAAACCAUUAUGUGCUGCUA and CGCCAAUAUUUACGUGCUGCUA), miR-15a and miR-16 mimics (GUGUUUGGUAAUACACGACGAU and GCGGUUAUAAAUGCACGACGAU), and nonspecific controls were purchased from RiboBio Co., Ltd. (Guangzhou, China). The following reagents and antibodies were obtained as indicated: rabbit anti-human poly(ADP-ribose) polymerase 1 (anti-PARP1) antibody was from Cell Signaling, rabbit anti-human caspase 9 antibody was from Cell Signaling, mouse anti-human Bcl-2 antibody was from Santa Cruz Biotechnology, rabbit anti-human actin antibody was from Santa Cruz Biotechnology, and horseradish peroxidase-conjugated secondary antibodies were from Jackson Laboratory.
Cell culture and transfection.
The human hepatoma cell lines HepG2 and HepG2.2.15 and the human normal liver cell line L-02 were obtained from the ATCC (Manassas, VA). Transfections were performed using Lipofectamine 2000 reagent (Invitrogen). Cells in 6-well plates were transfected with 50 nM miR-15a/16 inhibitor or miR-15a/16 mimic or the indicated amounts of plasmids.
RNA extraction and real-time PCR.
Total RNA was extracted from cells or frozen tissues by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Real-time PCR for Bcl-2 was performed using SYBR green premix reagent (Toyobo, Japan). β-Actin was used as an internal control. Real-time PCR analysis of miR-15a/16 was also performed using SYBR green premix reagent, with U6 as the internal control. Relative expression was quantified using the comparative threshold cycle (CT) method.
Dual-luciferase reporter activity assay.
HepG2 cells were cotransfected with a dual-luciferase reporter for HBV (UTR-HBV-wt or UTR-HBV-mut) and miR-15a or miR-16 for 24 h. The cell lysates were harvested for dual-luciferase assay according to the manufacturer's instructions. Three independent experiments were performed.
Immunoblotting.
Protein samples were subjected to SDS-PAGE and blotted with the rabbit anti-human PARP, caspase 9, or β-actin or mouse anti-human Bcl-2 antibody. After being washed with phosphate-buffered saline plus Tween 20 (PBST) three times, a horseradish peroxidase-conjugated secondary antibody was added for 1 h. Protein bands were visualized using Enlight Western blotting reagents (Engreen Biosystems, China).
Detection of HBsAg by ELISA.
HepG2 cells were transfected with pHBV1.3 or pHBV1.3-mut. The supernatants were collected at the indicated time points and subjected to enzyme-linked immunosorbent assay (ELISA) using a diagnostic kit for the hepatitis B virus surface antigen (Shanghai Kehua Bio-Engineering, China).
Southern blot analysis.
HepG2 cells were transfected with pHBV1.3 or pHBV1.3-mut for 48 h. The DNAs extracted from cells were subjected to Southern blot analysis using alkaline phosphatase (AP)-labeled HBV DNA as the probe (Amersham AlkPhos direct labeling and detection system; GE Healthcare).
Caspase activity assay.
HepG2 cells were plated in 96-well plates, treated as indicated, and then subjected to caspase activity assay with Caspase-Glo3/7 reagents (Promega). The luminescence of each sample was measured in a plate-reading luminometer.
Statistical analysis.
Differences between groups were determined using Student's t test. P values of <0.05 were considered significant. The degree of association between variables was determined by Spearman's nonparametric correlation.
RESULTS
HBV inhibits the apoptosis of hepatoma cells induced by the chemotherapy drug etoposide.
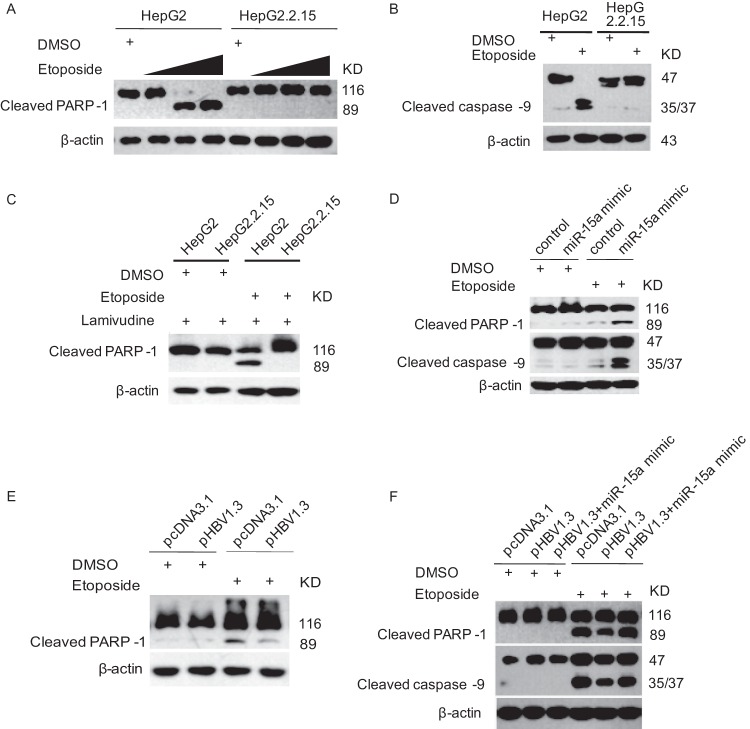
It has been reported that HBV has both proapoptotic and antiapoptotic effects in different experimental systems, but its contradictory functions have not yet been elucidated fully. In this study, we treated HepG2 and HepG2.2.15 cells, which are HBV-transgenic cells, with etoposide as indicated, and we examined the PARP-1 and caspase 9 cleavages by immunoblotting. As shown in Fig. 1A and B, PARP-1 and caspase 9 cleavages occurred dramatically in HepG2 but not HepG2.2.15 cells. These data indicated that HepG2.2.15 cells were resistant to apoptosis induced by etoposide. In order to determine if viral replication affected the HepG2.2.15 cells resisting apoptosis, we treated the cells with lamivudine and performed a similar experiment. The data showed that lamivudine treatment did not change the pattern of PARP-1 cleavage in HepG2.2.15 cells (Fig. 1C).
