Abstract
We identified the domains of CD26 involved in the binding of Middle East respiratory syndrome coronavirus (MERS-CoV) using distinct clones of anti-CD26 monoclonal antibodies (MAbs). One clone, named 2F9, almost completely inhibited viral entry. The humanized anti-CD26 MAb YS110 also significantly inhibited infection. These findings indicate that both 2F9 and YS110 are potential therapeutic agents for MERS-CoV infection. YS110, in particular, is a good candidate for immediate testing as a therapeutic modality for MERS.
TEXT
A novel coronavirus, Middle East respiratory syndrome coronavirus (MERS-CoV), was identified in patients with severe lower respiratory tract infections with almost 50% of cases resulting in lethal lower respiratory tract infections (1–5). Initially, MERS-CoV infection occurred sporadically; however, horizontal infection among human patients has been demonstrated and has potential pandemic ramifications. While MERS-CoV was reported to be sensitive to alpha interferon or cyclosporine treatment (6, 7), there are no vaccines or effective therapies currently available for clinical cases of MERS-CoV infection.
A recent report showed that the spike (S) protein of MERS-CoV mediates infection (8) using dipeptidyl peptidase IV (DPPIV; EC 3.4.14.5) as a functional receptor (9). This receptor is conserved among different species, such as bats and humans, which partially explains the large host range of MERS-CoV. DPPIV is also known as CD26, which is a 110-kDa cell surface glycoprotein with dipeptidase activity in its extracellular domain (10). CD26/DPPIV is a multifunctional cell surface protein that is widely expressed in most cell types, including T lymphocytes, bronchial mucosa, and the brush border of proximal tubules. This distribution of CD26 may play a role in the systemic dissemination of MERS-CoV infection in humans (11–13). Therefore, an effective therapy for MERS-CoV infection is needed not only to block the entry of MERS-CoV into such CD26-expressing organs as the respiratory system, kidney, liver, or intestine but also to eliminate circulating MERS-CoV. More recently, crystal structure analysis revealed the CD26–MERS-CoV binding regions (14, 15), and manipulation of CD26/DPPIV levels or the development of inhibitors that target the interaction between the MERS-CoV S domain and its receptor may provide therapeutic opportunities to combat MERS-CoV infection. In the present study, we mapped MERS-CoV S protein binding regions in human CD26 molecules and demonstrated that anti-CD26 monoclonal antibodies (MAbs) that were developed in our laboratory effectively blocked the interaction between the spike protein and CD26, thereby neutralizing MERS-CoV infectivity.
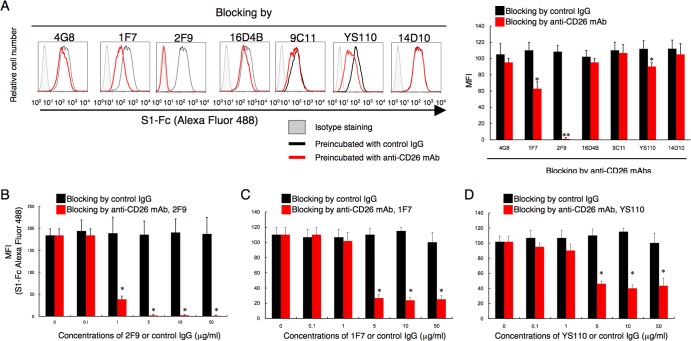
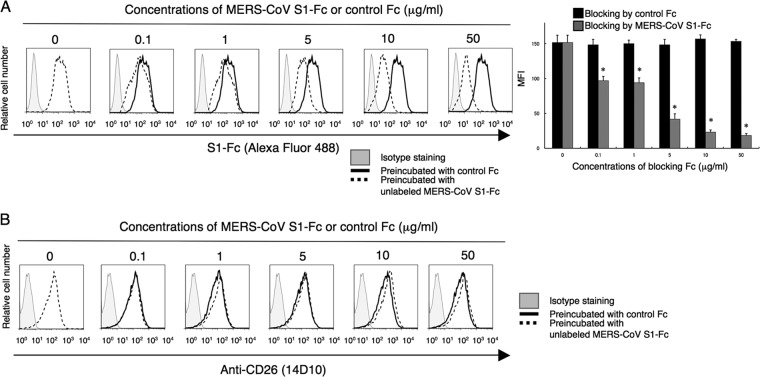
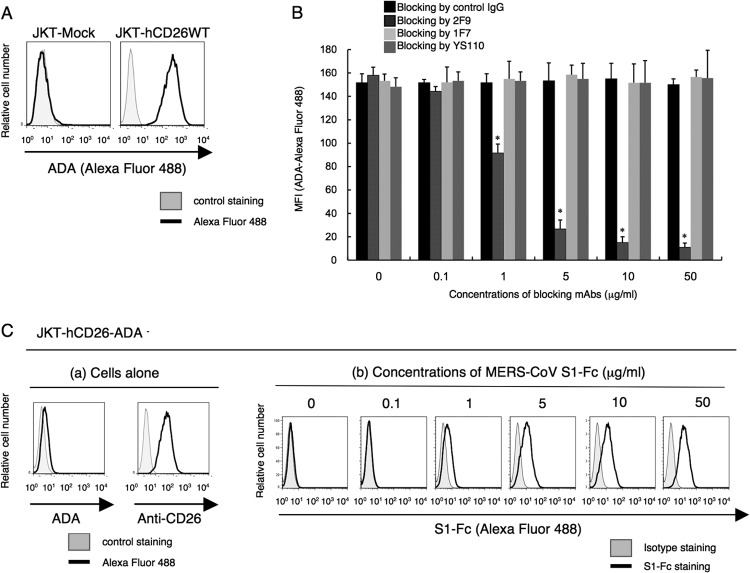