Fig 1.
HBV inhibits etoposide-induced apoptosis of hepatoma cells. (A) HepG2 and HepG2.2.15 cells were treated with etoposide (0.08 μM, 0.4 μM, and 2 μM) or dimethyl sulfoxide (DMSO) as a control for 24 h. The cell lysates were harvested for immunoblotting with PARP-1 antibody. (B) HepG2 and HepG2.2.15 cells were treated with etoposide (0.08 μM) for 24 h. The cell lysates were harvested for immunoblotting with caspase 9 antibody. (C) HepG2 and HepG2.2.15 cells were treated with etoposide (0.08 μM) and lamivudine (100 nM) for 24 h. The cell lysates were harvested for immunoblotting with PARP-1 antibody. (D) HepG2.2.15 cells were transfected with miR-15a mimic or a randomized oligonucleotide as a control for 24 h and were then treated with etoposide (0.08 μM) for 24 h. The cell lysates were harvested for immunoblotting with the indicated antibodies. (E) L-02 cells were transfected with the HBV replication plasmid pHBV1.3, using the empty plasmid pcDNA3.1 as a control, for 24 h and then treated with etoposide (0.08 μM) for 24 h. The cell lysates were harvested for immunoblotting with PARP-1 antibody. (F) HepG2 cells were transfected with the HBV replication plasmid pHBV1.3, with or without miR-15a mimic, or with the empty plasmid pcDNA3.1 as a control, for 24 h and then treated with etoposide (0.08 μM) for 24 h. The cell lysates were harvested for immunoblotting with PARP-1 and caspase 9 antibodies.
It has been reported that some microRNAs, including miR-15a/16, are differentially expressed in HepG2.2.15 compared with HepG2 cells by microarray analysis. The level of miR-15a/16 is about 10-fold lower in HepG2.2.15 cells than in HepG2 cells (24). Since miR-15a/16 has been reported to target Bcl-2, which is an important antiapoptotic protein, we wonder whether there is any correlation between miR-15a/16 and apoptosis of HepG2 cells. In order to address this question, HepG2.2.15 cells were transfected with a miR-15a mimic and then treated with etoposide. As shown in Fig. 1D, HepG2.2.15 cells transfected with the miR-15a mimic showed 4-fold more PARP-1 and caspase 9 cleavages than control cells treated with etoposide. We then transfected L-02 cells with the HBV replication plasmid pHBV1.3 and performed similar experiments. The data showed that PARP-1 cleavages were significantly lower in pHBV1.3-transfected cells than in control cells treated with etoposide (Fig. 1E). We also transfected HepG2 cells with the HBV replication plasmid pHBV1.3, with or without the miR-15a mimic, and performed experiments similar to those described above. The data showed that both PARP-1 and caspase 9 cleavages were 2-fold lower in pHBV1.3-transfected cells than in control cells treated with etoposide, while the miR-15a mimic could recover the PARP-1 and caspase 9 cleavages when it was cotransfected with pHBV1.3 (Fig. 1F). This suggested that HBV and miR-15a have opposite effects on apoptosis of hepatoma cells induced by etoposide.
HBV causes a reduction of miR-15a/16 in HepG2 cells.
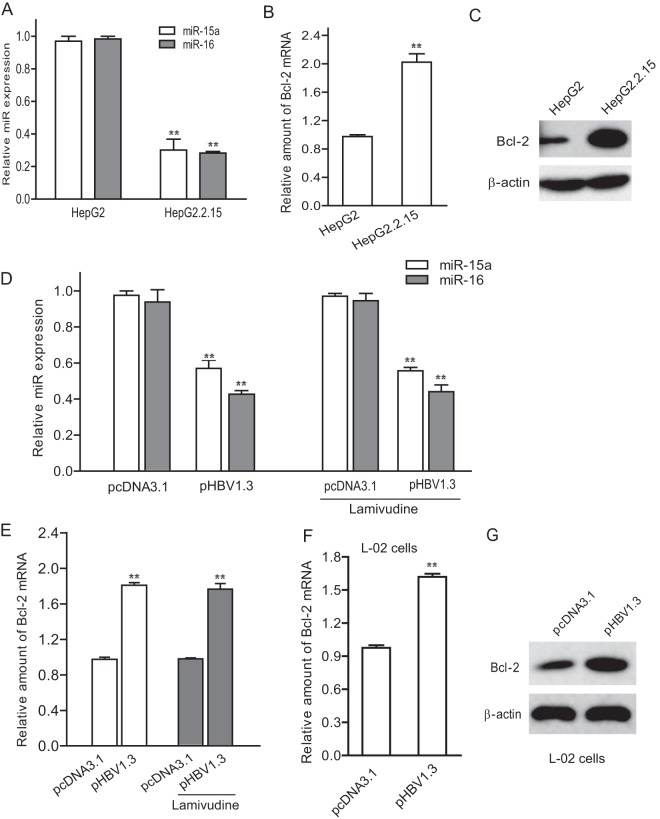
In order to understand how HBV inhibits miR-15a/16-related apoptosis of HepG2 cells, we first asked whether HBV affected miR-15a/16 expression. We examined the levels of miR-15a/16 in HepG2 and HepG2.2.15 cells by real-time PCR. The data showed that miR-15a/16 expression was decreased 60 to 70% in HepG2.2.15 cells compared to that in HepG2 cells (Fig. 2A). We also detected the expression level of Bcl-2, which is known to be one of the targets of miR-15a/16 in HepG2 and HepG2.2.15 cells. The results showed that both Bcl-2 mRNA and protein levels were significantly higher in HepG2.2.15 cells than in HepG2 cells (Fig. 2B and C). Next, we transfected pHBV1.3 into HepG2 cells and analyzed the miR-15a/16 expression. As shown in Fig. 2D, the level of miR-15a/16 in HepG2 cells transfected with pHBV1.3 was significantly reduced compared with that in control HepG2 cells (P < 0.01). In addition, we treated the HepG2 cells transfected with pHBV1.3 with lamivudine to inhibit HBV replication and examined the effect of HBV on miR-15a/16 and Bcl-2. The data indicated that cells transfected with pHBV1.3 could still reduce miR-15a/16 and upregulate Bcl-2 in lamivudine-treated cells (Fig. 2D and E). Furthermore, we performed a similar experiment in L-02 cells. Consistently, Bcl-2 mRNA and protein levels in L-02 cells transfected with pHBV1.3 were significantly elevated compared with those in control cells (Fig. 2F and G). Taking the data together, we propose that HBV transcripts can sponge miR-15a/16 and upregulate Bcl-2 expression.
Fig 2.
HBV causes a reduction of miR-15a/16 in HepG2 cells. (A and B) Total RNAs from HepG2 and HepG2.2.15 cells were extracted and subjected to real-time PCR for detection of miR-15a/16 expression (A) or Bcl-2 mRNA (B). The data for miR-15a/16 were normalized with endogenous U6. The data for Bcl-2 mRNA were normalized to β-actin mRNA. (C) The cell lysates from HepG2 and HepG2.2.15 cells were harvested and subjected to immunoblotting with Bcl-2 antibody and with β-actin antibody as a control. (D) HepG2 cells were transfected with pHBV1.3 or pcDNA3.1 (as a control) for 24 h and then treated with or without lamivudine (100 nM) for 24 h. The total RNA was extracted for real-time PCR to detect miR-15a/16 expression. (E) HepG2 cells were transfected with pHBV1.3 for 24 h and then treated with or without lamivudine (100 nM) for 24 h. The total RNA was extracted for real-time PCR to detect Bcl-2 mRNA levels. (F and G) L-02 cells were transfected with pHBV1.3 or pcDNA3.1 (as a control). The total RNA was extracted for real-time PCR to detect miR-15a/16 expression (F) and Bcl-2 mRNA (G). *, P < 0.05; **, P < 0.01.
HBV causes a reduction of the miR-15a/16 cluster in vivo.