In a recent study by Raj et al., anti-CD26 polyclonal antibody (pAb), but not DPPIV inhibitors, was used to inhibit in vitro MERS-CoV infection (9). Moreover, Mou et al. demonstrated that pAbs to the MERS-CoV S1 domain efficiently neutralize MERS-CoV infection (8). To determine the specific CD26 domain involved in MERS-CoV infection, we chose six different clones of anti-CD26 MAbs (4G8, 1F7, 2F9, 16D4B, 9C11, and 14D10) and the humanized anti-CD26 MAb YS110, which recognize six distinct epitopes of the CD26 molecule (16, 17), to conduct MERS-CoV S1-Fc (where S1-Fc is the S1 domain of MERS-CoV fused to the Fc region of human IgG) binding-inhibition assays. For this purpose, we used a CD26-negative Jurkat cell line stably transfected with full-length human CD26 (JKT-hCD26WT) or a pcDL-SRα296 vector control (JKT-Mock) (10). As shown in Fig. 1A, expression of CD26 was confirmed in JKT-hCD26WT cells but not in JKT-Mock cells, and binding of MERS-CoV S1-Fc to CD26 in JKT-hCD26WT cells was also confirmed (Fig. 1B). As shown in Fig. 2A, 2F9 inhibited full binding of MERS-CoV S1-Fc to JKT-hCD26WT, while other anti-CD26 MAbs demonstrated some inhibition (1F7 and YS110) or no significant inhibition (4G8, 16D4B, 9C11, and 14D10). The blocking effect of 2F9 was dose dependent (Fig. 2B). Since downmodulation of CD26 expression by anti-CD26 MAbs has been observed under certain experimental conditions (18), we evaluated surface expression of CD26, but expression levels of CD26 were not affected by changes in 2F9 concentration (data not shown). Moreover, MERS-CoV S1-Fc binding to JKT-hCD26WT was considerably inhibited by 1F7 or YS110 at concentrations of 5 to 10 μg/ml or greater, but complete blocking of MERS-CoV S1-Fc binding was not achieved even at a concentration of 50 μg/ml (Fig. 2C and D, respectively). These results suggest that 2F9 as well as 1F7 and YS110 inhibited binding of MERS-CoV S1-Fc to CD26 and that the binding regions of MERS-CoV S1-Fc are fully covered by 2F9 and partially overlap with the epitopes recognized by 1F7 or YS110. On the other hand, in the presence of unlabeled MERS-CoV S1-Fc at concentrations of 10 μg/ml or greater, MERS-CoV S1-Fc binding to JKT-hCD26WT was significantly inhibited (Fig. 3A), with no change in CD26 expression levels (Fig. 3B). However, complete blocking of MERS-CoV S1-Fc binding was not achieved even at a concentration of 50 μg/ml of preincubated MERS-CoV S1-Fc (Fig. 3A). These results strongly suggest that the anti-CD26 MAb 2F9 has greater therapeutic potential than recombinant MERS-CoV S1-Fc to prevent viral entry into susceptible cells and that 1F7 or YS110 also blocks MERS-CoV infection.
Fig 1.
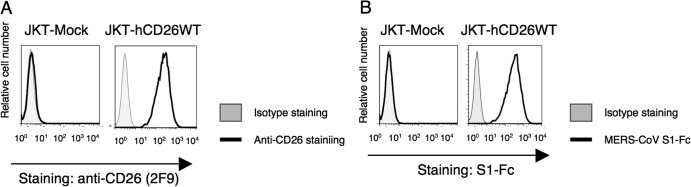
CD26 expression and binding of MERS-CoV S1-Fc in parental Jurkat cells and CD26 Jurkat transfectants. (A) Representative histograms showing results of staining of Jurkat cells stably transfected with the full-length human CD26 (JKT-hCD26WT) or vector control (JKT-Mock) with Alexa Fluor 488-labeled anti-CD26 MAb 2F9 (5 μg/ml; black).Gray histograms show results of staining with an isotype control (Alexa Fluor 488-labeled mouse IgG [msIgG-488]; 5 μg/ml). Results representative of three different experiments are shown. (B) Representative histograms showing results of staining with Alexa Fluor-labeled MERS-CoV S1-Fc (5 μg/ml; black) using JKT-Mock or JKT-hCD26WT. Gray histograms show staining with Alexa Fluor 488-labeled recombinant human Fc (Fc-488) as an isotype control. Results representative of three different experiments are shown.
Fig 2.