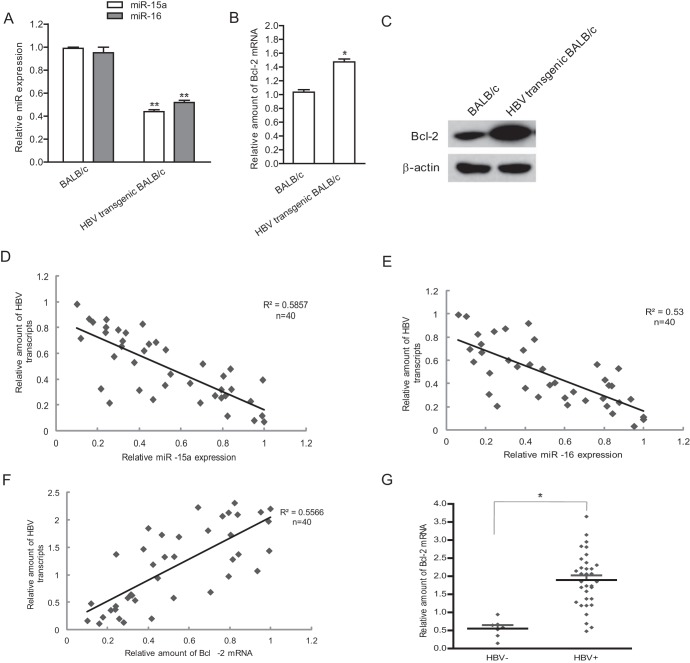
To further confirm that HBV can reduce the levels of miR-15a/16 in vivo, we examined the miR-15a/16 expression in HBV-transgenic BALB/c mice by real-time PCR. The data showed that the levels of miR-15a/16 in HBV-transgenic BALB/c mouse liver tissues were lower than those in control mice (P < 0.01) (Fig. 3A). Correspondingly, the expression of Bcl-2 mRNA (Fig. 3B) and protein (Fig. 3C) in liver tissues from HBV-transgenic mice was 2 (mRNA level)- and 3 (protein level)-fold higher than that in control mice (P < 0.05).
Fig 3.
HBV causes a reduction of miR-15a/16 in vivo. (A to C) Total RNAs extracted from liver tissues of HBV-transgenic BALB/c mice (n = 3) or control BALB/c mice (n = 3) were subjected to real-time PCR for detection of miR-15a/16 expression (A) and Bcl-2 mRNA (B) and to immunoblotting for detection of Bcl-2 protein (C). (D to F) Total RNAs from liver specimens of HBV-positive HCC patients were extracted and subjected to real-time PCR for miR-15a (D), miR-16 (E), and Bcl-2 mRNA (F), as well as HBV transcripts (D to F). Correlations between miR-15a/16, Bcl-2, and HBV mRNA levels in HBV-infected patient samples were analyzed by the Spearman rank test. Values for the correlation coefficient (R2) are shown. (G) Liver specimens from HBV-negative HCC patients were collected, and the total RNA was extracted and subjected to real-time PCR for Bcl-2 mRNA. The Bcl-2 mRNA levels were compared with those from HBV-positive patients. *, P < 0.05; **, P < 0.01.
We next examined the correlation between miR-15a/16 expression and HBV transcripts in liver tissues from HCC patients. As shown in Fig. 3D and E, there was a negative correlation between miR-15a/16 and HBV mRNA levels. We further examined the levels of Bcl-2 mRNA and HBV transcripts in liver tissues from the HCC patients by real-time PCR. The data indicated that there was a positive correlation between HBV mRNAs and Bcl-2 mRNA in tissues of HCC patients (Fig. 3F). We also compared the Bcl-2 expression levels in HCC with and without HBV infection. The results showed that HBV-positive HCC tissues displayed more expression of Bcl-2 protein than HBV-negative HCC tissues (Fig. 3G).
HBV mRNAs with a binding site complementary to miR-15a/16 sequester endogenous miR-15a/16.
It has been reported that microRNAs can be regulated by direct sponging (26). To address whether HBV downregulates miR-15a/16 through direct sponging, we first searched for putative miR-15a/16-binding sites in the HBV genomic sequence by using TargetScan at the miRBase website (http:www.mirbase.org). A putative miR-15a/16-complementary region (spanning nucleotides 1368 to 1383 of the HBV genome; GenBank accession no. EU562217) was found in HBV mRNAs, including pre-C/C (pgRNA), pre-S, and the S 3′-UTR (Fig. 4A). This predicted miR-15a/16-binding site is highly conserved among different HBV genotypes (Fig. 4B). We generated a 3′-UTR luciferase reporter for wild-type HBV (UTR-HBV-wt) or its mutant (UTR-HBV-mut), in which mutations in the miR-15a/16-binding site were made as indicated in Fig. 4A. HepG2 cells were transfected with the above-described 3′-UTR luciferase reporters and miR-15a/16 mimics. The cell lysates were harvested for luciferase assay. The data showed that miR-15a/16 mimics can reduce the reporter activity of UTR-HBV-wt but not UTR-HBV-mut (Fig. 4C).
Fig 4.
HBV can sponge the miR-15a/16 cluster. (A) The predicted miR-15a/16-binding sequences are located at nt 1362 to 1383 of the HBV genome. Perfect matches are indicated by vertical lines. Mutations in HBV were made in the predicted seed region of miR-15a/16-binding sites (UTR-HBV-mut) (upper panel; the mutated nucleotides are shown in bold italics). A putative miR-15a/16-complementary region was found in the HBV mRNAs and is indicated with gray boxes (lower panel). (B) miR-15a/16-binding sequences in different genotypes of HBV were pulled from the database, and the homology among them was determined. The seed region of miR-15a/16 is shown in bold italics. (C) HepG2 cells were cotransfected with a miR-15a/16 mimic, or a randomized oligonucleotide as a control, and the dual-luciferase reporter plasmid UTR-HBV-wt or UTR-HBV-mut. The cell lysates were harvested for firefly luciferase and Renilla luciferase activity assays. The relative luciferase activity was quantified. (D) HepG2 cells were transfected with pHBV1.3 or pHBV1.3-mut for 48 h. The total RNA was prepared for real-time PCR to measure miR-15a/16 levels. (E to H) HepG2 cells were transfected with pHBV1.3 or pHBV1.3-mut. Forty-eight hours after transfection, the total RNA was extracted and subjected to real-time PCR for HBV transcripts (E) and miR-15a/16 levels (H). (F) The supernatants were collected at the indicated time points for ELISA to detect HBV surface antigen. (G) The DNAs extracted from cells were subjected to Southern blotting for HBV DNA. RC and DL, relaxed circular and dendrimer DNAs, respectively. (I) HepG2 cells were transfected with the indicated amounts of pHBV1.3. The total HBV transcripts (left) and miR-15a/16 expression (right) were quantified by real-time PCR. *, P < 0.05; **, P < 0.01.
To further determine whether HBV UTR-HBV-wt could act as a “sponge” to absorb miR-15a/16, we transfected HepG2 cells with UTR-HBV-wt and its mutant, UTR-HBV-mut, and then analyzed the miR-15a/16 expression by real-time PCR. The results showed that miR-15a/16 expression was dramatically reduced in HepG2 cells transfected with UTR-HBV-wt compared to that in control cells and the cells transfected with UTR-HBV-mut (Fig. 4D).
Next, in order to test whether full-length HBV has the same effect on miR-15a/16 as UTR-HBV-wt, we made the same mutations in full-length HBV (pHBV1.3-mut) as those shown in Fig. 4A. HepG2 cells were transfected with pHBV1.3 or pHBV1.3-mut, and the levels of HBV transcripts were comparable in pHBV1.3- and pHBV1.3-mut-transfected cells (Fig. 4E). The data from ELISA and Southern blotting also indicated that HBV surface antigen in supernatants and HBV DNA in pHBV1.3- or pHBV1.3-mut-transfected cells were present at similar levels (Fig. 4F and G). We then examined the miR-15a/16 levels by real-time PCR. As shown in Fig. 4H, miR-15a/16 levels were decreased in pHBV1.3-transfected cells, but not in pHBV1.3-mut-transfected cells, compared with that in control cells. In addition, we performed a similar experiment with cells transfected with different amounts of pHBV1.3 and examined the HBV mRNA and miR-15a/16 levels by real-time PCR. The data showed that miR-15a/16 decreased more dramatically as the HBV mRNAs increased, indicating that HBV caused the reduction of miR-15a/16 in a dose-dependent manner (Fig. 4I). All these results suggested that HBV was able to sequester endogenous miR-15a/16 by direct binding.
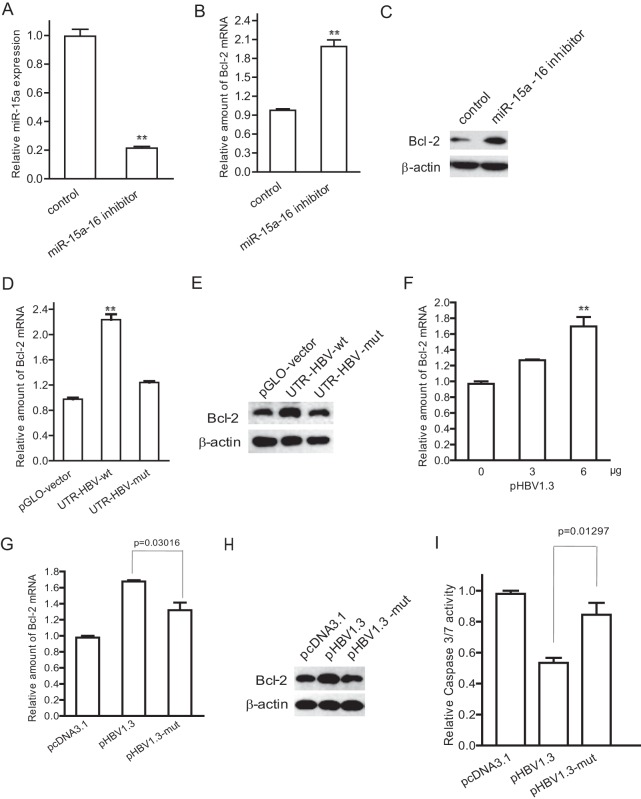
Reduction of miR-15a/16 by HBV leads to increased expression of its Bcl-2 target.