Anti-CD26 MAb 2F9 inhibits binding of MERS-CoV S1-Fc. (A) Representative histograms showing results of staining with MERS-CoV S1-Fc in the presence of various clones of anti-CD26 MAbs or control mouse IgG (left). JKT-hCD26WT cells were incubated with the indicated anti-CD26 MAb (mouse MAb 4G8, 1F7, 14D10, 2F9, 16D4B, or 9C11 or humanized MAb YS110) (red) or control IgG (black) (each 10 μg/ml) for 30 min at 4°C. After being washed, cells were stained with Alexa Fluor 488-labeled MERS-CoV S1-Fc (5 μg/ml). Gray histograms show results of staining with Fc-488 as an isotype control. Mean fluorescence intensities (MFI) of Alexa Fluor 488-labeled MERS-CoV S1-Fc are indicated in the bar graph (right). Results representative of three different experiments are shown as mean MFI. Error bars indicate standard errors of the means (SEMs) (two-tailed Student's t test; * or **, P < 0.05 versus control IgG). (B to D) MFI of staining with Alexa Fluor 488-labeled MERS-CoV S1-Fc in the presence of various concentrations of the anti-CD26 MAb 2F9 (B), 1F7 (C), or YS110 (D) (red) or control msIgG (black). JKT-hCD26WT cells were incubated with the indicated concentrations of the anti-CD26 MAbs or control IgG for 30 min at 4°C. After being washed, cells were stained with Alexa Fluor 488-labeled MERS-CoV S1-Fc (5 μg/ml). Results of three different experiments are shown as mean MFI ± SEMs (two-tailed Student's t test; *, P < 0.05 versus corresponding control IgG). The black and red bars at 0 μg/ml of preincubated control IgG or anti-CD26 MAbs were plotted using the same data.
Fig 3.
Preincubation with MERS-CoV S1-Fc partially inhibits binding of MERS-CoV S1-Fc. (A) Representative histograms showing results of staining with MERS-CoV S1-Fc in the presence of various concentrations of unlabeled MERS-CoV S1-Fc or control Fc (left). JKT-hCD26WT cells were incubated with the indicated concentrations of unlabeled MERS-CoV S1-Fc (dashed) or control Fc (black) for 30 min at 4°C. After being washed, cells were stained with Alexa Fluor 488-labeled MERS-CoV S1-Fc (5 μg/ml). Gray histograms show staining with the isotype control (Fc-488). MFI of Alexa Fluor 488-labeled MERS-CoV S1-Fc are indicated in the bar graph (right). Results representative of three different experiments are shown as mean MFI. Error bars indicate SEMs (two-tailed Student's t test; *, P < 0.05 versus corresponding control Fc). The black and dark-gray bars at 0 μg/ml of preincubated MERS-CoV S1-Fc or control Fc were plotted using the same data. (B) Representative histograms showing staining with the anti-CD26 MAb 14D10 in the presence of various concentrations of MERS-CoV S1-Fc or control Fc. The experiments were conducted as for panel A. Gray histograms show staining with the isotype control (msIgG-488).
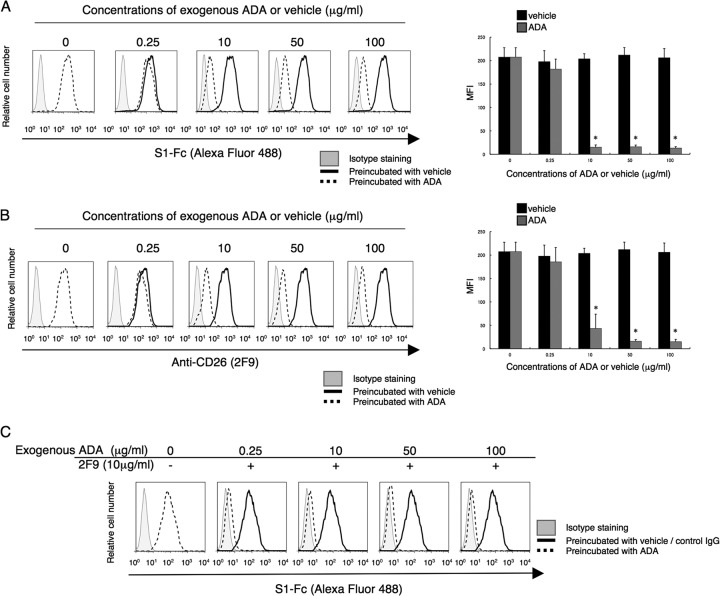
Human CD26 is known as the adenosine deaminsae 1 (ADA) binding protein (19–22). The epitope of the anti-human CD26 MAb 2F9 was estimated to be located near the ADA binding region of CD26, whereas the epitopes of the other anti-CD26 MAbs tested, including 1F7 and YS110, did not involve the ADA binding region (16). Moreover, the epitopes defined by 1F7 and YS110 were almost identical and binding of either antibody cross-blocked the other. Consistent with our previous work demonstrating CD26 binding to ADA (19), binding of exogenous ADA was detected on JKT-hCD26WT but not on CD26-negative parental Jurkat cells (Fig. 4A). Although 2F9 almost completely blocked binding of ADA to CD26, 1F7 or YS110 did not block binding of ADA to CD26 (Fig. 4B). However, as shown in Fig. 2C and D, 1F7 or YS110 considerably inhibited MERS-CoV binding to CD26. These observations suggest that MERS-CoV S1-Fc binding to CD26 involves ADA recognition sites of CD26 along with other potential CD26 domains. To further define the role of ADA recognition sites in MERS-CoV S1-Fc binding, we conducted binding assays using JKT-hCD26-ADA-negative (JKT-hCD26-ADA−) cells, which are Jurkat cells with the ADA binding regions of human CD26 mutated to prevent ADA binding (amino acid [aa] residues 340 to 344 of human CD26 replaced with those of mouse CD26) (20). While JKT-hCD26-ADA− cells expressed CD26, as determined by the anti-CD26 MAb 14D10, they did not bind to ADA (Fig. 4C, panels a). Importantly, binding of MERS-CoV S1-Fc to JKT-hCD26-ADA− was clearly observed at concentrations of 5 μg/ml or greater (Fig. 4C, panels b), but the binding intensity appeared to be lower than that observed with JKT-hCD26WT (Fig. 1B), suggesting that the region where CD26 binds to MERS-CoV S1-Fc partially overlaps with its ADA binding regions. Taken together, these results strongly suggest that the regions where CD26 binds