It was reported that Bcl-2 is the target of miR-15a/16. We also showed that a miR-15a/16 inhibitor transfected into HepG2 cells caused an obvious decrease of miR-15a/16 (Fig. 5A), a 2-fold increase of Bcl-2 mRNA (Fig. 5B), and a 3.2-fold increase of Bcl-2 protein (Fig. 5C). As we found that HBV mRNAs can sponge miR-15a/16, we therefore asked whether HBV can increase the expression of Bcl-2. To address this question, we transfected HepG2 cells with either UTR-HBV-wt or UTR-HBV-mut and examined the expression of Bcl-2. The data showed that both Bcl-2 mRNA and protein were about 2-fold higher in UTR-HBV-wt-transfected cells than in control and UTR-HBV-mut-transfected cells (Fig. 5D and E). The Bcl-2 mRNA was increased in pHBV1.3-transfected cells in a dose-dependent manner (Fig. 5F). We then transfected HepG2 cells with pHBV1.3 or its mutant, pHBV1.3-mut, and examined the Bcl-2 expression. The data showed that Bcl-2 mRNA and Bcl-2 protein were increased in pHBV1.3-transfected cells but not in pHBV1.3-mut-transfected cells (Fig. 5G and H). We further detected the caspase 3/7 activity of HepG2 cells transfected with pHBV1.3 and pHBV1.3-mut and treated with etoposide. As shown in Fig. 5I, caspase 3/7 activity was decreased in pHBV1.3- but not pHBV1.3-mut-transfected cells compared with that in control cells.
Fig 5.
Reduction of miR-15a/16 by HBV leads to increased expression of its target, Bcl-2. (A to C) HepG2 cells were transfected with a miR-15a/16 inhibitor or a randomized oligonucleotide as a control for 48 h. The total RNA was extracted and quantified by real-time PCR for miR-15a/16 expression (A) and Bcl-2 mRNA (B). (C) The cell lysates were harvested for immunoblotting with Bcl-2 antibody. (D and E) HepG2 cells were transfected with UTR-HBV-wt or UTR-HBV-mut for 48 h. (D) The Bcl-2 mRNA was quantified by real-time PCR. (E) The protein level of Bcl-2 was detected by immunoblotting analysis. (F) HepG2 cells were transfected with the indicated amounts of pHBV1.3. The amount of Bcl-2 mRNA was analyzed by real-time PCR. (G and H) HepG2 cells were transfected with pHBV1.3, pHBV1.3-mut, or pcDNA3.1 (as a control) for 48 h. The amount of Bcl-2 mRNA was quantified by real-time PCR (G), and the protein level of Bcl-2 was detected by immunoblotting (H). (I) HepG2 cells were transfected with pHBV1.3, pHBV1.3-mut, or pcDNA3.1 for 24 h and then treated with etoposide (0.08 μM) for 24 h. The cell lysates were harvested and subjected to caspase 3/7 activity assay. *, P < 0.05; **, P < 0.01.
Taking the data together, we made the conclusion that HBV can directly sponge miR-15a/16 and consequently upregulate the expression of Bcl-2 and then eventually inhibit the downstream cascade of apoptosis in hepatoma cells.
DISCUSSION
Liver cancer is the third leading cause of cancer mortality worldwide, with an annual death toll of 700,000 (27). More and more evidence indicates that chronic HBV infection is related to the occurrence and development of hepatoma (28). In the present study, we investigated the mechanism of how HBV inhibits etoposide-induced apoptosis of hepatoma cells and revealed that miR-15a/16 is involved in HBV-mediated inhibition of apoptosis. We found a negative correlation between HBV transcripts and miR-15a/16 in HBV-infected samples from HCC patients. Our further investigation showed that HBV mRNAs contain a binding site for miR-15a/16 and act as a sponge to bind and sequester endogenous miR-15a/16. Consequently, as the target of miR-15a/16, Bcl-2 mRNA was increased in HBV-infected cells, suggesting that HBV inhibits apoptosis of hepatoma cells by increasing expression of the antiapoptotic protein Bcl-2. Therefore, we proposed that the reduction of miR-15a/16 may contribute to chronic HBV infection, and perhaps the risk of development of HCC.