to MERS-CoV S1-Fc are located at the ADA recognition sites as well as additional regions distinct from the ADA binding region. Since ADA is found in the sera or body fluids, it is possible that existing ADA interferes with binding of MERS-CoV or 2F9 to CD26 in patients. We then examined whether MERS-CoV S1-Fc binding was inhibited by ADA. In the presence of ADA at concentrations of 10 μg/ml or greater, MERS-CoV S1-Fc binding to JKT-hCD26WT was inhibited (Fig. 5A) at a level similar to the level of inhibition seen with 2F9 (Fig. 5B). Meanwhile, CD26 expression was unchanged in the presence of exogenous ADA, as detected by the anti-CD26 MAb 14D10, which recognizes a distinct epitope (data not shown). Under this condition, adding 2F9 to exogenous ADA resulted in complete inhibition of MERS-CoV S1-Fc binding (Fig. 5C). These results strongly suggest that the regions where CD26 binds to MERS-CoV and 2F9 overlap with each other and include the ADA binding region and additional regions and that 2F9 inhibits MERS-CoV binding to CD26 with greater efficiency than ADA.
Fig 4.
Binding regions of ADA (adenosine deaminase 1) in CD26 are involved in the binding of MERS-CoV S1-Fc to human CD26. (A) Representative histograms showing results of the binding of ADA to JKT-Mock (left) or JKT-hCD26WT (right). JKT-Mock or JKT-hCD26WT was incubated with 10 μg/ml of Alexa Fluor 488-labeled ADA or ADA2 (CECR1) as a fluorescence control. Data are representative of three independent experiments, with similar results being obtained. (B) MFI for staining with Alexa Fluor 488-labeled ADA in the presence of various concentrations of the anti-CD26 MAb 2F9 (dark gray), 1F7 (light gray), YS110 (gray), or control msIgG (black). JKT-hCD26WT cells were incubated with the indicated concentrations of anti-CD26 MAbs or control IgG for 30 min at 4°C. After being washed, cells were stained with Alexa Fluor 488-labeled ADA (10 μg/ml). Alexa Fluor 488-labeled ADA2 was used as a fluorescence control, with MFI being <10. Results representative of three different experiments are shown as mean MFI. Error bars indicate SEMs (two-tailed Student's t test; *, P < 0.05 versus corresponding control IgG). (C, panel a) Representative histograms showing results for binding of ADA (left) or the anti-CD26-MAb 14D10 (right) to Jurkat cells stably transfected with human CD26 with a deletion of the ADA binding region (JKT-hCD26-ADA−). Gray histograms show Alexa Fluor 488-labeled ADA2 or msIgG-488 as a fluorescence control. (b) Representative histograms showing results for staining with MERS-CoV S1-Fc of JKT-hCD26-ADA−. JKT-hCD26-ADA− cells were stained with Alexa Fluor 488-labeled MERS-CoV S1-Fc (black) at the indicated concentrations. Gray histograms show results for staining with Fc-488 as an isotype control. Data are representative of three independent experiments, with similar results being obtained.
Fig 5.
2F9 fully inhibits binding to MERS-CoV S1-Fc of CD26 in the presence of exogenous ADA. (A and B) Representative histograms showing results for staining with MERS-CoV S1-Fc (A) or 2F9 (B) in the presence of various concentrations of exogenous ADA (dashed) or PBS (black) as a solvent control (vehicle) (left). JKT-hCD26WT cells were incubated with the indicated concentrations of exogenous ADA or corresponding concentrations of diluted PBS for 30 min at 37°C. After being washed, cells were stained with Alexa Fluor 488-labeled MERS-CoV S1-Fc or 2F9 (each 5 μg/ml). Gray histograms show results for staining with Fc-488 or msIgG-488 as an isotype control. MFI for Alexa Fluor 488-labeled MERS-CoV S1-Fc or 2F9 are indicated in the bar graphs (right). Results representative of three different experiments are shown as mean MFI. Error bars indicate SEMs (two-tailed Student's t test; *, P < 0.05 versus corresponding vehicle). The black and gray bars at 0 μg/ml of preincubated ADA or vehicle were plotted using the same data. (C) Representative histograms showing results for staining with MERS-CoV S1-Fc in the presence of various concentrations of exogenous ADA with the addition of 2F9 (dashed). JKT-hCD26WT cells were incubated with the indicated concentrations of exogenous ADA or corresponding concentrations of diluted PBS for 30 min at 37°C, followed by additional incubation with 2F9 (10 μg/ml) or control msIgG (10 μg/ml) for 30 min at 4°C. After being washed, cells were stained with Alexa Fluor 488-labeled MERS-CoV S1-Fc (5 μg/ml). The dashed histogram in the left panel shows results for staining with MERS-CoV S1-Fc in the presence of PBS with the addition of control msIgG. Gray histograms show staining with Fc-488 as an isotype control. Results representative of three different experiments are shown.