The major point of our work is to demonstrate that HBV inhibits apoptosis of hepatoma cells by sponging miR-15a/16. Direct HBV-mediated downregulation of miR-15a/16 was reported recently (29). In this study, we identified a novel region in HBV (nt 1362 to 1383) which can sponge miR-15a/16. This finding suggested that HBV possesses multiple ways to regulate the level of miR-15a/16. It has been reported that MCMV can produce highly abundant transcripts to downregulate cellular miR-27 for its efficient replication (21). Based on the evidence that miR-15a/16 is directly sponged by HBV mRNAs, we propose that HBV may downregulate miR-15a/16 to favor its replication and persistent infection. To support this hypothesis, we provided direct evidence to show that there is a negative correlation between HBV and miR-15a/16 and a positive correlation between HBV transcripts and Bcl-2 mRNA in HBV-infected patients. It has been reported that Bcl-2 is upregulated in liver tissues of HCC patients and that a Bcl-2 inhibitor can induce apoptosis in hepatocellular carcinoma cells (30). However, the detailed mechanism of Bcl-2 being regulated in hepatocytes has not been elucidated yet. Our results may explain the mechanism of upregulation of Bcl-2 expression in HBV-infected cells by HBV mRNAs sponging miR-15a/16.
Etoposide was selected as an apoptosis inducer for hepatoma cells in this study because a previous study showed that the Bcl-2 protein can inhibit etoposide-induced apoptosis (31). Since apoptosis plays an important role in the progress of liver diseases through various extrinsic or intrinsic pathways, with activation of caspases and the possible involvement of mitochondrial alteration (32), many efforts have been made to understand the role of HBV, mainly HBx, in the regulation of apoptosis. To date, the reported effects of HBx on apoptosis are controversial. The discrepancy of the role of HBx in apoptosis may be due to the different cell lines and experimental systems used in the studies. Nevertheless, the majority of these studies demonstrated that HBx can inhibit cell apoptosis (9, 33–35). In this report, we provide evidence that HBV mRNAs can inhibit hepatoma cell apoptosis by upregulating Bcl-2 protein expression.
In conclusion, our study provides a novel insight into the mechanism of HBV-mediated antiapoptosis of hepatoma cells by upregulating Bcl-2 expression through sponging miR-15a/16. Our data suggest that downregulation of miR-15a/16 during HBV infection causes increased expression of its targets, which gives a hint that another novel target(s) of miR-15a/16 may play important roles in chronic HBV infection or HCC development. Given the HBV mRNA–miR-15a/16–Bcl-2 regulatory pathway, our data may expand the knowledge of how HBV is involved in inhibiting apoptosis and provide a rational explanation for how HBV contributes to facilitating hepatoma cell survival and CHB progression.
ACKNOWLEDGMENTS
This work was supported by Ministry of Science and Technology of China (grants 2012CB519003, 2013ZX10004611, 2011CB504705, and 2012ZX10004501-001-003), National Natural Science Foundation of China (grant 81272272), and Chinese Academy of Sciences (grant KSCX2-EW-J-6) innovation projects. Xin Ye is a principal investigator of the Innovative Research Group of the National Natural Science Foundation of China (grant 81021003).
Footnotes
Published ahead of print 2 October 2013
REFERENCES
- 1.Neuveut C, Wei Y, Buendia MA. 2010. Mechanisms of HBV-related hepatocarcinogenesis. J. Hepatol. 52:594–604 [DOI] [PubMed] [Google Scholar]
- 2.Rehermann B, Nascimbeni M. 2005. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 5:215–229 [DOI] [PubMed] [Google Scholar]
- 3.Kew MC. 2010. Epidemiology of chronic hepatitis B virus infection, hepatocellular carcinoma, and hepatitis B virus-induced hepatocellular carcinoma. Pathol. Biol. 58:273–277 [DOI] [PubMed] [Google Scholar]
- 4.Beasley RP, Hwang LY, Lin CC, Chien CS. 1981. Hepatocellular carcinoma and hepatitis B virus. Lancet ii:1129–1133 [DOI] [PubMed] [Google Scholar]
- 5.De Mitri MS, Cassini R, Bernardi M. 2010. Hepatitis B virus-related hepatocarcinogenesis: molecular oncogenic potential of clear or occult infections. Eur. J. Cancer 46:2178–2186 [DOI] [PubMed] [Google Scholar]
- 6.Du J, Liang X, Liu Y, Qu Z, Gao L, Han L, Liu S, Cui M, Shi Y, Zhang Z, Yu L, Cao L, Ma C, Zhang L, Chen Y, Sun W. 2009. Hepatitis B virus core protein inhibits TRAIL-induced apoptosis of hepatocytes by blocking DR5 expression. Cell Death Differ. 16:219–229 [DOI] [PubMed] [Google Scholar]
- 7.Wang XW, Gibson MK, Vermeulen W, Yeh H, Forrester K, Sturzbecher HW, Hoeijmakers JH, Harris CC. 1995. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 55:6012–6016 [PubMed] [Google Scholar]
- 8.Su F, Schneider RJ. 1997. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor alpha. Proc. Natl. Acad. Sci. U. S. A. 94:8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diao J, Khine AA, Sarangi F, Hsu E, Iorio C, Tibbles LA, Woodgett JR, Penninger J, Richardson CD. 2001. X protein of hepatitis B virus inhibits Fas-mediated apoptosis and is associated with up-regulation of the SAPK/JNK pathway. J. Biol. Chem. 276:8328–8340 [DOI] [PubMed] [Google Scholar]
- 10.Krol J, Loedige I, Filipowicz W. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11:597–610 [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 12.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, 2nd, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. 2007. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13:613–618 [DOI] [PubMed] [Google Scholar]
- 13.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. 2009. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 69:1135–1142 [DOI] [PubMed] [Google Scholar]
- 14.Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG, Jia JH. 2011. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 411:586–592 [DOI] [PubMed] [Google Scholar]
- 15.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294:853–858 [DOI] [PubMed] [Google Scholar]
- 16.Aqeilan RI, Calin GA, Croce CM. 2010. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 17:215–220 [DOI] [PubMed] [Google Scholar]
- 17.Janumyan YM, Sansam CG, Chattopadhyay A, Cheng N, Soucie EL, Penn LZ, Andrews D, Knudson CM, Yang E. 2003. Bcl-xL/Bcl-2 coordinately regulates apoptosis, cell cycle arrest and cell cycle entry. EMBO J. 22:5459–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reed JC. 1994. Bcl-2 and the regulation of programmed cell death. J. Cell Biol. 124:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaux DL, Cory S, Adams JM. 1988. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 335:440–442 [DOI] [PubMed] [Google Scholar]
- 20.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. 1984. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 226:1097–1099 [DOI] [PubMed] [Google Scholar]
- 21.Marcinowski L, Tanguy M, Krmpotic A, Radle B, Lisnic VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, Babic M, Zimmer R, Trgovcich J, Koszinowski UH, Jonjic S, Pfeffer S, Dolken L. 2012. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 8:e1002510. 10.1371/journal.ppat.1002510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Wang Y, Wang S, Wu B, Hao J, Fan H, Ju Y, Ding Y, Chen L, Chu X, Liu W, Ye X, Meng S. 2013. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J. Virol. 87:2193–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu G, Yu F, Xiao Z, Xu K, Xu J, Tang W, Wang J, Song E. 2011. Hepatitis B virus X protein downregulates expression of the miR-16 family in malignant hepatocytes in vitro. Br. J. Cancer 105:146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Zhao JJ, Wang CM, Li MY, Han P, Wang L, Cheng YQ, Zoulim F, Ma X, Xu DP. 2009. Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin. Med. J. 122:10–14 [DOI] [PubMed] [Google Scholar]
- 25.Chen A, Wang L, Zhang J, Zou L, Jia Z, Zhou W, Wan Y, Wu Y. 2005. H-2 Kd-restricted hepatitis B virus-derived epitope whose specific CD8+ T lymphocytes can produce gamma interferon without cytotoxicity. J. Virol. 79:5568–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebert MS, Sharp PA. 2010. MicroRNA sponges: progress and possibilities. RNA 16:2043–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Yang L, Liu X, Chen W, Chang L, Chen L, Loera S, Chu P, Huang WC, Liu YR, Yen Y. 2013. MicroRNA-657 promotes tumorigenesis in hepatocellular carcinoma by targeting transducin-like enhancer protein 1 through nuclear factor kappa B pathways. Hepatology 57:1919–1930 [DOI] [PubMed] [Google Scholar]
- 28.Whittaker S, Marais R, Zhu AX. 2010. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 29:4989–5005 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu XD. 2013. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J. Biol. Chem. 288:18484–18493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X, Ogunwobi OO, Liu C. 2011. Survivin inhibition is critical for Bcl-2 inhibitor-induced apoptosis in hepatocellular carcinoma cells. PLoS One 6:e21980. 10.1371/journal.pone.0021980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamesaki S, Kamesaki H, Jorgensen TJ, Tanizawa A, Pommier Y, Cossman J. 1993. Bcl-2 protein inhibits etoposide-induced apoptosis through its effects on events subsequent to topoisomerase II-induced DNA strand breaks and their repair. Cancer Res. 53:4251–4256 [PubMed] [Google Scholar]
- 32.Liu Q, Chen J, Liu L, Zhang J, Wang D, Ma L, He Y, Liu Y, Liu Z, Wu J. 2011. The X protein of hepatitis B virus inhibits apoptosis in hepatoma cells through enhancing the methionine adenosyltransferase 2A gene expression and reducing S-adenosylmethionine production. J. Biol. Chem. 286:17168–17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elmore LW, Hancock AR, Chang SF, Wang XW, Chang S, Callahan CP, Geller DA, Will H, Harris CC. 1997. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc. Natl. Acad. Sci. U. S. A. 94:14707–14712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marusawa H, Matsuzawa S, Welsh K, Zou H, Armstrong R, Tamm I, Reed JC. 2003. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 22:2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo TC, Chao CC. 2010. Hepatitis B virus X protein prevents apoptosis of hepatocellular carcinoma cells by upregulating SATB1 and HURP expression. Biochem. Pharmacol. 80:1093–1102 [DOI] [PubMed] [Google Scholar]