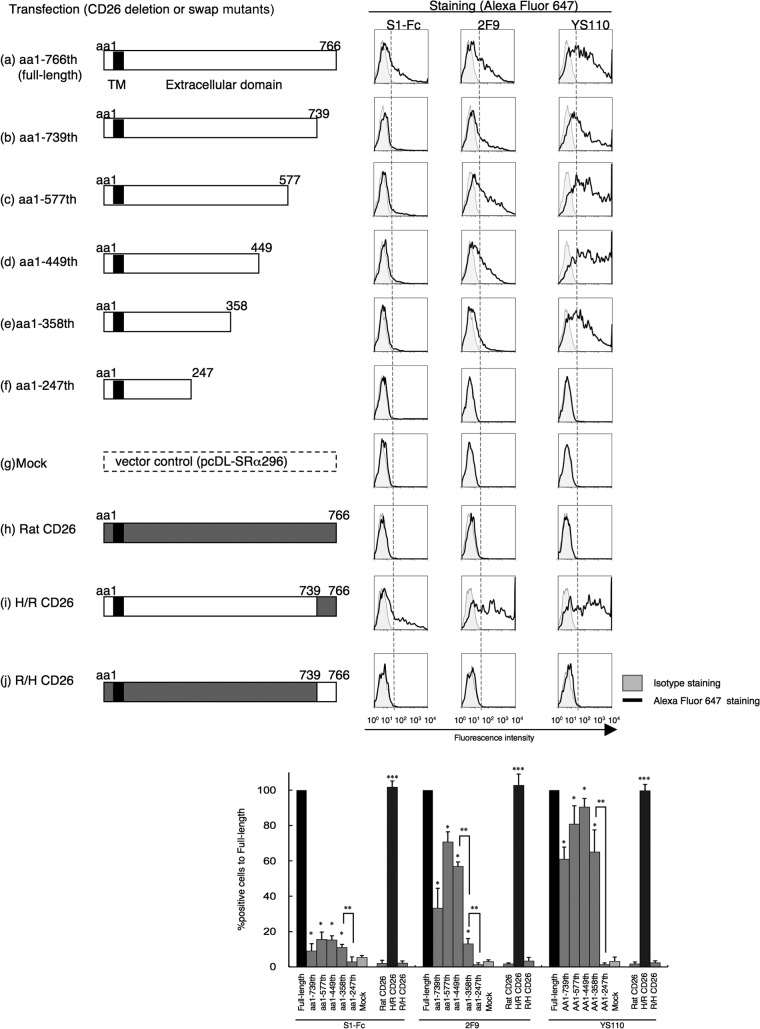
We next characterized the CD26 epitope involved in MERS-CoV S1-Fc binding to CD26 through the use of various CD26 mutants with a deletion in the C-terminal extracellular region, since CD26 is a type II transmembrane protein (23, 24). As shown in Fig. 6, the constructs containing the N-terminal aa residues 1 to 739 (Fig. 6b), 1 to 577 (Fig. 6c), 1 to 449 (Fig. 6d), or 1 to 358 (Fig. 6e) did not abrogate MERS-CoV S1-Fc binding (Fig. 6, bar graph, S1-Fc columns with one asterisk [*]), although binding efficiency was significantly decreased in comparison with that of the full-length human CD26 (Fig. 6a and black bars in graph). In contrast, MERS-CoV S1-Fc binding to CD26 mutants was not detected in aa 1 to 247 (Fig. 6f and bar graph, S1-Fc columns with two asterisks [**]), which was at levels similar to those in mock-transfected cells (Fig. 6g and bar graph, gray columns of S1-Fc). Furthermore, the results of Fig. 6 show that the epitopes defined by 2F9 or YS110 (which recognizes an epitope similar to 1F7) were located between aa 248 and 449 (Fig. 6, bar graph, 2F9 columns with two asterisks) or aa 248 to 358 (Fig. 6, bar graph, YS110 columns with two asterisks), respectively. These observations suggest that the CD26 epitope involved in binding to MERS-CoV S1-Fc might be located around aa 358, similar to the location of the 2F9 epitope. Our biological experiments above strongly indicate that binding of MERS-CoV S1-Fc to CD26 involves the ADA binding regions of CD26 as well as the additional sites distinct from the ADA binding regions, conclusions that are consistent with the results obtained in Fig. 2 and 4 and with those obtained using crystal structure analysis (14, 15). Moreover, observation using the CD26 mutant with a deletion of aa 740 to 766 indicated that the C-terminal region may be necessary for binding of MERS-CoV S1-Fc to CD26 (Fig. 6b). Since CD26 exists as homodimers and the C-terminal regions are located near the N-terminal membrane region to form the gate for grooves between each monomer, the deletion of the C-terminal region may disrupt the holding grooves of the dimeric CD26/DPPIV (25), thus disrupting the binding of MERS-CoV S1-Fc to the mutated CD26. To examine the role of this C-terminal region in maintaining the conformational structure, we generated a swap mutant using rat CD26, which has been estimated not to bind to MERS-CoV (9). The full-length rat CD26 or rat-human swap mutant consisting of aa 1 to 739 of rat CD26 with aa 740 to 766 of human CD26 showed no binding to MERS-CoV S1-Fc (Fig. 6h and j, respectively). On the other hand, the human-rat swap mutant consisting of aa 1 to 739 of human CD26 with aa 740 to 766 of rat CD26 (Fig. 6i) showed binding intensities to MERS-CoV S1-Fc, 2F9, or YS110 similar to those observed with the full-length human CD26 (Fig. 6a, bar graph, bars with three asterisks [***]). These results strongly suggest that maintaining the conformation of CD26 is important for its binding to MERS-CoV S1-Fc as well as to 2F9 or YS110. Taken together with the data that binding of MERS-CoV to CD26 was fully inhibited by 2F9 (Fig. 2B) and considerably inhibited by 1F7 or YS110 (Fig. 2C and D), these results strongly suggest that the main regions of CD26 for binding to MERS-CoV appear to be close to aa 358, recognized by 2F9, and that the region of CD26 defined by 1F7 and YS110 (aa 248 to 358) was also partially involved in MERS-CoV binding. Our biological experiments on the regions of CD26 that bind to MERS-CoV using MAbs show results comparable to those obtained from crystal structure analysis (14, 15), which are summarized in a schematic diagram of human CD26 at aa 1 to 449 (Fig. 7). Overall, our results indicate that targeting by 2F9 as well as 1F7 and YS110 may be an effective therapeutic strategy for MERS-CoV infection.
Fig 6.
Characterization of regions of CD26 that bind to MERS-CoV S1-Fc through the use of CD26 deletion or human/rat swap mutants. Representative histograms show results for staining for MERS-CoV S1-Fc, 2F9, or YS110. CD26 cDNAs with full-length human CD26 (a), the indicated deletion (b through f), human/rat (H/R) swap mutants (h through j), or vector control (g) were cotransfected with GFP-expressing plasmids to COS-1 cells. After 24 h of transfection, cells were stained with Alexa Fluor 647-labeled MERS-CoV S1-Fc, 2F9, or YS110 (each 5 μg/ml). Gray histograms show results of staining with Fc-647 or msIgG-647 as an isotype control. The histograms for Alexa Fluor 647 were obtained by gating for GFP-positive cells among all acquired cells. Results of three different experiments are shown as percent positive cells in full-length CD26 (bar graphs, bottom). Error bars indicate SEMs (two-tailed Student's t test; *, P < 0.05 versus mock; **, P < 0.05 versus corresponding mutants; ***, P < 0.05 versus corresponding rat full-length CD26). Vertical dashed lines in the histograms indicate borders between negative and positive intensities. In the diagrams of CD26, TM indicates a transmembrane region of human CD26, and amino acids (aa) derived from human (H) or rat (R) are represented in white or dark-gray boxes, respectively.
Fig 7.
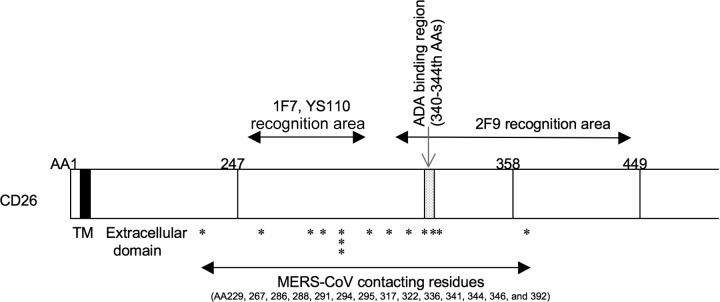
Schematic diagram of human CD26 profiling the predicted contact areas of anti-CD26 MAbs 2F9, 1F7, YS110, and MERS-CoV S1. 2F9 recognizes between 248 and 449 aa, including the ADA binding regions, and 1F7 and YS110 recognize between 248 and 358 aa, excluding the ADA binding regions. MERS-CoV-contacting residues of human CD26 are indicated by asterisks; information was obtained from recently published data (17, 18). TM indicates a transmembrane region of human CD26 (black box), and the extracellular domain of CD26 is located at the C-terminal residues of TM.
To determine whether treatment with anti-CD26 MAb 2F9 as well as 1F7 and YS110 could inhibit MERS-CoV infection, we preincubated susceptible cells with various anti-CD26 MAbs prior to inoculation with the virus. As shown previously, anti-CD26 goat pAb inhibited MERS-CoV infection of Huh-7 cells (Fig. 8). In this experimental system, infection was almost completely blocked by 2F9 (Fig. 8) but not by control IgG or several other anti-CD26 MAbs recognizing other epitopes (4G8, 16D4B, 14D10, or 9C11) (Fig. 8). Moreover, the anti-CD26 MAbs 1F7 and YS110 considerably inhibited MERS-CoV infection of Huh-7 cells (Fig. 8). These results demonstrate that 2F9 inhibits MERS-CoV entry and therefore can potentially be developed as a preventive or therapeutic agent for MERS-CoV infection in the clinical setting. More importantly, the humanized anti-CD26 MAb YS110 is being evaluated currently in patients with CD26-expressing cancers in an ongoing phase I clinical trial. Since no apparent adverse effects of YS110 have been reported besides the injection reaction, YS110 may be an immediate therapeutic candidate for clinical use as a treatment for MERS-CoV infection.
Fig 8.
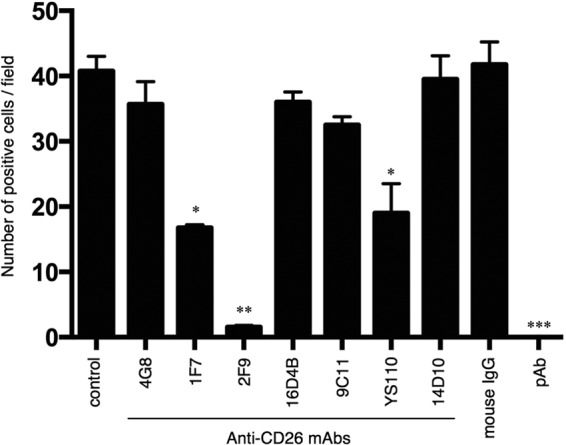
Inhibition of MERS-CoV infection by the anti-CD26 MAb 2F9. Huh-7 cells were preincubated with normal mouse IgG, various anti-CD26 MAbs (4G8, 1F7, 2F9, 16D4B, 9C11, or 14D10), humanized anti-CD26 MAb (YS110), or anti-CD26 goat polyclonal antibody (pAb) at a concentration of 40 μg/ml for 0.5 h prior to MERS-CoV virus inoculation (1 h), all at room temperature. Mock-incubated cells (control) were used as controls. Following incubation at 37°C for 8 h, infected cells were detected by immunofluorescence using anti-SARS-CoV NSP4 antibodies that are cross-reactive for MERS-CoV, and infection was quantified as the number of anti-SARS-CoV NSP4-positive cells. Two independent experiments were performed, and data from one representative experiment are shown. Error bars indicate SEMs (two-tailed Student's t test; *, **, or ***, P < 0.05 versus control.
On September 17, 2013, WHO was informed of a new confirmed case of MERS-CoV infection, resulting in a global total of 114 cases, including 54 deaths (5). A recent report of health care workers with MERS-CoV infections suggests it has person-to-person transmissibility (11, 13, 26), elevating concerns for a potential severe acute respiratory syndrome (SARS)-like pandemic (27). Given the potential for MERS-CoV to develop into a global health care crisis (4), development of effective and safe therapies is urgently needed. Our current results with anti-CD26 MAbs suggest the strategy of using CD26-specific reagents to inhibit MERS-CoV entry into susceptible cells and subsequent infections. The humanized anti-CD26 MAb YS110 is especially ready for testing as a potential treatment for MERS-CoV infection in the present outbreaks.
ACKNOWLEDGMENTS
We thank Haruna Otsuka and Aya Miwa for excellent assistance with experiments.
This work was supported by a grant-in-aid of the Ministry of Education, Science, Sports (K.O. and C.M.) and Culture, Ministry of Health, Labor, and Welfare, (C.M.) Japan. K.O. and C.M. are inventors on a patent application related to this work. The authors declare no competing financial interests.
K.O. and C.M. designed and coordinated the study. K.O., V.S.R, H.M., B.J.B., and B.L.H conducted the experiments. C.M., B.J.B., and B.L.H supervised part of the experiments. All authors contributed to the interpretations and conclusions presented. K.O. and C.M. wrote the manuscript, and B.J.B., B.L.H., R.H., and N.H.D. participated in editing it.
Footnotes
Published ahead of print 25 September 2013
REFERENCES
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367:1814–1820 [DOI] [PubMed] [Google Scholar]
- 2.Enserink M. 2013. New coronavirus reveals some of its secrets. Science 340:17–18 [DOI] [PubMed] [Google Scholar]
- 3.de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish Z, Perlman S, Poon LL, Snijder EJ, Stephens GM, Woo PC, Zaki AM, Zambon M, Ziebuhr J. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 87:7790–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC 2013. Update: severe respiratory illness associated with Middle East respiratory syndrome coronavirus (MERS-CoV)—worldwide, 2012-2013. MMWR Morb. Mortal. Wkly. Rep. 62:480–483 [PMC free article] [PubMed] [Google Scholar]
- 5.WHO 2013. Novel coronavirus infection—update. http://www.who.int/csr/don/2013_09_17_ncov/en/index.html
- 6.de Wilde AH, Ray VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens RW, Posthuma CC, van der Meer Y, Barcena M, Haagmans BL, Snijder EJ, van den Hoogen BG. 2013. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 94:1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falzarano D, de Wit E, Rasmussen AL, Feldmann F, Okumura A, Scott DP, Brining D, Bushmaker T, Martellaro C, Baseler L, Benecke AG, Katze MG, Munster VJ, Feldmann H. 8 September 2013, posting date Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV–infected rhesus macaques. Nat. Med. 10.1038/nm.3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mou H, Raj VS, van Kuppeveld FJM, Rottier PJM, Haagmans BL, Bosch BJ. 2013. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 87:9379–9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka T, Camerini D, Seed B, Torimoto Y, Dang NH, Kameoka J, Dahlberg HN, Schlossman SF, Morimoto C. 1992. Cloning and functional expression of the T cell activation antigen CD26. J. Immunol. 149:481–486 [PubMed] [Google Scholar]
- 11.Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM. 2013. Family cluster of Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 368:2487–2494 [DOI] [PubMed] [Google Scholar]
- 12.Drosten C, Seilmaier M, Corman VM, Hartmann W, Scheible G, Sack S, Guggemos W, Kallies R, Muth D, Junglen S, Müller MA, Haas W, Guberina H, Röhnisch T, Schmid-Wendtner M, Aldabbagh S, Dittmer U, Gold H, Graf P, Bonin F, Rambaut A, Wendtner C-M. 2013. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 13:745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guery B, Poissy J, El Mansouf L, Sejourne C, Ettahar N, Lemaire X, Vuotto F, Goffard A, Behillil S, Enouf V, Caro V, Mailles A, Che D, Manuguerra JC, Mathieu D, Fontanet A, van der Werf S. 2013. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet 381:2265–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF. 2013. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen YH, Zhang L, Wang X. 2013. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 23:986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong RP, Tachibana K, Hegen M, Scharpe S, Cho D, Schlossman SF, Morimoto C. 1998. Correlation of the epitopes defined by anti-CD26 mAbs and CD26 function. Mol. Immunol. 35:13–21 [DOI] [PubMed] [Google Scholar]
- 17.Inamoto T, Yamada T, Ohnuma K, Kina S, Takahashi N, Yamochi T, Inamoto S, Katsuoka Y, Hosono O, Tanaka H, Dang NH, Morimoto C. 2007. Humanized anti-CD26 monoclonal antibody as a treatment for malignant mesothelioma tumors. Clin. Cancer Res. 13:4191–4200 [DOI] [PubMed] [Google Scholar]
- 18.Dang NH, Torimoto Y, Sugita K, Daley JF, Schow P, Prado C, Schlossman SF, Morimoto C. 1990. Cell surface modulation of CD26 by anti-1F7 monoclonal antibody. Analysis of surface expression and human T cell activation. J. Immunol. 145:3963–3971 [PubMed] [Google Scholar]
- 19.Kameoka J, Tanaka T, Nojima Y, Schlossman SF, Morimoto C. 1993. Direct association of adenosine deaminase with a T cell activation antigen, CD26. Science 261:466–469 [DOI] [PubMed] [Google Scholar]
- 20.Dong RP, Tachibana K, Hegen M, Munakata Y, Cho D, Schlossman SF, Morimoto C. 1997. Determination of adenosine deaminase binding domain on CD26 and its immunoregulatory effect on T cell activation. J. Immunol. 159:6070–6076 [PubMed] [Google Scholar]
- 21.Ludwig K, Fan H, Dobers J, Berger M, Reutter W, Bottcher C. 2004. 3D structure of the CD26-ADA complex obtained by cryo-EM and single particle analysis. Biochem. Biophys. Res. Commun. 313:223–229 [DOI] [PubMed] [Google Scholar]
- 22.Weihofen WA, Liu J, Reutter W, Saenger W, Fan H. 2004. Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface. J. Biol. Chem. 279:43330–43335 [DOI] [PubMed] [Google Scholar]
- 23.Ohnuma K, Dang NH, Morimoto C. 2008. Revisiting an old acquaintance: CD26 and its molecular mechanisms in T cell function. Trends Immunol. 29:295–301 [DOI] [PubMed] [Google Scholar]
- 24.Ohnuma K, Uchiyama M, Yamochi T, Nishibashi K, Hosono O, Takahashi N, Kina S, Tanaka H, Lin X, Dang NH, Morimoto C. 2007. Caveolin-1 triggers T-cell activation via CD26 in association with CARMA1. J. Biol. Chem. 282:10117–10131 [DOI] [PubMed] [Google Scholar]
- 25.Rasmussen HB, Branner S, Wiberg FC, Wagtmann N. 2003. Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analog. Nat. Struct. Biol. 10:19–25 [DOI] [PubMed] [Google Scholar]
- 26.WHO 2013. Middle East respiratory syndrome coronavirus (MERS-CoV)—update. http://www.who.int/csr/don/2013_05_31_ncov/en/index.html [DOI] [PMC free article] [PubMed]
- 27.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen M, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TCT, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Günther S, Osterhaus ADME, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399 [DOI] [PubMed] [Google Scholar]